Abstract
To express its structural proteins, the arterivirus Equine arteritis virus (EAV) produces a nested set of six subgenomic (sg) RNA species. These RNA molecules are generated by a mechanism of discontinuous transcription, during which a common leader sequence, representing the 5′ end of the genomic RNA, is attached to the bodies of the sg RNAs. The connection between the leader and body parts of an mRNA is formed by a short, conserved sequence element termed the transcription-regulating sequence (TRS), which is present at the 3′ end of the leader as well as upstream of each of the structural protein genes. With the exception of RNA3, only one body TRS was previously assumed to be used to join the leader and body of each EAV sg RNA. Here we show that for the synthesis of two other sg RNAs, RNA4 and RNA5, alternative leader-body junction sites that differ substantially in transcriptional activity are used. By site-directed mutagenesis of an EAV infectious cDNA clone, the alternative TRSs used to generate RNA3, -4, and -5 were inactivated, which strongly influenced the corresponding RNA levels and the production of infectious progeny virus. The relative amounts of RNA produced from alternative TRSs differed significantly and corresponded to the relative infectivities of the virus mutants. This strongly suggested that the structural proteins that are expressed from these RNAs are limiting factors during the viral life cycle and that the discontinuous step in sg RNA synthesis is crucial for the regulation of their expression. On the basis of a theoretical analysis of the predicted RNA structure of the 3′ end of the EAV genome, we propose that the local secondary RNA structure of the body TRS regions is an important factor in the regulation of the discontinuous step in EAV sg mRNA synthesis.
The arterivirus Equine arteritis virus (EAV) is a plus-stranded RNA virus belonging to the order Nidovirales, which includes coronaviruses and arteriviruses (4, 11). The family Arteriviridae consists of EAV, Porcine reproductive and respiratory syndrome virus (PRRSV), murine Lactate dehydrogenase-elevating virus (LDV), and Simian hemorrhagic fever virus (SHFV) (see reference 34 for a recent review).
The EAV genome is a single 12.7-kb RNA molecule (11). Two large replicase polyproteins, the ORF1a and ORF1ab proteins, are encoded in the 5′ three-fourths of the genome. Translation of the ORF1b-encoded part of the ORF1ab protein involves a ribosomal frameshift (7). These polypeptides are proteolytically cleaved to yield 12 nonstructural proteins (36, 37, 44, 45, 48). In addition to genome replication, these proteins function in the production of a 3′-coterminal nested set of subgenomic (sg) RNAs, from which the viral structural proteins are translated (Fig. 1A).
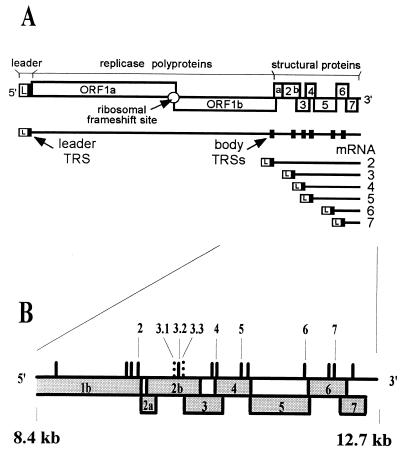
FIG. 1.
(A) Schematic diagram of the EAV genome organization and expression. The regions of the genome specifying the leader (L) sequence, the replicase gene (ORF1a and -1b), and the structural protein genes are indicated. The nested set of EAV mRNAs (genome and sg mRNA2 to -7) is depicted below. Black boxes, positions of leader and major body TRSs. (B) Close-up of the 3′-terminal one-third of the EAV genome. Numbers, previously identified body TRSs (see text); solid vertical lines, UCAAC sequences; dashed vertical lines, previously identified noncanonical leader-body junction sites.
EAV contains six or seven structural proteins (9, 10, 16, 35): the nucleocapsid protein N, the nonglycosylated, triple-spanning membrane protein M, the small envelope protein E, and four glycoproteins, GP2b, GP3, GP4, and GP5. It is unclear whether GP3 is a structural component of the arterivirus particle in view of the conflicting data reported for PRRSV GP3 (see reference 34 for a review). The genes encoding the EAV structural proteins (ORF2a to -7; Fig. 1) are located in the 3′-proximal 3 kb of the genome. From 5′ to 3′, the genes encode proteins in the following order: E-GP2b-GP3-GP4-GP5-M-N. mRNA2 is presumed to be bicistronic and is thought to encode both E and GP2b. The other five genes (encoding GP3 to N) are expressed from sg mRNA3 to -7.
The EAV sg RNAs are synthesized in different but constant molar ratios in the infected cell. The smallest sg RNA contains only the 3′-terminal open reading frame (ORF) of the genome (the N gene), while each next-larger sg RNA, with the exception of mRNA2, contains one additional ORF. Thus, all sg RNAs, except for the smallest RNA, are structurally polycistronic. However, with the exception of mRNA2, only the 5′-proximal ORF of each sg mRNA is assumed to be translated. Consequently, the expression of the EAV structural proteins can be primarily regulated at the level of sg mRNA synthesis.
The most characteristic feature of nidovirus sg mRNA transcription is that the sequences of the sg mRNAs are noncontiguous in the genome. At their 5′ ends, they include a common leader sequence (156 to 221 nucleotides [nt] for arteriviruses and 65 to 98 nt for coronaviruses), which is identical to the 5′ end of the genomic RNA (see references 19 and 34 for recent reviews). Thus, all nidovirus-specific RNAs, including the genome, are both 5′ and 3′ coterminal (Fig. 1A). The leader and body parts of the sg RNAs are connected by a short, conserved sequence element termed the transcription-regulating sequence (TRS), or intergenic sequence for coronaviruses. This sequence is present at the 3′ end of the leader (leader TRS) as well as upstream of (almost) every structural protein gene (body TRSs). The body TRS has been shown to be necessary for sg RNA synthesis (17, 22, 42, 46). A number of models have been put forward to explain the discontinuous step in nidovirus sg RNA synthesis (2, 31, 33, 38). Our recent data (46) are most consistent with the model of Sawicki and Sawicki (32), in which the discontinuous step occurs during minus-strand synthesis, producing minus-strand sg-sized templates for the synthesis of sg mRNAs in a process that resembles similarity-assisted RNA recombination (3, 27, 46).
The consensus sequence of the EAV TRS is 5′ UCAAC 3′ (6, 8). Recently, by site-directed mutagenesis of TRSs in the infectious cDNA clone of EAV, we have shown that base pairing between the complement of the RNA7 body TRS in the minus strand and the leader TRS in the plus strand is absolutely required for sg mRNA7 synthesis (46). However, at least for EAV, the mere presence of a UCAAC sequence in the genome is not sufficient for a leader-body junction event to occur (6, 7). The EAV genome contains multiple UCAAC sequences that appear to be transcriptionally silent. Upstream of ORF4, -5, and -7, two UCAAC boxes are present, whereas ORF2a and -2b are preceded by three UCAAC sequences (Fig. 1B) (11). Furthermore, the replicase gene contains a number of internal 5′ UCAAC 3′ sequences. In our previous study (6), RNA3 was shown to use three alternative leader-body junction sites, TRSs 3.1, 3.2, and 3.3, of which TRSs 3.1 and 3.3 are noncanonical junction sites. However, for the other EAV sg mRNAs mentioned above only a single active TRS was identified. The situation is different in PRRSV, where the use of two alternative junction sites has been documented for sg RNA4, -5, and -7 (isolate VR-2332 [28]) and RNA3 (isolate ISU-79 and some other isolates [24]).
Here, during the extension of our body TRS mutagenesis studies, two additional TRSs were discovered: one upstream of ORF4 and one upstream of ORF5. We show that, despite the fact that their primary sequences are almost identical, these alternative TRSs differ a lot in transcriptional activity. By mutagenizing the alternative TRSs of RNA3, -4, and -5, a strict correlation between individual RNA transcription levels and production of infectious progeny virus was established. Moreover, our results suggest that the glycoproteins that are expressed from these mRNAs are limiting factors during the viral life cycle and that the discontinuous step in sg mRNA synthesis is a crucial point in the regulation of structural protein expression.
MATERIALS AND METHODS
Cells and virus.
Baby hamster kidney (BHK-21; ATCC CCL10) cells were maintained and used for RNA transfection experiments as described previously (40, 43). They were also used for infections with wild-type (wt) EAV (Bucurus strain) (12) and mutant viruses.
Mutagenesis of the EAV full-length cDNA clone.
Translationally silent TRS mutations were introduced into a full-length cDNA clone of EAV (43) with the help of PCR mutagenesis (20). The oligonucleotides used for mutagenesis (purchased from Eurogentec, Seraing, Belgium) are listed in Table 1. Using standard recombinant DNA techniques (30), PCR fragments containing the mutations were cloned into shuttle vectors containing EAV sequences and were sequenced completely. PCR-based sequencing was carried out using the GeneAmp PCR system 2400 (Perkin-Elmer) and the ABI PRISM kit (Perkin-Elmer). Sequence reactions were analyzed using the ABI PRISM 310 genetic analyzer (Perkin-Elmer). To facilitate cloning, novel restriction sites were engineered in the EAV sequence by the introduction of translationally silent mutations. Subsequently, mutations were put into the full-length clone.
TABLE 1.
Oligonucleotides used in this study
| Primer | Polarity | Sequence | Genome position | Purpose |
|---|---|---|---|---|
| E239 | − | ACACGGGCCCAATGACTGAACCAATCTGCAGGAAATTAAACAGAGGTTTAC | 9699–9749 | RNA2 TRS mutagenesis |
| E241 | + | CACAAGAGCTGCAGCTCGAAGCCATTAATTGTAAATTGC | 10203–10241 | RNA3 TRS mutagenesis |
| E242 | − | CCGTAGATCTTCATCTACAAAGGAAAAAGACARCTGCCCCAAGCC | 10669–10713 | RNA4 TRS mutagenesis |
| E243 | − | CTCTTCCAAATTCACAATGACGTCGCCAGCC | 11011–11041 | RNA5 TRS mutagenesis |
| E244 | + | GCCTACGGTTCGAAAGTGAATCTTGTGAGGTTG | 11854–11886 | RNA6 TRS mutagenesis |
| E245 | + | ACCTTCCGGACCTGTTCCCATCCCTAGGAGTACAACTCAGGTAG | 12224–12267 | RNA7 TRS mutagenesis |
| E240 | − | GCTGGAGCTCTTGTGGATACTGCGACAAAATTGC | 10184–10217 | RNA3.1 TRS mutagenesis |
| E353 | + | TGGTTTGTAGCCATACGAATAGTACTACTGGCT | 10640–10672 | RNA4.1 TRS mutagenesis |
| E354 | + | TACGTTGGGCCCAGAGATGTTATCT | 11130–11154 | RNA5.2 TRS mutagenesis |
| E154 | − | TTGGTTCCTGGGTGGCTAATAACTACTT | 12679–12706 | Northern blot hybridization |
| E372 | − | ATAGTAGTATTATTGCTGCC | 10381–10400 | mRNA3 specific; RT and primer extension |
| E359 | − | GGTGGGATTAAGATA | 10886–10900 | mRNA4 specific; RT and primer extension |
| E122 | − | CATAAACACATCCAACACAACTATGC | 11495–11520 | mRNA5 specific; RT and primer extension |
| E371 | − | TGTAGGCACGACCCATGTGG | 10302–10321 | mRNA3 specific; RT and primer extension |
| E380 | − | GGAATTCGCATGCTTACGAGCCTCTGCAG | 10781–10797 | mRNA4 specific; PCR |
| E370 | − | CCGCGATCCGTCAGCATACA | 11246–11265 | mRNA5 specific; PCR and primer extension |
| EU153 | − | GGCCCAATCCATGAC | 10813–10827 | mRNA4 specific; primer extension |
| E157 | + | CTTGTGGGCCCCTCTCGGTAATCC | 63–89 | Leader specific; PCR |
| E373 | + | CTGGGATATGCTCTGTCGAT | 10001–10020 | mRNA3 specific; PCR |
| E374 | + | TTTGTAGTGCACACGGGTTA | 10501–10520 | mRNA4 specific; PCR |
| E375 | + | ACTCCCACTGCGCCGGCTAT | 10901–10920 | mRNA5 specific; PCR |
| E400 | + | TGGAGTCTTCTAGCTATGCT | 10140–10169 | mRNA3 specific; sequencing of virion RNA |
| E401 | + | TAATGCTTCTTGCGTGCAAG | 10620–10639 | mRNA4 specific; sequencing of virion RNA |
| E402 | + | GCTACTATCTGGCTGCAGCT | 10970–10988 | mRNA5 specific; sequencing of virion RNA |
| EL037 | + | GTAAATCCTAGAGGGCTTTC | 81–100 | Leader specific; sequencing of leader-body RT-PCR products |
In vitro RNA transcription, transfection, wt EAV infection, RNA isolation, and Northern blots.
Methods for in vitro RNA transcription from XhoI-linearized wt or mutant full-length EAV cDNA clones, as well as the procedures for transfection of BHK-21 cells, have been described previously (43). Infections with wt EAV were carried out as described previously (9) with a multiplicity of infection of 10 to 20 PFU/cell. For first-cycle RNA analysis, cells were kept at 39.5°C for 12 to 14 h and RNA was isolated using acidic phenol as described previously (47) and separated in denaturing agarose-formaldehyde gels. Gels were hybridized with 32P-labeled oligonucleotide E154, which is complementary to the 3′ end of the genome and which recognizes all plus-strand EAV RNAs (8) (Table 1). After hybridization, gels were analyzed using a Personal FX molecular imager and Quantity One software (both from Bio-Rad).
RT-PCR.
For reverse transcription-PCR (RT-PCR) analysis, poly(A)-containing RNA was isolated directly from the cell lysates using oligo(dT)-carrying Dynabeads (Dynal, Oslo, Norway) according to the manufacturer's instructions. cDNA synthesis was carried out at 42°C for 1.5 h using Moloney murine leukemia virus reverse transcriptase (Life Technologies Inc.) and antisense primers E372, E359, and E122, which map to the regions downstream of the RNA3, -4, and -5 body TRSs, respectively. Subsequently, the cDNA was used as the template for an sg RNA-specific PCR (leader-body PCR). Oligonucleotides E371, E380, and E370, which are located downstream of the RNA3, -4, and -5 body TRSs, respectively, were used as antisense primers, and oligonucleotide E157, located in the EAV leader sequence, was used as the sense primer (Table 1). The PCR consisted of 35 cycles, each comprising 45 s of denaturation at 95°C, 45 s of annealing at a temperature calculated to be 5°C below the lowest melting point of the primers involved, and 2.5 min of extension at 72°C. The 35 cycles were followed by a 5-min incubation at 72°C. Products were analyzed in 2% agarose gels and photographed using a GelDoc apparatus (Bio-Rad) and Quantity One software. For sequence analysis, PCR products were isolated from gel using a GeneClean kit (BIO 101 Inc.) and used directly as a template for the sequencing reaction.
Primer extension analysis.
Poly(A)-containing RNA was incubated with 10 pmol of 32P-labeled primer (E371 and E372 for the detection of RNA3-specific extension products, EU153 and E359 for RNA4, and E370 and E122 for RNA5) in the presence of 40 mM methylmercury hydroxide in a total volume of 10 μl for 10 min at room temperature. After the incubation, methylmercury hydroxide was neutralized by adding 4 μl of a 1.4 M β-mercaptoethanol solution. Primer extension reactions were performed in a total volume of 30 μl containing 5 μl of 6× first-strand buffer (Life Technologies Inc.), 1 mM deoxynucleoside triphosphates, 10 mM dithiothreitol, 40 U of RNase Out (Life Technologies Inc.), and 200 U of Superscript reverse transcriptase (Life Technologies Inc.) for 1.5 h at 42°C. Subsequently, reaction products were applied to an 8 M urea–6% acrylamide gel. A 33P-labeled sequence reaction performed with an M13mp18 single-stranded DNA and M13 forward primer using the T7 sequencing kit (Pharmacia), as well as a 32P-labeled 100-bp ladder (Life Technologies Inc.), were used as markers. Gels were analyzed and individual bands were quantitated as described above. All calculations were based on two or three experiments with two different primers.
IFAs.
Transfected cells were seeded on coverslips and fixed with 3% paraformaldehyde in phosphate-buffered saline (PBS) at 12, 24, or 36 h posttransfection. Immunofluorescence assays (IFAs) were carried out as described previously (41). Rabbit antisera directed against nonstructural protein 2 (nsp2) (37) and nsp3 (29) were used to monitor virus replication. Rabbit antisera recognizing the E (35), GP2b (9), and M (9) proteins and mouse monoclonal antibodies 6D10 (anti-GP5 [1]) and 3E2 (anti-N [21]) were used as primary antibodies to detect EAV structural proteins. As secondary antibodies, a Cy3-conjugated donkey anti-rabbit immunoglobulin G (IgG) antibody and a fluorescein isothiocyanate-conjugated donkey anti-mouse IgG antibody (both from Jackson ImmunoResearch Laboratories) were used. The samples were examined with an Olympus fluorescence microscope equipped with a digital camera and Qfluoro software (Leica).
Infectious-center assays and plaque assays.
For infectious-center assays, transfected cells were mixed, in different proportions, with untransfected BHK-21 cells directly after transfection. Cells were allowed to attach for 4 h, and then the medium was replaced with an overlay of medium containing 1.5% agarose and 2% fetal calf serum. For plaque assays, medium was collected from transfected cells at 24 h posttransfection and used to infect fresh BHK-21 cells. After a 1-h infection, cells were washed, and an agarose overlay was applied. At 3 to 6 days postinfection (p.i.), cells were fixed with 10% formaldehyde in PBS and stained with crystal violet.
Direct RT-PCR analysis of plaque-derived RNA.
Plaques were picked and resuspended in PBS, incubated for 1 h at 37°C with 0.5 mg of proteinase K/ml–0.5% sodium dodecyl sulfate, extracted twice with phenol-chloroform-isoamyl alcohol, ethanol precipitated in the presence of 10 μg of yeast RNA as the carrier, and resuspended in 8 μl of water to which 10 pmol of the appropriate RT primer was added. cDNA synthesis, followed by PCR, was carried out as described above, except that oligonucleotides E373, E374, and E375 were used as the sense PCR primers to amplify the RNA3, -4, and -5 TRS regions in the genome, respectively (Table 1). PCR products were purified from gel and sequenced as described above, using oligonucleotides E400, E401, and E402 as the sequencing primers.
RNA structure predictions.
The RNA structure of the 3′-terminal region of the EAV genome (GenBank accession no. Y07862) was predicted by using a previously described genetic algorithm implemented in the program STAR (15, 39).
RESULTS
Effects of body TRS mutagenesis on the transcription of the corresponding sg RNA and production of infectious progeny virus.
Previously, the absolute requirement of a body TRS for EAV sg RNA synthesis was shown for RNA7 only (46). To extend this study to the other sg RNA species, mutations were introduced in the EAV full-length cDNA clone in all canonical-body TRSs (UCAAC boxes) (6, 8), which were previously shown to serve as leader-body junction sites for the transcription of sg RNA2 to -7 (Table 2, mutants M2 to M7). Because all EAV structural protein genes are overlapping, every body TRS is also a part of the coding sequence for an upstream structural protein. In order to affect only sg RNA transcription, not the sequences of structural proteins encoded by upstream ORFs, the mutations introduced into TRSs were designed to be translationally silent.
TABLE 2.
Body TRS mutants
| Mutant | Mutated TRS location | Mutant TRS sequencea | Titer (PFU/ml) | Plaque diam after 3 days p.i. (mm) | TRS sequence of plaque-isolated RNA | No. of plaques analyzed | Reversion |
|---|---|---|---|---|---|---|---|
| M2 | 9711–9715 | UuAAu | N/ab | N/a | N/a | N/a | N/a |
| M3.2 | 10227–10231 | UuAAu | 6 × 107 | 4 | UuAAu | 4 | No |
| M4 (M4.2) | 10677–10681 | gCAgC | 1 × 107 | 2.5 | gCAgC | 5 | No |
| M5 (M5.1) | 11028–11032 | UgAAu | 6 × 104 | 4 | UCAAC | 4 | Yes |
| M6 | 11870–11874 | UgAAu | N/a | N/a | N/a | N/a | N/a |
| M7 | 12252–12256 | aguAC | N/a | N/a | N/a | N/a | N/a |
| M3.1 | 10195–10199 | gCAgU | 5.5 × 107 | 3.5 | gCAgU | 4 | No |
| M4.1 | 10660–10664 | aguAC | 3 × 108 | 4 | aguAC | 4 | No |
| M5.2 | 11140–11144 | cCAga | 2 × 108 | 4 | cCAga | 2 | No |
| M3+3 | 10195–10199, 10227–10231 | gCAgU + UuAAu | 4 × 106 | 2 | gCAgU + UuAAu (1 pl.); g/UCAg/AU + Uu/CAAu/C (3 pl.)d | 4 | No (1 pl.); mixe (3 pl.) |
| M4+4 | 10660–10664, 10677–10681 | aguAC + gCAgC | ∼1.5 × 105 | N/dc | N/d | N/d | N/d |
| M5+5 | 11028–11032, 11140–11144 | UgAAu + cCAga | 6 × 103 | 2.5 | UCAAC + UCAAC | 3 | Yes |
| wt | N/a | N/a | 2.5 × 108 | 3.5 | N/a | N/a | N/a |
Mutated nucleotides are in lowercase.
N/a, not applicable.
N/d, not detected.
pl., plaques.
Mix, mixture of mutant and wild-type nucleotides.
Cells were transfected with in vitro-transcribed full-length RNA containing these mutations, as well as with RNA transcribed from the wt EAV cDNA clone pEAV030, which served as a control. At 12 h posttransfection (p.t.) (after one EAV replication cycle), cells were stained with a replicase antiserum to monitor virus replication and, for mutants M2, M5, M6, and M7, with an antiserum recognizing the structural protein of which the expression was expected to be affected by the TRS mutation. Unfortunately, antisera specific for EAV GP3 and GP4 are not available. For all six mutant constructs tested, genome replication was not affected (data not shown). However, IFAs specific for structural proteins E and GP2 (M2), GP5 (M5), M (M6), or N (M7) were negative at 12 h (data not shown), providing an initial indication that the TRS mutations were deleterious for transcription of the corresponding sg RNA.
To study sg RNA transcription directly, total intracellular RNA was isolated at 12 h p.t. and analyzed by hybridization. With the exception of M3.2, mutagenesis of all TRSs studied abolished the transcription of the corresponding RNA (Fig. 2), thereby proving that a body TRS is absolutely required for transcription of each of the EAV sg mRNAs. The continued synthesis of RNA3 by M3.2 can be explained by the previously described use of two additional noncanonical leader-body junction sites upstream of the ORF3 initiation codon (3.1 and 3.3 [6]).
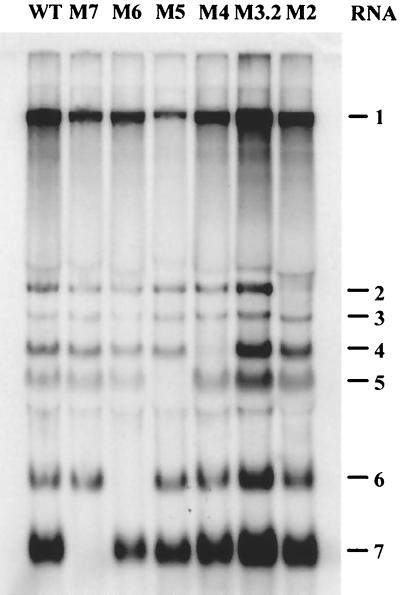
FIG. 2.
Northern blot analysis of virus-specific RNA isolated from cells transfected with the wt EAV cDNA construct and with the constructs in which major TRSs used for synthesis of sg mRNA2 to -7 were disrupted by mutations (see Table 2).
To assess whether any of the mutants was able to produce progeny virus, which should result in spread of the infection to initially untransfected cells in the monolayer, IFAs were done at 12, 24, and 36 h p.t. The EAV-specific signal produced by mutants M2, M6, and M7 did not spread to neighboring cells, even at 36 h p.t. (three EAV infection cycles) (Fig. 3 and data not shown). In contrast, M3.2, M4, and M5 mutant viruses had clearly spread at 24 h p.t., although for M5 spread was delayed (Fig. 3). Since we have recently shown that all EAV structural proteins are required for the production of infectious progeny virus (26), these observations suggested either leaky RNA transcription from mutant TRSs, rapid reversion of TRS mutations, or the use of alternative body TRSs for the transcription of RNA4 and -5, in addition to RNA3.
FIG. 3.
Immunofluorescence analysis of BHK-21 cells transfected with TRS mutants M3.2, M4, M5, and M7. At 12 and 24 h p.t., cells were fixed and stained for nsp3 (EAV replication marker) to test for spread of the EAV-specific signal.
EAV uses alternative body TRSs for transcription of sg RNA3, -4, and -5.
The observations described above prompted us to analyze the TRS mutants for RNA3, -4, and -5 in more detail. To discriminate between leaky transcription from mutated TRSs and the use of alternative TRSs, a sg mRNA-specific RT-PCR approach was designed, similar to that used by den Boon et al. (6) and van Marle et al. (47) (Fig. 4A). First, cDNA was synthesized using an RT primer located downstream of a body TRS region in an sg mRNA. Due to the nested structure of EAV mRNAs, such an RT primer also binds to the corresponding positions in all larger viral mRNAs, including the genome. Next, a PCR specific for the leader-body junction region of the desired mRNA was performed, using an internal antisense primer located just upstream of the RT primer in the body of the mRNA and a sense primer located in the leader sequence. This approach allowed us to obtain and separate by size RT-PCR products corresponding to the different mRNA species originating from all the potential TRSs upstream of the binding site of the antisense PCR primer. To make sure that such alternative mRNAs with different leader-body junctions would be detected with this method, we selected antisense primers that were located within the coding sequence of the corresponding ORF. After separation in agarose gel, PCR products were purified, and leader-body junctions were sequenced directly using an internal primer located in the leader.
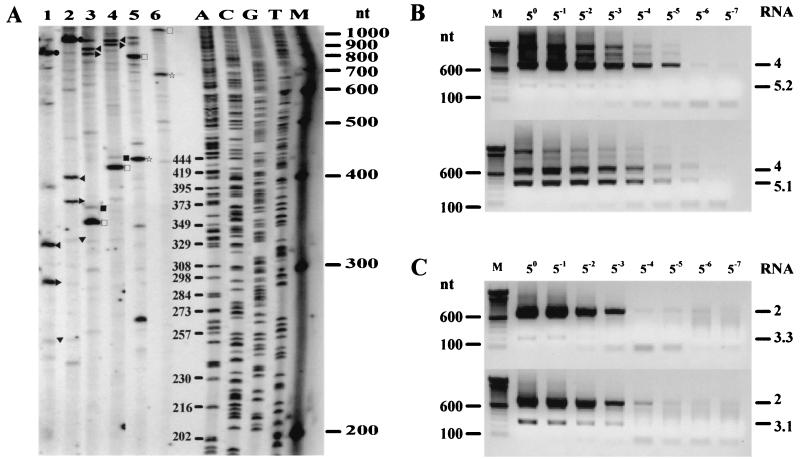
FIG. 4.
(A) Scheme of the mRNA-specific RT-PCR strategy. For clarity, a hypothetical gene with two upstream TRSs (A and B) (triangles) is depicted. Black and white arrows, RT and PCR (sense and antisense) primers, respectively. The genomic RNA (g RNA) and two sg RNAs originating from TRSs A and B are shown. Double lines, PCR products corresponding to sg RNAs A and B. (B) RT-PCR specific for mRNA3 (lanes M3.2 and WT3), mRNA4 (lanes M4 and WT4), and mRNA5 (lanes M5 and WT5). WT lanes, reactions performed on RNA from the wt EAV constructs; lanes M3.2, M4, and M5, RNA from M3.2, M4, and M5 constructs, respectively. ●, sg RNA2; ◂, sg RNA3.1; ▸, sg RNA3.2; ▾, sg RNA3.3; ■, sg RNA4.1; □, sg RNA4.2; ⋆, sg RNA5.1; ★, sg RNA5.2. A 100-bp ladder was used as a size marker.
Lane WT3 in Fig. 4B represents the products of the mRNA3-specific RT-PCR on poly(A)-containing RNA isolated at 12 h p.i. from wt EAV-infected cells. The PCR products in this lane corresponded to RNA species originating from TRSs 3.1 (269-nt band) and 3.2 (237-nt band), which were described by den Boon et al. (6), as well as from the more upstream RNA2 TRS (753 nt). As expected, RNA isolated from cells transfected with M3.2 RNA lacked the 237-nt band, but a minor band of 194 nt appeared; this band corresponded to the previously described alternative, noncanonical TRS 3.3 (6).
The mRNA4-specific RT-PCR on wt EAV RNA (lane WT4, Fig. 4B) showed that, besides a major product (275 nt) corresponding to TRS 4, a slightly larger minor product was formed. Direct sequence analysis of this product revealed that it represented a novel mRNA4 subspecies (mRNA4.1), which was derived from an alternative body TRS located 17 nt upstream (at nt 10660 to 10664) of TRS 4. This TRS 4.1 was previously thought to be silent. Consequently, we refer to the previously identified “major” RNA4 TRS as TRS 4.2. The analysis of M4 RNA showed that, while the generation of the major product was indeed prevented by the mutations, the minor TRS 4.1 was still active. In addition, bands of 725, 757, and 1,241 nt, corresponding to mRNA subspecies 3.2, 3.1, and 2, respectively, could be observed in both RNA4 RT-PCR lanes.
In contrast to RNA3 and -4, the RNA5-specific RT-PCR on wt EAV RNA (lane WT5; Fig. 4B) produced only one RNA5-specific product of 381 nt, corresponding to the use of TRS 5. However, when this TRS was mutated (lane M5), this major product was replaced with a smaller and much less abundant product. Sequencing revealed that it corresponded to a novel mRNA5 subspecies (mRNA5.2) originating from a TRS (TRS 5.2) located 112 nt downstream of TRS 5 (referred to as TRS 5.1) and directly upstream of the ORF5 initiation codon (nt 11140 to 11144). RNA4-specific products were also observed in both lanes.
Taken together, these observations indicated that not only for RNA3 but also for RNA4 and -5 alternative body TRSs are used for the joining of leader and body. The data also clearly showed the deleterious effects of TRS mutations on transcriptional activity. It should be noted, however, that these RT-PCR data are by no means quantitative because of the amplification step involved and the competition between cDNA molecules with overlapping sequences. For example, the fact that products corresponding to minor RNA species (3.3 and 5.2) could not be amplified from wt RNA but “appeared” when the synthesis of major RNA species was abolished did not necessarily imply up-regulation of the activity of these minor TRSs, but rather reflected competition at the amplification stage. Thus, a more direct method had to be applied in order to determine the relative abundance of these RNA subspecies in infected cells.
Relative abundances of RNA3 and -4 subspecies.
To measure the relative use of alternative TRSs, we developed primer extension assays specific for EAV sg RNAs similar to those used by de Vries et al. (8). Because the primer was always used in a large molar excess compared to the template RNA, the relative abundances of individual products in the same reaction could be assumed to be directly proportional to the relative abundances of the corresponding RNA subspecies. A radioactively labeled oligonucleotide was hybridized to poly(A)-containing RNA isolated at 12 h p.i. from wt EAV-infected cells. This step was followed by cDNA synthesis, and the reaction products were separated in a denaturing polyacrylamide gel (Fig. 5A). For each TRS region, a pair of antisense primers located in the corresponding ORF was used, one proximal to the TRSs and one more distal, so that sg mRNAs containing each of the potential leader-body junctions would be detected.
FIG. 5.
Quantitation of the relative amounts of RNA3, -4, and -5 subspecies. (A) Primer extension analysis specific for RNA3 (lanes 1 and 2), RNA4 (lanes 3 and 4), and RNA5 (lanes 5 and 6) subspecies. For each mRNA-specific reaction pair, RNA from the cells infected with wt EAV was hybridized with an antisense primer, which was either close to (uneven lanes) or more downstream of (even lanes) the TRSs. ●, sg RNA2; ◂, sg RNA3.1; ▸, sg RNA3.2; ▾, sg RNA3.3; ■, sg RNA4.1; □, sg RNA4.2; ⋆, sg RNA5.1. A 100-bp ladder as well as the products of a sequencing reaction were used as size markers. (B) Semiquantitative RT-PCR analysis aimed at estimating the relative amounts of the RNA5.1 and -5.2 subspecies. Eight fivefold serial dilutions of RNA from M5.1 (top) or M5.2 (bottom) were subjected to RNA5-specific RT-PCR analysis. Indicated are the positions of PCR products corresponding to RNA species 4 and 5.2 (top) or 4 and 5.1 (bottom). (C) The same approach was used to estimate the relative amounts of RNA3.1 and -3.3 subspecies. RNA is from M3+3 (top) or M3.2 (bottom). Indicated are the positions of PCR products corresponding to RNA species 2 and 3.3 (top) or 2 and 3.1 (bottom). A 100-bp ladder was used as a size marker for both panels B and C.
Lanes 1 and 2 in Fig. 5A correspond to RNA3-specific reactions. Both primers recognized RNA subspecies 3.1 and 3.2 (332- and 300-nt products in lane 1 and 411- and 379-nt products in lane 2), which were present in almost equimolar amounts (concentration of 3.1 [C3.1]/C3.2 = 1.1 ± 0.2), as well as RNA2 (816 or 895 nt). In addition, a minor band, corresponding to the RNA3.3 subspecies, could be detected (257 nt in lane 1 or 336 nt in lane 2). The average ratio between the intensities of this band and the band corresponding to RNA3.1 was approximately 1:10 (C3.1/C3.3 = 10.5 ± 0.6). Lanes 3 and 4 correspond to RNA4-specific primer extension reactions. The major subspecies RNA4.2 and the minor subspecies RNA4.1 could be detected using both primers (373 and 356 nt in lane 3; 446 and 429 nt in lane 4). The average molar ratio between these subspecies was determined to be approximately 40:1 (C4.2/C4.1 = 40.4 ± 9.1). Larger products corresponding to RNA subspecies 3.1 and 3.2 were also seen in these lanes (838 and 806 nt in lane 3; 911 and 879 nt in lane 4). Lanes 5 and 6 correspond to RNA5-specific primer extension reactions. In this case, only the RNA5.1 (major) subspecies was detected using both primers (444 or 691 nt), presumably because the level of the RNA5.2 subspecies in infected cells is extremely low. Larger products (795 nt in lane 5 and 1,042 nt in lane 6) correspond to RNA4.
Effects of single and double knockouts of alternative RNA3, -4, and -5 TRSs on mRNA synthesis.
To study the importance of the use of alternative body TRSs for viral mRNA synthesis and production of infectious progeny virus, additional mutant constructs were designed (Table 2, M3.1 to M5+5) to inactivate TRSs 3.1, 4.1, and 5.2. For each RNA, we also constructed double mutants (TRS 3.1 plus 3.2 in M3+3; TRS 4.1 plus 4.2 in M4+4; TRS 5.1 plus 5.2 in M5+5). These novel mutants were transfected and analyzed as described above for the initial set of TRS mutants.
The RT-PCR analysis of RNA3, -4, and -5 synthesis by all single and double TRS knockout mutants is shown in Fig. 6. In each case, TRS mutagenesis had a clear deleterious effect on its use for the leader-body junction. While M3+3 still produced a small amount (Fig. 6A) of RNA3 from TRS 3.3 (which was not mutagenized), RNA4 or -5 synthesis could not be detected for M4+4 or M5+5, respectively (Fig. 6B and C). In a number of lanes novel products or larger amounts of certain products were observed when the generation of one of the major products was abrogated. Again, this observation did not necessarily imply up-regulation of the transcription of other mRNA (sub)species when a major (sub)species was not generated; most likely, it reflected competition effects during the RT-PCR amplification stage.
FIG. 6.
RT-PCR specific for sg RNA3 (A), RNA4 (B), and RNA5 (C). For each set of RNA-specific reactions, RT-PCR was performed on poly(A)-containing RNA from single as well as double TRS mutants and on RNA from a wt EAV construct. A 100-bp ladder was used as a size marker.
The additional mutants described above were used to resolve another important issue. Because the primer extension assays described in the previous section were not sensitive enough to detect extension products corresponding to sg RNA subspecies 5.2, we used a semiquantitative RT-PCR method similar to the one described by van Marle et al. (47) to estimate the relative abundance of this minor subspecies. As shown in Fig. 5B, RT-PCR was performed on fivefold serial dilutions (undiluted to 5−7) of poly(A)-containing RNA isolated from an equal number of cells transfected with either M5.1 (transcribing only RNA5.2) or M5.2 (transcribing only RNA5.1). In both cases, the mRNA5-specific RT-PCR was performed using the same set of primers as that described above. Assuming that the RT-PCRs were equally efficient, we estimated the ratio of RNA5.2 to RNA5.1 to be between 1:125 and 1:625. This estimation was based on the fact that the intensity of the RNA5.2 band in lane 5−3 (which corresponded to the highest dilution used with which we could still see this signal) was lower than the intensity of the RNA5.1 band in lane 5−6 but higher than that of the RNA5.1 band in lane 5−7 (Fig. 5B). As a control, the same approach was used to determine the relative abundance of the RNA3.3 subspecies (Fig. 5C), which could be compared to the values obtained in the primer extension experiments (Fig. 5A). mRNA3-specific RT-PCRs were done on fivefold serial dilutions of intracellular RNA from transfections with M3.2 (transcribing predominantly RNA3.1) or M3+3 (transcribing RNA3.3 only). In this case, by an approach similar to that described above, the RNA3.3 amount was estimated to be 5 to 25 times lower than the amount of RNA3.1, a value that corresponded nicely to the approximately 1:10 ratio determined by primer extension. The important assumption behind these experiments is that mutagenizing a particular body TRS does not dramatically influence the activities of adjacent body TRSs. Though we were unable to directly prove this point for the alternative TRSs for RNA3, -4, and -5, we have recently established this for all major EAV sg RNAs (Fig. 1 and unpublished data).
Effects of single and double knockouts of alternative RNA3, -4, and -5 TRSs on production of infectious progeny virus.
Cells transfected with the six additional constructs described in the previous paragraph were also used for IFAs at 12 and 24 h p.t. using an anti-nsp3 antiserum. The phenotype of M3.1, as revealed by this assay, was similar to the phenotype of M3.2 described above. Both mutant viruses were infectious, and the infection was found to spread through the monolayer rapidly. Surprisingly, this was also true for M3+3 (data not shown), which was assumed to rely on ORF3 expression from a single, minor RNA3 subspecies, RNA3.3.
Remarkably, for the TRS 4 and TRS 5 mutant series not only all single TRS knockout viruses were infectious and able to spread to neighboring cells but also both double-knockout mutants were able to do so, although much less efficiently than the wt control (data not shown). Due to the lack of GP3 and GP4 antibodies, we could only monitor the expression of the corresponding structural protein for the TRS5 mutants. Cells were stained with an antibody recognizing GP5, which is translated from RNA5, as well as with the anti-nsp3 antiserum, as an internal control on virus replication. All TRS 5 mutants and the wt control produced similar amounts of nsp3 (data not shown). M5.2, in which only a minor portion of RNA5 synthesis should be abolished, appeared to produce wt amounts of GP5 already at 12 h p.t., as well as at 24 h. However, the GP5 signal of M5.1, which should lack most of the RNA5 synthesis, and that of M5+5 were detectable only at 24 h p.t. and were much fainter than that of the wt control (data not shown).
Taken together, these data implied that expression of just a minor RNA subspecies (3.3, 4.1, or 5.2) may be enough for production of infectious progeny virus (M3+3, M4.2, and M5.1, respectively). Moreover, the results obtained with M4+4 and M5+5 suggested that infectious progeny virus can be produced without any detectable amounts of RNA4 or RNA5 (Fig. 6B and C). However, we have recently established that both GP4 and GP5 are absolutely required for the production of infectious progeny virus (26). This implied either the rapid reversion of TRS mutations or residual expression of the corresponding proteins by M4+4 and M5+5. The latter situation might occur on the basis of leaky transcription from mutated TRSs or could be due to the internal initiation of translation from larger sg mRNAs (e.g., GP4 would then be expressed from mRNA2 or -3).
Virus titers and plaque phenotypes of single- and double-knockout TRS 3, 4, and 5 mutants.
To characterize the TRS mutants in more detail, we first determined the virus titers produced upon transfection of these constructs. Cells were transfected with wt or mutant RNAs, and infectious-center assays and/or IFAs were done in order to determine the transfection efficiencies (data not shown). At 24 h p.t., the medium of transfected cultures was harvested and used for triplicate plaque assays. Cells were fixed after 3, 4, 5, or 6 days. Table 2 shows the titers of the mutant viruses and the wt control at 24 h p.t. normalized to their individual transfection efficiencies, as well as the average plaque diameters at 3 days p.i. For all TRS mutants analyzed, there was a clear difference in virus production between mutants using major RNA subspecies (3.1, 3.2, 4.2, or 5.1) for the production of corresponding structural proteins and mutants using minor RNA subspecies (3.3, 4.1, or 5.2). Furthermore, the differences in titers between M3.1 and M3.2 (1:1.1), M3.1 and M3+3 (15:1), and M4.1 and M4.2 (30:1) corresponded very well to the differences in molar amounts between the RNA subspecies produced from the active TRSs in these mutants (see above).
Surprisingly, double mutant M4+4, though capable of spread on the basis of IFAs, did not form detectable plaques even at 6 days p.i. To study this mutant more carefully, we performed a transfection experiment in which we compared M4+4 to a construct in which ORF4 was disrupted by a 129-nt internal deletion in the region which does not overlap with ORF3 or ORF5 (Fig. 1). This ORF4 mutant does not produce any infectious progeny virus, presumably due to the lack of a functional GP4 protein (S. Greve and E. J. Snijder, unpublished data). Medium from both transfections was harvested at 24 h p.t., diluted 10- to 1,000-fold, and used to infect fresh cells, which were seeded on coverslips. IFAs specific for nsp3 were done at 12, 24, and 36 h p.i. While there were no positive cells after infection with medium from the transfection of the ORF4 deletion construct, even after 36 h of incubation and at the lowest dilution, the sample from the M4+4 transfection yielded positive cells already at 12 h p.i. At later time points their number slowly increased (data not shown). Based on this infection experiment, we were able to determine an approximate titer of this mutant virus (Table 2). The comparison with the ORF4 deletion mutant suggested that M4+4 still expressed small amounts of GP4, most likely by internal initiation of translation from a larger sg mRNA or by leaky transcription of RNA4 from either of the mutated TRSs (or an undetected alternative TRS). On the basis of the titer of this mutant and that of the wt control, we estimated the amount of GP4 produced in this manner to be approximately 1,000-fold reduced. If this protein was produced from sg RNA4, then the amount of RNA4 may also be 1,000-fold reduced, which might explain why we could not detect its synthesis by RT-PCR. Reversion of the TRS mutations in this case is unlikely, since a revertant with a functional TRS 4.1 or TRS 4.2 would probably produce enough RNA4 to allow for the formation of plaques (Table 2, mutants 4.1 and 4.2). For M5.1 and M5+5, however, reversion was the most logical explanation for our observations, since they produced plaques with almost wt diameters, despite the dramatically reduced virus titers detected in the medium (Table 2). To clarify this issue, we performed sequence analysis of plaque-purified progeny of the various TRS mutants.
Sequence analysis of the progeny of TRS mutants.
At 3 or 4 days p.i., plaques of TRS mutants were picked, and virion RNA was isolated as described in Materials and Methods. The relevant body TRS regions in the viral genome were amplified by RT-PCR, and PCR products were sequenced directly (Table 2).
Both TRS 3 single mutants were stable, i.e., four out of four plaque-purified viruses retained the mutant TRS. This implied that the amount of RNA3 produced from either of the major RNA3 body TRSs (3.1 and 3.2) sufficed for the virus to replicate. Apparently there was no selective pressure, at least not within this passaging experiment, favoring the reversion of either of these TRS knockout mutations, although the titers of both mutant viruses were about fourfold reduced. Additional experiments are needed to study the long-term fitness of these mutant viruses. For double mutant M3+3, the situation was different. Virion RNA from one plaque contained both mutant TRSs, confirming the IFA result that the generation of the minor RNA3.3 species is sufficient for the virus to survive. However, each of the other three plaques analyzed contained a mixed population of nucleotides at all mutated positions in TRS 3.1 and 3.2, with one nucleotide still being mutant and one nucleotide having reverted to the wt sequence. Most likely, the initial virus particle which produced such a plaque contained both mutant TRSs, and reversion occurred during plaque growth. Since reversion was not observed for both TRS3 single mutants, it is unlikely that a genome in which both mutant TRSs of the double mutant had reverted simultaneously was present in one of the progeny viruses. More likely, there was a mixed population of TRS 3.1 revertants, TRS 3.2 revertants, and particles containing mutations in both TRSs. The reversion of the double mutant suggested that the virus requires at least one major RNA3 body TRS.
Both TRS 4 single mutants had remained stable in all plaques analyzed. This was surprising in view of the only fourfold difference in titer between the major RNA4 TRS mutant (TRS 4.2) and M3+3, which was unstable in the long run, and also because equivalent nucleotide substitutions had been introduced to generate M3.1 and M4.2 (Table 2). Moreover, even after 6 days, double mutant M4+4 apparently did not revert, in spite of its 1,000-fold-reduced titer. The absence of reversion may be due to differences in local RNA structure or flanking sequences, which may modulate the reversion frequency by influencing the error rate of the viral polymerase. Clearly, the possibility that reversion of M4.2 and M4+4 may occur upon prolonged passaging of these viruses cannot be ruled out.
As hypothesized above, the infectivity of M5.1 and M5+5 was completely due to reversion of the mutated nucleotides to the wt sequence. The minor RNA5 subspecies (RNA5.2), which was several hundred times less abundant than the major RNA5.1 subspecies, can apparently not support translation of sufficient GP5 protein to ensure production of infectious progeny virus. It is not clear whether the GP5 protein produced by M5.1 at 24 h p.t. was expressed from the minor TRS 5.2 or from the reverted TRS 5.1. Surprisingly, plaques isolated from the infection with M5+5 contained wt nucleotides in both TRSs, while reversion of the TRS 5.2 mutations in single mutant M5.2 was not observed.
DISCUSSION
Heterogeneity of arterivirus sg mRNAs.
To express their structural proteins, arteriviruses generate a 3′-coterminal nested set of at least six sg mRNAs (reviewed in references 11 and 34). Generally, only the most 5′-proximal ORF is assumed to be expressed from each of these sg mRNAs. However, there are some notable exceptions: mRNA2 of EAV and probably the corresponding mRNA of all other arteriviruses (35) have been postulated to be functionally bicistronic. Moreover, SHFV generates two extra sg mRNA species, one of which is again presumed to be functionally bicistronic, in connection with the postulated duplication of three structural protein genes (14).
Clearly, a single sg mRNA species would suffice to express each of the arterivirus structural proteins. Nevertheless, two types of sg mRNA heterogeneity have been documented for various arteriviruses and coronaviruses: differences in mRNA sequence, in particular in the region of the leader-body junction site, and differences in mRNA length. Heterogeneity of the first type is attributed to an imprecise leader-body junction mechanism, reflected in sequence differences in the leader-body junction region of sg mRNAs. This phenomenon was first described for the mouse hepatitis coronavirus (23) and was also described for arteriviruses. It appears to be caused by the use of different junction sites within or directly upstream of a body TRS. Variant mRNA subspecies, which can be explained in this manner, were described for PRRSV (25) and EAV (6) but not for LDV (5) and SHFV (14). The Lelystad strain of PRRSV in particular shows considerable heterogeneity of this type. In fact, only mRNA4 appears to contain a homogeneous leader-body junction sequence; all other mRNAs consist of two or even three subspecies (25). For EAV, this type of heterogeneity is much less pronounced: two subspecies, which differ in a single nucleotide immediately upstream of the body TRS, were detected for mRNA3.2 only (6).
The second type of sg mRNA heterogeneity, differences in mRNA length, was again described for EAV and PRRSV. Both these viruses generate mRNA subspecies, which originate from the use of alternative body TRSs for the joining of leader and body but which can still be used for the translation of the same structural protein. For EAV, only mRNA3 was previously reported to be derived from the use of more than one body TRS. Two major mRNA3 subspecies were detected, mRNA3.1 and -3.2, their body TRSs being separated by 32 nt in the genome (6). Furthermore, there is a third variant of mRNA3, mRNA3.3, which is produced by the leader-body junction at a sequence (43 nt downstream of TRS 3.2) that bears no resemblance to the canonical 5′ UCAAC 3′ box. However, in this case extended base pairing possibilities with the leader TRS region immediately downstream of the RNA3.3 body TRS exist, and these appear to compensate for the absence of the TRS consensus sequence (6).
For PRRSV, this kind of size heterogeneity is more pronounced. Meng et al. (24) reported the use of a second body TRS for RNA3 (referred to as RNA3-1) for some PRRSV isolates. In these isolates, a single U-to-C point mutation led to the activation of a cryptic body TRS and the synthesis of an additional mRNA3 subspecies. This additional TRS is, however, located downstream of the ORF3 translation initiation codon, and thus a full-length GP3 protein cannot be expressed from this RNA. A small alternative ORF (ORF3-1) was identified in the 5′-terminal region of mRNA3.1, but whether this ORF is actually expressed or whether RNA3.1 can be used to express GP4 remains to be determined. Recently, the use of alternative body TRSs was also reported for three sg RNA species (mRNA4, -5, and -7) of the North American VR-2332 isolate of PRRSV (28). However, the European PRRSV prototype, Lelystad virus, does not appear to use more than one body TRS were observed for each of these sg RNAs. Especially interesting differences in the utilization of potential RNA7 body TRSs by these two strains (28). While both viruses contain three potential RNA7 TRSs, conserved in position relative to the ORF7 translation initiation codon, Lelystad virus uses only the most downstream box to join leader and body, while VR-2332 preferentially uses the most upstream site and to a limited extent also the most downstream site. Interestingly, neither strain uses the middle potential TRS, in spite of its perfect match with the leader TRS in both isolates. Finally, it is worth mentioning that also some strains of LDV generate an extra sg mRNA (mRNA1-1), which is derived from a “body TRS-like” sequence located in the 3′-terminal region of the replicase gene (5, 18). The functionality of this mRNA remains to be investigated.
The use of alternative body TRSs for EAV mRNA3, -4, and -5.
In this report, we have described novel alternative body TRSs that are used for the generation of alternative subspecies of EAV mRNA4 and -5. In both cases, as well as for the previously described alternative body TRSs for RNA3, all TRSs are located upstream of the translation initiation codons of the respective ORFs. Thus, sg mRNAs produced from these TRSs can be used to express the corresponding structural proteins (Fig. 7A). While there are no alternative AUG codons downstream of the most-upstream body TRSs for RNA3 and -4 (TRS 3.1 and 4.1), there is an AUG codon between TRSs 5.1 and 5.2 that is followed by a potential ORF of 59 codons, which is in the −1 frame relative to the ORF that encodes the GP5 protein. However, this start codon is in an unfavorable context, and in this study, the GP5 protein has been shown to be translated from RNA5.1. Thus, all subspecies of EAV mRNA3, -4, and -5 most likely express the same glycoprotein, i.e., GP3, GP4, or GP5, respectively.
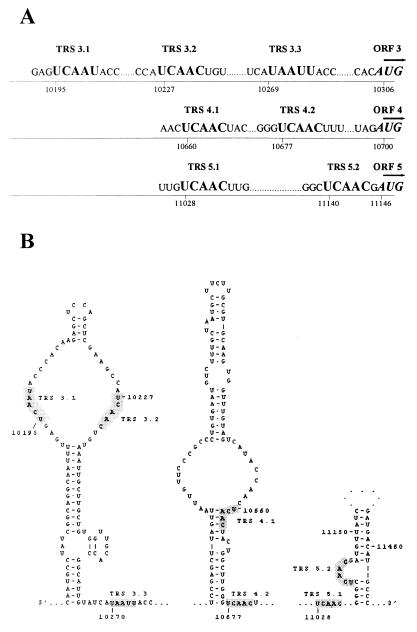
FIG. 7.
(A) RNA3, -4, and -5 body TRS regions. The sequences are aligned with respect to the translation initiation for the corresponding structural protein. (B) Putative secondary structure of the RNA3, -4, and -5 body TRS regions. The predicted structures are shown only for the regions immediately adjacent to TRSs; other regions are indicated by dots. Additional 3-bp segments at positions 10193 to 10195 and 10230 to 10232 (not shown), possible in the loop containing TRS 3.1 and TRS 3.2, provide a gain in free energy of only −1.3 kcal/mol and therefore are estimated to be rather labile. TRSs are shaded.
The fact that the mRNA4.1 and -5.2 subspecies went unnoticed in previous studies can be explained by their low abundance in infected cells and, for TRS 5.2, by the fact that an antisense primer located upstream of this TRS was used for a previous mRNA5-specific RT-PCR analysis (6). The activity of TRS 5.2 was recently also reported by de Vries et al. (10) but was not quantitated in any detail. Our semiquantitative RT-PCR experiments (Fig. 5B) now revealed that RNA5.2 is 125- to 625-fold less abundant than RNA5.1. de Vries et al. (10) further suggested that TRS 5.2 may be less active in leader-body joining because it contains a G residue at the sixth position. We have recently shown that nucleotides at position 6 of the body and leader TRSs are indeed important for discontinuous transcription (A. O. Pasternak, E. van den Born, J. C. Dobbe, W. J. M. Spaan, and E. J. Snijder, unpublished data). In general, however, sequence differences between individual TRSs cannot explain the differences in molar amounts between different EAV mRNA (sub)species, including those described in this report. For example, body TRS 4.1 has a perfect 8-nt match with the leader TRS but is nevertheless 40 times less active than TRS 4.2, which has a match of only 6 nt. Sequence identity at positions 7 and 8 of the leader and body TRSs also can be important for body TRS activity (A. O. Pasternak et al., unpublished data). Therefore, if the primary sequence of a body TRS was the principal determinant of its activity, RNA4.1 would be more abundant than RNA4.2. In fact, as observed in this study, TRS 4.1, in spite of its 8-nt match with the leader TRS, is approximately as active as the noncanonical TRS 3.3 (data not shown). Furthermore, the EAV genome, as well as the genomes of other arteriviruses, contains several potential body TRSs (5′ UCAAC 3′ sequences for EAV) that match the leader TRS but that are not used, at least not at detectable levels, for leader-body joining. Thus, the mere presence of sequence 5′ UCAAC 3′ in the EAV genome is clearly not sufficient to direct the discontinuous transcription of an sg mRNA. Our results, as well as other studies on nidovirus mRNA transcription (28, 42), suggest that, although the TRS primary sequence is very important for leader-body joining, differences in body TRS activities cannot be explained by just the number of nucleotides that match the leader TRS. Other factors, such as flanking sequences and/or RNA secondary structure, are likely to influence or determine the activities of different body TRSs.
RNA structure of the plus-strand template may determine body TRS activity during discontinuous minus-strand transcription.
It has been proposed that nidovirus discontinuous transcription resembles similarity-assisted RNA recombination (3, 27, 46). During this process, a nascent minus strand is translocated from a donor (body) template to the acceptor (leader) template, a process that may be guided by sequence similarity between the leader and body TRSs and by the secondary structure of the TRS regions. The leader TRS in the genome was predicted to be located in a loop “presented” by a striking hairpin structure (46), a mechanism that would resemble a number of antisense RNA-regulated control mechanisms that are based on interactions between single-stranded tails and hairpin loops (reference 46 and references therein). The 3′ part of the stem of the leader TRS hairpin is located downstream of the TRS, and therefore it is only present in the genomic plus strand. Consequently, the formation of similar, alternative leader TRS hairpins in one or multiple subgenomic mRNAs is considered unlikely. Therefore, if the leader TRS hairpin indeed directs the reinitiation of transcription, the genomic leader in the (plus) sense would be the sole acceptor molecule during discontinuous minus-strand synthesis. The frequency with which the nascent strand is released from the body TRS, a factor that is likely to be a major determinant of the transcriptional activity of a TRS, may be determined by the local secondary structure of the plus-strand template, from which the minus-strand body is transcribed. To investigate this issue, we generated RNA structure predictions for all EAV body TRS regions, including those described in this report. An intriguing relationship was revealed: body TRSs that are used to produce major sg RNA species tend to be located in single-stranded regions of the predicted structure of the genomic template, while less-active and inactive potential TRSs (5′ UCAAC 3′ boxes) are fully or partially base paired. This suggests that the RNA structure of the donor template may regulate the discontinuous step in minus-strand RNA synthesis. A single-stranded body TRS may be more “open” for interactions that can promote attenuation of minus-strand synthesis and release of the nascent strand. Clearly, the possibility that the structure of the nascent minus strand itself may also play a role in this process cannot be excluded. Experiments to test this hypothesis in the EAV system are currently under way.
To illustrate the principle outlined above, Fig. 7B shows the RNA structure predictions for the RNA3, -4, and -5 body TRS regions. TRSs 3.1 and 3.2 are symmetrically located at opposite sides of a large internal loop, a finding that seems to correspond to their comparable transcriptional activities. TRS 3.3 resides in a large multibranch loop (which is not shown in Fig. 7B), which may explain its activity in spite of the reduced sequence similarity with the leader TRS. The difference in activity between TRSs 4.1 and 4.2 can be explained by the fact that the latter is fully open, whereas the former is partially closed (except for nt 1 and 2). For RNA5, TRS 5.1 lies in a large single-stranded region, whereas TRS 5.2 is located in a bulge and is probably shielded by base pairs in immediately flanking sequences. In this case, however, the reduced activity of this TRS can also be explained by a mismatch with the leader at the sixth position.
Further supporting this hypothesis, TRSs of other arteriviruses, in particular LDV and PRRSV, were also predicted to be mostly single stranded (data not shown). Interestingly, structure predictions of the context of the RNA7 body TRSs of two PRRSV strains, Lelystad virus and VR-2332, can at least partially explain the observations of Nelsen et al. (28) (data not shown).
Role of alternative body TRSs in the viral replication cycle.
In this paper, we have functionally tested the significance of the use of alternative body TRSs for the transcription of EAV RNA3, -4, and -5 using both biochemical and virological assays. We have been able to show a strict quantitative correlation between the levels of these individual sg mRNAs and production of infectious progeny virus. Recently, we have reported that the expression of all EAV proteins encoded by ORF2a to -7 is required for production of infectious progeny virus (26). The present study not only confirmed this for at least three of these proteins, GP3, GP4, and GP5, but also revealed that they are produced by the virus in optimized quantities. For example, for mutants M3.1 and M3.2, a twofold reduction in the amount of mRNA3 (and that of the corresponding structural protein) resulted in a corresponding reduction of the titer of infectious progeny virus (Table 2, M3.1 and M3.2). This suggests that the expression of EAV structural proteins is tightly regulated at the level of sg RNA synthesis. By determining the rate of minus-strand release at a body TRS, the RNA structure of the plus-strand template may be part of such a regulatory mechanism.
Having determined the relative transcriptional activities of alternative body TRSs, we observed that TRSs 3.3, 4.1, and 5.2 do not contribute much to the transcription of the corresponding sg mRNA species. This raises the question of why these minor body TRSs are maintained in the viral genome when the major body TRS(s) suffices to produce 90 to 99% of the amount of a given sg mRNA species. Maybe this feature has evolved as a defense mechanism against spontaneous mutations in body TRSs, which would otherwise kill the virus and which may now have a chance to revert more easily. It is unclear why mRNA6 and -7, which express major EAV structural proteins, have not been provided with such a mechanism. In contrast to that of EAV, RNA7 of PRRSV isolate VR-2332 is made from two alternative body TRSs, one of which appears to produce much less mRNA7 than the other, but in PRRSV only one TRS appears to be used for mRNA3 transcription (28).
Alternatively, once a structural protein gene is provided with one “strong” TRS (or, for EAV RNA3, two TRSs of medium strength) upstream of its translation initiation codon, the presence of additional leader-body junction sites in this region may be a chance event. If “TRS-like” (or even noncanonical) sequences in this region are situated in a favorable structural context, they can serve as additional leader-body junction sites; if not, they remain silent. The finding of a number of functional leader-body junction sites (none of which was a canonical TRS) in the heterologous green fluorescent protein gene, which was inserted in the genome of coronavirus mouse hepatitis virus (13), suggests that sometimes the presence of a leader-body junction site may be evolutionarily uncoupled from the presence of a translation initiation site for a structural protein and may be determined solely by an appropriate structural context of this site. In contrast, no leader-body junction site is present between the initiation codons of the structural proteins E and GP2b of EAV (encoded by ORF2a and -2b), and therefore the latter protein is assumed to be expressed from the second cistron of EAV mRNA2 (35). This indicates that sometimes selection for certain codons in the upstream ORF may prevent the formation of a body TRS for the next sg mRNA.
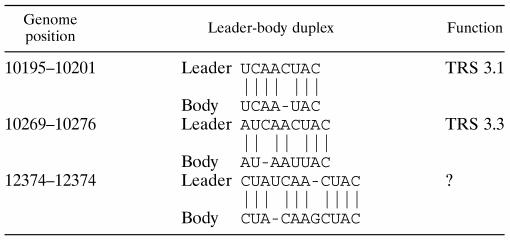
The results presented above prompted us to search for other possible leader-body junction events in the 3′ end of the genome. By using sensitive RT-PCR techniques similar to those described above (data not shown), we have detected four additional minor sg RNA species produced from TRSs situated in the 3′ end of the replicase reading frame (nt 8646 to 8648 and 9623 to 9627) and close to the RNA7 TRS region (nt 12184 to 12188 and 12374 to 12384). The latter TRS does not contain a canonical UCAAC; instead, it has extended possibilities for duplex formation with the leader TRS region at flanking nucleotides. Interestingly, no leader-body junction events involving the UCAAC boxes located at position 9557 to 9561 and 12489 to 12493 could be detected. Though RNA transcribed from the TRSs located at positions 9623 to 9627 and 12184 to 12188 can, in principle, be used for translation of E and GP2 or N proteins, respectively, no infectious progeny virus was detected when the major RNA2 and RNA7 body TRSs were knocked out, meaning that these UCAAC boxes apparently do not have a significant biological role.
Table 3 combines the data on the usage of EAV TRS-like sequences. The EAV genome contains 18 5′ UCAAC 3′ boxes, 13 of which are located in the 3′-terminal one-third of the genome. Interestingly, only two of these 13 UCAAC boxes are apparently silent (or produce undetectable amounts of sg RNAs). In addition, Table 4 includes leader-body duplex possibilities for three noncanonical junction points. Note that in each case, base pairing possibilities extend beyond the usual 5-nt duplex and involve flanking sequences. Together, the number of leader-body junction sites in the 3′ one-third of the genome is 14, meaning a twofold redundancy relative to the number of structural protein genes.
TABLE 3.
EAV leader-to-body junction sites (UCAAC boxes)
| Genome position | Sequence | Function | Used for leader-to body junction? |
|---|---|---|---|
| 207–211 | UCAACU | Leader TRS | Yes |
| 1567–1571 | UCAACA | Not tested | Not tested |
| 2437–2441 | UCAACA | Not tested | Not tested |
| 2760–2764 | UCAACU | Not tested | Not tested |
| 7911–7915 | UCAACA | Not tested | Not tested |
| 8646–8650 | UCAACC | ? | Yes |
| 9557–9561 | UCAACU | No function | No |
| 9623–9627 | UCAACA | ? | Yes |
| 9711–9715 | UCAACU | TRS 2 | Yes |
| 10227–10231 | UCAACU | TRS 3.2 | Yes |
| 10660–10664 | UCAACU | TRS 4.1 | Yes |
| 10677–10671 | UCAACU | TRS 4.2 | Yes |
| 11028–11032 | UCAACU | TRS 5.1 | Yes |
| 11140–11144 | UCAACG | TRS 5.2 | Yes |
| 11870–11874 | UCAACC | TRS 6 | Yes |
| 12184–12188 | UCAACC | ? | Yes |
| 12252–12256 | UCAACU | TRS 7 | Yes |
| 12489–12493 | UCAACA | No function | No |
TABLE 4.
Noncanonical leader-to-body junction sites
The data described in this and other studies have modified our understanding of arterivirus transcription. We and others have now firmly established that many structural proteins can be translated from more than one RNA species. Eleven out of 13 5′ UCAAC 3′ boxes in the 3′ end of the EAV genome and also three noncanonical junction sites, are used for the synthesis of sg RNA molecules, some of which are fully redundant for the viral life cycle. Our findings suggest that the amount of a structural protein and production of infectious progeny virus are determined by the combined activity of all TRSs situated upstream of its translation initiation codon. We propose that the RNA structure of the donor template (or nascent strand) and leader-body duplex formation possibilities (not necessarily involving the actual pentanucleotide box) regulate this activity by determining the relative portion of nascent minus strands which are joined to the antileader sequence in a recombination-like strand transfer reaction. The absolute TRS activity would, of course, also depend on the molar amount of the substrate of this reaction (nascent minus strands), and thus on the position of the TRS relative to the other TRSs.
ACKNOWLEDGMENTS
We are grateful to Udeni Balasuryia, James Maclachlan, Twan de Vries, and Peter Rottier for providing monoclonal antibodies and rabbit antisera. We thank Kees Pleij, Marieke Tijms, and Richard Molenkamp for helpful discussions.
A.O.P. was supported by grant 700-31-020 from the Council for Chemical Sciences of The Netherlands Organization for Scientific Research.
REFERENCES
- 1.Balasuriya U B, Patton J F, Rossitto P V, Timoney P J, McCollum W H, MacLachlan N J. Neutralization determinants of laboratory strains and field isolates of equine arteritis virus: identification of four neutralization sites in the amino-terminal ectodomain of the G(L) envelope glycoprotein. Virology. 1997;232:114–128. doi: 10.1006/viro.1997.8551. [DOI] [PubMed] [Google Scholar]
- 2.Baric R S, Stohlman S A, Lai M M. Characterization of replicative intermediate RNA of mouse hepatitis virus: presence of leader RNA sequences on nascent chains. J Virol. 1983;48:633–640. doi: 10.1128/jvi.48.3.633-640.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brian D A, Spaan W J M. Recombination and coronavirus defective interfering RNAs. Semin Virol. 1997;8:101–111. doi: 10.1006/smvy.1997.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 5.Chen Z, Kuo L, Rowland R R, Even C, Faaberg K S, Plagemann P G. Sequences of 3′ end of genome and of 5′ end of open reading frame 1a of lactate dehydrogenase-elevating virus and common junction motifs between 5′ leader and bodies of seven subgenomic mRNAs. J Gen Virol. 1993;74:643–659. doi: 10.1099/0022-1317-74-4-643. [DOI] [PubMed] [Google Scholar]
- 6.den Boon J A, Kleijnen M F, Spaan W J, Snijder E J. Equine arteritis virus subgenomic mRNA synthesis: analysis of leader-body junctions and replicative-form RNAs. J Virol. 1996;70:4291–4298. doi: 10.1128/jvi.70.7.4291-4298.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.den Boon J A, Snijder E J, Chirnside E D, de Vries A A, Horzinek M C, Spaan W J. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Vries A A, Chirnside E D, Bredenbeek P J, Gravestein L A, Horzinek M C, Spaan W J. All subgenomic mRNAs of equine arteritis virus contain a common leader sequence. Nucleic Acids Res. 1990;18:3241–3247. doi: 10.1093/nar/18.11.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Vries A A, Chirnside E D, Horzinek M C, Rottier P J. Structural proteins of equine arteritis virus. J Virol. 1992;66:6294–6303. doi: 10.1128/jvi.66.11.6294-6303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Vries A A, Glaser A L, Raamsman M J, de Haan C A, Sarnataro S, Godeke G J, Rottier P J. Genetic manipulation of equine arteritis virus using full-length cDNA clones: separation of overlapping genes and expression of a foreign epitope. Virology. 2000;270:84–97. doi: 10.1006/viro.2000.0245. [DOI] [PubMed] [Google Scholar]
- 11.de Vries A A, Horzinek M C, Rottier P J M, de Groot R J. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin Virol. 1997;8:33–47. doi: 10.1006/smvy.1997.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doll E R, Bryans J T, McCollum W H, Crowe M E W. Isolation of a filterable agent causing arteritis of horses and abortion by mares. Its differentiation from the equine abortion (influenza) virus. Cornell Vet. 1957;47:3–41. [PubMed] [Google Scholar]
- 13.Fischer F, Stegen C F, Koetzner C A, Masters P S. Analysis of a recombinant mouse hepatitis virus expressing a foreign gene reveals a novel aspect of coronavirus transcription. J Virol. 1997;71:5148–5160. doi: 10.1128/jvi.71.7.5148-5160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godeny E K, de Vries A A, Wang X C, Smith S L, de Groot R J. Identification of the leader-body junctions for the viral subgenomic mRNAs and organization of the simian hemorrhagic fever virus genome: evidence for gene duplication during arterivirus evolution. J Virol. 1998;72:862–867. doi: 10.1128/jvi.72.1.862-867.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gultyaev A P, van Batenburg F H, Pleij C W. The computer simulation of RNA folding pathways using a genetic algorithm. J Mol Biol. 1995;250:37–51. doi: 10.1006/jmbi.1995.0356. [DOI] [PubMed] [Google Scholar]
- 16.Hedges J F, Balasuriya U B, MacLachlan N J. The open reading frame 3 of equine arteritis virus encodes an immunogenic glycosylated, integral membrane protein. Virology. 1999;264:92–98. doi: 10.1006/viro.1999.9982. [DOI] [PubMed] [Google Scholar]
- 17.Joo M, Makino S. Mutagenic analysis of the coronavirus intergenic consensus sequence. J Virol. 1992;66:6330–6337. doi: 10.1128/jvi.66.11.6330-6337.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo L, Chen Z, Rowland R R, Faaberg K S, Plagemann P G. Lactate dehydrogenase-elevating virus (LDV): subgenomic mRNAs, mRNA leader and comparison of 3′-terminal sequences of two LDV isolates. Virus Res. 1992;23:55–72. doi: 10.1016/0168-1702(92)90067-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai M M, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landt O, Grunert H P, Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990;96:125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- 21.MacLachlan N J, Balasuriya U B, Hedges J F, Schweidler T M, McCollum W H, Timoney P J, Hullinger P J, Patton J F. Serologic response of horses to the structural proteins of equine arteritis virus. J Vet Diagn Investig. 1998;10:229–236. doi: 10.1177/104063879801000302. [DOI] [PubMed] [Google Scholar]
- 22.Makino S, Joo M, Makino J K. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J Virol. 1991;65:6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makino S, Soe L H, Shieh C K, Lai M M. Discontinuous transcription generates heterogeneity at the leader fusion sites of coronavirus mRNAs. J Virol. 1988;62:3870–3873. doi: 10.1128/jvi.62.10.3870-3873.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng X J, Paul P S, Morozov I, Halbur P G. A nested set of six or seven subgenomic mRNAs is formed in cells infected with different isolates of porcine reproductive and respiratory syndrome virus. J Gen Virol. 1996;77:1265–1270. doi: 10.1099/0022-1317-77-6-1265. [DOI] [PubMed] [Google Scholar]
- 25.Meulenberg J J, de Meijer E J, Moormann R J. Subgenomic RNAs of Lelystad virus contain a conserved leader-body junction sequence. J Gen Virol. 1993;74:1697–1701. doi: 10.1099/0022-1317-74-8-1697. [DOI] [PubMed] [Google Scholar]
- 26.Molenkamp R, van Tol H, Rozier B C D, van der Meer Y, Spaan W J, Snijder E. The arterivirus replicase is the only viral protein required for genome replication and subgenomic RNA transcription. J Gen Virol. 2000;81:2491–2496. doi: 10.1099/0022-1317-81-10-2491. [DOI] [PubMed] [Google Scholar]
- 27.Nagy P D, Simon A E. New insights into the mechanisms of RNA recombination. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- 28.Nelsen C J, Murtaugh M P, Faaberg K S. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol. 1999;73:270–280. doi: 10.1128/jvi.73.1.270-280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pedersen K W, van der Meer Y, Roos N, Snijder E J. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J Virol. 1999;73:2016–2026. doi: 10.1128/jvi.73.3.2016-2026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Sawicki S G, Sawicki D L. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J Virol. 1990;64:1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawicki S G, Sawicki D L. Coronavirus use discontinuous extension for synthesis of subgenome-length negative strands. Adv Exp Med Biol. 1995;380:499–506. doi: 10.1007/978-1-4615-1899-0_79. [DOI] [PubMed] [Google Scholar]
- 33.Sethna P B, Hung S L, Brian D A. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc Natl Acad Sci USA. 1989;86:5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snijder E J, Meulenberg J J. The molecular biology of arteriviruses. J Gen Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- 35.Snijder E J, van Tol H, Pedersen K W, Raamsman M J, de Vries A A. Identification of a novel structural protein of arteriviruses. J Virol. 1999;73:6335–6345. doi: 10.1128/jvi.73.8.6335-6345.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snijder E J, Wassenaar A L, Spaan W J. Proteolytic processing of the N-terminal region of the equine arteritis virus replicase. Adv Exp Med Biol. 1993;342:227–232. doi: 10.1007/978-1-4615-2996-5_36. [DOI] [PubMed] [Google Scholar]
- 37.Snijder E J, Wassenaar A L, Spaan W J. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J Virol. 1994;68:5755–5764. doi: 10.1128/jvi.68.9.5755-5764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spaan W, Delius H, Skinner M, Armstrong J, Rottier P, Smeekens S, van der Zeijst B A, Siddell S G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2:1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Batenburg F H, Gultyaev A P, Pleij C W. An APL-programmed genetic algorithm for the prediction of RNA secondary structure. J Theor Biol. 1995;174:269–280. doi: 10.1006/jtbi.1995.0098. [DOI] [PubMed] [Google Scholar]
- 40.van Berlo M F, Horzinek M C, van der Zeijst B A. Equine arteritis virus-infected cells contain six polyadenylated virus-specific RNAs. Virology. 1982;118:345–352. doi: 10.1016/0042-6822(82)90354-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Meer Y, van Tol H, Locker J K, Snijder E J. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J Virol. 1998;72:6689–6698. doi: 10.1128/jvi.72.8.6689-6698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Most R G, de Groot R J, Spaan W J. Subgenomic RNA synthesis directed by a synthetic defective interfering RNA of mouse hepatitis virus: a study of coronavirus transcription initiation. J Virol. 1994;68:3656–3666. doi: 10.1128/jvi.68.6.3656-3666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Dinten L C, Den Boon J A, Wassenaar A L, Spaan W J, Snijder E J. An infectious arterivirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc Natl Acad Sci USA. 1997;94:991–996. doi: 10.1073/pnas.94.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Dinten L C, Rensen S, Gorbalenya A E, Snijder E J. Proteolytic processing of the open reading frame 1b-encoded part of arterivirus replicase is mediated by nsp4 serine protease and is essential for virus replication. J Virol. 1999;73:2027–2037. doi: 10.1128/jvi.73.3.2027-2037.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Dinten L C, Wassenaar A L, Gorbalenya A E, Spaan W J, Snijder E J. Processing of the equine arteritis virus replicase ORF1b protein: identification of cleavage products containing the putative viral polymerase and helicase domains. J Virol. 1996;70:6625–6633. doi: 10.1128/jvi.70.10.6625-6633.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Marle G, Dobbe J C, Gultyaev A P, Luytjes W, Spaan W J, Snijder E J. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc Natl Acad Sci USA. 1999;96:12056–12061. doi: 10.1073/pnas.96.21.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Marle G, van Dinten L C, Spaan W J, Luytjes W, Snijder E J. Characterization of an equine arteritis virus replicase mutant defective in subgenomic mRNA synthesis. J Virol. 1999;73:5274–5281. doi: 10.1128/jvi.73.7.5274-5281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wassenaar A L, Spaan W J, Gorbalenya A E, Snijder E J. Alternative proteolytic processing of the arterivirus replicase ORF1a polyprotein: evidence that NSP2 acts as a cofactor for the NSP4 serine protease. J Virol. 1997;71:9313–9322. doi: 10.1128/jvi.71.12.9313-9322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]