Abstract
Transformers are a powerful subclass of neural networks catalyzing the development of a growing number of computational methods for RNA structure modeling. Here, we conduct an objective and empirical study of the predictive modeling accuracy of the emerging transformer-based methods for RNA structure prediction. Our study reveals multi-faceted complementarity between the methods and underscores some key aspects that affect the prediction accuracy.
Introduction
In silico modeling of RNA three-dimensional (3D) structures is undergoing rapid transformation, inspired, in part, by the unprecedented success of DeepMind’s AlphaFold2 method for protein 3D structure prediction [1]. The recently concluded collaborative effort between the 15th Critical Assessment of Structure Prediction (CASP15) and RNA-Puzzles [2], the first of its kind double-blind assessment of RNA structure prediction, revealed an emerging trend of deep learning-based methods for RNA 3D structure prediction, with over 40% of the participating groups (17 out of 42) using deep learning. Given such intense interest, a natural question arises: Are there similarities or differences between the new generation of deep learning-based methods for modeling RNA 3D structures, in terms of the underlying methodology and/or the resulting accuracy?
Methodologically, a number of new deep learning-based methods for RNA 3D modeling [3–5, 7, 8] rely on attention-powered transformers as the core neural architecture, which is also a key ingredient of the AlphaFold2 framework [1]. However, there are fundamental differences among the transformer-based models of RNA 3D structure prediction in terms of architectural design and information flow. Some methods, such as DeepFoldRNA [3] and trRosettaRNA [4], are based on geometric restraints prediction using transformers, followed by gradient-based optimization. Other approaches, such as RoseTTAFoldNA [5], employ end-to-end learning leveraging the SE(3)-transformer [6] architecture. Yet other methods, such as DRfold [7] and RhoFold [8], adopt a hybrid approach by integrating end-to-end learning with geometrical restraints. The architectural diversity of the RNA 3D modeling transformers naturally leads to different learning paradigms with varied training efficiencies and/or inference modalities. It is worth mentioning here that although the recently published AlphaFold 3 method [9] can predict 3D structures of a diverse set of biomolecules including RNA, we did not incorporate it in this comparison due to the lack of publicly available source code.
Beyond the realm of neural architecture, there are intrinsic differences in terms of the representation of RNA conformational space and the cardinalities of the input and output. For instance, DeepFoldRNA adopts a coarse-grained representation specified by the phosphate P, ribose C4′, and glycosidic N atoms of the nucleobase, and the predicted geometric restraints involving these three atoms lead to 3-bead models with the remaining atoms subsequently added and refined. trRosettaRNA uses a finer-grained 5-bead representation by including C1′ and C3′ atoms in addition to P, C4′, and N atoms with direct optimization of all-atom models using the predicted finer-grained geometric restraints. In contrast, RoseTTAFoldNA uses a coordinate frame representation where each nucleotide-specific coordinate frame captures the position and orientation of the phosphate group and a set of 10 torsion angles (6 backbone, 3 ribose ring, and 1 nucleoside) that can build all-atom nucleic acid models. DRfold relies on a reduced 3-bead representation with P, C4′, and N atoms, both for predicting geometric restraints and for defining the coordinate frames, with the all-atom models recovered from the 3-vector virtual bond system. RhoFold, on the other hand, uses C4′, C1′, and N atoms with a set of four torsion angles to reconstruct all atom coordinates except for P, C4′, and N atoms for pairwise distance prediction. It is worth noting that DRfold is a single-sequence RNA structure prediction method with the only required input being the nucleotide sequence itself, whereas all the other methods depend on multiple sequence alignment (MSA) as an additional input, typically generated using the rMSA pipeline [10] with Rhofold additionally incorporating information from RNA language model (RNA-FM). As such, the nature of the evolutionary information encoded in the MSA partly affects the MSA-dependent methods.
Notwithstanding the multifaceted methodological differences, there is no clear consensus on which method performs objectively better than the others. This is partially due to the lack of robust and comprehensive evaluation metrics, as well as limited head-to-head comparisons on an independent dataset. Why is it important? First, to help the practitioners choose a better method from a growing ecosystem of approaches for RNA 3D structure prediction powered by transformers. Second, to make progress towards better learning paradigms and/or inference modalities, it is useful to objectively assess how certain design choices affect the resulting accuracy. Third, to help understand how the existing deep learning-based predictors fare against fully automated deep learning-free methods. To this end, we have chosen RNAComposer [11], a fragment-assembly-based fully automated pipeline designed for RNA 3D structure prediction with secondary structure as the only input information.
While the CASP15 offers an excellent testbed to evaluate these emerging methods over the same set of targets, it only contains 12 RNA targets that may be insufficient for meaningful statistical significance tests. To address this issue, we curated an in-house benchmarking set containing 60 additional non-redundant RNA targets deposited into the Protein Data Bank (PDB) [12] after the preparation of the training set of the individual methods wherever known and after the release of the respective open-source code/data repositories. On the combined set of 72 RNA targets, we benchmarked the predictive modeling performance of DeepFoldRNA, trRosettaRNA, DRfold, RoseTTAFoldNA, RhoFold, and RNAComposer using a number of complementary evaluation metrics. Our study underscores the mutual complementarity between different methods and highlights some key aspects that affect prediction accuracy.
Results
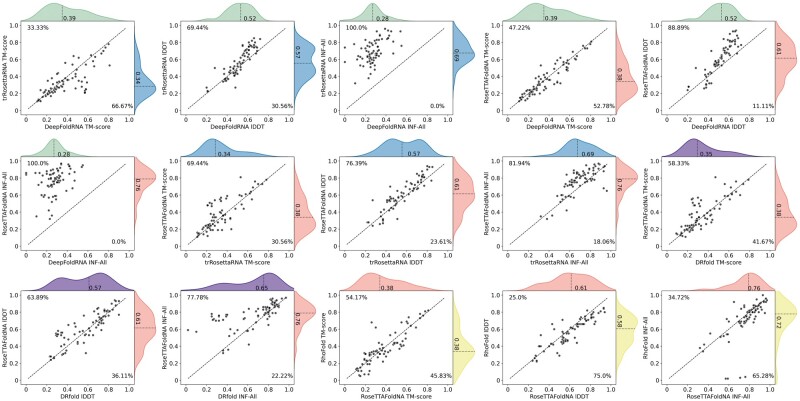
Figure 1 shows the prediction accuracy of different methods and their head-to-head comparisons in terms of the correctness of global fold prediction measured by TM-score [13], distance-based structural similarity measured by lDDT [14], and environment-dependent local quality measured by INF [15]. We have only shown a subset of comparisons in Fig. 1 as described below for brevity. The full set of comparisons is available in Supplementary Figs S1–S3 and the target-by-target results are available in Supplementary Tables S3–S8. First, the comparison between the two geometric restraints prediction methods shows that despite using a coarser-grained 3-bead representation, DeepFoldRNA is comparable to finer-grained 5-bead trRosettaRNA in terms of global fold prediction (TM-score). However, when both structural similarity and local quality are considered, trRosettaRNA convincingly outperforms DeepFoldRNA for 69.4% and 100% of cases with respect to lDDT and INF respectively, with the difference being statistically significant at a 95% confidence level for both lDDT (p = 4.4 × 10−6) and INF (p = 1.6 × 10−13). Next, we compare the end-to-end method RoseTTAFoldNA with both geometric restraints prediction methods trRosettaRNA and DeepFoldRNA. RoseTTAFoldNA strikes a balance between complementary aspects of modeling accuracy by being comparable to trRosettaRNA (or DeepFoldRNA) in terms of TM-score and much better (more than 75% and 80% of cases) than trRosettaRNA with respect to lDDT and INF respectively, resulting in statistically significant performance difference for both lDDT (p = 2.5 × 10−6) and INF (p = 2.4 × 10−7). Performance comparison between the pure end-to-end method RoseTTAFoldNA and the single-sequence hybrid method DRfold reveals a narrower accuracy margin with RoseTTAFoldNA still consistently outperforming DRfold across all three metrics, although the performance difference is not statistically significant in terms of TM-score but significant in terms of lDDT (p = 0.01) and INF (p = 3.9 × 10−7). Finally, a similar trend can be observed when comparing two end-to-end methods RoseTTAFoldNA and RhoFold, where RoseTTAFoldNA is similar to RhoFold in terms of TM-score but outperforms in terms of lDDT and INF for more than 75% and 65% of cases, respectively.
Figure 1.
Partial head-to-head comparison of five transformer-based RNA 3D modeling methods on a benchmark set of 72 targets (The full set of comparisons is available in Supplementary Figs S1–S3). Scatterplots showing comparisons between two competing methods. Percentage numbers reported in the top-left quadrant correspond to targets where the method shown in the Y-axis outperforms the competing method shown in the X-axis, and vice versa for the bottom-right quadrant. The distributions of the scores color-coded for different methods for ease of visualization are demonstrated for both axes, with the dashed line and the numbers next to it indicating the mean values.
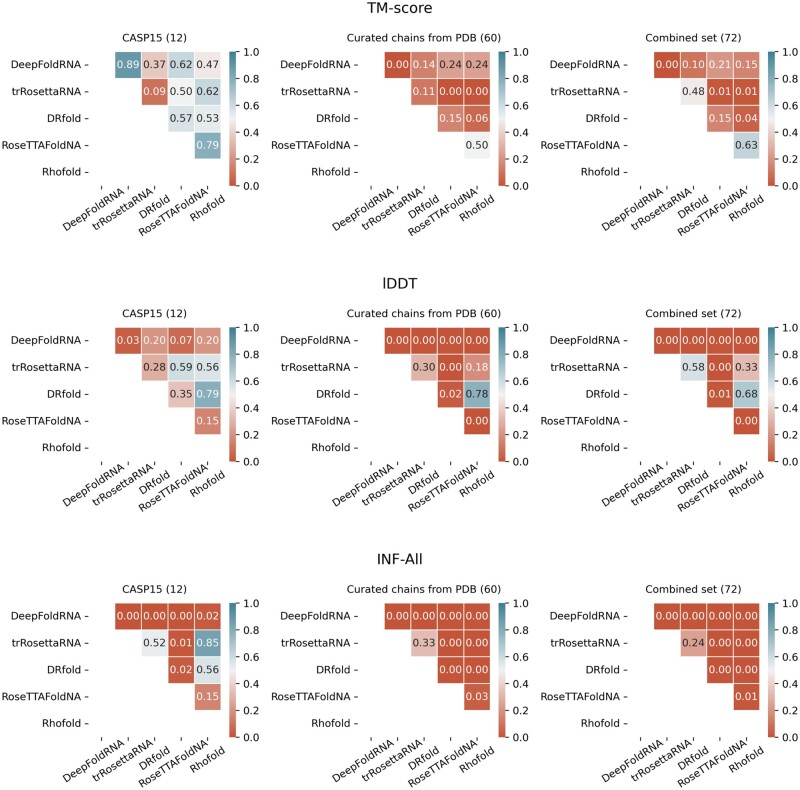
What is the contribution of our in-house benchmarking set of 60 nonredundant RNA targets above and beyond the 12 CASP15 RNA targets during head-to-head performance comparisons between different methods? As shown in Fig. 2, the fraction of times the pairwise statistical tests between the competing methods leads to statistically significant performance difference at a 95% confidence level (i.e. p < 0.05) is substantially higher for our in-house benchmarking set of 60 nonredundant RNA targets than for the CASP15 set having only 12 RNA targets across all three different performance evaluation metrics. The trend in statistical significance is preserved even when the two sets are combined having a total of 72 RNA targets, demonstrating that our in-house benchmarking set is the main driver in bringing out meaningful statistically significant performance differences between various methods during head-to-head comparisons.
Figure 2.
Results of pairwise statistical tests (p-values) of the performance difference between all five deep learning-based methods in terms of three different performance evaluation metrics for three partitions of the benchmark set: (i) CASP15 set having only 12 RNA targets (CASP15); (ii) our in-house benchmarking set of 60 non-redundant RNA targets (TS60); and (iii) the combination of the two sets having a total of 72 RNA targets (TS72).
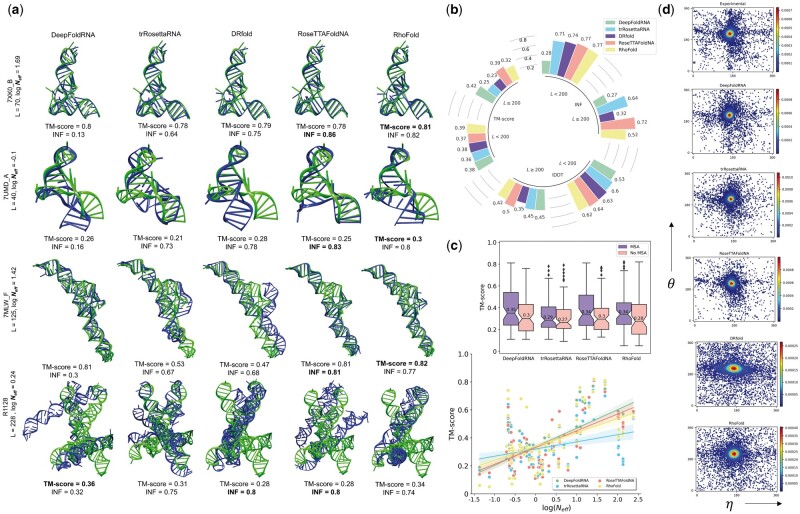
Four representative examples are shown in Fig. 3a. For the first target 7XK0_B having sufficient evolutionary information, measured by the logarithm of the normalized number of effective sequences (Neff) in the MSA, with a log(Neff) value of 1.69, all methods yield high accuracy across all evaluation metrics. For the second target 7UMD_A with a shallow evolutionary profile having a low log(Neff) value of −0.1, none of the methods could predict the global fold accurately with the single-sequence method DRfold achieving comparatively higher accuracy than most of the MSA-based methods. The third target, 7MLW_F, a comparatively larger RNA of 125 nucleotides having reasonable evolutionary information with a log(Neff) value of 1.42 shows that the relatively more MSA-dependent methods DeepFoldRNA, RoseTTAFoldNA, and RhoFold led to noticeably better prediction accuracy than the relatively less MSA-dependent method trRosettaRNA and the single-sequence method DRfold. That is, the availability of evolutionary information (or the lack thereof) directly or indirectly affects the predictive modeling accuracy of the MSA-based methods regardless of the architectural design or modeling paradigms. Finally, for the CASP15 target R1128 having a length of 238 nucleotides, none of the methods could predict the global fold correctly with the best TM-score of 0.36 achieved by DeepFoldRNA, revealing the limitations of these methods for larger targets. It is interesting to note that the incorporation of language model information in RhoFold appears to have reduced the dependency on evolutionary information, achieving the highest TM-score for the first three targets despite varying log(Neff) values.
Figure 3.
Predictive modeling accuracy of five transformer-based RNA 3D modeling methods on a benchmark set of 72 targets. (a) Four representative targets shown with the predicted structural models colored in blue superimposed on the experimental structures in green and the TM-score and INF scores shown below with bold numbers indicating the best performance. (b) Polar bar plot showing the mean accuracies of the methods annotated on top of each bar along with the impact in accuracy with the change in sequence length (L). (c) Notched box plots showing the TM-score distributions with and without using MSA as input for the MSA-based methods with numbers indicating the median values (top); and scatterplot between predicted TM-scores and MSA depths (log(Neff) (bottom) where the solid lines represent tendency lines constructed by linear fit to the data. (d) Joint angular distribution of the pseudo torsion angles, with color code ramping from blue to red for low to high density.
Do the aforementioned characteristics of the RNA targets, such as length and evolutionary profile, have any systematic effect on the prediction accuracy? As shown in Fig. 3b, the accuracies of all methods in terms of TM-score, lDDT, and INF are somewhat lower for sequences having length ≥ 200. One potential reason for this prediction discrepancy can be the presence of a higher number of noncanonical base pairings in larger RNAs, such as ribosomal RNAs (rRNAs) and long non-coding RNAs (lncRNAs); and accurate RNA 3D structure prediction relies heavily on correctly predicting both noncanonical and canonical base pairings [16]. The choice of a length threshold of 200 for this analysis is based on two reasons: first, this threshold is used in the existing literature [17], and second, DRfold was trained on sequences shorter than 200 nucleotides. Thus, our analysis allows for a fair comparison of DRfold against the other methods. The results demonstrate that DeepFoldRNA and RoseTTAFoldNA exhibit relatively less accuracy decline for targets with length ≥200, whereas DRfold suffers from a drastic accuracy drop of 50% or even more when switching from shorter to longer sequences. The noticeably reduced accuracy of DRfold for longer sequences represents a serious weakness of this single-sequence method. On the other hand, MSA-based methods DeepFoldRNA, trRosettaRNA, RoseTTAFoldNA, and RhoFold are somewhat dependent on the quality of evolutionary information as measured by log(Neff) values. As shown in Fig. 3c, DeepFoldRNA, RoseTTAFoldNA, and RhoFold are more MSA-dependent than trRosettaRNA, and running these MSA-based methods in single sequence mode leads to a consistent decline in global fold prediction accuracy (i.e. TM-score). The single-sequence method DRfold, on the other hand, remains naturally unaffected by the nature of evolutionary information, demonstrating its unique advantage.
With respect to the stereochemistry of the predicted structures, there are noticeable differences between different methods. As shown in Fig. 3d, the joint distribution of the two pseudo-torsion angle pairs η (defined by C4′i−1, Pi, C4′i, and Pi+1 atom coordinates) and θ (defined by Pi, C4′i, Pi+1, and C4′i+1 atom coordinates), where i represents the ith nucleotide in a specific RNA sequence, are closer to experimental observations for DeepFoldRNA, trRosettaRNA, and RoseTTAFoldNA than the other two methods. In terms of clash score metric computed from the MolProbity package [18] quantifying the steric violations, trRosettaRNA achieves the best performance (with the lowest mean clash score of 27.4), followed by RoseTTAFoldNA (mean clash score of 45.9), whereas DeepFoldRNA, DRfold, and RhoFold exhibit much inferior stereochemical qualities with a mean clash score of 167.1, 175.2, and 498.36, respectively.
How does the prediction accuracy of the five deep learning-based methods compare against the deep learning-free method RNAComposer chosen in this study? Supplementary Table S2 presents the average performance of all six methods on the common set of 70 targets excluding R1138 and 7QDU_Q due to RNAComposer’s prediction limit of targets having lengths up to 500 nucleotides. RNAComposer lags behind most of the deep learning-based methods across all three performance evaluation metrics, TM-score, lDDT, and INF-All, while outperforming all methods in terms of clash score. The mean prediction accuracy of RNAComposer in terms of TM-score is 0.22 compared to 0.39 of the best performing deep learning-based method DeepFoldRNA with a p-value of 1.7e × 10−11. In terms of lDDT and INF-All, once again RNAComposer shows lower mean values of 0.49 and 0.61, respectively, compared to the best-performing method RoseTTAFoldNA in both cases having mean values of 0.62 and 0.77 with statistically significant p-values of 1.2 × 10−10 and 3.1 × 10−9, respectively. Regarding clash score, RNAComposer achieves the lowest mean score of 22.66 compared to the best-performing method trRosettaRNA's mean score of 27.25, even though the performance difference is not statistically significant at 95% confidence level (p = 0.1). It is worth noting that despite our study showing evidence that deep learning-based methods attain comparable or better accuracy than the fully-automated deep learning-free counterparts, a combination of methods such as RNAComposer with expert human input [19] still outperforms the deep learning-based methods participating in CASP15 [2], highlighting the potential for further improvement in deep learning-based RNA 3D structure prediction.
Discussion
Amid the growing interest in the development of transformer-powered RNA 3D structure prediction methods, our neutral, multi-faceted, and empirical study reveals some interesting insights. First, some fundamental methodological differences exist between these emerging methods. These differences often lead to accuracy complementarity in terms of nucleotide orientation, global and local environment, and stereochemistry of the predicted structures. Furthermore, certain characteristics of the input itself such as length and availability of evolutionary information have more influence on the predictive modeling accuracy than some of the other subtle differences like the granularity of RNA conformational space representation. Second, leveraging the latest advances in RNA language models may help in reducing the dependency on length and/or evolutionary information as shown in our case study leading to a potentially improved generalizability. Third, even with the same input information, there exists a great deal of performance variability between different approaches, with certain methods having unique strengths in certain aspects of the evaluation measures. This suggests that further improvement may be possible by combining these methods. Finally, it is worth noting that despite the interest, the accuracies of the deep learning-based RNA structure predictors, including some of the methods discussed here, are still lagging behind the top-notch groups in CASP15, who did not use deep learning [2]. Progress on exploiting knowledge- and/or physics-guided deep learning may be beneficial for improving the state-of-the-art of deep learning enabled RNA 3D structure prediction.
Materials and methods
We used a benchmark set of 72 RNA targets consisting of 12 targets from the CASP15 website and 60 additional nonredundant RNA targets collected from the PDB repository. Although the recently published RNA structural dataset RNA3DB [20] is available for evaluating deep learning-based RNA modeling tasks, we chose not to use it due to the potential overlap with training sequences used by competing methods. Since none of the competing methods has made their training dataset public except for trRosettaRNA, it is impossible to curate a truly non-redundant benchmark set in terms of sequence and/or structural similarity. As such, we collected sequences from PDB that were released between 1 January 2022 and 6 July 2023 to minimize any overlap between our benchmark set and the datasets used for training or hyperparameter optimization in different methods wherever known (e.g. trRosettaRNA used structures released before 1 January 2022, RoseTTAFoldNA used structures released before May 2020, and DRfold used structures released before 2021) and after the release of the respective open-source code/data repositories. To curate our in-house benchmarking set of 60 RNAs, we downloaded the targets based on the above-mentioned date filtering from the PDB and performed several preprocessing steps on the collected sequences including discarding sequences with one or more unknown characters, removing fully duplicate sequences, and separating monomers from complexes. Finally, we ran CD-HIT [21] with default parameter settings to ensure that the curated set of additional RNA targets is nonredundant with respect to the set of 12 CASP15 targets. This yielded a set of 60 RNA targets of which 9 were extracted from protein–RNA complexes and the rest were RNA monomers (Supplementary Table S9). The final set of 72 targets represents a diverse array of structures characterized by a mean pairwise TM-score of 0.25, indicating minimal fold similarity among the chains and pairwise RMSD distribution spanning a wide range from 1 to 6 Å (Supplementary Fig. S4).
We predicted the structures of 72 sequences in the combined benchmark set using publicly available versions of DeepFoldRNA, trRosettaRNA, DRfold, RoseTTAFoldNA, RhoFold, and RNAComposer. Except for RoseTTAFoldNA, all four methods leverage AlphaFold2-inspired 48-block transformer architecture to update the sequence, pairwise and MSA representation of the input. These transformer architectures differ from each other in terms of channel dimension, number of attention heads, and attention-based hyperparameters such as number of query and value points (Supplementary Table S1). RoseTTAFoldNA uses SE(3)-equivariant transformer network consisting of 36 blocks that exploits 1D, 2D, and 3D tracks to update the input feature representations. We downloaded the open-source software implementations of DeepFoldRNA from https://github.com/robpearc/DeepFoldRNA/ (downloaded on 27 September 2022), trRosettaRNA from https://yanglab.nankai.edu.cn/trRosettaRNA/ (downloaded on 11 November 2022), RoseTTAFoldNA from https://github.com/uw-ipd/RoseTTAFold2NA/ (downloaded on 13 May 2023), DRfold from https://zhanggroup.org/DRfold/ (downloaded on 11 June 2023), and RhoFold from https://github.com/ml4bio/RhoFold (downloaded on 13 May 2023). All methods were run in our in-house 64-bit Linux cluster using an 80-GB NVIDIA A100 GPU with their default parameter settings without any post-prediction optimization or refinement. All MSA-based methods used the same MSA as input generated by the rMSA [10] pipeline. For methods that output multiple predicted structural models, we only considered the top-ranked models based on the method’s internal ranking (or the first model, in the case of DeepFoldRNA, which does not rank models) for a fair performance evaluation. For RNAComposer, we predicted the structures using a publicly available web server at https://rnacomposer.cs.put.poznan.pl with CentroidFold [22] as secondary structure input.
The predicted structural models were evaluated against the experimental coordinates provided by the CASP organizers for targets cleared for public access as of 20 December 2022 from https://predictioncenter.org/download_area/CASP15/targets/ (Supplementary Table S10) or obtained from PDB. We evaluated the predictive modeling accuracy using various metrics that include the correctness of global fold using the template modeling score (TM-score) [13], distance-based structural similarity using a pared-down RNA-specific version of local distance difference test lDDT [14] that does not penalize stereochemical violation, environment-dependent local quality using the interaction network fidelity (INF) [15], and clash score from the MolProbity package [18].
Author contributions
Sumit Tarafder (Data curation [equal], Formal analysis [equal], Investigation [equal], Resources [equal], Software [equal], Validation [equal], Visualization [equal], Writing—original draft [equal], Writing—review & editing [equal]), Rahmatullah Roche (Resources [supporting], Validation [supporting], Writing—review & editing [supporting]), and Debswapna Bhattacharya (Conceptualization [equal], Data curation [equal], Formal analysis [equal], Funding acquisition [equal], Investigation [equal], Methodology [equal], Project administration [equal], Resources [equal], Software [equal], Supervision [equal], Validation [equal], Visualization [equal], Writing—original draft [equal], Writing—review & editing [equal])
Supplementary Material
Contributor Information
Sumit Tarafder, Department of Computer Science, Virginia Tech, Blacksburg, VA 24061, United States.
Rahmatullah Roche, Department of Computer Science, Virginia Tech, Blacksburg, VA 24061, United States.
Debswapna Bhattacharya, Department of Computer Science, Virginia Tech, Blacksburg, VA 24061, United States.
Supplementary data
Supplementary data is available at Biology Methods and Protocols online.
Conflict of interest statement. None declared.
Funding
This work was partially supported by the National Institute of General Medical Sciences (R35GM138146 to D.B.) and the National Science Foundation (DBI2208679 to D.B.). Funding for open access charge: National Institute of General Medical Sciences [R35GM138146 to D.B.]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data availability
Predicted RNA structural models and metrics calculated to benchmark the predictive modeling performance have been deposited to https://github.com/Bhattacharya-Lab/RNAmark/ (https://doi.org/10.5281/zenodo.12533828).
References
- 1. Jumper J, Evans R, Pritzel A. et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021;596:583–89. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Das R, Kretsch RC, Simpkin AJ. et al. Assessment of three-dimensional RNA structure prediction in CASP15. Proteins: Struct Funct Bioinf 2023;91:1747–70. 10.1002/prot.26602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pearce R, Omenn GS, Zhang Y.. De Novo RNA tertiary structure prediction at atomic resolution using geometric potentials from deep learning. Preprint at bioRxiv, 2022. 10.1101/2022.05.15.491755. [DOI] [Google Scholar]
- 4. Wang W, Feng C, Han R. et al. trRosettaRNA: automated prediction of RNA 3D structure with transformer network. Nat Commun 2023;14:7266. 10.1038/s41467-023-42528-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baek M, McHugh R, Anishchenko I. et al. Accurate prediction of protein–nucleic acid complexes using RoseTTAFoldNA. Nat Methods 2023;21:117–21. 10.1038/s41592-023-02086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fuchs F, Worrall D, Fischer V. et al. SE(3)-transformers: 3D roto-translation equivariant attention networks. In: Advances in Neural Information Processing Systems. Curran Associates, Inc., 2020, 1970–81. https://proceedings.neurips.cc/paper_files/paper/2020/file/15231a7ce4ba789d13b722cc5c955834-Paper.pdf
- 7. Li Y, Zhang C, Feng C. et al. Integrating end-to-end learning with deep geometrical potentials for ab initio RNA structure prediction. Nat Commun 2023;14:5745. 10.1038/s41467-023-41303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shen T, Hu Z, Peng Z. et al. E2Efold-3D: End-to-end deep learning method for accurate de novo RNA 3D structure prediction. Preprint at arXiv: 2207.01586, 2022. 10.48550/arXiv.2207.01586. [DOI]
- 9. Abramson J, Adler J, Dunger J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024;630:493–500. 10.1038/s41586-024-07487-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang C, Zhang Y, Pyle AM.. rMSA: a sequence search and alignment algorithm to improve RNA structure modeling. J Mol Biol 2023;435:167904. 10.1016/j.jmb.2022.167904. [DOI] [PubMed] [Google Scholar]
- 11. Popenda M, Szachniuk M, Antczak M. et al. Automated 3D structure composition for large RNAs. Nucleic Acids Res 2012;40:e112. 10.1093/nar/gks339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Berman HM, Westbrook J, Feng Z. et al. The protein data bank. Nucleic Acids Res 2000;28:235–42. 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang C, Shine M, Pyle AM. et al. US-align: universal structure alignments of proteins, nucleic acids, and macromolecular complexes. Nat Methods 2022;19:1109–15. 10.1038/s41592-022-01585-1. [DOI] [PubMed] [Google Scholar]
- 14. Mariani V, Biasini M, Barbato A. et al. lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics 2013;29:2722–28. 10.1093/bioinformatics/btt473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Parisien M, Cruz JA, Westhof É. et al. New metrics for comparing and assessing discrepancies between RNA 3D structures and models. RNA 2009;15:1875–85. 10.1261/rna.1700409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kryshtafovych A, Antczak M, Szachniuk M. et al. New prediction categories in CASP15. Proteins: Struct Funct Bioinf 2023;91:1550–57. 10.1002/prot.26515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ma L, Bajic VB, Zhang Z.. On the classification of long non-coding RNAs. RNA Biol 2013;10:925–33. 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen VB, Arendall WB, Headd JJ. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 2010;66:12–21. 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sarzynska J, Popenda M, Antczak M, Szachniuk M.. RNA tertiary structure prediction using RNAComposer in CASP15. Proteins: Struct. Funct. Bioinf. 2023;91:1790–99. 10.1002/prot.26578. [DOI] [PubMed] [Google Scholar]
- 20. Szikszai M, Magnus M, Sanghi S. et al. RNA3DB: a structurally-dissimilar dataset split for training and benchmarking deep learning models for RNA structure prediction. J Mol Biol 2024;168552:168552. 10.1016/j.jmb.2024.168552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fu L, Niu B, Zhu Z. et al. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 2012;28:3150–12. 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hamada M, Kiryu H, Sato K. et al. Prediction of RNA secondary structure using generalized centroid estimators. Bioinformatics 2009;25:465–73. 10.1093/bioinformatics/btn601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Predicted RNA structural models and metrics calculated to benchmark the predictive modeling performance have been deposited to https://github.com/Bhattacharya-Lab/RNAmark/ (https://doi.org/10.5281/zenodo.12533828).