Abstract
Amelogenesis imperfecta (AI) is an inherited developmental enamel defect affecting tooth masticatory function, esthetic appearance, and the well-being of patients. As one of the major enamel matrix proteins (EMPs), enamelin (ENAM) has three serines located in Ser-x-Glu (S-x-E) motifs, which are potential phosphorylation sites for the Golgi casein kinase FAM20C. Defects in FAM20C have similarly been associated with AI. In our previous study of EnamRgsc514 mice, the Glu57 in the S55-X56-E57 motif was mutated into Gly, which was expected to cause a phosphorylation failure of Ser55 because Ser55 cannot be recognized by FAM20C. The severe enamel defects in ENAMRgsc514 mice reminiscent of Enam-knockout mouse enamel suggested a potentially important role of Ser55 phosphorylation in ENAM function. However, the enamel defects and ENAM dysfunction may also be attributed to distinct physicochemical differences between Glu57 and Gly57. To clarify the significance of Ser55 phosphorylation to ENAM function, we generated two lines of Enam knock-in mice using CRISPR-Cas9 method to eliminate or mimic the phosphorylation state of Ser55 by substituting it with Ala55 or Asp55 (designated as S55A or S55D), respectively. The teeth of 6-day or 4-week-old mice were subjected to histology, micro-CT, SEM, TEM, immunohistochemistry, and mass spectrometry analyses to characterize the morphological, microstructural and proteomic changes in ameloblasts, enamel matrix and enamel rods. Our results showed that the enamel formation and EMP expression in S55D heterozygotes (Het) were less disturbed than those in S55A heterozygotes, while both homozygotes (Homo) had no mature enamel formation. Proteomic analysis revealed alterations of enamel matrix biosynthetic and mineralization processes in S55A Hets. Our present findings indicate that Asp55 substitution partially mimics the phosphorylation state of Ser55 in ENAM. Ser55 phosphorylation is essential for ENAM function during amelogenesis.
Keywords: Enamelin, FAM20C, Amelogenesis, Phosphorylation, Kinase, Biomineralization
Introduction
Amelogenesis imperfecta (AI) is a heterogeneous group of conditions characterized by inherited developmental enamel defects. The prevalence of AI in human varies widely depending upon the gene pool. Values of 1:14,000 in the USA to 1:700 in Sweden have been reported [1,2]. AI enamel is abnormally thin, soft, fragile, pitted and/or discolored, causing patients to suffer from functional and psychological problems such as early tooth loss, eating difficulties, embarrassment, and pain. Normal enamel formation results from highly orchestrated extracellular processes involving matrix proteins, proteases, and mineral ion fluxes [3,4]. Three principal EMPs that ameloblasts secrete, amelogenin (AMEL), ameloblastin (AMBN), and enamelin (ENAM), are considered to control multiple steps in the crystallization of hydroxyapatite [5]. Matrix metallopeptidase 20 (MMP20) secreted by ameloblasts at the secretory stage and kallikrein-related peptidase 4 (KLK4) at the maturation stage are two key proteinases within the enamel matrix that proteolytically degrade EMPs [3]. Each of these key EMPs has been shown to be essential for proper enamel formation using knockout mouse models [6–10]. During amelogenesis, the secretion and post-translational modification processes of EMPs are tightly controlled. Disturbance of any of these procedures may cause AI.
The EMPs belong to the secreted calcium-binding phosphoproteins (SCPPs) superfamily, in which each member has one or several Ser-x-Glu/pSer (S-x-E/pS) motifs, wherein Ser phosphorylation is considered to promote binding of the peptide to hydroxyapatite (HA) in the regulation of skeletal and dental mineralization [11–14]. FAM20C is the primary kinase phosphorylating the serine residue within the S-x-E/pS motif in a broad spectrum of secretory proteins, including SCPPs [15,16]. Conditional ablation of FAM20C from ameloblasts or mutations in FAM20A, a FAM20C co-factor, caused severe enamel and ameloblasts defects reminiscent of EMP knockout mice [17,18], highlighting the importance of EMP phosphorylation for enamel formation. Indeed, studies have shown that phosphorylation of the single phosphorylation site in AMEL greatly enhanced the capacity of AMEL to stabilize amorphous calcium phosphate and prevented apatite crystal formation both in vitro and in vivo [19–21]. In line with this, mutations of the phosphorylation sites in AMEL and several other EMPs resulted in AI in mice and humans [22–24].
In comparison with the knowledge about the role of AMEL phosphorylation in amelogenesis [21], the functional significance of Ser phosphorylation in ENAM has not been well understood. Three highly conserved putative pSer sites have been identified in the ENAM of pigs (Ser53, Ser191, Ser216), mice (Ser 55, Ser196, Ser219), and humans (Ser54, Ser191, and Ser216) [22]. The S216L mutation in humans was reported to cause minor pitting or localized enamel hypoplasia in heterozygotes and severe enamel malformation with little or no mineral covering dentin in compound heterozygotes [24]. N-ethyl-N-nitrosourea- (ENU-) induced S55I or E57G mutations in the S55-x-E57 motif of murine ENAM resulted in AI-like enamel breakage [22,25]. It is anticipated that both S55I and E57G mutations cause phosphorylation failure on Ser55 by either preventing phosphorylation at this site or making this site unrecognizable for FAM20C kinase after changing the S-x-E motif into S-x-G. However, it is controversial whether or not both S>I and E>G mutations significantly change the physicochemical properties of the original amino acids and that such changes may have an impact on the biological outcomes. To clarify the functional significance of phosphorylated Ser55 in murine ENAM, we have generated knock-in mice to mutate Ser55 into S55A or S55D to either eliminate or mimic the phosphorylation state of this residue in vivo.
Materials and methods
Animals
All animal procedures were approved by the Institutional Animal Care and Use Committee of Texas A&M University College of Dentistry (Dallas, TX, USA) and performed following the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
S55A and S55D mice were generated by homology-directed repair using CRISPR-Cas9 technology (Fig. 1A). The exons and splicing junctions of the Enam gene were sequenced to exclude off-target mutations (Fig. 1B). Genomic DNA was extracted from tail lysates by standard protocol [26] and subjected to PCR genotyping using primers: Forward, 5’- CTAAGGCTCTGCATTTCTCCCTCAG-3’, Reverse, 5’- AACACTGAAACACATAGCAGGCTCG-3’. The wild-type PCR amplicon was 274 bp with no restriction enzyme site at the Ser55 location. The S55A mutation created an AluI restriction site, and digestion of the S55A PCR products with AluI created 206 bp and 68 bp fragments; The S55D mutation created a BtsCI restriction site, and digestion of the S55D PCR products with BtsCI gave rise to 201 bp and 73 bp fragments. The PCR and digestion products were examined on 3% agarose gel (Fig. 1C). All animals were weaned at 21 days and provided with gel food. Gross pictures of mouse incisors at 4-week-old were taken under a stereomicroscope (Olympus SZX2).
Fig. 1.
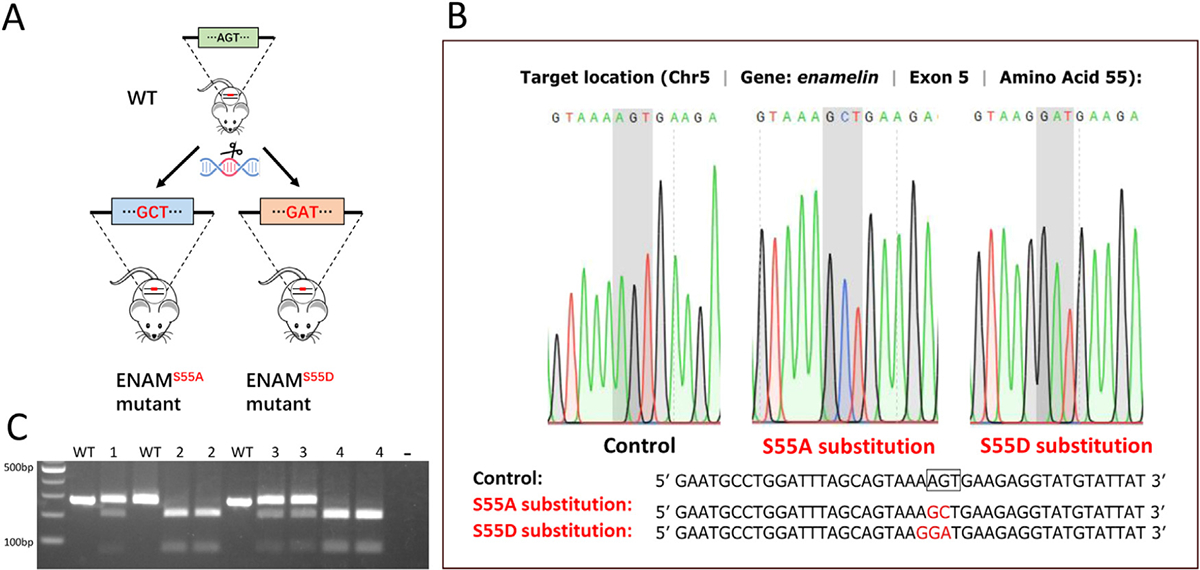
Generation of ENAM S55A and S55D knock-in mice. (A) Strategy of mutating ENAM S55 into S55A or S55D by CRISPR-Cas9 technique. (B) Sequencing chromatograms of WT mice, ENAM S55A, and ENAM S55D mutant mice. The shaded codons are Ser in control mice, Ala in S55A knock-in mice, and Asp in S55D knock-in mice. (C) AluI digestion of Enam PCR products from the S55A mice produced 206 bp and 68 bp fragments. BtsCI digestion of Enam PCR products from the S55D mice produced 201bp and 73 bp fragments. The 274 bp PCR products from WT mice cannot be cleaved by AluI or BtsCI restriction enzymes. Lanes: 1, S55A Het; 2, S55A Homo; 3, S55D Het; 4, S55D Homo.
Micro-computed tomography (micro-CT) analysis
Micro-CT analysis was performed as previously described [26]. Briefly, mandibles were dissected from 4-week-old mice and fixed in 4% paraformaldehyde (PFA), then stored in 0.5% PFA. Serial tomographic images were obtained by Micro-CT35 imaging system (Scanco Medical) at an energy level of 55 kV and intensity of 145 mA with a scanning interval of 7μm. Enamel structures were manually plotted for evaluation of the total volume and density. Scanco ISQ files were transformed into ids/ics files using ImageJ, and 3-D images were generated with iMaris (9.0.1, Bitplane). Virtual sections were made along the sagittal axial of lower incisors or first molars. Pseudo-colors were rendered onto the 3-D structures using the same parameters across samples.
Scanning electron microscopy (SEM)
Gross SEM and cross-section SEM analysis were performed as previously described [22,27]. Briefly, mandibles collected from postnatal 6 days (P6), P14 and 4-week-old mice in each group were fixed in 4% PFA at 4°C overnight and dehydrated through ethanol gradients. For cross-section SEM analysis, samples were embedded in methyl methacrylate resin, and were cross-sectioned using a slow-speed diamond saw at the level 8 site (located 8 mm from the apical end of the incisor and at the crest of alveolar bone near gingival margin) [27]. The dissected surface was made perpendicular to the enamel surface with minimal apical-to-incisal and mesial-to-lateral angulation. The surface of the section was then ground using 1200 grade silicon carbide abrasive paper and finely polished using a 0.5 μm diamond paste. Samples were further washed in distilled water and subjected to ultrasonic cleaning. After drying overnight, both gross and cross-sectioned mandible samples were coated with carbon. Gross mandible samples were examined using scanning electron microscope in field emission mode (JSM-6010LA, JEOL, Japan). Cross-sectioned incisor samples were observed under backscatter mode. Overview of the dissected surfaces were taken at × 250 magnification and overlapping images of the entire enamel layer from mesial to lateral CEJ were taken at × 800 magnification. The montage map showing the alignment of enamel rods were manually drew and colored in Adobe Photoshop (http://www.adobe.com).
Transmission electron microscopy (TEM)
The impact of the ENAM S55A and S55D mutations on the development of enamel mineral structures during amelogenesis was assessed by TEM at defined intervals as previously described [21]. Maxillary incisors from 4-week-old mice of each genotype were isolated by dissection and immediately fixed with 70% ethanol followed by dehydration through graded ethyl alcohol treatment. The incisors were then infused with a 1:1 solution of 100% ethyl alcohol and LR White resin (Hard Grade, Electron Microscopy Sciences) overnight, followed by two repeated two-hour infusions with the LR White solution. Treated incisors were embedded in the LR White resin using a cold cure method. Ultra-thin incisor sections (70–100 nm thick) were cut in the sagittal plane with an ultramicrotome (Leica, Buffalo Grove, IL) or using a diamond knife (DiATOME, Hatfield, PA) and floated into a pool of distilled water pre-saturated with HA with a few added drops of ethanol. The sections were immediately mounted on 200-mesh hexagonal carbon-coated copper TEM grids (EMS, Hatfield, PA). Developing enamel structures from each genotype were examined using TEM (JEM 1400-PLUS, JEOL, Peabody, MA) in bright-field mode at 100 kV. At least three maxillary incisors of each genotype were analyzed. A series of sequential TEM micrographs starting from the beginning of enamel secretion to the maturation stage were taken from each section (Supplementary Fig. 1).
Microhardness test
Mandibles from three 4-week-old mice per genotype were collected and embedded in resin as described above for TEM. Resin blocks were sectioned above the alveolar bone ridge using a diamond saw in a transverse plane. The erupted portions of the incisors were then polished with a series of SiC papers. The micro indentation tests were performed using IdentaMet 1105 microhardness tester (Buehler, Lake Bluff, Il) equipped with a CCD camera, connected to a computer equipped with OmniMet software (Buehler, Lake Bluff, Il). The tests were conducted using a Vickers hardness diamond tip with a load of 25 gf and dwell time of 5 sec. A standardized set of testing points of outer, middle and inner enamel were selected according to previously reported method [28]. The averages of measurements were used as the Vickers hardness numbers (HV) of individual specimens.
H&E staining and immunohistochemistry
Mandibles collected from 6-day-old mice were fixed in 4% PFA at 4°C for 16 h and then decalcified in 10% ethylenediaminetetraacetic acid (EDTA)/PBS at 4°C for five days, followed by dehydration and paraffin embedding. Serial sections with 4 μm thickness were prepared for H&E and immunohistochemistry staining as described previously [22,29]. The immunohistochemistry or immunofluorescence analyses were performed using primary antibodies against ENAM C-terminus (1:50, Santa Cruz Biotechnology Cat# sc-33107, RRID: AB_2098749), ENAM N-terminus (1:500) [30], AMBN (1:50, Santa Cruz Biotechnology Cat# sc-33100, RRID: AB_2226389) and AMEL (1:500, Santa Cruz Biotechnology Cat# sc-32892, RRID: AB_2226455). Primary antibodies were detected using biotinylated secondary antibodies (Vector Laboratories Cat# BA-5000, RRID:AB_2336126; Cat# BA-1000, RRID:AB_2313606) and VECTASTAIN ABC kit (Vector Laboratories Cat# PK-7100, RRID: AB_2336827) for color development with DAB substrate, or AlexaFluor488 conjugated secondary antibody (Thermo Fisher Scientific Cat# A-11034, RRID:AB_2576217) and AlexaFluor633 conjugated secondary antibody (Molecular Probes Cat# A-21082, RRID:AB_141493) for immunofluorescence. Slides were counterstained with methyl green or DAPI.
Mass spectrometry (MS) and proteomic profiling
Mandibular incisors were dissected from 6-day-old wild-type (WT), S55D, and S55A heterozygous mice under the stereomicroscope. Soft enamel matrix and outer enamel epithelium ameloblasts were dissected under stereomicroscope and lysed in RIPA buffer containing a proteinase inhibitor cocktail (ThermoFisher, 78429). After quantitation by bicinchoninic acid (BCA) protein assay, the lysates containing equal amounts of total protein from each group were loaded on a sodium dodecyl sulfate-polyacrylamide gel for electrophoresis (SDS-PAGE). Protein bands were stained with Coomassie Blue (ThermoFisher, 24594), cut off, and diced into ~1 mm3 pieces.
Samples were digested overnight with trypsin (Pierce), followed by reduction and alkylation with DTT and iodoacetamide (Sigma–Aldrich). The samples were then subject to solid-phase extraction cleanup with an Oasis HLB plate (Waters) and dried and reconstituted into 10 μl 2% CAN and 0.1% TFA. 2 μl of each sample was examined on a QExactive HF mass spectrometer coupled to an Ultimate 3000 RSLC-Nano liquid chromatography system. Samples were injected onto a 75 μm i.d., 15-cm long EasySpray column (Thermo) and eluted with a gradient of 0–28% buffer B over 90 min with a flow rate of 250 nL/min. Buffer A contained 2% (v/v) ACN and 0.1% formic acid in water. Buffer B contained 80% (v/v) ACN, 10% (v/v) trifluoroethanol and 0.1% formic acid in water. The mass spectrometer operated in positive ion mode with a source voltage of 2.4 kV and an ion transfer tube temperature of 275°C. MS scans were acquired at 120,000 resolution in the Orbitrap, and up to 20 MS/MS spectra were obtained for each full spectrum acquired using higher-energy collisional dissociation (HCD) for ions with charges 2–8. Dynamic exclusion was set for 20 s after an ion was selected for fragmentation.
Bioinformatics and data analysis
Raw MS data files were analyzed using Proteome Discoverer v2.4 (Proteome Discoverer, RRID: SCR_014477), with peptide identification performed using Sequest HT searching against the mouse protein database from UniProt (downloaded on Jan. 8, 2021). Fragment and precursor tolerances of 10 ppm and 0.02 Da were specified, and three missed cleavages were allowed. Carbamidomethylation of Cys was set as a fixed modification, with oxidation of Met set as a variable modification. The false-discovery rate (FDR) cutoff was 1% for all peptides. Proteins that were identified with only 1 PSM (peptide-spectrum match), or identified but not quantified, or with potential contaminants were listed but not included in the further analysis.
The data analysis was performed following the workflow of the DEP package [31]. Briefly, the Summarized Experiment object was generated, and proteins that were identified in all replicates of at least one group were filtered. The data was background corrected and normalized by variance stabilizing transformation (vsn). Missing values were imputed with “MiniProb”. Protein-wise linear models combined with empirical Bayes statistics were used for the differential expression analysis. Stacked bar plots were employed to visualize the distribution of significant conditions per protein and the overlap between conditions. The correlation matrix of all groups and significant proteins were plotted as heat-maps. Differential expressed proteins (DEPs) were plotted as volcano plots. Gene Ontology (GO) enrichment of DEPs was performed using GOnet [32] and visualized with GOplot [33].
Statistical analysis
Data were expressed as mean ± SD of at least three independent samples. Unpaired Student’s t-test was used to analyze data sets with two groups. When comparing more than two groups, P values were determined by one-way ANOVA followed by Tukey’s multiple comparison tests. Statistical calculations were performed using the Prism software package (GraphPad Prism7). *P<0.05, **P<0.01, ***P< 0.001, and ****P<0.0001 indicated a significant difference.
Results
ENAM Ser55 substitutions caused AI in a dosage-dependent manner
The amelogenesis of incisors and molars was compared between 4-week-old WT, ENAM S55A/D Hets and Homos. At the gross level, the enamel of WT mice showed light brown color with semi-translucency. Both incisor and molar enamel presented smooth surfaces with high density to resist the occlusal forces (Figs. 2A, 2F, 2K, 3A, and 3B).
Fig. 2.
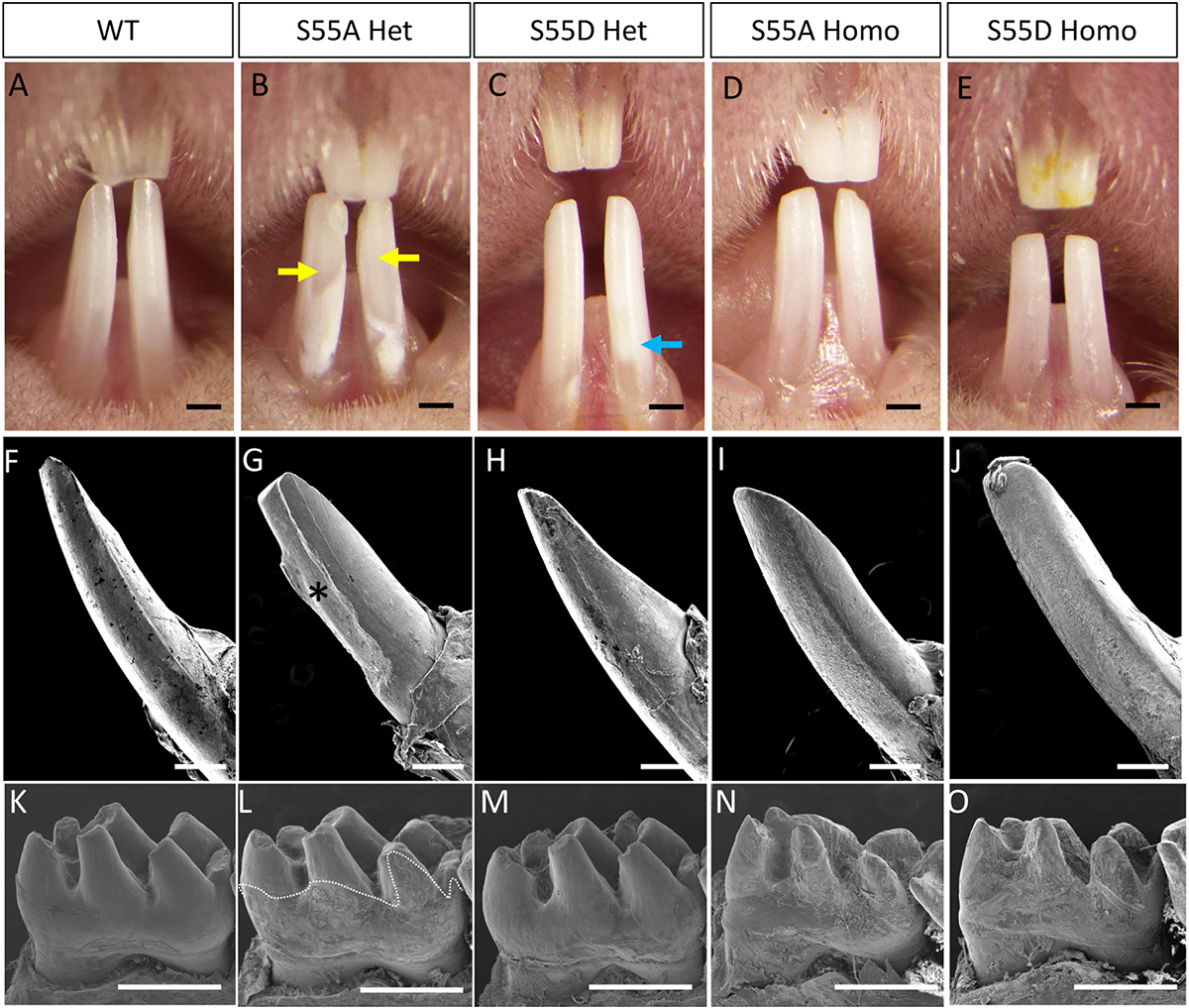
ENAM S55A/D substitution caused AI in a dosage-dependent manner. (A–E) Gross pictures of incisors in 4-week-old WT and ENAM S55A/D mutant mice. The yellow arrow indicates enamel breakage. The Blue arrow indicates white spots on the enamel. (F–J) SEM images of lower incisors of WT and ENAM S55A/D mutant mice. Asterisk indicates enamel crack off and dentin exposure. (K–O) SEM images of mandibular first molars of WT and ENAM S55A/D mutant mice. The dotted line shows the boundary between smooth surface of enamel and rough surface of dentin. Note that WT and S55D Het in (K) and (M) showed normal or nearly normal enamel with smooth surfaces, while S55A Het in (L) showed extensive enamel defects in the plotted areas. Both S55A and S55D Homo mutants in (N) and (O) did not form any mature enamel on dentin surfaces. Scale bars: 500 μm inA–O.
Fig. 3.
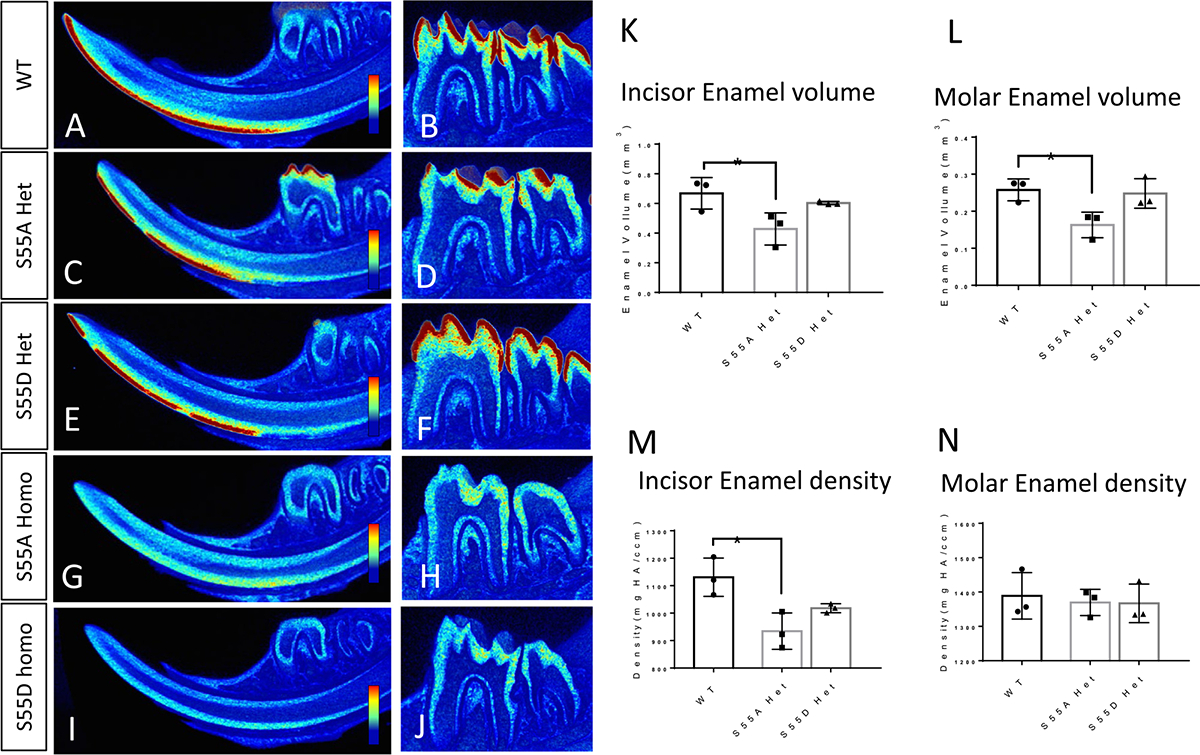
Micro-CT analyses of enamel in 4-week-old WT and ENAM S55A/D mutant mice. (A, C, E, G, and I) Virtual sagittal sections of lower incisors. Blue to red colors indicates the density of tissues from low to high. (B, D, F, H, and J) Virtual sagittal sections of mandibular first molar. (K and L) S55A Het showed less enamel volume than WT. (M) S55A Het showed lower enamel density than WT. (N) Molar enamel density did not show significant differences among WT, S55A, and S55D Het. *P<0.05, n=3.
Amelogenesis imperfecta was identified in both ENAM S55A and S55D Hets and Homos. In Het mice, the enamel defects were localized, while the severity and extent varied between S55A and S55D. The incisors of S55A Hets showed a chalky white color with large areas of enamel cracked off (Fig. 2B, G). In contrast, the S55D Hets had a nearly normal appearance of enamel in incisors with minor color and translucency changes (Fig. 2C, H). In S55A Het molars, the crown surfaces below molar cusps were rough. Less evident hypoplasia was only detected on molar cusps (Fig. 2L). In contrast, the molar enamel of S55D Hets was nearly intact, with a smooth surface covering the entire molar crown. The enamel defects were localized to the enamel-cementum junction shown as small rough areas (Fig. 2M). We scored the enamel defects based on the severity and extent of the defects. The results further indicated that the enamel defects of S55D Hets were more localized than that of S55A Hets (Supplementary Fig. 2).
In S55A and S55D Homos, the enamel defects were much more severe than those inHets that no mature enamel was detected on their incisors and molars (Figs. 2D, E, I, J, N, O, and 3G–J). However, small amounts of radiopaque materials along the incisal dentin surfaces and below the alveolar bone indicate immature enamel formation (Supplementary Fig. 3). The dentin of incisors and molars were exposed to the oral cavity to resist occlusal forces, making the dentin surfaces rough and incisal tips blunt. The more severe enamel phenotypes in ENAM S55 Homos than Hets indicated dosage-dependent effects of the mutations.
S55D substitution partially mimicked pSer55 function in ENAM
The volume and density of mature enamel were compared between S55A and S55D Hets by micro-CT analysis (red color in Fig. 3). The mature enamel of S55A Hets was discontinuous in the incisors (Fig. 3C), which is consistent with the enamel breakage defects observed at gross level (Fig. 2). The enamel volume of S55A Hets was significantly reduced in both incisors and molars compared to WT, while that of S55D Hets did not show significant differences from WT (Fig. 3A–F, K and L). The enamel volume of S55D Hets was slightly higher than S55A Hets but not statistically significant. The incisal tips of S55A Hets lost enamel, resulting in a blunter shape than WT and S55D Het (Fig. 3A, C, and E). The incisor enamel density of S55A Hets was significantly decreased compared to WT (Fig. 3M). S55D Hets showed a enamel density similar to WT and higher than S55A Hets, but no significant differences were identified between S55A and S55D Hets.
To evaluate the microstructural changes of enamel rods in S55A/D Hets, we performed scanning electron microscopy on the cross-sections of lower incisors cut at the crest of alveolar bone near gingival margin of mandibular first molars from 4-week-old mice (Fig. 4). In S55A Hets, the enamel rods aligned sparsely and discontinued, especially in the middle and outer enamel (Fig. 4B). The length of enamel rods and inter-rod spaces were increased compared to WT, and enamel rods detachment was observed (Fig. 4B1–3). In comparison, the enamel structures of S55D Hets were less affected (Fig. 4C), although their inter-rod spaces and rod length were slightly increased in some severe cases (Fig. 4C1–3). The enamel defects in S55A Hets were further indicated by the fabric changes of enamel matrix after acid etching of the enamel sections (Fig. 5).
Fig. 4.
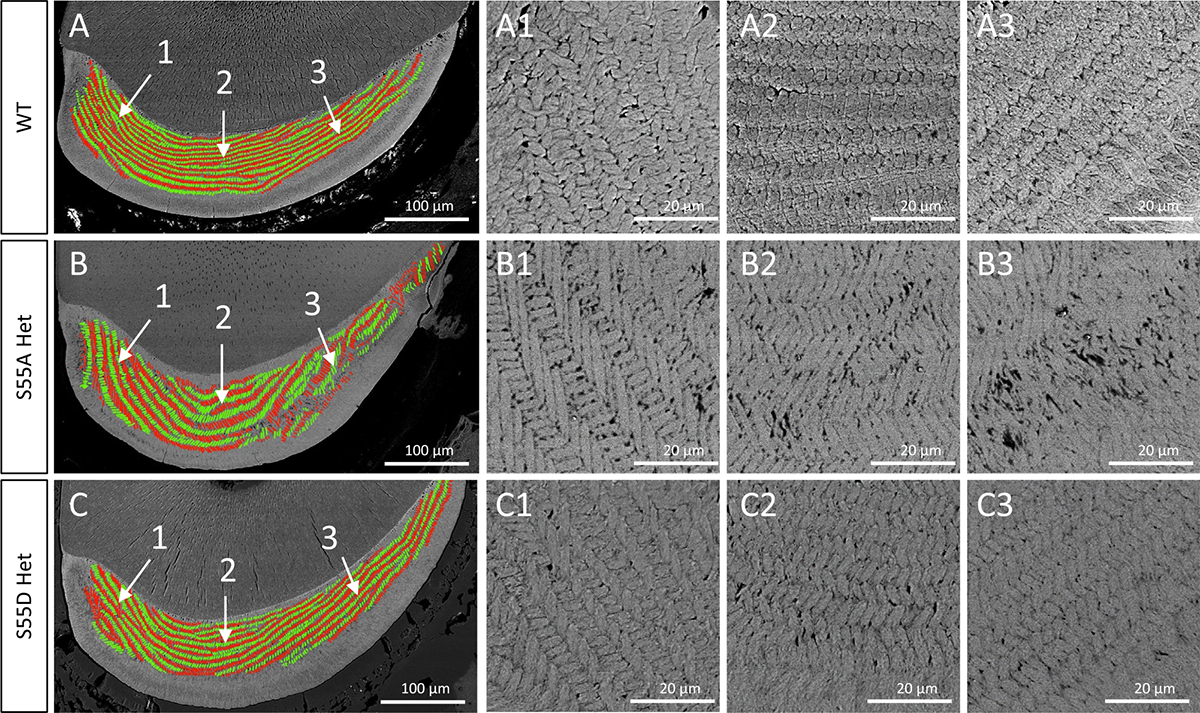
SEM analyses on the transections of lower incisors cut at the crest of alveolar bone near gingival margin of mandibular first molars from 4-week-old mice. In the left panel, enamel rods were color-coded green for rows having a medial tilt and red for rows having a lateral tilt. Sites 1,2,3 represent the medial, middle, and distal regions of incisor section. The panels on right side are higher resolution images (× 800) of the indicated regions in left panels.
Fig. 5.
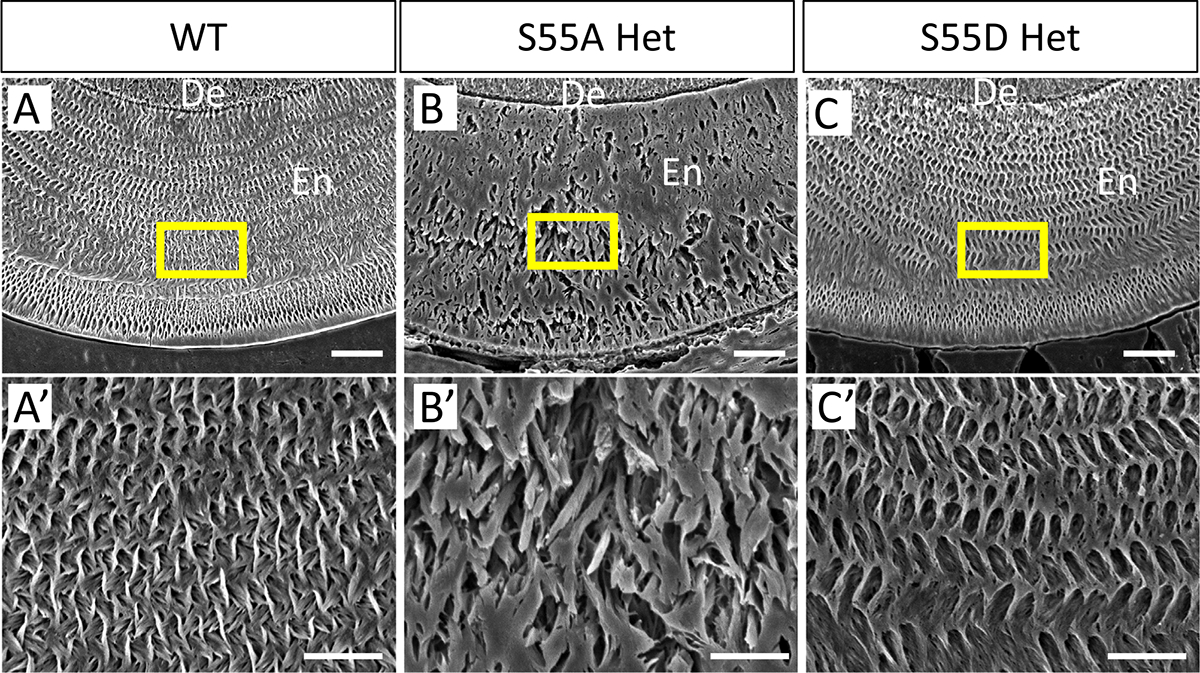
SEM analyses of the fabric pattern of enamel matrix in the lower incisors of 4-week-old mice after acid etching of transections cut at the mesial root level of the first lower molars. (A’–C’) Enlarged images of boxed areas in (A–C). The enamel matrix of S55A Hets showed disorganized fabric compared to WT and S55D Hets. De, dentin. En, enamel. Scale bars: 20um in A, B, and C; 10um in A’,B’, and C’.
To clarify if post-eruption changes such as attrition or chipping caused the enamel defects in S55A Hets (all mice were fed gel food after wean), we also performed micro-CT and SEM analyses on 2-week-old mice. The results are consistent with those obtained from 4-week-old mice (Supplementary Fig. 4), indicating that the enamel defects in S55A Hets were intrinsic.
To determine if the structural changes have an impact on the physical properties of enamel, we performed micro-hardness tests on the transections of lower incisors (Fig. 6). Vickers hardness number (HV) of erupted incisal enamel in WT was similar to the previously reported values [34,35]. The hardness of erupted incisal enamel was decreased in both S55A and S55D Hets compared to WT; S55A Het HV was significantly lower than S55D Hets throughout the enamel thickness. The micro-hardness results are consistent with those of microscopy, histology, SEM and micro-CT analyses.
Fig. 6.
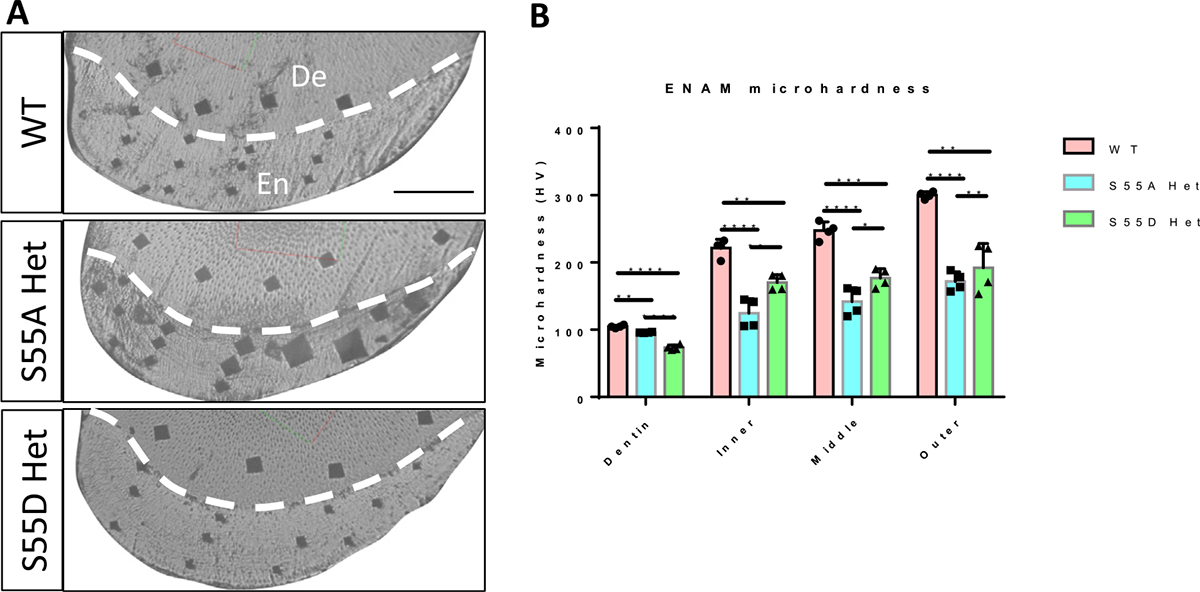
Micro-hardness test of incisors of 4-week-old mice. (A) Light micrographs taken from the indentation sites in the incisal enamel of WT, S55A Het, and S55D Het mice. De, dentin. En, enamel. Scale bar, 100um. (B) Statistic comparison of micro-hardness of incisal enamel at inner, middle and outer locations between WT, S55A Hets and S55D Hets. S55A Hets showed significantly lower enamel hardness than S55D Hets and WT. S55D Hets also showed decreased enamel hardness compared to WT. Both S55A and S55D Hets showed lower dentin hardness than WT. * P<0.05. ** P<0.01. *** P<0.005. **** P<0.001.
To confirm and further assess differences in enamel organization at the submicron scale, we conducted a TEM study on developing incisal enamel from the early-secretory stage to the maturation stage by taking a sequential series of TEM micrographs along the dentin-enamel junction (DEJ). The micrographs were taken in 11 open areas (grids) starting at the beginning of enamel deposition (Supplementary Fig. 1). In the WT, the normal decussating rod pattern was observed throughout the secretory stage of amelogenesis and across the enamel thickness (Fig. 7A, D, and G). At higher magnification, well-organized elongated mineral crystals were observed within each rod and the inter-rod matrix (Fig. 8A). In S55A Hets, enamel at the early-secretory stage rods and interrod were detected. However, they were less organized than WT enamel at the early-secretory stage (Fig. 7B). The analysis of the mid-secretory enamel in S55A Het revealed increased variability of enamel phenotypes and further deterioration of enamel organization (Fig. 7E). At the late-secretory stage, S55A Het enamel presented with a significantly distorted decussating pattern of the inner enamel and with outer enamel consisting of randomly organized short mineral crystals (Figs. 7H and 8C), and high-density mineral aggregates stated to appear in S55A Het enamel, both inside the rods and in spaces between the rods (Fig. 8C–E). Importantly, the thickness of the enamel layer of S55A Hets was significantly lower than in WT and S55D Hets (Fig. 7D–I). S55D Het enamel appeared similar to the WT (Fig. 7C, F, and I). However, at high magnification, the rods and inter rods appeared less organized (Fig. 8B).
Fig. 7.
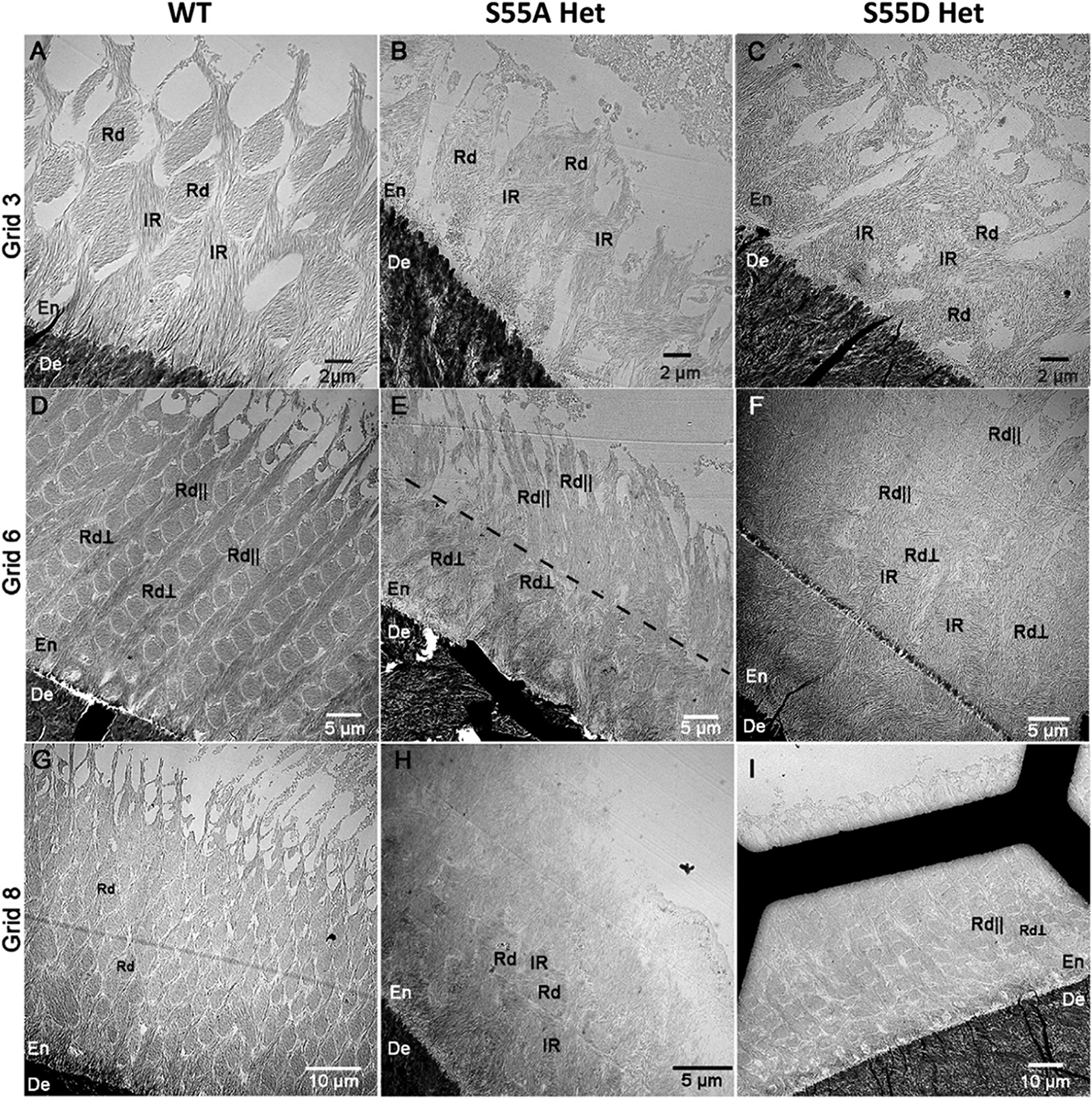
TEM micrographs of fully mineralized early (grid 6), mid (grid 6), and late (grid 8) secretory stage enamel. (A) Early-secretory stage enamel of WT mice. The enamel rods were sectioned in an oblique plane. Note the well-defined out-lines of the rods (Rd) surrounded by contiguous interrod matrix (IR). (B) Early-secretory stage of enamel in S55A Het mouse. The rods and interrod were less organized than in WT. (C) Early-secretory stage of enamel in S55D Het mouse. Similar to S55A Het, the rods and interrod were less organized than in WT. (D) Mid-secretory stage enamel of WT mice. Two sets of rods - one oriented in the section plane (Rd||) and another oriented perpendicular to the section plane (Rd⊥) were presented. (E) Mid-secretory stage enamel in S55A Het mouse. The section contains both Rd⊥ and Rd||. Importantly, the direction of the in-plane rods (Rd||) changed in this sample at roughly 15 um from the DEJ (dotted line). It is also worth noting that S55A Het enamel was significantly thinner than WT and S55D Het enamel at this stage. (F) Mid-secretory stage of enamel in S55D Het mouse. Enamel organization was similar to WT enamel at the mid-secretory stage. (G) Late-secretory enamel of WT mice with the rods sectioned in an oblique plane as in (A). (H) Late-secretory enamel of S55A Het mice. Note that the enamel layer was very thin, and the surface layer lacked enamel rods and was composed of short disorganized crystals. (I) Late-secretory enamel of S55D Het mice. The organization of enamel was similar to WT enamel in (D). En, enamel; De, dentin.
Fig. 8. TEM micrographs of the late-secretory stage enamel.
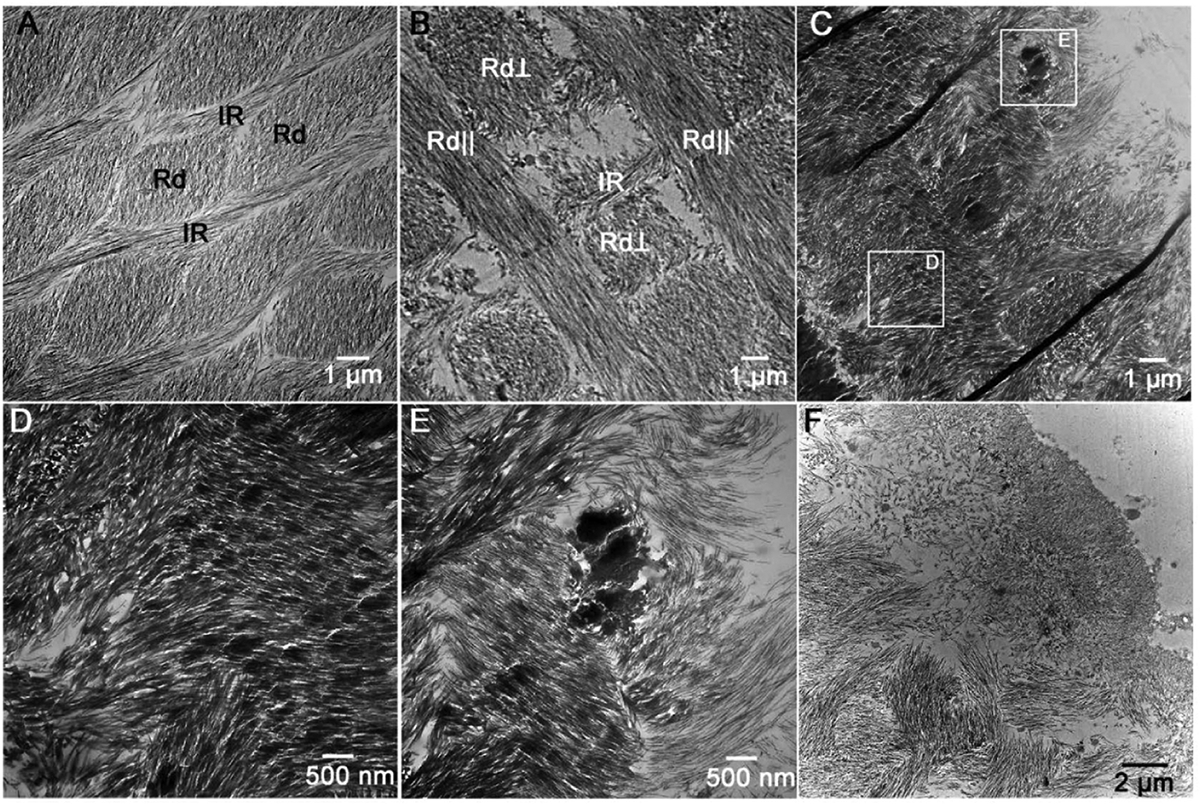
(A) WT enamel with well-defined obliquely sectioned discrete rods (Rd) and a contagious interrod matrix (IR). (B) S55D Het enamel with alternating rods oriented in the section plane (Rd||) and perpendicular to the section plane (Rd⊥). The structural organization of enamel was similar to WT, although individual enamel rods were less organized. (C) S55A Het enamel. Note that the decussating pattern could not be obscured by nodules of highly mineralized nodules between the rods (square E) and in the rod regions (square D). (D and E) Closeups of the regions E and D in (C). (F) Outer layer of S55A Het enamel containing short disorganized mineral crystals.
Histological changes in the ameloblasts of ENAM S55A and S55D mutant mice
To evaluate if ameloblast morphology and matrix secretion were affected in S55 mutant mice, we characterized mandibular incisors and molars in 6-day-old WT, S55A, and S55D mice at three well-defined amelogenesis stages known as the secretary, transition, and maturation phases. The ameloblasts alignment in S55A Hets started showing evident disorganization at the mid-secretory stage. (Fig. 9, Supplementary Fig. 5G and H). The interface between Tomes’ process and enamel matrix showed micro-bubbles (Supplementary Fig. 5I). Some ameloblasts detached from the enamel matrix at the late-secretory stage, changed into the stratified low-column epithelium, and secreted amorphous enamel matrix into the vacuolated spaces (Fig. 9C–D’, Supplementary Fig. 5I and J). In contrast, the ameloblasts in S55D Hets did not show apparent changes throughout the process of amelogenesis (Fig. 9E and F’, Supplementary Fig. 5K–O). However, early onset of severe defects in the ameloblasts and enamel matrix occurred in both S55A and S55D Homos (Fig. 9G–J’).
Fig. 9.
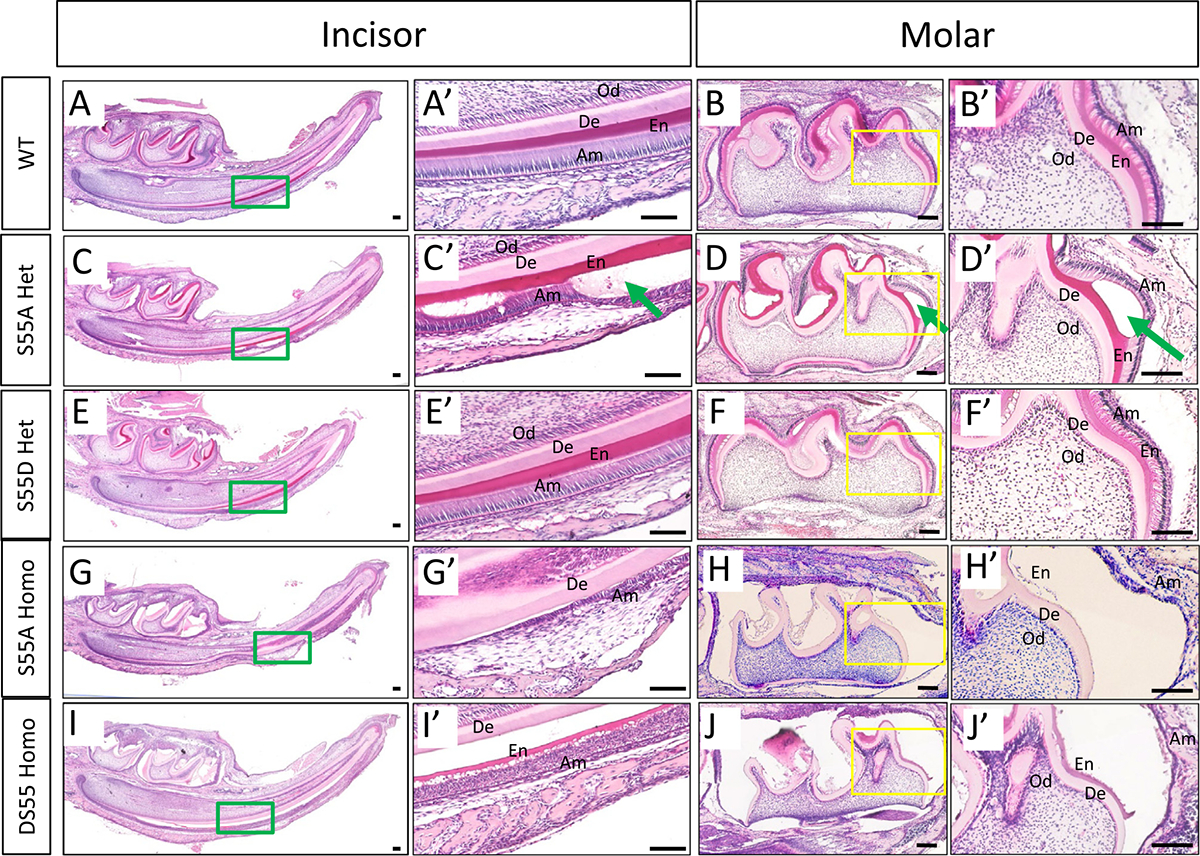
Morphological changes of ameloblasts and enamel matrix in P6 ENAM mutant mice. (A-B’) The ameloblasts in WT mice showed polarized high-column morphology at the late-secretory stage. The enamel matrix displayed smooth surfaces with an incremental thickness along the maturation axis. (C-D’) The ameloblasts in S55A Het mice detached from the dentin surface and started secreting amorphous substance (green arrow) at the late-secretory stage. (E-F’) The ameloblasts and enamel matrix in S55D Het mice showed nearly normal morphology. (G-H’) The ameloblasts in S55A Homo mice detached from the dentin surface and showed disorganized morphology. The enamel matrix remained in an amorphous state and could not form any tangible morphology. (I-J’) The ameloblasts and enamel matrix in S55D Homo mice showed severe defects similar to those in S55A Homo mice. De, dentin. Od, odontoblasts. En, enamel matrix. Am, ameloblasts. Scale bars: 100um.
At the secretary stage, columnar-shaped ameloblasts secrete enamel matrix proteins into enamel matrix, which act as scaffolds for enamel crystallization. After that, the full-length ENAM is cleaved into intermediate fragments by MMP20 and transported to specific locations in the enamel matrix [22]. Using anti-N- and anti-C-ENAM antibodies, we detected an accumulation of ENAM N-terminal fragments in the amorphous matrix secreted by ameloblasts detached from the enamel in S55A Hets (Fig. 10B, B’, E and E’). The boundary between aprismatic and prismatic enamel became blurry in S55A Hets (Fig. 10B’ and E’) compared to a clear one in WT (Fig. 10A’ and D’) and S55D Hets (Fig. 10C’ and F’). ENAM C-terminal fragments did not show apparent differences in the distribution patterns between WT and S55A/D Hets (Supplementary Fig. 6A). As S55A and S55D Homos did not form enamel on the dentin surfaces, both antibodies only detected amorphous matrix in cyst-like bulges (Supplementary Fig. 6B).
Fig. 10.
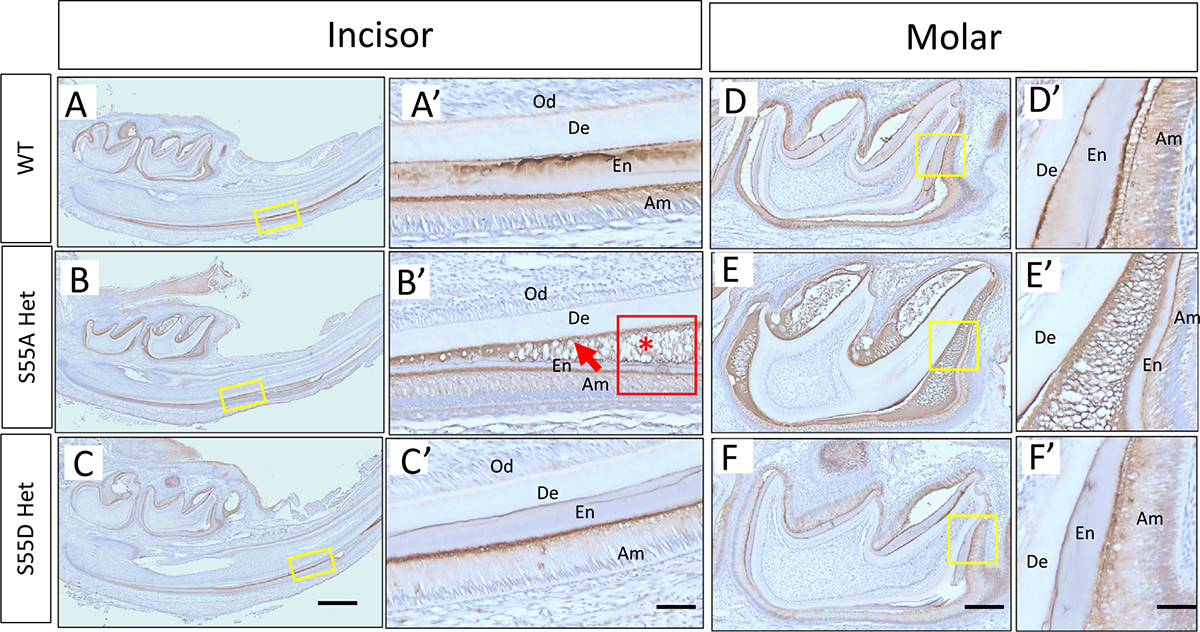
The distribution pattern of ENAM N-terminus in incisors and molars of 6-days old WT and ENAM S55 A/D Het mice. (A-C) ENAM was observed at the aprismatic enamel layer, at the mineralization front, and along with the enamel prisms in WT and S55D Het incisors. While in S55A Het incisor, it lacked clear boundaries between aprismatic enamel and prismatic enamel (red arrow in B’). The ENAM was also observed to accumulate in the amorphous matrix (red asterisk in B’) where ameloblasts lost their well-aligned columnar shape and bulged up. Clear prismatic structures in the enamel layer were not seen where ameloblasts detached from the underlying enamel matrix (red boxed area in (B’). (D–F) Consistent with incisors, ENAM N-terminus distributed well along with enamel prisms in WT and S55D Het molars, while in S55A Het, the ENAM distributed sparsely and loosely along with prisms. De, dentin. Od, odontoblasts. En, enamel matrix. Am, ameloblasts. Scale bars: 500 μm in A, B and C; 50 μm in A’, B’ and C’; 200 μm in D, E and F; 50 μm in D’, E’ and F’.
EMPs were found to co-localize and cooperate to control crystal nucleation and structural hierarchy during amelogenesis [36–39]. In S55A Het mice, the distribution and colocalization patterns of ENAM, AMBN and AMEL were dramatically changed at the secretory stage in the ameloblasts and enamel matrix in comparison to WT and S55D Het mice (Fig. 11C and D’, Supplementary Fig. 7); large amounts of these EMPs accumulated in the amorphous matrix encompassed in the epithelial bulges formed by detached ameloblasts (Fig. 11C’ and D’, Supplementary Fig. 7). AMEL in S55D Hets appeared to be less secreted into the matrix and retained in ameloblasts (Fig. 11F and F’, Supplementary Fig. 7). Both S55A and S55D Homo showed a disorganized distribution of EMPs in the amorphous matrix and malformed ameloblasts (Supplementary Fig. 8), which is consistent with the aforementioned histological defects.
Fig. 11.
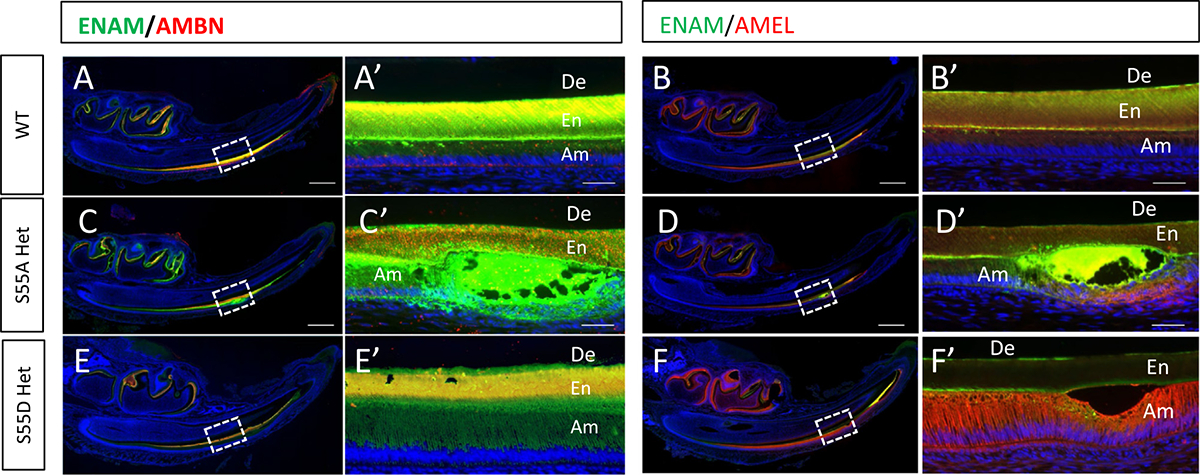
The distribution and colocalization pattern of ENAM with AMBN or AMEL. (A-B’) The distribution and colocalization pattern of ENAM with AMBN and AMEL in WT mice. (C-D’) In S55A Het mice, ENAM was accumulated in the amorphous matrix in the vacuolated spaces formed by detached ameloblasts. AMBN and AMEL showed different distribution patterns from WT. (E-F’) In S55D Het mice, ENAM and AMBN showed a similar distribution pattern with WT. AMEL appeared to be less secreted into the matrix and more retained in ameloblasts. Scale bars: 500 μm in A-J; 50 μm in A’-J’.
Potential biological process and function associated with ENAM Ser55 phosphorylation
As the hypoplastic AI in S55A Het mice may result from the functional changes of ameloblasts during the secretory stage, we employed label-free quantitation mass spectrometry, which is especially suitable for identifying differences between proteins of low abundance [40], to compare the proteomic profiles among S55A Hets, S55D Hets and WT. The triplicates in each group displayed good reproducibility, while both S55A Hets and S55D Hets showed distinct proteomic profiles from WT (Fig. 12A). Differential expressed proteins (DEPs) were screened in S55A Hets and S55D Hets compared to WT (Supplementary Table 1). S55A Hets and S55D Hets shared 55 DEPs, while 42 DEPs in S55A Het and 46 DEPs in S55D Het were distinct from one another (Fig. 12B). Similar GO items were enriched when DEPs in S55A Het or S55D Het to WT were input, which indicated the alterations of the enamel matrix biosynthetic process for biological process, alterations of enamel matrix mineralization and ameloblasts maturation for molecular function, and potential involvement of ER stress for cellular component (Supplementary Tables 2 and 3).
Fig. 12.
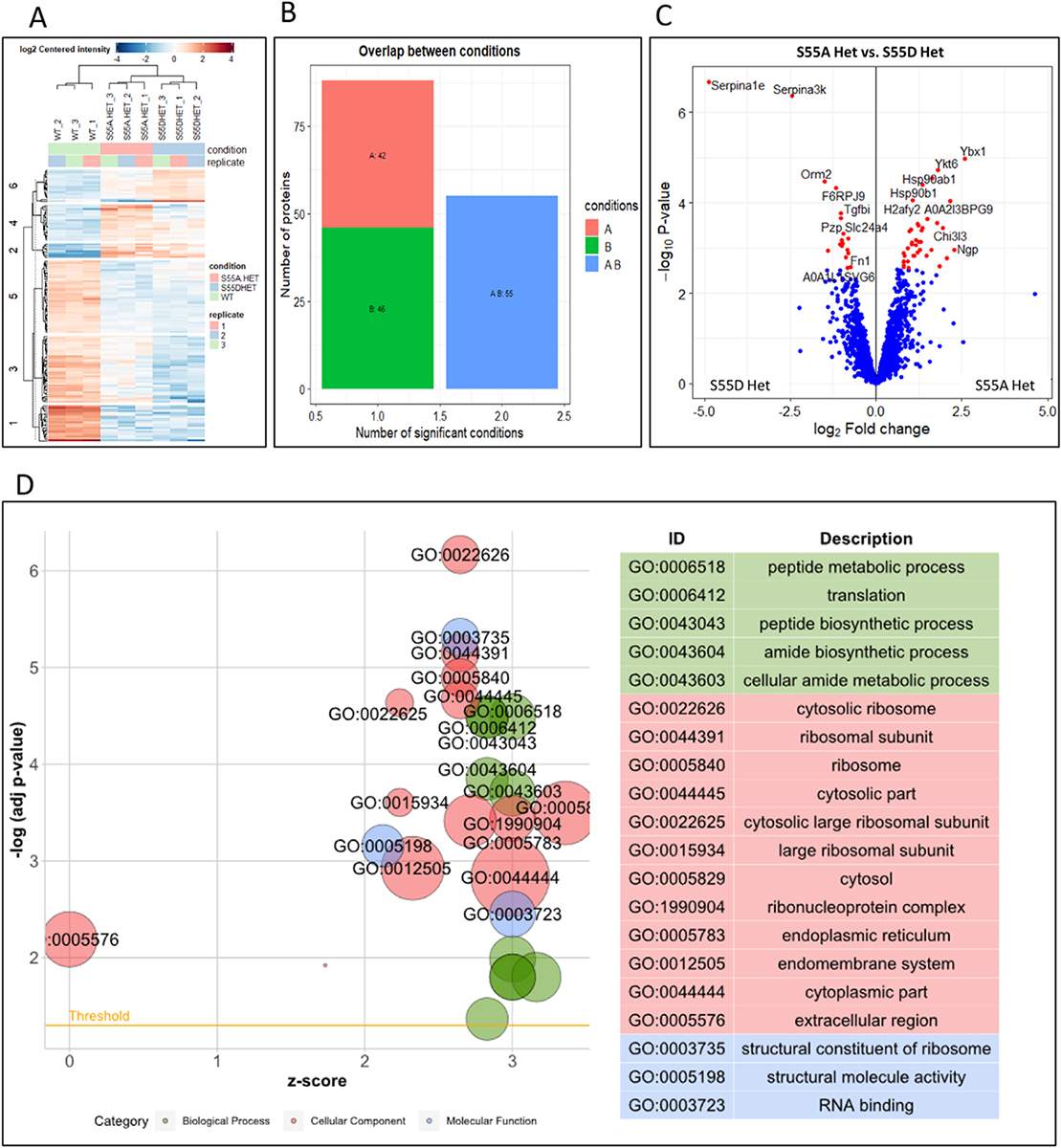
Proteomic profiling associated with ENAM S55A or S55D mutation. (A) Heatmap of significant proteins in all tested samples compared to WT. The rows represented different proteins and were clustered by k-means (numbers on the left). The columns depicted the different samples. Rows and columns were hierarchically clustered on Euclidean distance. Colors represented log2 transformed protein content of each sample vs. WT (red: high; blue: low). (B) The distribution of significant proteins per condition and the overlap between conditions. A: S55A Het vs. WT. B: S55D Het vs. WT. (C) The volcano plot showed the DEPs between S55A Het and S55D Het. (D) GO enrichment of DEPs between S55A Het and S55D Het. The left panel showed the plot of enriched GO ID, and the right panel showed the enriched GO terms on top.
Given that the S55A or S55D mutations were designed to represent different states of Ser55, and different severity of AI has been detected between S55A Hets and S55D Hets, we compared the proteomic profiles between S55A Hets and S55D Hets. The results revealed that 9 proteins were downregulated and 38 proteins upregulated in S55A Hets. The biological processes enriched in GO analysis suggest an association with peptide metabolic process and peptide biosynthetic process, etc., which may relate to the disturbances on the orchestrated enamel matrix protein regulation. The endoplasmic reticulum was enriched in cellular components, hinting a potential association with ER stress that was reported in a previous study of ENAMS55I mice [41,42] (Fig. 12D and Supplementary Table 4). However, we did not identify significant changes in ER stress markers Heat Shock 70 kDa Protein 5a (HSPA5a), the C/EBP homologous protein (CHOP), UPR signal activator (IRE1), PRKR-like ER kinase (PERK), ER-localized protein (ATF6) and X-box-binding protein 1 (XBP1) by Q-PCR analyses (Supplementary Fig. 9).
Discussion
The discovery of protein kinases VLK and FAM20C (also known as Golgi casein kinase) has changed the old dogma that restricted the concept of phosphorylation process occurring just in the intracellular milieu. These kinases have been shown to be involved in the classical ER-Golgi secretory pathway to recognize and phosphorylate the serine in S-x-E motifs in substrates like SCPPs [43]. The specific role of FAM20C in amelogenesis was evidenced by the enamel defects when FAM20C was specifically inactivated from the epithelial cells using Cre-loxP system mediated by K14-Cre [17]. Previous studies found that ENU-induced S55I and E57G mutations in the S55-x-E57 motif of murine ENAM resulted in AI-like enamel defects [22,25]. However, it remains controversial if the ENAM dysfunction arose from phosphorylation failure of Ser55 or the physicochemical changes when Ser55 and Glu57 were substituted by Ile and Gly, respectively. To clarify the essential role of Ser55 phosphorylation for ENAM function, we generated knock-in mice to replace Ser55 with alanine or aspartic acid.
Substitution of Ser with Ala, Asp or Glu has been used as a standard algorithm for phosphorylation site replacement [44,45]. Ala eliminates the side chain beyond the βcarbon and yet does not alter the main-chain conformation nor does it impose extreme electrostatic or steric effects [46–48]. In contrast, the physicochemical property of Asp or Glu is very different from serine, but their negative charge is often used to mimic phosphorylation state [45,49–52]. In addition, it has been shown that phospho-serine sites evolve from Asp/Glu residues [53].
Our S55A Het mice recapitulated the AI phenotypes as in ENU-induced S55I and E57G Het mutant mice [42]. The reduced enamel volume and disorganized ameloblasts conform to a diagnosis of hypoplastic AI [39,54]. The ameloblasts of S55A Het mice secreted an initial layer of enamel that appeared normal with a prismatic structure, but subsequent layers displayed multiple abnormities, including a slowdown of appositional growth, loss of enamel rod structure, a smaller size of enamel crystals and hypermineralization. In comparison, the enamel formation in S55D Het mice was less disturbed and showed a generally well-organized enamel rod structure similar to the WT. These results indicate that the phosphorylation of Ser55 is essential for ENAM function and that mutations changing the phosphorylation state of ENAM Ser55 could be a potential etiology factor of AI.
The enamel defects in molars were mainly located at the cervical and middle third of the crown while the cusps were less defective. This is consistent with previous observations in human permanent dentition [55]. Crown development and morphological factors, such as enamel rod length and direction, may influence the development and expression of enamel surface defects [55]. Kierdorf et al. identified a marked reduction of enamel extension rates in cervical compared with more cuspal crown portions of molars, with formation of the cervical 25% of the crown taking about the same time as that of the upper 75% of the crown, which explains the more frequent occurrence of enamel hypoplasia in cervical compared with upper and middle crown portions [56].
Although S55D Hets showed generally normal enamel, both S55A and S55D Homo mice displayed similarly severe hypoplastic AI. The distinct phenotypes between S55D Hets and Homos may be associated with several lines of facts. First, the severity of enamel defects associated with ENAM dysfunction is dosage-dependent, which has been well documented in Enam-knockout mice [57]. In humans, individuals with Enam mutations on both alleles developed more severe enamel hypoplasia than those affected with only one allele. The former exhibited generalized hypoplastic AI and anterior open bite, while the latter showed localized defects and/or horizontal grooves on their teeth [57–59]. Second, the ENAM expression level is tightly controlled during amelogenesis. Hu et al. found that introducing various dosages of ENAM transgenes into the Enam−/− mice cannot fully rescue the enamel defects and that WT mice developed AI when ENAM was overexpressed with the lowest expression level of transgenic ENAM. This suggests a precise control of ENAM level is required in order to achieve proper enamel formation [57]. It is conceivable that a precise control may also apply to the post-translational modifications of ENAM, such as phosphorylation. Our proteomic data showed that the phosphorylation state of Ser55 is required for the precise control of EMPs’ biosynthetic and secretary processes. Third, the amino acid substitution strategy has its intrinsic limitations for mimicking natural phosphorylation. Natural phosphorylation should be dynamic and often transient in response to cellular events, with the level of phosphorylation being regulated by its removal (e.g., dephosphorylation by a resident phosphatase) to modify protein function. The balance of phosphorylation and dephosphorylation is a key factor for phosphorylation-associated regulatory events, which is generally employed by not only intracellular signaling molecules but also extracellular matrix proteins [16,60–63]. However, Asp substitution creates a sustained state of phosphorylation. In heterozygotes, the mutant and WT alleles may partially mimic the phosphorylation/dephosphorylation modality, while in homozygotes, this balance was completely broken. This is further supported by the observation that there was a differentiation issue of ameloblasts related to ENAM phosphorylation: in the homozygotes, ameloblasts did not develop into high-column cells like in WT and heterozygotes.
Our proteomic profiling revealed that the phosphorylation of ENAM Ser55 is associated with enamel matrix biosynthesis, mineralization, and maturation processes. This is consistent with our TEM and immunofluorescence data, and mutually in agreement with previous studies in bones and dentin, where phosphorylation failure interferes with the processing, folding, transport, secretion, release, and degradation of matrix proteins, as well as their binding and interaction with other ECM components [64]. Our proteomic profiling also enriched the cellular component term endoplasmic reticulum, implying a potential involvement of ER stress, which has been suggested by a previous study with ENAM S55I mutant mice [42]. However, our further investigation did not detect significant changes in Hspa5a, Chop, Ire1, Perk, Atf6 and Xbp1.
We also noticed that ribosome, cytosol, endomembrane system and extracellular region were enriched for cellular component in GO analyses of proteomic profiling data, and structure constitutes of the ribosome, structural molecular activity, and RNA binding were enriched for molecular function (Fig. 9D, Supplementary Table S3). These data suggest that the ENAM dysfunction due to the phosphorylation loss of Ser55 may also lead to ameloblast pathology and the disruption of other biological processes and signaling pathways involved in amelogenesis.
Several previous studies have proved that proper phosphorylation of SIBLING proteins by secreted kinase is essential for the biomineralization in bones and dentin. The phospho-serines phosphorylated by FAM20C (at the sequence motif S-x-E) bind calcium and regulate the formation of calcium phosphate-containing hydroxyapatite (HA) crystals [65]. On the other hand, the inhibitory regulation of osteopontin (OPN) on HA formation also relies on the degree of phosphorylation [66]. In this study, we characterized the function of pSer55 in ENAM by eliminating or mimicking the phosphorylation state in vivo. The phosphorylation of Ser55 appears to be critical for the function of ENAM during several intracellular and extracellular processes associated with amelogenesis. Understanding the mechanism and function of EMPs phosphorylation may gain novel insights for AI etiology and disease treatment.
Supplementary Material
Abbreviations:
- Fam20C
Family with sequence similarity 20, member C
Footnotes
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.matbio.2022.07.001.
References
- [1].Bäckman B, Holm AK, Amelogenesis imperfecta: prevalence and incidence in a northern Swedish county, Community Dent. Oral Epidemiol 14 (1) (1986) 43–47. [DOI] [PubMed] [Google Scholar]
- [2].Gadhia K, McDonald S, Arkutu N, Malik K, Amelogenesis imperfecta: an introduction, Br. Dent. J 212 (8) (2012) 377–379. [DOI] [PubMed] [Google Scholar]
- [3].Simmer JP, Hu JCC, Expression, structure, and function of enamel proteinases, Connect. Tissue Res 43 (2–3) (2002) 441–449. [DOI] [PubMed] [Google Scholar]
- [4].Margolis H, Beniash E, Fowler C, Role of macromolecular assembly of enamel matrix proteins in enamel formation, J. Dent. Res 85 (9) (2006) 775–793. [DOI] [PubMed] [Google Scholar]
- [5].Uchida T, Tanabe T, Fukae M, Shimizu M, Immunocytochemical and immunochemical detection of a 32kDa nonamelogenin and related proteins in porcine tooth germs, Arch. Histol. Cytol 54 (5) (1991) 527–538. [DOI] [PubMed] [Google Scholar]
- [6].Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, Krebsbach PH, Nanci A, Kulkarni AB, Yamada Y, Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts, J. Cell Biol 167 (5) (2004) 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lu Y, Papagerakis P, Yamakoshi Y, Hu JCC, Bartlett JD, Simmer JP, Functions of KLK4 and MMP-20 in dental enamel formation, Biol. Chem 389 (6) (2008) 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington R, Amelogenin-deficient mice display an amelogenesis imperfecta phenotype, J. Biol. Chem 276 (34) (2001) 31871–31875. [DOI] [PubMed] [Google Scholar]
- [9].Smith CE, Wazen R, Hu Y, Zalzal SF, Nanci A, Simmer JP, Hu JCC, Consequences for enamel development and mineralization resulting from loss of function of ameloblastin or enamelin, Eur. J. Oral Sci 117 (5) (2009) 485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Caterina JJ, Skobe Z, Shi J, Ding Y, Simmer JP, Birkedal-Hansen H, Bartlett JD, Enamelysin (matrix metalloproteinase 20)-deficient mice display an amelogenesis imperfecta phenotype, J. Biol. Chem 277 (51) (2002) 49598–49604. [DOI] [PubMed] [Google Scholar]
- [11].Kawasaki K, Weiss KM, Mineralized tissue and vertebrate evolution: the secretory calcium-binding phosphoprotein gene cluster, Proc. Natl. Acad. Sci. USA 100 (7) (2003) 4060–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Addison WN, Nakano Y, Loisel T, Crine P, McKee MD, MEPE-ASARM peptides control extracellular matrix mineralization by binding to hydroxyapatite: an inhibition regulated by PHEX cleavage of ASARM, J. Bone Miner. Res 23 (10) (2008) 1638–1649. [DOI] [PubMed] [Google Scholar]
- [13].Tagliabracci VS, Engel JL, Wen J, Wiley SE, Worby CA, Kinch LN, Xiao J, Grishin NV, Dixon JE, Secreted kinase phosphorylates extracellular proteins that regulate biomineralization, Science 336 (6085) (2012) 1150–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kiselnikova L, Vislobokova E, Voinova V, Dental manifestations of hypophosphatasia in children and the effects of enzyme replacement therapy on dental status: a series of clinical cases, Clin. Case Rep 8 (5) (2020) 911–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tagliabracci VS, Wiley SE, Guo X, Kinch LN, Durrant E, Wen J, Xiao J, Cui J, Nguyen KB, Engel JL, A single kinase generates the majority of the secreted phosphoproteome, Cell 161 (7) (2015) 1619–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yang X, Yan W, Tian Y, Ma P, Opperman LA, Wang X, Family with sequence similarity member 20C is the primary but not the only kinase for the small-integrin-binding ligand N-linked glycoproteins in bone, FASEB J. 30 (1) (2016) 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang X, Jung J, Liu Y, Yuan B, Lu Y, Feng J, Qin CJ, The specific role of FAM20C in amelogenesis, J. Dent. Res 92 (11) (2013) 995–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].O’Sullivan J, Bitu CC, Daly SB, Urquhart JE, Barron MJ, Bhaskar SS, Martelli-Júnior H,dos Santos Neto PE, Mansilla MA, Murray JC, Whole-exome sequencing identifies FAM20A mutations as a cause of amelogenesis imperfecta and gingival hyperplasia syndrome, Am. J. Hum. Genet 88 (5) (2011) 616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kwak SY, Wiedemann-Bidlack FB, Beniash E, Yamakoshi Y, Simmer JP, Litman A, Margolis HC, Role of 20-kDa amelogenin (P148) phosphorylation in calcium phosphate formation in vitro, J. Biol. Chem 284 (28) (2009) 18972–18979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Le Norcy E, Kwak SY, Wiedemann-Bidlack F, Beniash E, Yamakoshi Y, Simmer J, Margolis H, Leucine-rich amelogenin peptides regulate mineralization in vitro, J. Dent. Res 90 (9) (2011) 1091–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shin NY, Yamazaki H, Beniash E, Yang X, Margolis SS, Pugach MK, Simmer JP, Margolis HC, Amelogenin phosphorylation regulates tooth enamel formation by stabilizing a transient amorphous mineral precursor, J. Biol. Chem 295 (7) (2020) 1943–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yan WJ, Ma P, Tian Y, Wang JY, Qin CL, Feng JQ, Wang XF, The importance of a potential phosphorylation site in enamelin on enamel formation, Int. J. Oral Sci 9 (11) (2017) e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ma P, Yan W, Tian Y, He J, Brookes S, Wang X, The importance of serine phosphorylation of ameloblastin on enamel formation, J. Dent. Res 95 (12) (2016) 1408–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chan HC, Mai L, Oikonomopoulou A, Chan H, Richardson A, Wang SK, Simmer J, Hu JC, Altered enamelin phosphorylation site causes amelogenesis imperfecta, J. Dent. Res 89 (7) (2010) 695–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Masuya H, Shimizu K, Sezutsu H, Sakuraba Y, Nagano J, Shimizu A, Fujimoto N, Kawai A, Miura I, Kaneda HJ, Enamelin (Enam) is essential for amelogenesis: ENU-induced mouse mutants as models for different clinical subtypes of human amelogenesis imperfecta (AI), Hum. Mol. Genet 14 (5) (2005) 575–583. [DOI] [PubMed] [Google Scholar]
- [26].Rangiani A, Cao ZG, Liu Y, Rodgers AV, Jiang Y, Qin CL, Feng J.Q.J.I.j.o.o.s., Dentin matrix protein 1 and phosphate homeostasis are critical for postnatal pulp, dentin and enamel formation, Int. J. Oral Sci 4 (4) (2012) 189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Smith CE, Hu Y, Hu JC, Simmer JP, Characteristics of the transverse 2D uniserial arrangement of rows of decussating enamel rods in the inner enamel layer of mouse mandibular incisors, J. Anat 235 (5) (2019) 912–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wang SK, Hu Y, Smith CE, Yang J, Zeng C, Kim JW, Hu JC, Simmer JP, The enamel phenotype in homozygous FAM83h truncation mice, Mol. Genet. Genom. Med 7 (6) (2019) e724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wu J, Tian Y, Han L, Liu C, Sun T, Li L, Yu Y, Lamichhane B, D’Souza RN, Millar SE, FAM20B-catalyzed glycosaminoglycans control murine tooth number by restricting FGFR2b signaling, BMC Biol 18 (1) (2020) 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Brookes SJ, Kingswell NJ, Barron MJ, Dixon MJ, Kirkham JJ, Is the 32-kDa fragment the functional enamelin unit in all species, Eur. J. Oral Sci 119 (2011) 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhang X, Smits AH, van Tilburg GB, Ovaa H, Huber W, Vermeulen M, Proteome-wide identification of ubiquitin interactions using UbIA-MS, Nat. Protoc 13 (3) (2018) 530–550. [DOI] [PubMed] [Google Scholar]
- [32].Pomaznoy M, Ha B, Peters B, GOnet: a tool for interactive gene ontology analysis, BMC Bioinform. 19 (1) (2018) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Walter W, Sánchez-Cabo F, Ricote M, GOplot: an R package for visually combining expression data with functional analysis, Bioinformatics 31 (17) (2015) 2912–2914. [DOI] [PubMed] [Google Scholar]
- [34].Bartlett J, Beniash E, Lee D, Smith C, Decreased mineral content in MMP-20 null mouse enamel is prominent during the maturation stage, J. Dent. Res 83 (12) (2004) 909–913. [DOI] [PubMed] [Google Scholar]
- [35].Tian Z, Lv X, Zhang M, Wang X, Chen Y, Tang P, Xu P, Zhang L, Wu B, Zhang L, Deletion of epithelial cell-specific Cdc42 leads to enamel hypermaturation in a conditional knockout mouse model, Biochim. Biophys. Acta (BBA) Mol. Basis Dis 1864 (8) (2018) 2623–2632. [DOI] [PubMed] [Google Scholar]
- [36].Zalzal SF, Smith CE, Nanci AJ, Ameloblastin and amelogenin share a common secretory pathway and are co-secreted during enamel formation, Matrix Biol. 27 (4) (2008) 352–359. [DOI] [PubMed] [Google Scholar]
- [37].Gallon V, Chen L, Yang X, Moradian-Oldak JJ, Localization and quantitative co-localization of enamelin with amelogenin, J. Struct. Biol 183 (2) (2013) 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mazumder P, Prajapati S, Lokappa SB, Gallon V, Moradian-Oldak JJ, Analysis of co-assembly and co-localization of ameloblastin and amelogenin, Front. Physiol 5 (2014) 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mazumder P, Prajapati S, Bapat R, Moradian-Oldak JJ, Amelogenin-ameloblastin spatial interaction around maturing enamel rods, J. Dent. Res 95 (9) (2016) 1042–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Dowle AA, Wilson J, Thomas JR, Comparing the diagnostic classification accuracy of iTRAQ, peak-area, spectral-counting, and emPAI methods for relative quantification in expression proteomics, J. Proteome Res 15 (10) (2016) 3550–3562. [DOI] [PubMed] [Google Scholar]
- [41].Brookes SJ, Barron MJ, Dixon MJ, Kirkham JJ, The unfolded protein response in amelogenesis and enamel pathologies, Front. Physiol 8 (2017) 653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Brookes SJ, Barron MJ, Smith CE, Poulter JA, Mighell AJ, Inglehearn CF, Brown CJ, Rodd H, Kirkham J, Dixon MJ, Amelogenesis imperfecta caused by N-terminal enamelin point mutations in mice and men is driven by endoplasmic reticulum stress, Hum. Mol. Genet 26 (10) (2017) 1863–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Klement E, Medzihradszky KF, Extracellular protein phosphorylation, the neglected side of the modification, Mol. Cell Proteom 16 (1) (2017) 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Chen Z, Cole PA, Synthetic approaches to protein phosphorylation, Curr. Opin. Chem. Biol 28 (2015) 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Thorsness PE, Koshland D, Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by negative change of the phosphate, J. Biol. Chem 262 (22) (1987) 10422–10425. [PubMed] [Google Scholar]
- [46].Cunningham BC, Wells JA, High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis, Science 244 (4908) (1989) 1081–1085. [DOI] [PubMed] [Google Scholar]
- [47].Fabrice L, Marie-Hélène R, Jean-Michel M, Alanine-stretch scanning mutagenesis: a simple and efficient method to probe protein structure and function, Nucleic Acids Res 2 (1997) 447–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Morrison KL, Weiss GA, Combinatorial alanine-scanning, Curr. Opin. Chem. Biol 5 (3) (2001) 302–307. [DOI] [PubMed] [Google Scholar]
- [49].Guerra-Castellano A, Díaz-Moreno I, Velázquez-Campoy A, Miguel A, Díaz-Quintana A, Structural and functional characterization of phosphomimetic mutants of cytochrome c at threonine 28 and serine 47, Biochim. Biophys. Acta Bioenerg 1857 (4) (2016) 387–395. [DOI] [PubMed] [Google Scholar]
- [50].Sanchez CP, Cubel SM, Nyboer B, Jankowska-Döllken M, Schaeffer-Reiss C, Ayoub D, Planelles G, Lanzer M, Phosphomimetic substitution at Ser-33 of the chloroquine resistance transporter PfCRT reconstitutes drug responses in Plasmodium falciparum, J. Biol. Chem 294 (34) (2019) 12766–12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cooper JA, Sefton BM, Hunter T, Detection and quantification of phosphotyrosine in proteins, Methods Enzymol 99 (1983) 387–402. [DOI] [PubMed] [Google Scholar]
- [52].Lé J, Kempf M, Lee G, Brandt R, conversion of serine to aspartate imitates phosphorylation- induced changes in the structure and function of microtubule- associated protein tau*, J. Biol. Chem 272 (13) (1997) 8441–8446. [DOI] [PubMed] [Google Scholar]
- [53].Pearlman S, Serber Z, Ferrell J, A mechanism for the evolution of phosphorylation sites, Cell 147 (4) (2011) 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Smith CE, Poulter JA, Antanaviciute A, Kirkham J, Brookes SJ, Inglehearn CF, Mighell AJ, Amelogenesis imperfecta; genes, proteins, and pathways, Front. Physiol 8 (2017) 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Goodman AH, Armelagos GJ, Factors affecting the distribution of enamel hypoplasias within the human permanent dentition, Am. J. Phys. Anthropol 68 (4) (1985) 479–493. [DOI] [PubMed] [Google Scholar]
- [56].Kierdorf H, Witzel C, Upex B, Dobney K, Kierdorf U, Enamel hypoplasia in molars of sheep and goats, and its relationship to the pattern of tooth crown growth, J. Anat 220 (5) (2012) 484–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hu JCC, Hu Y, Lu Y, Smith CE, Lertlam R, Wright JT, Suggs C, McKee MD, Beniash E, Kabir M.E.J.P.o., Enamelin is critical for ameloblast integrity and enamel ultrastructure formation, PloS ONE 9 (3) (2014) e89303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ozdemir D, Hart P, Firatli E, Aren G, Ryu O, T.J.J.o.d.r. Hart, Phenotype of ENAM mutations is dosage-dependent, J. Dent. Res 84 (11) (2005) 1036–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Wright JT, Hart TC, Hart PS, Simmons D, Suggs C, Daley B, Simmer J, Hu J, Bartlett JD, Li YJ, Human and mouse enamel phenotypes resulting from mutation or altered expression of AMEL, ENAM, MMP20 and KLK4, Cells Tissues Organs 189 (1–4) (2009) 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Prince CW, Oosawa T, Butler W, Tomana M, Bhown A, Bhown M, Schrohenloher R, Isolation, characterization, and biosynthesis of a phosphorylated glycoprotein from rat bone, J. Biol. Chem 262 (6) (1987) 2900–2907. [PubMed] [Google Scholar]
- [61].Kurosawa M, Phosphorylation and dephosphorylation of protein in regulating cellular function, J. Pharmacol. Toxicol. Methods 31 (3) (1994) 135–139. [DOI] [PubMed] [Google Scholar]
- [62].Fuchs SY, Tappin I, Z.e. Ronai, Stability of the ATF2 transcription factor is regulated by phosphorylation and dephosphorylation, J. Biol. Chem 275 (17) (2000) 12560–12564. [DOI] [PubMed] [Google Scholar]
- [63].Curtin P, McHugh KP, Zhou HY, Fluckiger R, Goldhaber P, Oppenheim FG, Salih E, Modulation of bone resorption by phosphorylation state of bone sialoprotein, Biochemistry 48 (29) (2009) 6876–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Yalak G, Olsen BR, Proteomic database mining opens up avenues utilizing extracellular protein phosphorylation for novel therapeutic applications, J. Transl. Med 13 (1) (2015) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].George A, Veis A, Phosphorylated proteins and control over apatite nucleation, crystal growth, and inhibition, Chem. Rev 108 (11) (2008) 4670–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Hunter GK, Kyle CL, Goldberg HA, Modulation of crystal formation by bone phosphoproteins: structural specificity of the osteopontin-mediated inhibition of hydroxyapatite formation, Biochem. J 300 (Pt 3) (1994) 723–728 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


