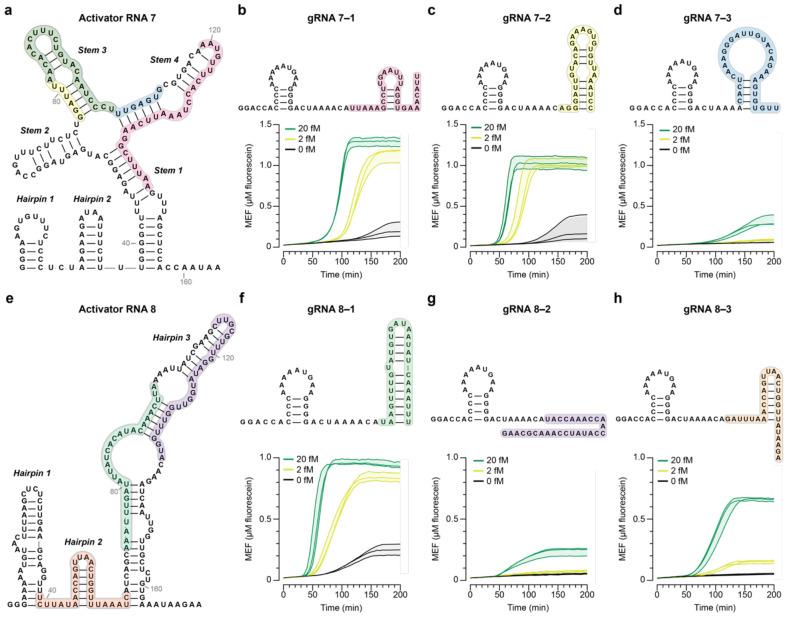
Fig. 2 ∣. Screening of LbuCas13a gRNAs identifies factors that could impact cleavage efficiency.
a, Predicted secondary structure of activator RNA 7 generated from NASBA with Primer Set 7. Regions targeted by gRNAs are shaded in different colors. Fluorescence kinetics from NASBA-Cas13a at varying concentrations of synthetic SARS-CoV-2 genome with b, gRNA 7–1, c, gRNA 7–2, or d, gRNA 7–3 with predicted secondary structures of each gRNA shown above. e, Predicted secondary structure of activator RNA 8 generated from NASBA with Primer Set 8. Regions targeted by gRNAs are shaded in different colors. Fluorescence kinetics from NASBA-Cas13a at varying concentrations of synthetic SARS-CoV-2 genome with f, gRNA 8–1, g, gRNA 8–2, or h, gRNA 8–3 with predicted secondary structures of each gRNA shown above. Data are n=3 independent biological replicates, each plotted as a line with raw fluorescence standardized to MEF. Shading indicates the average of the replicates ± standard deviation.