Abstract
Vaccinia virus gene F12L is shown to encode a 65-kDa protein that is synthesized early and late during infection and that is not modified by glycosylation. Computational sequence comparison revealed that related proteins are encoded by all sequenced chordopoxviruses. A virus deletion mutant lacking the F12L gene (vΔF12L) and a revertant virus with the F12L gene reinserted into the deletion mutant (vF12L-rev) were constructed and analyzed. A version of the F12L gene with a C-terminal amino acid tag derived from the influenza virus hemagglutinin and that is recognized by a monoclonal antibody was also inserted into the F12L locus of vΔF12L. Loss of the F12L protein reduced the formation of IMV 2-fold, but there was a dramatic (99.5%) reduction in actin tail formation, and the levels of cell-associated enveloped virus and extracellular enveloped virus were reduced 8- to 11-fold and 7-fold, respectively. Consistent with the lack of actin tail formation, vΔF12L produced a very small plaque. The vΔF12L virus was severely attenuated in vivo, such that a dose of vΔF12L 10,000-fold greater than the dose of wild-type virus that induced severe disease was unable to induce disease in mice infected intranasally.
Vaccinia virus (VV) is a large DNA virus that replicates in the cytoplasm of infected cells (34). Like all poxviruses, the VV particle is large and complex, and more than 100 polypeptides have been identified in the purified virion (15). The virus genome encodes approximately 200 genes and has been sequenced for VV strains Copenhagen (18), modified virus Ankara (2), and most of the Western Reserve (WR) strain (see reference 46 for additional references).
VV produces two infectious forms of virus called intracellular mature virus (IMV) and extracellular enveloped virus (EEV). EEV was shown to be antigenically distinct from IMV (3), and an immune response against the EEV-specific antigens is necessary for protection against orthopoxvirus challenge (6, 51). Virus morphogenesis begins in cytoplasmic factories that are largely devoid of cellular organelles (11, 28). The first visible structures within these factories are virus crescents that are composed of lipid and virus protein, but the origin and composition of these crescents is disputed. Early studies proposed that crescents contain a single lipid membrane that was synthesized de novo (11). Later these structures were proposed to contain a double lipid bilayer that was derived from and was continuous with intermediate compartment membranes between the endoplasmic reticulum and the Golgi complex (47). More recently, additional electron microscopic evidence reported that there was only a single lipid bilayer without continuity with cellular membranes (22). After their formation, lipid crescents extend into ovals called immature virus that lack infectivity. These mature into electron-dense IMV particles by condensation of the core and proteolytic processing of several core proteins.
IMV particles represent the majority of infectious progeny and most remain within the infected cell until cell lysis. However, some IMV particles are transported away from the virus factory in a process that is dependent upon the A27L protein and microtubules (41) to sites where they become wrapped by two additional cellular membranes (25). These wrapping membranes are derived from the tubular endosomes (50) or trans-Golgi network (20, 42) that have been modified by the inclusion of virus-encoded proteins that ultimately become part of the EEV outer envelope. This wrapping process produces an intracellular enveloped virus (IEV). IEV particles move to the cell surface where the outer membrane fuses with the plasma membrane, exposing a virion the cell surface. If this virion is retained on the cell surface or is released and then reattaches, it is called cell associated enveloped virus (CEV) (4), but if it is released it is called EEV. VV infection induces the polymerization of actin tails that protrude from the cell surface with an enveloped virion at their tip (9). Virus mutants that are unable to induce the polymerization of actin form a small plaque due to inefficient cell-to-cell spread (see below).
Six VV genes were reported to encode EEV-specific proteins. These are F13L (p37) (21), A33R (gp22-28) (37), A34R (gp22-24) (12), A36R (p45-50) (36), A56R (gp86, the virus hemagglutinin [HA]) (45), and B5R (gp42) (13, 27). Recently, however, the A36R protein was found not to be present in the CEV or EEV particle, despite copurifying with EEV in density gradients (52). The roles of these proteins have been investigated by the analysis of virus mutants in which the individual genes are mutated, repressed, or deleted. These studies have shown that none of the EEV proteins have a major effect on the production or infectivity of IMV, but they have different effects on the subsequent stages of morphogenesis. F13L (4), B5R (14, 56), and A34R (12, 32, 57) are needed for the wrapping of IMV particles since the formation of IEV is reduced or abolished in their absence. Consequently, the formation of actin tails is also greatly inhibited or abolished with these mutants (9, 19, 31, 39, 40, 57, 58). A56R is not required for either IEV or actin tail formation (40), and the plaque formed by the deletion mutant is of wild-type size but syncytial. In the absence of A33R, some IEV particles are made but an increased proportion of these have incomplete wrapping and actin tails are not formed (38). Lastly, A36R is not required for IEV formation but is required for production of actin tails (17, 36, 39, 40, 58). A36R has a type Ib membrane topology with the N terminus and the majority of the protein in the cytosol (39, 52). It has an unusual distribution and is present on the outer IEV membrane and the plasma membrane beneath CEV particles (52).
The production of EEV by these mutants is variable. Without F13L (4) and B5R (14, 56), EEV formation is reduced by 10- to 100-fold, and without A36R EEV it is reduced 3- to 5-fold (36). In contrast, loss of the HA did not affect EEV formation or infectivity (G. L. Smith, unpublished data) and loss of A33R (38) and A34R (32) increased EEV formation 2- to 4-fold and 25-fold, respectively. However, the EEV formed in the absence of A34R had a fivefold-reduced specific infectivity (32).
In this report we have characterized the F12L gene product and studied the properties of a virus lacking this gene. This deletion mutant formed normal amounts of IMV but was defective in actin tail formation, plaque size, CEV and EEV production, and virulence.
MATERIALS AND METHODS
Cells and viruses.
Monkey kidney BS-C-1 and CV-1 cells, rabbit kidney (RK)13 cells, human osteosarcoma TK−143B cells and HeLa D980R cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Gibco). VV strain WR was used throughout.
Plasmids.
A plasmid that could be used to construct a VV F12L deletion mutant (vΔF12L) was constructed as follows. The 5′ (353 bp) and 3′ (349 bp) ends of the F12L gene were amplified by PCR using VV WR DNA as a template. Oligonucleotides 5′-CCCGGATCCATGTTAAACAGGGTACAA (forward) and 5′-CCCGGATCCGTGCTATATCTCCCTGTT (reverse) were used for the 5′ fragment and included terminal BamHI sites (underlined). Oligonucleotides 5′-CCCGTCGACCTGCTATCAGAGCAGGAT (forward) and 5′-CCCGTCGACTTATAATTTTACCATCTG (reverse) were used for the 3′ fragment and included terminal SalI sites (underlined). The initiation codon and termination codon (complement of) are shown in boldface type. The PCR fragment representing the 5′ end of the gene was digested with BamHI and cloned into pSJH7 (24) that had been digested with BamHI. pSJH7 contains the Escherichia coli guanine xanthine phosphoribosyl transferase (Ecogpt) gene linked to the VV 7.5K early-late promoter (7). The resultant plasmid was then digested with SalI and ligated with the PCR fragment representing the 3′ end of the open reading frame (ORF) that has also been digested with SalI. The resulting plasmid, pΔF12L, contained the F12L gene with a 1,207-bp internal deletion.
To express the F12L ORF in an inducible manner in VV-infected cells, the entire F12L ORF flanked by 5′ NdeI and 3′ SalI sites (underlined) was amplified by PCR using VV DNA as template and oligonucleotides 5′-GGGAATTCCATATGTTAAACAGGGTACAAATCTTG (forward) and 5′-CCCGTCGACTTATAATTTTACCATCTG (reverse) as primers. Emboldened nucleotides indicate the translation initiation codon and termination codon (complement of). The PCR product was digested with NdeI and SalI and inserted into plasmid pVOTE.2 (54), forming pVOTE.2-F12L.
To attach the influenza virus HA epitope YPYDVPDYA at the C terminus of F12L, two PCR fragments were generated using virus DNA as a template. One fragment contained the entire F12L ORF, together with a 5′ HindIII site and the C-terminal HA tag (primers 5′-GCGAAGCTTATGTTAAACAGGGTACAAATCTTGATG [forward] and 5′-agcgtaatcaggcacgtcgtaaaggtaTAATTTTACCATCTGACTCATG [reverse]). The second fragment contained 501 bp downstream from the F12L termination codon, together with the 5′ HA tag and the 3′ XhoI restriction site (primers 5′-tacgacgtgcctgattacgctTAAAAAGTGAAAAACAATATTATTTTT [forward] and 5′-GCGCTCGAGCTCATTTTTTAAGCAGATTGTTGC [reverse]). Boldface, underlined, or lowercase nucleotides represent the translation initiation and termination codons, restriction sites or HA tag, respectively. These PCR fragments were then assembled into a single DNA fragment by splicing by overlap extension (23). The assembled DNA fragment was digested with HindIII and XhoI and inserted into pBAC-1 (Clontech) that had been digested with the same enzymes. All DNA fragments that were generated by PCR and cloned into plasmid vectors were sequenced and shown to be correct.
Construction of recombinant VVs.
A recombinant VV lacking 1,207 bp (63%) of the F12L ORF was constructed by transient dominant selection (16). Briefly, plasmid pΔF12L was transfected with Lipofectin (Gibco-BRL) into CV-1 cells that had been infected with VV WR. A mycophenolic acid (MPA)-resistant virus was selected from the resultant progeny virus by plaque assay on BS-C-1 cells. This intermediate virus was then resolved by plaque assay on HeLa D980R cells (29) in the presence of 6-thioguanine (26) to produce the deletion mutant (vΔF12L) and a plaque purified wild-type virus (vF12L) derived from the same intermediate. A revertant virus was constructed by rescue of the small-plaque phenotype. CV-1 cells were infected with vΔF12L and transfected with a plasmid containing the entire HindIII F fragment cloned into pUC13. A virus forming a large plaque was selected from the progeny by plaque assay on BS-C-1 cells, plaque purified three times, and designated vF12L-rev. The recombinant virus expressing the HA-tagged version of the F12L protein was selected in a similar way after transfection of pBAC-1-F12LHA into vΔF12L-infected CV-1 cells. This virus was called vF12LHA. Lastly, a virus with an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible version of the F12L gene was constructed by infecting CV-1 cells with vT7LacOI (54), transfecting these cells with pVOTE.2-F12L, and selecting an MPA-resistant recombinant virus by plaque assay on BS-C-1 cells. After three cycles of plaque purification, this virus was amplified and designated vindF12L.
Antiserum to the F12L protein.
A rabbit polyclonal antibody was raised against synthetic peptide SYKDINESMSQMVK (F12L amino acids 621 to 634) coupled to keyhole limpet hemocyanin by intramuscular injection with the peptide emulsified in complete Freund adjuvant. Rabbits were boosted with the same antigen emulsified in incomplete Freund adjuvant.
Immunocytochemistry.
BS-C-1 cells were grown on glass coverslips and were infected with 0.1 PFU/cell. At 15 h postinfection (p.i.) the infected cells were washed with cold phosphate-buffered saline (PBS) and fixed in ice-cold acetone for 1 min at room temperature (RT). After incubation in blocking buffer (5% FBS–1% bovine serum albumin in PBS) for 1 h at RT, the cells were incubated at RT for 1 h with mouse monoclonal antibody (MAb) AB1.1 directed against the VV D8L protein (36). After extensive washing, the bound antibody was detected with fluorescein isothiocyanate-conjugated donkey anti-mouse (Stratech Scientific, Luton, United Kingdom) (diluted 1:100 in blocking buffer) for 45 min at RT. F-actin was stained with tetramethyl rhodamine isothiocyanate (TRITC)-phalloidin (Sigma, Poole, United Kingdom) for 1 h at RT. Samples were mounted in mowiol (87.5% glycerol, 10% PBS, 2.5% diazabicydo) and analyzed by a Bio-Rad MRC 1024 confocal microscope, and images were processed with Adobe Photoshop software.
Virulence assay.
Groups of five BALB/c mice were inoculated intranasally under general anaesthetic with doses of vF12L, vΔF12L, or vF12L-rev of between 104 and 108 PFU in 25 μl of PBS. Mice were weighed daily before and after infection, and the mean weights for each group of animals were calculated and compared with the mean weights of the same group of animals on day 0. Animals that had lost 30% of their body weight were sacrificed by cervical dislocation.
RESULTS
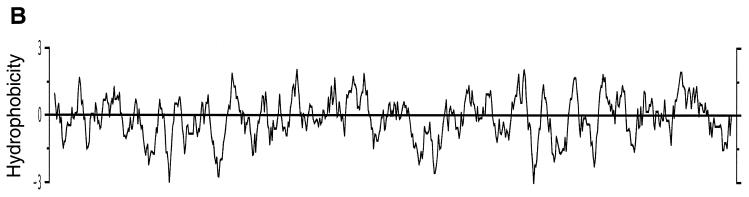
The VV F12L gene was selected for study because there is a conserved counterpart of this ORF in fowlpox virus (24% amino acid identity) and insertional mutagenesis of this fowlpox virus gene resulted in a small-plaque phenotype and sixfold-reduced production of extracellular virus (35). The phenotype of the fowlpox virus mutant is similar to some VV mutants that have lost genes encoding EEV-specific proteins (Introduction). In addition, there are related genes in all other sequenced chordopoxviruses, including variola virus (44), molluscum contagiosum virus (43), Shope fibroma virus (55), and myxoma virus (8), implying an important and conserved function. The F12L-like proteins encoded by all these viruses have similar lengths. The VV protein shares 95% amino acid identity with the comparable protein in variola virus strain Bangladesh, 34% identity and 55% similarity with Shope fibroma virus, 27% identity and 49% similarity with molluscum contagiosum virus, and 27% identity and 45% similarity with fowlpox virus. The F12L ORF in VV strain Copenhagen was predicted to encode a protein of 73.2 kDa (635 amino acid residues) (18). The sequence of the F12L ORF in VV WR was determined by J. Sisler and kindly provided by B. Moss (National Institutes of Health, Bethesda, Md.) and showed 16 nucleotide changes from the Copenhagen sequence out of 1,908 nucleotides (99.2% identity). Four of these resulted in amino acid substitutions H38R, S98P, I239M, and N545H (Copenhagen residue shown first), but the proteins were the same lengths. An alignment of the proteins from different chordopoxviruses is shown in Fig. 1A, and the hydropathy profile is shown in Fig. 1B. A computational search showed that VV F12L had six potential sites for attachment of N-linked carbohydrate, but no signal sequence was identified. Several regions of hydrophobic nature are present within F12L (Fig. 1B), but it is unclear if these would cause association of the protein with membranes of the infected cell or virus particles.
FIG. 1.
(A) Amino acid alignment of the VV F12L protein and related proteins from other chordopoxviruses. VAC, VV strain WR; VAR, variola virus strain Bangladesh-1975 (30); SFV, Shope fibroma virus (55); MCV, molluscum contagiosum virus (43); FPV, fowlpox virus (35). Positions showing conserved amino acid residues in five, four, or three sequences are highlighted in black, dark gray, or light gray, respectively. (B) Hydrophobicity profile of the VV F12L protein.
Characterization of the F12L protein.
To identify the F12L protein, a recombinant virus was constructed that contained the F12L ORF fused to a nine-amino-acid tag at the C terminus that is recognized by MAb HA.11 (Covalence, Luton, United Kingdom). This tagged version of the F12L gene was inserted into the VV mutant from which the majority of the F12L gene had been deleted (vΔF12L) (see Materials and Methods), taking advantage of the small plaque phenotype of vΔF12L (see below). By transfecting the plasmid with the HA-tagged F12L allele into cells infected with vΔF12L, a large plaque recombinant virus (vF12LHA) was selected. Southern blotting and PCR analysis of the genome of vF12LHA confirmed that it had the predicted genomic structure (data not shown). The plaque size and growth properties of this virus in cell culture were indistinguishable from the wild-type virus, indicating that the addition of the short C-terminal tag to the F12L protein was not deleterious.
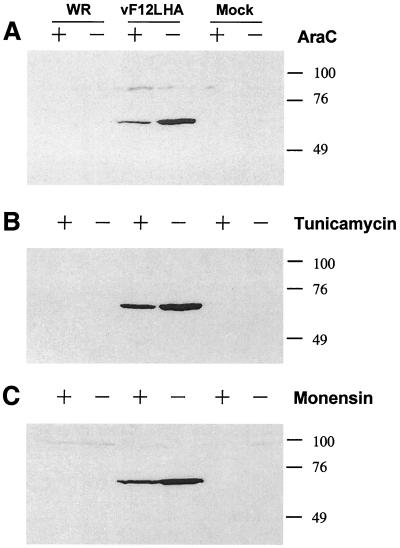
Cells were mock infected or were infected with WR or vF12LHA viruses, and extracts of these cells were analyzed by immunoblotting using MAb HA.11 (Fig. 2A). This antibody detected a protein of approximately 65 kDa in vF12LHA-infected cells that was absent from cells infected with WR and from mock-infected cells. In the presence of cytosine arabinoside (a drug that inhibits virus DNA replication and hence the expression of intermediate and late virus proteins), the amount of F12L protein detected by immunoblot was reduced greatly but was still detectable. This indicated that the protein is expressed early and late during infection, despite the lack of a TAAAT late transcriptional start motif near the 5′ end of the ORF. To determine if the F12L protein was modified by the addition of carbohydrate, parallel cultures were infected or mock infected in the presence of tunicamycin or monensin, drugs that inhibit the addition of N- or O-linked carbohydrate, respectively (Fig. 2B and C). Neither of these drugs affected the size of the F12L protein as far as could be determined from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), suggesting that the F12L protein was either not glycosylated or contained very little carbohydrate, despite containing six potential sites for addition of N-linked glycans. However, both drugs reduced the amount of F12L protein detected at late times in infection.
FIG. 2.
Immunoblot showing the identification of the F12L protein. BS-C-1 cells were infected with the indicated viruses at 10 PFU/cell. At 15 h p.i. the infected cells were harvested, and extracts of the infected cells were analyzed by SDS-PAGE and immunoblotting. Cells were infected in the presence or absence of 40 μg of cytosine arabinoside (AraC) (A), 10 μg of tunicamycin (B), or 10 μg of monensin (C) per ml, as indicated. Blots were incubated with MAb HA.11 (diluted 1:400 in blocking buffer), and bound antibody was detected by incubation with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G, followed by chemiluminescent reagents as directed by the manufacturer (Amersham). The positions of molecular weight markers are shown in kilodaltons.
The F12L protein copurifies with EEV.
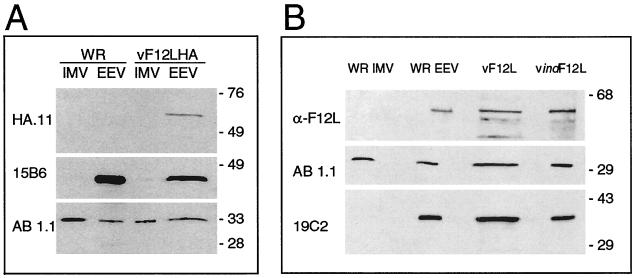
To determine if F12LHA is incorporated into virions, cells were infected with either vF12LHA or WR viruses, and IMV and EEV particles were purified from the cytoplasm of infected cells or from the cell culture supernatant by sucrose density gradient centrifugation. Equal amounts of IMV and EEV proteins were then analyzed by SDS-PAGE and immunoblotting using MAbs HA.11, 15B6 that is directed against the p37 EEV protein encoded by gene F13L (20), and AB1.1 that is directed against the 32-kDa protein of IMV encoded by gene D8L (36) (Fig. 3A). MAb AB1.1 detected protein in IMV and EEV from each virus, while MAb 15B6 detected the p37 protein only in EEV from each virus, as expected. However, MAb HA.11 detected protein only in EEV from vF12LHA. This indicated that the F12L protein copurified with EEV particles.
FIG. 3.
Immunoblots showing F12L associated with EEV. (A) IMV and EEV were purified from cells or culture supernatants of cells infected with WR or vF12LHA as described in Materials and Methods. Equivalent amounts (3 μg) of purified EEV or IMV were then analyzed by SDS-PAGE and immunoblotting with the MAbs HA.11 (anti-HA tag, diluted 1:400), 15B6 (anti-F13L, diluted 1:2,000), and AB1.1 (anti-D8L, diluted 1:2,000) overnight at 4°C for MAb HA.11 or at RT for 1 h for other antibodies. (B) WR IMV and EEV as in panel A or extracts from BS-C-1 cells infected with vF12L or vindF12L in the presence of 10 mM IPTG were analyzed as in panel A. A rabbit antiserum raised against the F12L protein and MAbs AB1.1 (anti-D8L) and 19C2 directed against the B5R protein (tissue culture supernatant diluted 1:8) (42) were used. Molecular weight markers are shown in kilodaltons.
To obtain independent evidence that the F12L protein was associated with EEV and that this was not due to the attachment of the HA tag at the C terminus of F12L, WR IMV and EEV were purified and immunoblotted with an antibody raised against the F12L protein (see Materials and Methods) (Fig. 3B). This detected a protein or approximately 65-kDa in EEV and not IMV, although the signal was weak. To determine if the protein detected in EEV was the same size as the F12L protein in infected cells, extracts of cells infected with a plaque-purified wild-type virus (vF12L) (see Materials and Methods) or a virus that expressed the F12L gene upon addition of IPTG (vindF12L) (see Materials and Methods) were analyzed in parallel (Fig. 3B). A protein of the same size was seen in cells infected with each virus. As controls, MAb AB1.1 detected the D8L protein in extracts from infected cells and each type of virion, and MAb 19C2, directed against the B5R gene product (42), detected the B5R protein in infected cells and EEV but not in IMV.
Phenotype of a mutant virus lacking F12L.
The role of the F12L protein in the virus life cycle was examined by the construction of a virus mutant lacking 63% of the F12L ORF (Materials and Methods). Five days after infection under semisolid overlay, the plaque size of this mutant was very small compared to a plaque purified wild-type virus (vF12L) isolated from the same intermediate (Fig. 4). A revertant virus in which the F12L gene was reinserted into the vΔF12L genome (vF12L-rev) was analyzed in parallel and showed a similar-sized plaque to that of the wild type. Incubation of infected cells under liquid overlay showed that the formation of secondary plaques was reduced, suggesting reduced release of EEV (data not shown). The genome structures of these viruses were analyzed by PCR and Southern blotting and shown to be as expected (data not shown).
FIG. 4.
Plaque size formed by viruses. BS-C-1 cells were infected with the indicated viruses and incubated under semisolid overlay (DMEM containing 2.5% FBS and 1.5% carboxymethylcellulose) for 5 days. The monolayers were stained with 0.1% (wt/vol) crystal violet in 15% ethanol.
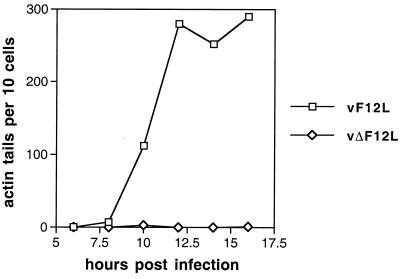
The small plaque size of vΔF12L was consistent with the phenotype of the fowlpox virus lacking this gene (35) and also with VV mutants that lacked the F13L, A33R, A34R, A36R, and B5R proteins (see the introduction). These other VV mutants had greatly reduced or abolished actin tail formation. Consequently, the ability of the vΔF12L mutant to induce actin tails was examined (Fig. 5). Infected cells were stained with MAb AB1.1 to detect all virus particles and costained with TRITC-phalloidin to detect F-actin. In cells infected with vF12L (Fig. 5) or vF12L-rev (data not shown), there were numerous actin tails with a virus particle at their tip. In contrast, in vΔF12L-infected cells the number of actin tails was reduced by 99.5% and the rare actin tails that were detected appeared thicker than those in vF12L-infected cells (data not shown).
FIG. 5.
Graph showing the number of virus-tipped actin tails in infected cells at different times p.i. BS-C-1 cells were infected with vF12L, vΔF12L, or vF12L-rev at 0.1 PFU/cell and at the indicated times p.i. were stained with MAb AB1.1 to reveal all VV particles and with TRITC-phalloidin to detect F-actin. Samples were analyzed by confocal microscopy and reconstructed z-series of images of different sections through the cell were examined. The number of virus-tipped actin tails was counted for each of 10 infected cells (as shown by reactivity with AB1.1), and the average number is shown. The number of virus-tipped actin tails in cells infected with vF12L and vF12L-rev were indistinguishable, and data are shown for only vF12L compared with vΔF12L.
Growth properties of vΔF12L.
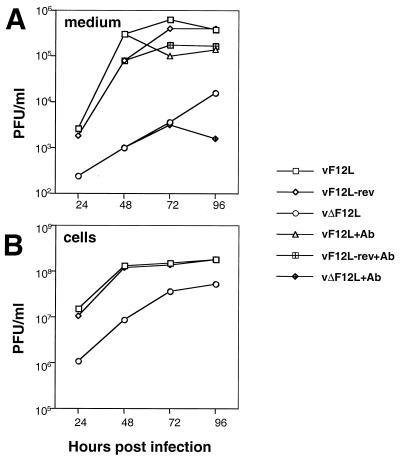
The growth properties of the virus deletion mutant were examined in cell culture (Fig. 6). After low-multiplicity infection (0.01 PFU/cell), the production of both IMV and EEV was reduced greatly. For IMV, this reduction was probably attributable partly to the reduced rate of spread of virus from the first infected cells (consistent with the small-plaque phenotype), but even after infection at 10 PFU/cell there was a twofold reduction in infectious IMV (data not shown). Notably, the difference between the titer of infectious IMV made by vΔF12L and vF12L or vF12L-rev decreased at later times after infection, presumably as the infection spread slowly to surrounding uninfected cells. For EEV, the difference in infectious titer between the deletion mutant and the controls after infection at 0.01 PFU/cell was approximately 100-fold by 48 and 72 h p.i., and a large difference remained even at 96 h p.i. To determine the proportion of this extracellular virus that was EEV, rather than IMV that had been released from cells at these late times p.i., virus in the supernatant was incubated with mouse MAb 5B4/2F2 directed against the IMV surface protein encoded by the A27L gene and that neutralizes IMV infectivity (10). Under these conditions the remaining infectivity of vΔF12L, representing EEV, was still approximately 100-fold lower than that of wild-type or revertant viruses. This indicated a specific defect in the production of EEV from cells infected with a virus lacking the F12L protein.
FIG. 6.
Growth of vΔF12L in tissue culture. BS-C-1 cells were infected with the indicated viruses at 0.01 PFU/cell. After incubation for 1 h at 37°C, nonabsorbed virus was washed away, and cells were incubated in minimal essential medium containing 1% FBS. (A) At the indicated times, culture supernatants were removed and, after centrifugation at 1,000 × g for 10 min to remove detached cells and cell debris, the virus infectivity was determined by plaque assay on fresh monolayers of BS-C-1 cells. Where indicated (+Ab), the infectivity of the diluted virus samples was determined after incubation with MAb 5B4/2F2 that neutralizes IMV (10), as described previously (53). (B) The infectivity associated with the infected cells was determined by scraping the cells into PBS, recovering the cells by centrifugation as described above, and combining the pellets with the pellets derived from centrifugation of the culture supernatant. The infectivity present was determined by plaque assay on fresh monolayers of BS-C-1 cells.
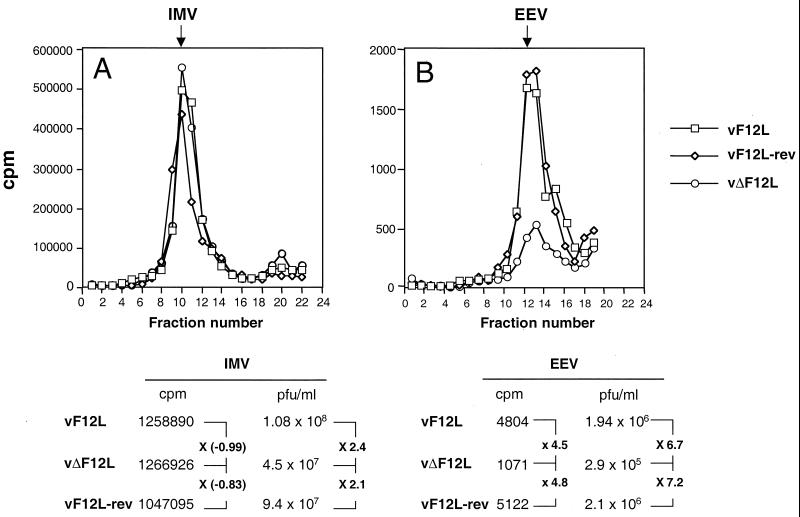
To examine further the formation of IMV and EEV, cells were infected with vF12L, vΔF12L, or vF12L-rev and labeled with [3H]thymidine from 1.75 to 24 h p.i. IMV within infected cells and EEV present in the culture supernatant were then purified by CsCl density gradient centrifugation (36). The radioactivity in the gradient fractions, determined by scintillation counting, showed similar-sized peaks for each virus that corresponded to the density of IMV (Fig. 7). However, when the titer of infectious virus present in these peaks was determined by plaque assay, there was a reduction in IMV titer for the deletion mutant of between 2.1- and 2.4-fold compared with revertant and wild-type viruses, respectively.
FIG. 7.
CsCl density gradient analysis of IMV and enveloped viruses produced by vF12L, vΔF12L and vF12L-rev. RK13 cells were infected with the indicated viruses at 10 PFU per cell and labeled with 100 μCi of [3H]thymidine (Amersham) from 1.75 h p.i. At 24 h p.i., the virus in the infected cells (A) or culture supernatant (B) was purified by CsCl density gradient centrifugation as described previously (36). The density of the fractions was determined by refractometry. The infectious virus present in peak fractions corresponding to IMV or EEV was titrated by plaque assay on BS-C-1 cells, and the amount of radioactivity in these fractions was determined by scintillation counting. These values are displayed underneath the graphs, and the relative differences between the deletion mutant and control viruses are displayed.
For EEV there was a more dramatic difference. The amount of radioactive EEV made by vΔF12L was reduced 4.5- to 4.8-fold compared to controls. The difference in the infectious virus present in these peaks was slightly greater, 6.7- to 7.2-fold, compared to vF12L and vF12L-rev, respectively. Thus, the loss of the F12L protein had a relatively minor effect on IMV production but reduced the production of EEV physical particles approximately fivefold and of EEV infectious particles approximately sevenfold.
The formation of CEV particles by the vΔF12L was also analyzed. To do this, BS-C-1 cells were infected with vF12L, vΔF12L, or vF12L-rev at 5 PFU/cell for 24 h p.i., the medium was removed, and the cells were incubated in PBS containing 1 μg of trypsin per ml for 1 h at 37°C. Infectious virus released from the cells during this time (predominately CEV) (5) was then measured by plaque assay in duplicate. The virus titers were as follows: vF12L, 5.2 × 105 PFU/ml; vΔF12L, 4.6 × 104 PFU/ml; and vF12L-rev, 3.6 × 105 PFU/ml. These results show that the level of infectious virus released from vΔF12L-infected cells by this treatment was approximately 8- to 11-fold lower than from cells infected with vF12L or vF12L-rev. This suggests that low levels of CEV particles are produced by vΔF12L and, taken together with the low levels of EEV made by this virus (Fig. 7), indicates that the F12L protein plays an important role in the formation of cell surface or released enveloped virus.
The vΔF12L virus is attenuated in vivo.
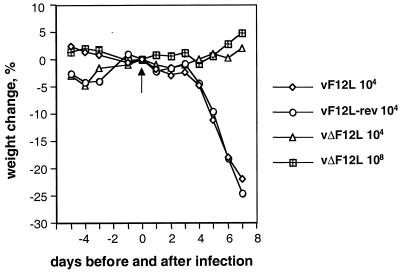
In several other cases, mutant viruses that have a decreased plaque size are attenuated in in vivo models. To determine if the loss of the F12L protein affected virus virulence, groups of 5 BALB/c mice were infected with vF12L, vΔF12L, or vF12L-rev, and their body weights were recorded (Fig. 8). After infection with 104 PFU of vF12L or vF12L-rev, all animals suffered rapid weight loss and were sacrificed by 8 days p.i. In contrast, animals infected with 104 PFU of vΔF12L showed no weight loss and continued to increase in weight similar to uninfected controls. Moreover, even at a 10,000-fold-greater dose of virus (108 PFU), the animals infected with the vΔF12L remained healthy and had only a very small (5% maximum), transient weight loss. The loss of the F12L gene thus reduces VV virulence profoundly.
FIG. 8.
Virulence assay. Groups of five BALB/c female mice were infected intranasally with the indicated doses of vF12L, vΔF12L, or vF12L-rev on day 0 (arrow). The mean weight of each group of animals compared to the weight of the same group on day zero are shown ± the standard error of the mean (n = 5).
DISCUSSION
This report shows that the VV F12L gene encodes a 65-kDa protein that is made early and late during infection and that copurifies with EEV but not IMV. Although, this might suggest that F12L is another EEV-specific protein, this is not certain, and additional data using immunoelectron microscopy and confocal microscopy are needed to examine this more carefully. These studies are in progress. Previously, the A36R protein was found to copurify with EEV on density gradients (36), but subsequent microscopic analysis found that the protein was not on CEV particles but copurified with membrane fragments attached to the WR EEV particles (52).
The roles of the F12L protein in virus replication, dissemination, and virulence were analyzed by using plaque-purified wild-type, deletion mutant, and revertant viruses that did or did not express the F12L gene. These analyses showed that F12L protein had only a minor (twofold) effect on the formation of IMV particles, but the formation of actin tails and CEV particles was greatly reduced, and the deletion mutant formed a small plaque. vΔF12L produced sevenfold-reduced levels of infectious EEV, and the virus was dramatically attenuated in vivo.
It remains to be determined where F12L is located in the infected cell and whether it is associated with cellular or virus membranes. For these purposes, the virus containing the F12L protein tagged at its C terminus with an epitope recognized by an MAb will be useful. Although the F12L protein has several regions of hydrophobic amino acids residues and several potential sites for attachment of carbohydrate, these seem not to be used since the protein was unaltered in electrophoretic mobility in the presence of inhibitors of N- or O-linked carbohydrate. This suggests that if the protein is associated with membranes, it has a topology that places the majority of the mass in the cytosol, so that it is not accessible to enzymes within vesicles that attach carbohydrate. In this regard, F12L might be similar to the F13L and A36R proteins that are predominantly in the cytosol rather than within the lumen of the wrapping membranes (21, 39, 52).
The loss of the F12L protein greatly reduced the formation of actin tails. Like other viruses that make fewer actin tails, the F12L mutant has a reduced plaque size, emphasizing the importance of actin tails for efficient cell-to-cell spread. The production of EEV by viruses that do not form actin tails is quite variable. In some cases where the formation of IEV particles is reduced greatly (vΔF13L and vΔB5R), the production of EEV is reduced 10- to 100-fold. However, with vΔA34R and vΔA33R, where IEV formation is also reduced or incomplete, the production of EEV is increased 25-fold and 2- to 4-fold, respectively. The vΔA36R mutant makes normal IEV particles but no actin tails and forms three- to fivefold-reduced EEV. In comparison with these phenotypes the vΔF12L mutant described here makes approximately sevenfold-less infectious EEV.
Lastly, the virulence of the vΔF12L virus was reduced dramatically. Doses of 104 PFU of wild-type and revertant virus caused severe illness in mice infected by the intranasal route, whereas this dose of vΔF12L had no noticeable effect. Moreover, even at 10,000-fold-higher doses of vΔF12L, the animals suffered little ill effect. Other mutants that are defective in EEV formation or cell-to-cell spread also showed dramatic attenuation. In comparison, mutant viruses that have lost specific immunomodulators, such as the biosynthetic steroid enzyme 3-β hydroxysteroid dehydrogenase (33), the type I interferon-binding protein (49), or the soluble interleukin-1 receptor (48), exhibited only modest attenuation or even enhanced virulence (1) in this model. Evidently, the ability of VV to spread efficiently from cell-to-cell and more widely by the release of EEV is far more important for virus virulence than the possession of these immunomodulators. The latter however, may have profound affects on the immune response to infection and the immunogenicity of recombinant viruses.
ACKNOWLEDGMENTS
This work was supported by a Program Grant from the United Kingdom Medical Research Council and an equipment grant from The Wellcome Trust.
We thank Henriette van Eijl for critical reading of the manuscript and Caroline Gubser and Han-Joo Lee for computational analysis.
REFERENCES
- 1.Alcamí A, Smith G L. A soluble receptor for interleukin-1 beta encoded by vaccinia virus: a novel mechanism of virus modulation of the host response to infection. Cell. 1992;71:153–167. doi: 10.1016/0092-8674(92)90274-g. [DOI] [PubMed] [Google Scholar]
- 2.Antoine G, Scheiflinger F, Dorner F, Falkner F G. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology. 1998;244:365–396. doi: 10.1006/viro.1998.9123. [DOI] [PubMed] [Google Scholar]
- 3.Appleyard G, Hapel A J, Boulter E A. An antigenic difference between intracellular and extracellular rabbitpox virus. J Gen Virol. 1971;13:9–17. doi: 10.1099/0022-1317-13-1-9. [DOI] [PubMed] [Google Scholar]
- 4.Blasco R, Moss B. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-dalton outer envelope protein. J Virol. 1991;65:5910–5920. doi: 10.1128/jvi.65.11.5910-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blasco R, Moss B. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J Virol. 1992;66:4170–4179. doi: 10.1128/jvi.66.7.4170-4179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulter E A, Zwartouw H T, Titmuss D H I, Maber H B. The nature of the immune state produced by inactivated vaccinia virus in rabbits. Am J Epidemiol. 1971;94:612–620. doi: 10.1093/oxfordjournals.aje.a121360. [DOI] [PubMed] [Google Scholar]
- 7.Boyle D B, Coupar B E H. A dominant selectable marker for the construction of recombinant poxviruses. Gene. 1988;65:123–128. doi: 10.1016/0378-1119(88)90424-6. [DOI] [PubMed] [Google Scholar]
- 8.Cameron C, Hota-Mitchell S, Chen L, Barrett J, Cao J X, Macaulay C, Willer D, Evans D, McFadden G. The complete DNA sequence of myxoma virus. Virology. 1999;264:298–318. doi: 10.1006/viro.1999.0001. [DOI] [PubMed] [Google Scholar]
- 9.Cudmore S, Cossart P, Griffiths G, Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 10.Czerny C P, Mahnel H. Structural and functional analysis of orthopoxvirus epitopes with neutralizing monoclonal antibodies. J Gen Virol. 1990;71:2341–2352. doi: 10.1099/0022-1317-71-10-2341. [DOI] [PubMed] [Google Scholar]
- 11.Dales S, Mosbach E H. Vaccinia as a model for membrane biogenesis. Virology. 1968;35:564–583. doi: 10.1016/0042-6822(68)90286-9. [DOI] [PubMed] [Google Scholar]
- 12.Duncan S A, Smith G L. Identification and characterization of an extracellular envelope glycoprotein affecting vaccinia virus egress. J Virol. 1992;66:1610–1621. doi: 10.1128/jvi.66.3.1610-1621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelstad M, Howard S T, Smith G L. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology. 1992;188:801–810. doi: 10.1016/0042-6822(92)90535-w. [DOI] [PubMed] [Google Scholar]
- 14.Engelstad M, Smith G L. The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology. 1993;194:627–637. doi: 10.1006/viro.1993.1302. [DOI] [PubMed] [Google Scholar]
- 15.Essani K, Dales S. Biogenesis of vaccinia: evidence for more than 100 polypeptides in the virion. Virology. 1979;95:385–394. doi: 10.1016/0042-6822(79)90493-8. [DOI] [PubMed] [Google Scholar]
- 16.Falkner F G, Moss B. Transient dominant selection of recombinant vaccinia viruses. J Virol. 1990;64:3108–3111. doi: 10.1128/jvi.64.6.3108-3111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frischknecht F, Moreau V, Röttger S, Gonfloni S, Rechmann I, Superti-Furga G, Way M. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature. 1999;401:926–929. doi: 10.1038/44860. [DOI] [PubMed] [Google Scholar]
- 18.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 19.Herrera E, del Mar Lorenzo M, Blasco R, Isaacs S N. Functional analysis of vaccinia virus B5R protein: essential role in virus envelopment is independent of a large portion of the extracellular domain. J Virol. 1998;72:294–302. doi: 10.1128/jvi.72.1.294-302.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiller G, Weber K. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J Virol. 1985;55:651–659. doi: 10.1128/jvi.55.3.651-659.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirt P, Hiller G, Wittek R. Localization and fine structure of a vaccinia virus gene encoding an envelope antigen. J Virol. 1986;58:757–764. doi: 10.1128/jvi.58.3.757-764.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollinshead M, Vanderplasschen A, Smith G L, Vaux D J. Vaccinia virus intracellular mature virions contain only one lipid membrane. J Virol. 1999;73:1503–1517. doi: 10.1128/jvi.73.2.1503-1517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horton R M, Cai Z L, Ho S N, Pease L R. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques. 1989;8:528–535. [PubMed] [Google Scholar]
- 24.Hughes S J, Johnston L H, de Carlos A, Smith G L. Vaccinia virus encodes an active thymidylate kinase that complements a cdc8 mutant of Saccharomyces cerevisiae. J Biol Chem. 1991;266:20103–20109. [PubMed] [Google Scholar]
- 25.Ichihashi Y, Matsumoto S, Dales S. Biogenesis of poxviruses: role of A-type inclusions and host cell membranes in virus dissemination. Virology. 1971;46:507–532. doi: 10.1016/0042-6822(71)90056-0. [DOI] [PubMed] [Google Scholar]
- 26.Isaacs S N, Kotwal G J, Moss B. Reverse guanine phosphoribosyltransferase selection of recombinant vaccinia viruses. Virology. 1990;178:626–630. doi: 10.1016/0042-6822(90)90367-z. [DOI] [PubMed] [Google Scholar]
- 27.Isaacs S N, Wolffe E J, Payne L G, Moss B. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J Virol. 1992;66:7217–7224. doi: 10.1128/jvi.66.12.7217-7224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joklik W K. The intracellular uncoating of poxvirus DNA. II. The molecular basis of the uncoating process. J Mol Biol. 1964;8:277–288. doi: 10.1016/s0022-2836(64)80137-6. [DOI] [PubMed] [Google Scholar]
- 29.Kerr S M, Johnston L H, Odell M, Duncan S A, Law K M, Smith G L. Vaccinia DNA ligase complements Saccharomyces cerevisiae cdc9, localizes in cytoplasmic factories and affects virulence and virus sensitivity to DNA damaging agents. EMBO J. 1991;10:4343–4350. doi: 10.1002/j.1460-2075.1991.tb05012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Massung R F, Liu L I, Qi J, Knight J C, Yuran T E, Kerlavage A R, Parsons J M, Venter J C, Esposito J J. Analysis of the complete genome of smallpox variola major virus strain Bangladesh-1975. Virology. 1994;201:215–240. doi: 10.1006/viro.1994.1288. [DOI] [PubMed] [Google Scholar]
- 31.Mathew E, Sanderson C M, Hollinshead M, Smith G L. The extracellular domain of vaccinia virus protein B5R affects plaque phenotype, extracellular enveloped virus release, and intracellular actin tail formation. J Virol. 1998;72:2429–2438. doi: 10.1128/jvi.72.3.2429-2438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McIntosh A A, Smith G L. Vaccinia virus glycoprotein A34R is required for infectivity of extracellular enveloped virus. J Virol. 1996;70:272–281. doi: 10.1128/jvi.70.1.272-281.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore J B, Smith G L. Steroid hormone synthesis by a vaccinia enzyme: a new type of virus virulence factor. EMBO J. 1992;11:1973–1980. doi: 10.1002/j.1460-2075.1992.tb05251.x. . (Erratum, 11:3490.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J, Monath T P, Roizman B, Straus S E, editors. Virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2637–2671. [Google Scholar]
- 35.Ogawa R, Calvert J G, Yanagida N, Nazerian K. Insertional inactivation of a fowlpox virus homologue of the vaccinia virus F12L gene inhibits the release of enveloped virions. J Gen Virol. 1993;74:55–64. doi: 10.1099/0022-1317-74-1-55. [DOI] [PubMed] [Google Scholar]
- 36.Parkinson J E, Smith G L. Vaccinia virus gene A36R encodes a Mr 43-50 K protein on the surface of extracellular enveloped virus. Virology. 1994;204:376–390. doi: 10.1006/viro.1994.1542. [DOI] [PubMed] [Google Scholar]
- 37.Roper R L, Payne L G, Moss B. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J Virol. 1996;70:3753–3762. doi: 10.1128/jvi.70.6.3753-3762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roper R L, Wolffe E J, Weisberg A, Moss B. The envelope protein encoded by the A33R gene is required for formation of actin-containing microvilli and efficient cell-to-cell spread of vaccinia virus. J Virol. 1998;72:4192–4204. doi: 10.1128/jvi.72.5.4192-4204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Röttger S, Frischknecht F, Reckmann I, Smith G L, Way M. Interactions between vaccinia virus IEV membrane proteins and their roles in IEV assembly and actin tail formation. J Virol. 1999;73:2863–2875. doi: 10.1128/jvi.73.4.2863-2875.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanderson C M, Frischknecht F, Way M, Hollinshead M, Smith G L. Roles of vaccinia virus EEV-specific proteins in intracellular actin tail formation and low pH-induced cell-cell fusion. J Gen Virol. 1998;79:1415–1425. doi: 10.1099/0022-1317-79-6-1415. [DOI] [PubMed] [Google Scholar]
- 41.Sanderson C M, Hollinshead M, Smith G L. The vaccinia virus A27L gene is needed for the microtubule-dependent transport of intracellular mature virus particles. J Gen Virol. 2000;81:47–58. doi: 10.1099/0022-1317-81-1-47. [DOI] [PubMed] [Google Scholar]
- 42.Schmelz M, Sodeik B, Ericsson M, Wolffe E, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Senkevich T G, Bugert J J, Sisler J R, Koonin E V, Darai G, Moss B. Genome sequence of a human tumorigenic poxvirus: prediction of specific host response-evasion genes. Science. 1996;273:813–816. doi: 10.1126/science.273.5276.813. [DOI] [PubMed] [Google Scholar]
- 44.Shchelkunov S N, Massung R F, Esposito J J. Comparison of the genome DNA sequences of Bangladesh-1975 and India-1967 variola viruses. Virus Res. 1995;36:107–118. doi: 10.1016/0168-1702(94)00113-q. [DOI] [PubMed] [Google Scholar]
- 45.Shida H. Nucleotide sequence of the vaccinia virus hemagglutinin gene. Virology. 1986;150:451–462. doi: 10.1016/0042-6822(86)90309-0. [DOI] [PubMed] [Google Scholar]
- 46.Smith G L, Chan Y S, Howard S T. Nucleotide sequence of the 42 kbp of vaccinia virus strain WR from near the right inverted terminal repeat. J Gen Virol. 1991;72:1349–1376. doi: 10.1099/0022-1317-72-6-1349. [DOI] [PubMed] [Google Scholar]
- 47.Sodeik B, Doms R W, Ericsson M, Hiller G, Machamer C E, van't Hof W, van Meer G, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spriggs M, Hruby D E, Maliszewski C R, Pickup D J, Sims J E, Buller R M L, Vanslyke J. Vaccinia and cowpox viruses encode a novel secreted interleukin-1 binding protein. Cell. 1992;71:145–152. doi: 10.1016/0092-8674(92)90273-f. [DOI] [PubMed] [Google Scholar]
- 49.Symons J A, Alcamí A, Smith G L. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell. 1995;81:551–560. doi: 10.1016/0092-8674(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 50.Tooze J, Hollinshead M, Reis B, Radsak K, Kern H. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur J Cell Biol. 1993;60:163–178. [PubMed] [Google Scholar]
- 51.Turner G S, Squires E J. Inactivated smallpox vaccine: immunogenicity of inactivated intracellular and extracellular vaccinia virus. J Gen Virol. 1971;13:19–25. doi: 10.1099/0022-1317-13-1-19. [DOI] [PubMed] [Google Scholar]
- 52.van Eijl H, Hollinshead M, Smith G L. The vaccinia virus A36R protein is a type Ib membrane protein present on intracellular but not extracellular enveloped particles. Virology. 2000;271:26–36. doi: 10.1006/viro.2000.0260. [DOI] [PubMed] [Google Scholar]
- 53.Vanderplasschen A, Smith G L. A novel virus binding assay using confocal microscopy: demonstration that the intracellular and extracellular vaccinia virions bind to different cellular receptors. J Virol. 1997;71:4032–4041. doi: 10.1128/jvi.71.5.4032-4041.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ward G A, Stover C K, Moss B, Fuerst T R. Stringent chemical and thermal regulation of recombinant gene expression by vaccinia virus vectors in mammalian cells. Proc Natl Acad Sci USA. 1995;92:6773–6777. doi: 10.1073/pnas.92.15.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willer D O, McFadden G, Evans D H. The complete genome sequence of shope (rabbit) fibroma virus. Virology. 1999;264:319–343. doi: 10.1006/viro.1999.0002. [DOI] [PubMed] [Google Scholar]
- 56.Wolffe E J, Isaacs S N, Moss B. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J Virol. 1993;67:4732–4741. doi: 10.1128/jvi.67.8.4732-4741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolffe E J, Katz E, Weisberg A, Moss B. The A34R glycoprotein gene is required for induction of specialized actin-containing microvilli and efficient cell-to-cell transmission of vaccinia virus. J Virol. 1997;71:3904–3915. doi: 10.1128/jvi.71.5.3904-3915.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolffe E J, Weisberg A S, Moss B. Role for the vaccinia virus A36R outer envelope protein in the formation of virus-tipped actin-containing microvilli and cell-to-cell virus spread. Virology. 1998;244:20–26. doi: 10.1006/viro.1998.9103. [DOI] [PubMed] [Google Scholar]