Abstract
Adenovirus codes for a DNA polymerase that is a member of the DNA polymerase α family and uses a protein primer for initiation of DNA synthesis. It contains motifs characteristic of a proofreading 3′-5′-exonuclease domain located in the N-terminal region and several polymerase motifs located in the C-terminal region. To determine the role of adenovirus DNA polymerase in DNA replication, 22 site-directed mutations were introduced into the conserved DNA polymerase motifs in the C-terminal region of adenovirus DNA polymerase and the mutant forms were expressed in insect cells using a baculovirus expression system. Each mutant enzyme was tested for DNA binding activity, the ability to interact with pTP, DNA polymerase catalytic activity, and the ability to participate in the initiation of adenovirus DNA replication. The mutant phenotypes identify functional domains within the adenovirus DNA polymerase and allow discrimination between the roles of conserved residues in the various activities carried out by the protein. Using the functional data in this study and the previously published structure of the bacteriophage RB69 DNA polymerase (J. Wang et al., Cell 89:1087–1099, 1997), it is possible to envisage how the conserved domains in the adenovirus DNA polymerase function.
Human adenoviruses, of which there are over 40 different serotypes, are similar in morphology and genome organization. The genomes of human adenoviruses are linear double-stranded DNA (dsDNA) molecules of approximately 36,000 bp with inverted terminal repeats of about 100 bp, the precise size depending on the serotype. Located within the inverted terminal repeats are the cis-acting sequences which define the origin of DNA replication. Covalently attached to each 5′ end of the genome DNA is a terminal protein (TP) which is likely to constitute an additional cis-acting component of the origin of DNA replication (reviewed in references 22 and 62).
Replication of the adenovirus genome is catalyzed by adenovirus DNA polymerase (Adpol) via a protein-priming mechanism (reviewed in references 22 and 62) in which the adenovirus preterminal protein (pTP) acts as the protein primer. Adpol and pTP form a stable heterodimer, and following the binding of pTP-Adpol to the core origin of replication, DNA synthesis is initiated by Adpol catalyzing the addition of dCMP to the hydroxyl group of serine580 of pTP. Initiation is enhanced by a virus-encoded single-stranded DNA (ssDNA) binding protein (DBP) and two cellular factors, nuclear factor I (NFI or CTF1) and nuclear factor III (NFIII or OCT1), are required for virus DNA replication (41, 42, 48, 61). The presence of the adenovirus TP, which is covalently attached to the 5′ end of the viral DNA, also stimulates initiation of DNA replication in vitro (46, 47). Replication initiates opposite the GTA at positions 4 to 6 in the genome. After addition of dCMP, dAMP, and dTMP, the initiation product generated (pTP-CAT) jumps back to occupy positions 1 to 3 and then Adpol catalyzes the synthesis of the elongation product via a strand displacement mechanism (22).
DNA polymerases (Pols) are a family of enzymes responsible for faithful maintenance, replication, and transmission of genetic information. Although individual Pols differ in size, overall structure, the requirement for accessory proteins, and their roles in DNA replication, their fundamental function is to catalyze the addition of nucleotides onto the 3′ end of the growing nucleic acid chain with high accuracy. This process may vary from the insertion of one to a few nucleotides in the case of human DNA Pol α (63) or β (54) to the replication of many thousands of bases of a complete genome, as in the case of bacteriophage T7 (57) or φ29 (50) DNA Pol. In 1988, Wong et al. (67) identified six highly conserved domains, designated Pol I to Pol VI, by their extent of similarity in the eukaryotic and prokaryotic DNA Pols with the deduced human Pol α amino acid sequence. The Pols that contain these six conserved regions are designated α-like Pols (or Pol α) (13). With the availability of new sequence data, the list of Pols that contain these six regions has grown rapidly. The seventh conserved region, designated Pol VII, was identified among the α-like Pols (24, 35). Four families, A, B, C, and D, of Pols have been designated based on amino acid sequences similar to Escherichia coli Pols I, II, and III and to the cellular repair enzyme Pol β, respectively (6, 26). Eukaryotic viral Pols are α-like Pols and are found exclusively in Pol family B, while most bacteriophage Pols are in families A and B (35).
During recent years, there has been a significant increase in our understanding of the catalytic mechanism of Pols. Amino acid residues involved in 3′-5′ exonuclease activity, metal binding, deoxynucleoside triphosphate (dNTP) binding, and DNA Pol activity were identified by sequence comparisons and site-directed mutagenesis using φ29 Pol (2, 3, 14, 15, 52), human Pol α (16–18), herpes simplex virus Pol (25), and bacteriophage RB69 Pol (68). Delarue et al. (13) revealed that the exonuclease (EXO) motifs (EXO I, II, and III) and Pol segments (or motifs) I, II, and III, or A, B, and C, were present in prokaryotic and eukaryotic Pols. Three EXO motifs (I, II, and III), which are responsible for the 3′-5′ exonuclease activity are located in the N-terminal peptide sequence, while three Pol motifs (A, B, and C) in the C-terminal peptide sequence were proposed to be important for the DNA Pol activity of all Pols. It is suggested that the conserved amino acid residues in these motifs are the components of the Pol catalytic site (for reviews, see references 1 and 30). X-ray crystallographic analysis of several Pols revealed that they are similar in having a structure that can be represented as a hand with subdomains corresponding to the palm, finger, and thumb. The catalytic center has been visualized, and most of the conserved residues are located at or near this center (12, 19, 27, 33, 34, 36, 37, 43, 53, 55), although the structural framework that supports this arrangement of conserved residues varies considerably. In 1997, significant insights into the mechanism of the Pol α family in DNA replication were provided with the structure of a Pol from bacteriophage RB69 (64). In RB69 Pol, three highly conserved motifs, A, B, and C, converge on the catalytic center from the palm, finger, and thumb base subdomains to produce a continuous conserved surface. In human Pol α (11) and φ29 DNA Pol (52), conserved aspartic acid residues in these motifs are essential for catalysis. Other conserved residues are located near the catalytic center (Fig. 1A) and play various roles in DNA synthesis (68). This information, together with the mutagenesis data, provides a structural framework for α-like DNA Pols, although diversion of each individual might exist.
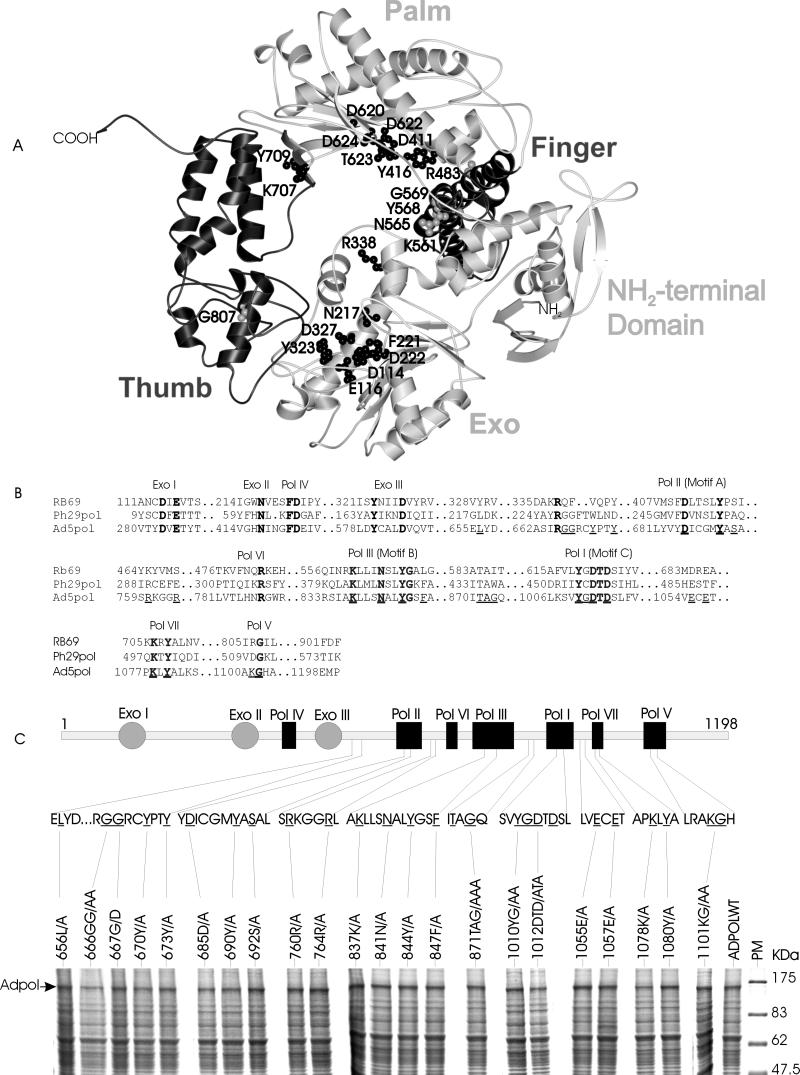
FIG. 1.
Locations of mutations in Adpol and expression in Sf9 cells. (A) Ribbon diagram of RB69 DNA Pol from Wang et al. (64) showing the relative locations of five domains (EXO, N terminus, finger, palm, and thumb) and the conserved residues in these domains. Each residue is marked with a letter (one-letter amino acid code) and a number indicating its position in the amino acid sequence of RB69 DNA Pol. (B) Multiple-sequence alignment of conserved domains in RB69 Pol, φ29 Pol, and Adpol. The conserved residues in each domain indicated in panel A are in boldface. Residues mutated in Adpol are underlined. (C) Locations of mutations in Adpol and expression in Sf9 cells. Filled circles represent the EXO domains, and rectangles represent the DNA Pol domains. Numbers at each end indicate the length of the Adpol peptide chain, and below is the one-letter amino acid sequence of the regions in which mutations are located. Mutated residues are underlined. The gel images show the expression of mutant Adpol in Sf9 insect cells. Total cell extract was fractionated by SDS–10% PAGE and stained with 0.25% Coomassie blue. The 140-kDa Adpol is indicated. Each lane is labeled with the mutant enzyme name showing its mutation site (numbers) in the Adpol peptide chain and the amino acid substitutions (one-letter code). PM, prestained protein molecular size markers.
Adpol is one of several protein-primed Pols included in the Pol α family (21, 22). Like all other Pols, its proofreading EXO domains are located in the N-terminal portion with DNA Pol domains in the C-terminal portion. In the C-terminal portion of Adpol, seven conserved domains (designated Pol I to Pol VII) have been identified (24, 35). Analysis of Adpol mutant forms demonstrated that two cycteine-histine-rich clusters in Adpol are required for DNA binding and initiation of DNA replication (28). Mutation of the conserved residues in Pol I decreased the Pol activities (29). By insertional mutagenesis, Chen and Horwitz (8) showed that Pol IV and V of Adpol are required for Pol and initiation activities and multiple regions of Adpol are essential for adenovirus DNA replication (49). However, the precise contribution of these conserved domains to Adpol catalytic activity has not been fully established. To address these points, we used the RB69 Pol structure as a framework into which conserved residues in Adpol could be fitted (Fig. 1A and B). Using this model as a guide, mutations were introduced into the C-terminal regions of Adpol suspected of having an important role in the activity of the enzyme. Each mutant enzyme was tested in a range of assays designed to differentiate between the different activities of Adpol in viral DNA replication. The mutant phenotypes identify functional domains within Adpol and suggest a model for the arrangement of the conserved domains within the enzyme.
MATERIALS AND METHODS
Nucleotides and DNA manipulation enzymes.
Unlabeled nucleotides were purchased from Pharmacia Biochemicals. [α-32P]dCTP (3,000 Ci/mmol) and [α-32P]dATP (3,000 Ci/mmol) were obtained from Amersham International Plc. All of the enzymes used for molecular cloning were purchased from New England Biolabs.
Cells and viruses.
Spodoptera frugiperda (Sf9) cells, cultured in TC-100 medium supplemented with 7% fetal calf serum, were used for protein expression. HeLa cells were maintained in suspension culture in Glasgow S-minimal essential medium supplemented with 7% newborn calf serum. Adenovirus was extracted from the infected cells with fluoroethane and purified on CsCl gradients, and adenovirus template DNA was prepared as previously described (39). Recombinant baculoviruses containing the genes for DBP, pTP, and Adpol have been described previously (41, 58). The procedures used for infection of Sf9 cells were previously described (65).
Site-directed mutagenesis of Adpol.
A full-length cDNA encoding Adpol was released from a pGEM construct (provided by Michael Stanglmaier and Ernst Winnacker) by EcoRI/SphI digestion. Plasmid pAd5FastBac was constructed by inserting the Adpol DNA coding sequence into the EcoRI/SphI-linearized pFastBac1 vector. Site-directed mutagenesis was performed on the Adpol-encoding gene in a pAd5FastBac clone based on PCR-generated mutagenesis (7). All of the primers used in this study were synthesized by Oswel Research Products Ltd. (University of Southampton). The mutation sites and the amino acid substitutions are detailed in Fig. 1C. For each mutation, two stages of PCR amplification were performed. To generate the 656L/A, 666GG/AA, 667G/D, 670Y/A, 673Y/A, 685D/A, 690Y/A, 692S/A, 760R/A, and 764R/A mutations, the first PCR was carried out using a forward mutagenesis primer with a backward primer (2541 to 2521, 5′ AAACGACCCGGCGAGGGCGTTG 3′) and the second (cloning) PCR was performed using the first PCR product as a megaprimer and an external primer (619 to 640, 5′ CTCTGCTTCCTTGTGCGCGGTC 3′), respectively. The first PCR of mutation 837K/A, 841N/A, 844Y/A, 847F/A, or 871TAG/AAA was carried out using a backward mutagenesis primer and a forward primer (2065 to 2085, 5′ ATGTACGCCGCCGCGCTCACC 3′), and the second (cloning) PCR was performed using a megaprimer of the first PCR product and an external primer (3411 to 3390, 5′ CTTGAGGCTGGTCCTGCTGGTG 3′), respectively. For mutation 1010YG/AA, 1012DTD/ATA, 1055E/A, 1057E/A, 1078K/A, 1080Y/A, or 1101KG/AA, the first PCR was carried out, respectively, using a backward primer (3411 to 3390, 5′ CTTGAGGCTGGTCCTGCTGGTG 3′) and a forward mutagenesis primer and the second (cloning) PCR was performed using a megaprimer of the first PCR product and an external primer (2065 to 2085, 5′ ATGTACGCCGCCGCGCTCACC 3′). To clone the mutations within the EcoNI-NotI region, the amplified PCR products were digested using EcoNI and NotI and inserted into EcoNI/NotI-cleaved plasmid pAd5FastBacpol. For mutations within the NotI-Eco47III region, the amplified PCR products were digested by NotI and Eco47III and ligated into NotI/Eco47III-cleaved plasmid pAd5FastBacpol, respectively. A lysine mutation (764R/A) was cloned using SmaI and Eco47III digestion and ligated into plasmid pAd5FastBacpol digested with the same enzymes. The mutations and the integrity of the DNA sequence of all regions of pAd5FastBacpol clones amplified by PCR and cloning junctions were confirmed by automatic sequencing (Alex Houston, St. Andrews DNA Sequence Service).
Expression of Adpol and pTP in Sf9 cells.
Wild-type Adpol and mutated Adpol were expressed in Sf9 cells using the Bac-To-Bac baculovirus expression system (Gibco BRL) and following the manufacturer's protocol. About 1 ng of donor vector pFastBac containing a mutated Adpol-encoding gene was transformed into DH10Bac cells, and colonies harboring a recombinant baculovirus genome were selected on plates containing kanamycin, gentamicin, tetracycline, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and isopropyl-β-d-thiogalactopyranoside (IPTG). The prepared baculovirus recombinant DNA was transfected into Sf9 cells using CellFectin. Recombinant viruses were amplified once before being used for protein expression. Protein expression with each recombinant virus was optimized using different multiplicities of infection (MOIs) and times postinfection. A nuclear extract of Sf9 cells expressing Adpol was prepared as follows. Cells from a 50-ml culture of Sf9 cells infected with a recombinant baculovirus containing the gene for Adpol were resuspended in 1 ml of cell resuspension buffer (25 mM HEPES-NaOH [pH 8.0], 5 mM KCl, 0.5 mM MgCl2, 0.5 mM dithiothreitol [DTT], protease inhibitors). Cells were disrupted using a Dounce homogenizer with a type B pestle. After centrifugation at 8,000 rpm for 10 min at 4°C in a microcentrifuge (Eppendorf 5040), the pellet was resuspended in 0.2 ml of cell resuspension buffer containing 0.2 M NaCl. After incubation on ice for 30 min and centrifugation at 60,000 rpm for 20 min at 4°C in a TL100 ultracentrifuge (Beckman), the supernatant was collect as nuclear extract. The amount of mutant Adpol in each nuclear extract preparation was determined using an indirect competitive enzyme-linked immunosorbent assay (ELISA) essentially as previously described (23) with purified wild-type Adpol as the standard. Immulon 4 (Dynatech) microtiter plates were coated with 50 μl of purified Adpol (10 μg/ml) in PBSN (phosphate-buffered saline [PBS] containing 0.05% NaN3) buffer and incubated for 2 h at 37°C and blocked with PBS containing 0.25% bovine serum albumin and 0.05% Tween 20. A mixture of 100 μl of serial dilutions of mutant Adpol, serial dilutions of purified wild-type Adpol (used to prepare a standard inhibition curve), or blocking buffer (used as an uninhibited control) and 100 μl of a 2 × 10−3 dilution of rabbit anti-Adpol serum was prepared, respectively, in a microtiter plate. After incubation for 1 h at room temperature, 50 μl of each mixture was transferred into the Adpol-coated plates and incubated for 2 h at room temperature. After washing of the plates with PBST (PBS containing 0.05% Tween 20), 50 μl of horseradish peroxidase–goat anti-rabbit conjugate (4 × 10−3 dilution) was added, and the plates were incubated for another hour. The plates were thoroughly washed with PBST and read at a wavelength of 450 nm on a microplate reader after addition of 100 μl of substrate (0.66 mg of o-phenylenediamine per ml, 0.03% H2O2). The protein concentration of each mutant Adpol was interpolated from the purified Adpol inhibited curve. Expression and purification of wild-type Adpol and pTP were done as previously described (58, 65). The protein concentration of the purified Adpol and pTP was measured by Bradford assay.
DNA binding assay.
To analyze the DNA binding activity of mutant and wild-type Adpol, an extract containing equal amounts of the expressed Adpol proteins was incubated with either ssDNA-Sepharose or dsDNA-Sepharose and the bound Adpol was measured by Western blotting. DNA-Sepharose was prepared as previously described (31). In each of triplicate assays, a total of 1 μg of Adpol protein (in 50 μl of nuclear extract, adjusted to 0.1 or 0.2 M NaCl) was pretreated with 10 μl (packed volume) of Sepharose. After incubation for 30 min at 4°C, beads were sedimented and the supernatant was mixed with 10 μl (packed volume) of ssDNA-Sepharose or dsDNA-Sepharose. After incubation for 30 min at 4°C, the DNA-Sepharose beads were washed twice with 100 μl of 0.1 or 0.2 M NaCl in 25 mM HEPES-NaOH (pH 8.0)–5 mM KCl–0.5 mM MgCl2–0.5 mM DTT plus protease inhibitors. After the final washing, the supernatant was carefully removed and the beads were resuspended in 50 μl of 1× Laemmli loading buffer (38) and boiled for 3 min. A 20-μl sample was loaded onto a 10% polyacrylamide gel containing sodium dodecyl sulfate (SDS), and the bound Adpol was fractionated by electrophoresis and measured by enhanced-chemiluminescence (ECL) Western blotting using the previously described Pol D antiserum (59). The amount of Adpol detected by Western blotting was then quantitated by densitometric analysis using a multianalysis system (Doc-2000; Bio-Rad).
Immunoprecipitation assay.
To analyze the interaction of mutant Adpol and pTP, immunoprecipitation assays (65) were carried out to detect the Adpol-pTP complex in SF9 cells coexpressing both proteins. Approximately 3 × 106 Sf9 cells were coinfected with an MOI of 3 PFU each of Adpol and pTP recombinant baculoviruses per cell, and infected cells were harvested at 60 h after infection. Nuclear extract was obtained as described above and clarified by centrifugation at 13,000 rpm for 20 min in an Eppendorf 5040 microcentrifuge. The amount of Adpol mutants and pTP in the supernatant was determined by competitive ELISA as described before using purified Adpol and pTP as the standards and equalized before being used in the assays. All of the following steps were carried out at 4°C. The supernatant was diluted fivefold with ice-cold extraction buffer supplemented with 2.5 mM MgCl2 (NEB-M). A total of 1 μg of Adpol and pTP (in 200 μl of diluted extract) was mixed with 20 μl (packed volume) of protein A-Sepharose (Sigma), incubated for 30 min with gentle agitation, and then centrifuged at 13,000 rpm for 5 min in an Eppendorf 5040 microcentrifuge. The supernatant was then transferred into a fresh tube with 50 μl of anti-pTP monoclonal antibody 16H1 (65) and 10 μl (packed volume) of protein A-Sepharose. The mixture was incubated overnight, and the protein A-Sepharose beads were collected by centrifugation. The beads were washed four times with NEB-M, and the bound Adpol was measured by ECL Western blotting using the previously described Pol D antiserum (59). The amount of Adpol in each assay was then quantitated by densitometric analysis as described before. All of the assays were done in triplicate, and parallel experiments were carried out in which the assays contained DNase I (20 U/ml) or ethidium bromide (2 μg/ml) (51) to confirm that the interaction of Adpol and pTP was direct and was not a consequence of both proteins binding to DNA.
DNA Pol assay.
The catalytic activity of wild-type and mutated versions of Adpol was determined in a DNA Pol assay (40) using activated calf thymus DNA (20) as the template. Each mutant was analyzed in triplicate, and a nuclear extract of Sf9 cells infected with a wild-type baculovirus was used as a control. For each assay, the incubation mixture contained 10 μg of activated calf thymus DNA, 0.1 μl of nuclear extract of Sf9 cells containing 5 ng of Ad5pol, 100 μM dTTP, 100 μM dGTP, 100 μM dCTP, 20 μM dATP, and 1 μCi of [α-32P]dATP in a total volume of 50 μl of reaction buffer (50 mM Tris-HCl (pH 8.0)–5 mM MgCl2–10 mM DTT). The mixture was incubated for 1 h at 37°C, and the reaction was stopped by addition of 5 ml of 10% trichloroacetic acid containing 0.5% pyrophosphate. Precipitated DNA was captured on Whatman GF/C discs by filtration under vacuum and washed twice with 5% trichloroacetic acid and once with ethanol, and the radioactivity was determined by liquid scintillation counting using a Tri-Carb Liquid Scintillation Analyzer (Packard Instrument Co.). The relative DNA Pol activity of the mutant enzymes was calculated after subtraction of the background incorporation obtained with an extract of Sf9 cells infected with a wild-type baculovirus.
Initiation assay.
The ability of wild-type and mutant versions of Adpol to participate in the initiation of adenovirus DNA replication was assayed essentially as previously described (66). Each mutant form was assayed in triplicate together with a control using a nuclear extract of Sf9 cells without Adpol. The incubation mixture contained 500 ng of DBP, 50 ng of pTP, 0.5 μl of nuclear extract of Sf9 cells containing 25 ng of Adpol, 20 ng of adenovirus DNA template, and 2.5 μCi of [α-32P]dCTP in a total of 10 μl of reaction buffer (25 mM Bicine-NaOH [pH 8.0], 2 mM DTT, 1 mM MnCl2, 0.15 mM dATP, 0.2 mg of bovine serum albumin per ml). After incubation for 1 h at 30°C, CaCl2 was added to a final concentration of 10 mM and 1 U of micrococcal nuclease was added to destroy the template and any DNA attached to pTP. After further incubation for 30 min at 37°C, the reaction was stopped by addition of disruption buffer (20% glycerol, 5% SDS, 570 mM 2-mercaptoethanol, 33 mM Tris-HCl [pH 6.8], 0.2% bromophenol blue) and boiling for 3 min. The reaction mixtures were fractionated in a 10% polyacrylamide gel containing SDS, and the dried gel was exposed to X-ray film. The pTP-dCMP complex was quantitated by densitometric analysis as described before.
RESULTS
Expression of mutant Adpol in Sf9 cells.
Using the structure of RB69 DNA Pol as a guide (Fig. 1A), the functions of conserved amino acids (Fig. 1B and C) in Adpol activity were predicted. To test the validity of these predictions, identified amino acids were altered by site-directed mutagenesis. In most cases, the amino acid was changed to alanine, representing a deletion of the functional group of the original amino acid. All mutated Adpol cDNAs were engineered into recombinant baculoviruses, and the proteins were expressed in Sf9 insect cells. Optimal expression was achieved at 68 h postinfection with an MOI of 4 PFU per cell. Analysis of whole-cell extracts by SDS-PAGE and Coomassie blue staining indicated that all of the mutant Adpols were expressed well, at a level comparable to that of the wild type (Fig. 1C). Cell fractionation indicated that the wild type and each of the mutant proteins were found predominantly in the nuclear extract. Thus, as predicted, none of the mutated residues affect the nuclear localization of Adpol.
Interaction of mutant Adpol with ssDNA and dsDNA.
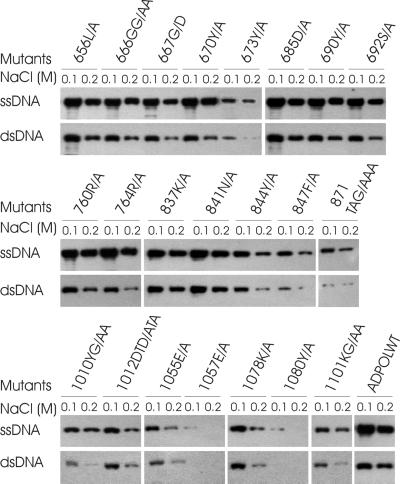
Interaction of Adpol with viral DNA is required for DNA replication, although the details of this requirement are not clear. It is suspected that the stability of this interaction will affect the efficiency of Adpol-catalyzed DNA synthesis. To ascertain the role of Adpol domains and the involvement of individual residues in DNA binding, all of the mutant enzymes were analyzed for DNA binding and their binding was compared in the presence of 0.1 and 0.2 M NaCl. Nuclear extracts containing equivalent amounts of the expressed Adpol and mutant enzymes were incubated with ssDNA- or dsDNA-Sepharose beads, and bound Adpol was detected by Western blotting. The results are shown in Fig. 2. Although the mechanism of Adpol-DNA interaction is not clear, increasing the concentration of NaCl in the reaction buffer decreased the amount of Adpol bound to both ssDNA and dsDNA. Mutation of 673Y/A, 844Y/A, 847F/A, 871TAG/AAA, 1010YG/AA, 1012 DTD/ATA, 1055E/A, 1057E/A, 1078 K/A, 1080Y/A, and 1101KG/AA caused a decrease in DNA binding activity, suggesting that these residues are required for efficient interaction of Adpol with DNA. Mutations 1057E/A and 1080Y/A, in particular, resulted in almost no DNA binding activity to either ssDNA or dsDNA, even at the lower NaCl concentration. Thus, the residues required for DNA binding of Adpol are clustered in the C-terminal region of the protein and are located both within conserved domains and between conserved domains. Interestingly, the conserved residues in Pol II of Adpol seem less important in Adpol-DNA interaction. Mutation of these residues did not affect Adpol binding to DNA.
FIG. 2.
DNA binding activity of mutant Adpol. Overexpressed Adpol in a nuclear extract of infected Sf9 cells was incubated with ssDNA- and dsDNA-Sepharose in the presence of either 0.1 or 0.2 M NaCl as described in the text. DNA-Sepharose beads were washed twice, and the bound proteins were fractionated by SDS-PAGE. Adpol was detected by Western blotting with Adpol-specific antiserum using ECL. The designation of each mutant enzyme is at the top of each lane.
Identification of residues required for formation of the Adpol-pTP heterodimer.
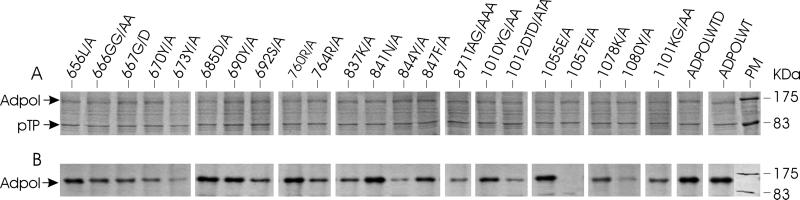
Adpol is primed by pTP in adenovirus DNA replication, and an Adpol-pTP complex is required prior to the initiation of DNA synthesis (22). Thus, mutations which disrupt or decrease formation of the Adpol-pTP complex could abolish or reduce the efficiency of the initiation reaction. To identify residues and domains involved in the formation of the Adpol-pTP heterodimer, pTP was coexpressed with wild-type or mutant Adpol in Sf9 cells and complex formation was tested in an immunoprecipitation assay. Both Adpol and pTP were expressed well in Sf9 cells using an MOI of 3 PFU each of Adpol and pTP recombinant baculoviruses per cell, although the expression level varied slightly (Fig. 3A). The Adpol-pTP complex formed in vivo was immunoprecipitated using anti-pTP monoclonal antibody 16H1, which has been shown to recognize pTP, as well as pTP engaged in a complex with Adpol (65). Adpol bound to immunoprecipitated pTP was detected by Western blotting (Fig. 3B). It is apparent that a number of mutations (673Y/A, 844Y/A, 1080Y/A, and 1057E/A) reduce the ability of Adpol to interact with pTP. Mutations 670Y/A, 871TAG/AAA, 1078K/A, and 1101KG/AA also reduce the ability of Adpol to interact with pTP. Thus, a collection of residues in the C-terminal portion of the Adpol molecule are important for interactions with pTP. In support of this conclusion, an N-terminal truncation mutant form of Adpol did not abolish the interaction of Adpol and pTP (data not shown). Control experiments in which the extracts containing Adpol and pTP were pretreated with DNase I or ethidium bromide indicated that the interaction between Adpol and pTP was direct and was not a consequence of both proteins binding to DNA in the cell extract.
FIG. 3.
Ability of mutant Adpol to interact with pTP. (A) Sf9 cells were coinfected with 3 PFU each of Adpol and pTP baculoviruses per cell and harvested after 70 h. SDS-PAGE shows the expression of Adpol and pTP in Sf9 cells stained with 0.25% Coomassie blue. Adpol and pTP are indicated. (B) Adpol-pTP complexes were immnoprecipitated using an anti-pTP monoclonal antibody, and Adpol was detected by Western blotting using a specific Adpol antiserum and ECL. The designation of each mutant enzyme is at the top of each lane, and ADPOLWTD indicates the detection of wild-type Adpol in the assay containing DNase. ADPOLWT, wild-type Adpol; PM, prestained protein molecular size markers.
DNA Pol activity of Adpol mutants.
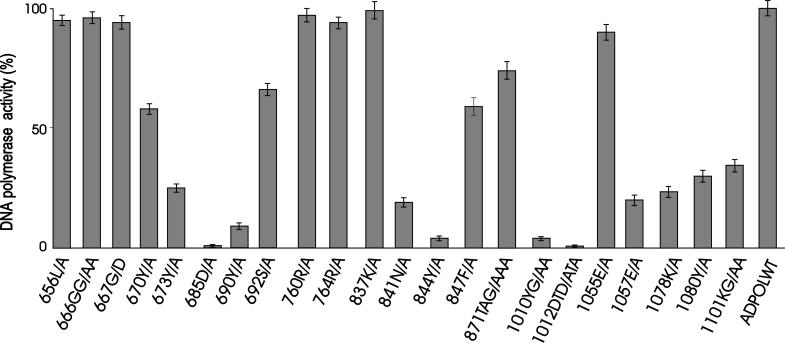
Using activated calf thymus DNA as a template, the DNA Pol activity of Adpol expressed in Sf9 cells was determined (Fig. 4). Mutation of an aspartic acid (685D/A) in Pol II or two aspartic acids (1012DTD/ATA) in Pol I to alanine completely abolished DNA Pol activity. Based on the conservation of these residues in all identified members of the Pol α family of DNA Pols (13, 35), it is likely that they are involved in metal-specific catalysis (4, 9, 11). Other residues, tyrosine and glycine (1010YG/AA) in Pol I, tyrosine (690Y/A) in Pol II, asparagine (841N/A) or tyrosine (844Y/A) in Pol III, a lysine and glycine (1101KG/AA) in Pol V, and lysine (1078K/A) and tyrosine (1080Y/A) in Pol VII, also play important roles in the DNA Pol activity of Adpol. Mutation of any of these residues substantially reduced the DNA Pol activity of Adpol, although mutation of conserved lysine (837K/A) in Pol III had little effect on the catalytic activity of Adpol. Interestingly, a nonconserved glutamic acid residue (1057E/A) is also required for DNA Pol activities. Other mutations (656L/A, 666 GG/AA, 667G/D, and 670Y/A) in the junction region between Exo III and Pol II had little effect on DNA Pol activity.
FIG. 4.
Catalytic activity of mutant Adpol. The DNA Pol activity of mutant Adpol present in nuclear extracts of infected Sf9 cells was determined using activated calf thymus DNA as the template. For each mutant enzyme, the incorporation of dAMP into acid-insoluble radioactivity was measured and compared to that of wild-type Adpol (21 pmol/60 min) after subtraction of the background incorporation (0.39 pmol/60 min) obtained with extract of Sf9 cells infected with a wild-type baculovirus. Experimental values are the means of triplicate determinations and are graphically represented relative to that of wild-type Adpol (ADPOLWT). Error bars represent the 95% confidence limits of the calculated means.
Adpol residues required for initiation of adenovirus DNA replication in vitro.
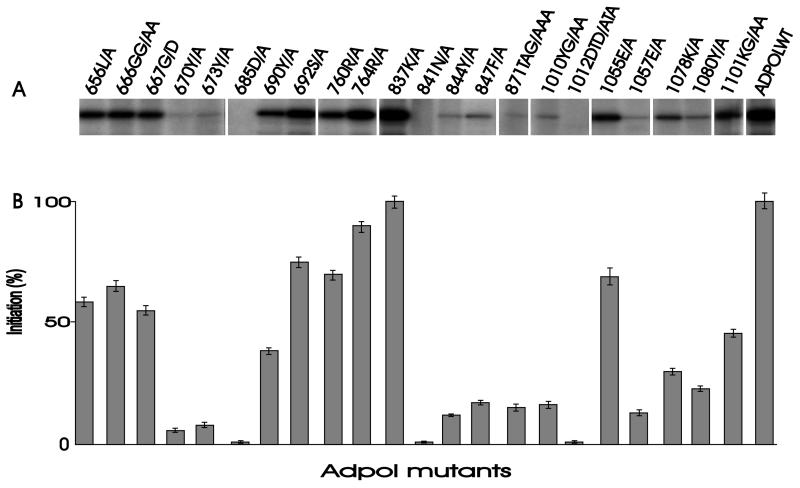
To analyze the effects of Adpol domains and residues on initiation of adenovirus DNA synthesis, each of the mutant enzymes was tested in an in vitro system containing adenovirus template DNA, purified DBP, purified pTP, and a nuclear extract containing equivalent amounts of the expressed Adpol and mutant enzymes. The template-directed transfer of [32P]dCMP onto pTP catalyzed by mutant Adpol, relative to that catalyzed by the wild type, is a measure of the ability of the mutant Adpol enzymes to participate in the initiation reaction. Variation in the ability of these mutant enzymes to participate in the initiation of DNA replication (Fig. 5) identified the residues that are important in the initiation reaction. Mutation of asparagine (841N/A) or aspartic acid (685D/A) residues in the Pol II domain or in the Pol I domain (1012DTD/ATA) to alanine abolished initiation. These residues are conserved in all members of the Pol α family and are also required for DNA Pol activity (Fig. 4; Table 1), suggesting that initiation of DNA synthesis by Adpol occurs at the same catalytic site as DNA chain elongation. Mutation of tyrosine (844Y/A), phenylalanine (847F/A), or a tyrosine and a glycine (1010YG/AA) also caused a substantial reduction in initiation. Mutation of glycine residues (666GG/AA, 667G/D) in a sequence conserved between DNA Pols which utilize protein primers (60) reduced but did not abolish the initiation of DNA synthesis (Fig. 5B). Interestingly, mutation of tyrosine residues to alanine (670Y/A or 673Y/A) near the GG sequence in Adpol greatly reduced the efficiency of initiation. As glutamic acid (1057E/A), tyrosine (673Y/A, 1080Y/A), or lysine and glycine (1101KG/AA) residues play important roles in the interaction of Adpol with pTP and DNA (Fig. 2 and 3 and Table 1), the decrease in initiation observed with these mutant enzymes is likely to be caused by a decrease in the ability of Adpol to interact with pTP and DNA.
FIG. 5.
Ability of mutant Adpol to participate in the initiation of adenovirus DNA replication in vitro. (A) Autoradiograph showing template-directed incorporation of dCMP onto pTP in vitro. (B) Graphical representation of incorporation of dCMP onto pTP for each mutant enzyme. Data presented are the mean of three independent experiments and are relative to the initiation observed with wild-type Adpol (ADPOLWT). Error bars represent the 95% confidence limits of the calculated means.
TABLE 1.
Phenotypes caused by mutations in Adpola
| Mutation(s) in Adpol | DNA bindingb
|
Adpol-pTP complexc | Pol activityd (%) | Initiatione (%) | |
|---|---|---|---|---|---|
| ssDNA | dsDNA | ||||
| None (wild type) | ++++ | ++++ | ++++ | ++++ | ++++ |
| 656L/A | +++ | +++ | +++ | ++++ | +++ |
| 666GG/AA | +++ | ++ | +++ | ++++ | +++ |
| 667G/D | +++ | +++ | +++ | ++++ | +++ |
| 670Y/A | ++++ | ++ | +++ | +++ | + |
| 673Y/A | + | + | + | ++ | + |
| 685D/A | ++++ | +++ | ++++ | − | − |
| 690Y/A | +++ | +++ | ++++ | + | ++ |
| 692S/A | +++ | ++ | +++ | +++ | +++ |
| 760R/A | ++++ | ++ | ++++ | ++++ | ++ |
| 764R/A | +++ | ++ | +++ | ++++ | ++++ |
| 837K/A | ++++ | ++++ | +++ | ++++ | ++++ |
| 841N/A | ++++ | ++++ | ++++ | ++ | + |
| 844Y/A | + | ++ | ++ | + | + |
| 847F/A | + | + | +++ | ++ | + |
| 871TAG/AAA | + | + | ++ | ++ | + |
| 1010YG/AA | + | + | +++ | + | + |
| 1012DTD/ATA | + | + | ++ | − | − |
| 1055E/A | + | + | ++++ | ++++ | ++ |
| 1057E/A | − | − | − | + | + |
| 1078K/A | + | + | +++ | ++ | ++ |
| 1080Y/A | − | − | + | ++ | + |
| 1101KG/AA | + | + | +++ | ++ | ++ |
Activities of wild-type Adpol and the mutant forms in the assays. ++++, activity of wild-type Adpol or the mutant forms showing 76 to 100% of wild-type activity, +++, ++, and +, mutant forms showing 51 to 75, 25 to 50, and <25% of wild-type activity, respectively; −, no activity detected.
Ability of mutant Adpol to bind ssDNA or dsDNA at 0.2 M NaCl. Adpol bound to ssDNA or dsDNA-Sepharose beads was determined by Western blotting with Adpol-specific antiserum. The scanned image of the ECL blot was used to determine the binding of each mutant Adpol relative to that of the wild type.
Ability of mutant Adpol to form the Adpol-pTP complex. The amount of Adpol bound by pTP was determined by immunoprecipitation of the Adpol-pTP complex using anti-pTP monoclonal antibody 16H1 and detection of Adpol by Western blotting as described in Materials and Methods.
Catalytic activity of Adpol on activated DNA was determined in a DNA Pol assay and is based on dAMP incorporation into wild-type Adpol.
The ability of mutant Adpol to participate in initiation of DNA replication was determined based on incorporation of the TP-dCMP complex.
DISCUSSION
Structural analysis of diverse DNA Pols bound to a template primer and incoming dNTP (19, 32, 45) has illustrated that nucleotide addition by polynucleotide Pols occurs by a common two-metal ion mechanism. It is proposed that the two metal ions correctly position the dNTP in the active site and participate in the chemistry of phosphoryl transfer. While one metal ion activates the 3′ hydroxyl of the primer, the other helps the departure of pyrophosphate and both stabilize the charge and structure of the predicted transition state (56). In all of the structures analyzed, the template primer and dNTP are in contact with the highly conserved residues which make up the active sites of the enzymes. No structure is available for a Pol α family member bound to a template primer and dNTP. The structure of Pol α-like RB69 DNA Pol has been determined in the absence of substrates and displays a very similar arrangement of conserved residues at the active site (64). These residues are conserved among RB69 DNA Pol, φ 29 DNA Pol, human DNA Pol α, herpes simplex virus DNA Pol, and adenovirus DNA Pol, and mutational and biochemical studies of these enzymes indicate that the conserved residues play similar roles in catalysis (25, 64; this study). In RB69 DNA pol, these residues are all within 10Å of the catalytic center and form a contiguous conserved surface. In the case of Adpol, this region would display three chemically distinct clusters of amino acids that would be expected to interact with the primer terminus and the incoming dNTP. These conserved clusters consist of exposed aromatic residues Y670, Y690, Y844, F847, Y1010, and Y1080; negatively charged residues D685, D1012, D1014, E1055, and E1057; and positively charged residues R787, R791, K837, and K1078. It is primarily these residues which have been tested for their role in protein-primed initiation of adenovirus DNA replication.
Inspection of Table 1 reveals the phenotypes of the mutated DNA Pols analyzed and indicates that a limited number either have no phenotype (764R/A and 837K/A) or affect all activities (673Y/A, 844Y/A, 1010YG/AA, 1012DTD/ATA, and 1057E/A) while the remaining mutant DNA Pols have selected effects on the different activities of the DNA Pol. It is this latter class of mutant enzymes which allows the role of individual residues to be ascribed to particular roles in adenovirus DNA replication. Since none of the mutant enzymes analyzed in this study has completely lost all of the activities tested, it seems likely that the mutations do not grossly alter the conformation of Adpol. This conclusion is strengthened by the observation that all of the mutated proteins were translocated to the nucleus and all were relatively undegraded when expressed in insect cells. In addition to its catalytic activity of DNA polymerization this enzyme has to carry out a number of other activities related to its complex function in adenovirus DNA replication. This enzyme binds to the origin of DNA replication, interacts with pTP, binds to nuclear factor I, transfers dCMP onto pTP, extends the pTP dCMP primer, separates from pTP, and catalyzes strand displacement DNA synthesis in the presence of the viral DBP. As not all of these activities have been tested, it is entirely possible that some of the mutant enzymes will have additional phenotypes in DNA replication.
As we chose to focus on the conserved residues that are expected to interact with the template primer, it is perhaps not surprising that a large number of the mutations reduce the DNA binding activity of Adpol (Table 1). In this respect, it is also worth noting that all of the mutant DNA Pols which are compromised for pTP binding also display reduced DNA binding activity (Table 1). This suggests that, as the β-OH of S580 in pTP is the initial primer for DNA synthesis, it is likely that pTP contacts a group of residues during initiation similar to that which the DNA primer contacts during elongation. However, a number of Adpol mutant enzymes are competent to bind pTP but fail to bind DNA (847F/A, 1010YG/AA, 1055E/A, and 1078K/A), suggesting that during elongation additional contacts with the template primer are required. There is not a strict correlation between DNA binding and DNA Pol activity, as Adpol with a number of mutations (847F/A, 871TAG/AAA, and 1055E/A) that cause substantially reduced DNA binding activity still retains substantial (>50%) DNA Pol activity. It is possible that the high primer template concentration in DNA Pol assays using activated DNA overcomes the relatively low affinity of these proteins for DNA. It is also worth noting that while DNA binding assays were performed in the presence of 0.1 or 0.2 M NaCl, DNA Pol assays are conducted in the absence of NaCl, thus stabilizing low-affinity interaction between Adpol and DNA. The importance of the C-terminal region in DNA binding suggested by the deleterious effect of mutations 1055E/A and 1057E/A is supported by previous work in which mutation of cysteine residues (C1060, C1063, C1087, and C1090), thought to ligate Zn2+ in a zinc finger domain, also had a deleterious effect on DNA binding, Pol activity, and initiation (28).
Like other members of the Pol α family, Adpol has a multidomain structure and noncontinuous regions of the polypeptide chains representing a large surface area are required for interaction with pTP (44, 49). Analysis of the Adpol mutants described here allows the identification of residues that are likely to participate in the interactions with pTP and an assessment of their role in the initiation of DNA synthesis (Table 1). Residues involved in Adpol-pTP interaction are located in different regions of the Adpol peptide chain, but since the structure of Adpol has yet to be determined, the positions of these residues in three-dimensional space are not known. As shown in Fig. 3, mutation of tyrosine (673Y/A, 844Y/A, or 1080Y/A) or a glutamic acid (1057E/A) residue reduced the Adpol-pTP interaction dramatically. Alignment of the Adpol sequence with that of RB69 DNA Pol, whose structure is known, indicates that tyrosines (673Y, 844Y) and glutamic acid (1057E) of Adpol have positions similar to those of tyrosines (391Y, 567Y) and glutamic acid (686E) near the catalytic center in RB69 Pol (64). This suggests that in addition to forming part of the catalytic site, these residues of Adpol also play roles in protein primer selection and template interaction as in φ29 DNA Pol (5). It has been reported that a YxGG/A motif of φ29 Pol is necessary for the formation of a stable TP-Pol complex (60). The same motif was also found in Adpol. However, mutation of the GG sequence in these mutant enzymes did not abolish Adpol-pTP interaction. Interestingly, two tyrosine residues (670Y/A, 673Y/A) close to the YxGG/A motif in Adpol play an important role in Adpol-pTP interaction and are also required for initiation of DNA synthesis.
Two mutant (670Y/A and 841N/A) Adpol enzymes have a particularly interesting phenotype in that they bind pTP and DNA with wild-type characteristics and are only moderately affected in DNA Pol activity but are severely compromised in the ability to participate in the initiation of DNA replication (Table 1). This indicates that residues Y670 and N841 have functions that are likely to be unique to protein-primed initiation of DNA synthesis. Sequence alignment shows that residue Y670 of Adpol is close to Y391 in RB69 Pol, which model building suggests may be involved in template binding by hydrophobic stacking interactions between the base and the aromatic side chain. Residues N841 and Y844 are conserved in both RB69 Pol (N564, Y567) and φ29 Pol (N387, Y390). In φ29 Pol, N387 and Y390 are involved in primer-template binding and dNTP selection. As in Adpol, mutation of these residues severely reduces protein-primed initiation of φ29 Pol (5). Interestingly, residue F847 in Adpol seems to play more roles in protein-primed initiation while the equivalent (F393) in φ29 DNA Pol is involved mainly in DNA primer-dependent polymerization (5). As reported previously (29), mutation of the highly conserved DTD motif in Adpol abolished DNA Pol and initiation activities. Studies of other DNA Pols have indicated that these residues are involved in metal-specific catalysis and metal-induced infidelity of DNA synthesis (10, 11, 52). Mutation of D685 in Adpol also abolished DNA Pol and initiation activities. This residue is absolutely conserved among all members of the Pol α family (16) and is one of the two acidic residues that ligate Mg2+, bind dNTP, and participate in catalysis.
RB69 DNA Pol has the overall shape of a disc with a hole in the center (64). Three deep grooves formed by five domains, converge on the central hole, and a number of residues surrounding the central hole are directly involved in catalytic activity (68). Similar to those of other members of the Pol α family, the catalytic site of Adpol is probably composed of the conserved motifs YGDTD in Pol I, Dx2SLYP in Pol II, and Kx3NSx2YG in Pol III. Residues within or between the conserved regions of Adpol are also involved in DNA Pol activity. However, elucidation of the exact roles of these residues or how these residues function in adenovirus DNA replication awaits the structural analysis of Adpol.
ACKNOWLEDGMENTS
We thank Catherine H. Botting for setting up the initiation assay and providing the anti-pTP monoclonal antibody and Michael H. Tatham for cloning the Adpol-encoding gene into the pFastBac1 vector.
This work was supported by the Wellcome Trust.
REFERENCES
- 1.Arnold E, Ding J, Hughes S H, Hostomsky Z. Structures of DNA and RNA polymerases and their interactions with nucleic acid substrates. Curr Opin Struct Biol. 1995;5:27–38. doi: 10.1016/0959-440x(95)80006-m. [DOI] [PubMed] [Google Scholar]
- 2.Bernad A, Blanco L, Lazaro J M, Martin G, Salas M. A conserved 3′-5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989;59:219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- 3.Bernad A, Zaballos A, Salas M, Blanco L. Structural and functional relationships between prokaryotic and eukaryotic DNA polymerases. EMBO J. 1987;6:4219–4225. doi: 10.1002/j.1460-2075.1987.tb02770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasco M A, Lazaro J M, Blanco L, Salas M. Phi 29 DNA polymerase active site. Residue ASP249 of conserved amino acid motif “Dx2SLYP” is critical for synthetic activities. J Biol Chem. 1993;268:24106–24113. [PubMed] [Google Scholar]
- 5.Blasco M A, Lazaro J M, Blanco L, Salas M. Phi 29 DNA polymerase active site. The conserved amino acid motif “Kx3NSxYG” is involved in template-primer binding and dNTP selection. J Biol Chem. 1993;268:16763–16770. [PubMed] [Google Scholar]
- 6.Braithwaite D K, Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brons-Poulsen J, Petersen N E, Horder M, Kristiansen K. An improved PCR-based method for site directed mutagenesis using megaprimers. Mol Cell Probes. 1998;12:345–348. doi: 10.1006/mcpr.1998.0187. [DOI] [PubMed] [Google Scholar]
- 8.Chen M, Horwitz M S. Dissection of functional domains of adenovirus DNA polymerase by linker-insertion mutagenesis. Proc Natl Acad Sci USA. 1989;86:6116–6120. doi: 10.1073/pnas.86.16.6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Copeland W C, Lam N K, Wang T S. Fidelity studies of the human DNA polymerase alpha. The most conserved region among alpha-like DNA polymerases is responsible for metalinduced infidelity in DNA synthesis. J Biol Chem. 1993;268:11041–11049. [PubMed] [Google Scholar]
- 10.Copeland W C, Wang T S. Enzymatic characterization of the individual mammalian primase subunits reveals a biphasic mechanism for initiation of DNA replication. J Biol Chem. 1993;268:26179–26189. [PubMed] [Google Scholar]
- 11.Copeland W C, Wang T S. Mutational analysis of the human DNA polymerase alpha. The most conserved region in alpha-like DNA polymerases is involved in metalspecific catalysis. J Biol Chem. 1993;268:11028–11040. [PubMed] [Google Scholar]
- 12.Davies J F, 2nd, Almassy R J, Hostomska Z, Ferre R A, Hostomsky Z. 2.3 A crystal structure of the catalytic domain of DNA polymerase beta. Cell. 1994;76:1123–1133. doi: 10.1016/0092-8674(94)90388-3. [DOI] [PubMed] [Google Scholar]
- 13.Delarue M, Poch O, Tordo N, Moras D, Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- 14.de Vega M, Blanco L, Salas M. phi29 DNA polymerase residue Ser122, a single-stranded DNA ligand for 3′-5′ exonucleolysis, is required to interact with the terminal protein. J Biol Chem. 1998;273:28966–28977. doi: 10.1074/jbc.273.44.28966. [DOI] [PubMed] [Google Scholar]
- 15.de Vega M, Lazaro J M, Salas M, Blanco L. Mutational analysis of phi29 DNA polymerase residues acting as ssDNA ligands for 3′-5′ exonucleolysis. J Mol Biol. 1998;279:807–822. doi: 10.1006/jmbi.1998.1805. [DOI] [PubMed] [Google Scholar]
- 16.Dong Q, Copeland W C, Wang T S. Mutational studies of human DNA polymerase alpha. Identification of residues critical for deoxynucleotide binding and misinsertion fidelity of DNA synthesis. J Biol Chem. 1993;268:24163–24174. [PubMed] [Google Scholar]
- 17.Dong Q, Copeland W C, Wang T S. Mutational studies of human DNA polymerase alpha. Serine 867 in the second most conserved region among alpha-like DNA polymerases is involved in primer binding and mispair primer extension. J Biol Chem. 1993;268:24175–24182. [PubMed] [Google Scholar]
- 18.Dong Q, Wang T S. Mutational studies of human DNA polymerase alpha. Lysine 950 in the third most conserved region of alpha-like DNA polymerases is involved in binding the deoxynucleoside triphosphate. J Biol Chem. 1995;270:21563–21570. doi: 10.1074/jbc.270.37.21563. [DOI] [PubMed] [Google Scholar]
- 19.Doublie S, Tabor S, Long A M, Richardson C C, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 20.Fansler B S, Loeb L A. Sea urchin nuclear DNA polymerase. Methods Enzymol. 1974;29:53–70. doi: 10.1016/0076-6879(74)29009-8. [DOI] [PubMed] [Google Scholar]
- 21.Field J, Gronostajski R M, Hurwitz J. Properties of the adenovirus DNA polymerase. J Biol Chem. 1984;259:9487–9495. [PubMed] [Google Scholar]
- 22.Hay R H. Adenovirus DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 699–719. [Google Scholar]
- 23.Hornbeck P. Direct competitive ELISA to detect soluble antigens. In: Coligan J E, Kruisbeek A M, Shevach D H, Strober W, editors. Current protocols in immunology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 2.1.6–2.1.11. [Google Scholar]
- 24.Hwang C B C, Ruffner K L, Coen D M. A point mutation within a distinct conserved region of the herpes simplex virus DNA polymerase gene confers drug resistance. J Virol. 1992;66:1774–1776. doi: 10.1128/jvi.66.3.1774-1776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang Y T, Liu B-Y, Coen D M, Hwang C B C. Effects of mutations in the Exo III motif of the herpes simplex virus DNA polymerase gene on enzyme activities, viral replication, and replication fidelity. J Virol. 1997;71:7791–7798. doi: 10.1128/jvi.71.10.7791-7798.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito J, Braithwaite D K. Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 1991;19:4045–4057. doi: 10.1093/nar/19.15.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joung I, Engler J A. Mutations in two cysteine-histidine-rich clusters in adenovirus type 2 DNA polymerase affect DNA binding. J Virol. 1992;66:5788–5796. doi: 10.1128/jvi.66.10.5788-5796.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joung I, Horwitz M S, Engler J A. Mutagenesis of conserved region 1 in the DNA polymerase from human adenovirus serotype 2. Virology. 1991;184:235–241. doi: 10.1016/0042-6822(91)90840-8. [DOI] [PubMed] [Google Scholar]
- 30.Joyce C M, Steitz T A. Function and structure relationships in DNA polymerases. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 31.Kadonaga J T, Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci USA. 1986;83:5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiefer J R, Mao C, Braman J C, Beese L S. Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature. 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- 33.Kiefer J R, Mao C, Hansen C J, Basehore S L, Hogrefe H H, Braman J C, Beese L S. Crystal structure of a thermostable Bacillus DNA polymerase I large fragment at 2.1 A resolution. Structure. 1997;5:95–108. doi: 10.1016/s0969-2126(97)00169-x. [DOI] [PubMed] [Google Scholar]
- 34.Kim Y, Eom S H, Wang J, Lee D S, Suh S W, Steitz T A. Crystal structure of Thermus aquaticus DNA polymerase. Nature. 1995;376:612–616. doi: 10.1038/376612a0. [DOI] [PubMed] [Google Scholar]
- 35.Knopf C W. Evolution of viral DNA-dependent DNA polymerases. Virus Genes. 1998;16:47–58. doi: 10.1023/a:1007997609122. [DOI] [PubMed] [Google Scholar]
- 36.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 37.Korolev S, Nayal M, Barnes W M, Di Cera E, Waksman G. Crystal structure of the large fragment of Thermus aquaticus DNA polymerase I at 2.5-A resolution: structural basis for thermostability. Proc Natl Acad Sci USA. 1995;92:9264–9268. doi: 10.1073/pnas.92.20.9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laemmli U K, Quittner S F. Maturation of the head of bacteriophage T4. IV. The proteins of the core of the tubular polyheads and in vitro cleavage of the head proteins. Virology. 1974;62:483–499. doi: 10.1016/0042-6822(74)90409-7. [DOI] [PubMed] [Google Scholar]
- 39.Leith I R, Hay R T, Russell W C. Adenovirus subviral particles and cores can support limited DNA replication. J Gen Virol. 1989;70:3235–3248. doi: 10.1099/0022-1317-70-12-3235. [DOI] [PubMed] [Google Scholar]
- 40.Monaghan A, Hay R T. Pyridoxal 5′-phosphate inhibition of adenovirus DNA polymerase. J Biol Chem. 1996;271:24242–24248. doi: 10.1074/jbc.271.39.24242. [DOI] [PubMed] [Google Scholar]
- 41.Monaghan A, Webster A, Hay R T. Adenovirus DNA binding protein: helix destabilising properties. Nucleic Acids Res. 1994;22:742–748. doi: 10.1093/nar/22.5.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mul Y M, Verrijzer C P, van der Vliet P C. Transcription factors NFI and NFIII/oct-1 function independently, employing different mechanisms to enhance adenovirus DNA replication. J Virol. 1990;64:5510–5518. doi: 10.1128/jvi.64.11.5510-5518.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ollis D L, Brick P, Hamlin R, Xuong N G, Steitz T A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature. 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 44.Parker E J, Botting C H, Webster A, Hay R T. Adenovirus DNA polymerase: domain organisation and interaction with preterminal protein. Nucleic Acids Res. 1998;26:1240–1247. doi: 10.1093/nar/26.5.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pelletier H, Sawaya M R, Kumar A, Wilson S H, Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 46.Pronk R, Stuiver M H, van der Vliet P C. Adenovirus DNA replication: the function of the covalently bound terminal protein. Chromosoma. 1992;102:S39–S45. doi: 10.1007/BF02451784. [DOI] [PubMed] [Google Scholar]
- 47.Pronk R, van der Vliet P C. The adenovirus terminal protein influences binding of replication proteins and changes the origin structure. Nucleic Acids Res. 1993;21:2293–2300. doi: 10.1093/nar/21.10.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pruijn G J, van Driel W, van der Vliet P C. Nuclear factor III, a novel sequence-specific DNA-binding protein from HeLa cells stimulating adenovirus DNA replication. Nature. 1986;322:656–659. doi: 10.1038/322656a0. [DOI] [PubMed] [Google Scholar]
- 49.Roovers D J, van der Lee F M, van der Wees J, Sussenbach J S. Analysis of the adenovirus type 5 terminal protein precursor and DNA polymerase by linker insertion mutagenesis. J Virol. 1993;67:265–276. doi: 10.1128/jvi.67.1.265-276.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salas M. Protein-priming of DNA replication. Annu Rev Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 52.Saturno J, Lazaro J M, Blanco L, Salas M. Role of the first aspartate residue of the “YxDTDS” motif of phi29 DNA polymerase as a metal ligand during both TP-primed and DNA-primed DNA synthesis. J Mol Biol. 1998;283:633–642. doi: 10.1006/jmbi.1998.2121. [DOI] [PubMed] [Google Scholar]
- 53.Sawaya M R, Pelletier H, Kumar A, Wilson S H, Kraut J. Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism. Science. 1994;264:1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- 54.Sobol R W, Horton J K, Kuhn R, Gu H, Singhal R K, Prasad R, Rajewsky K, Wilson S H. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 55.Sousa R, Chung Y J, Rose J P, Wang B C. Crystal structure of bacteriophage T7 RNA polymerase at 3.3 A resolution. Nature. 1993;364:593–599. doi: 10.1038/364593a0. [DOI] [PubMed] [Google Scholar]
- 56.Steitz T A. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 57.Tabor S, Richardson C C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci USA. 1987;84:4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Temperley S M, Hay R T. Recognition of the adenovirus type 2 origin of DNA replication by the virally encoded DNA polymerase and preterminal proteins. EMBO J. 1992;11:761–768. doi: 10.1002/j.1460-2075.1992.tb05109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Temperley S M, Hay R T. Replication of adenovirus type 4 DNA by a purified fraction from infected cells. Nucleic Acids Res. 1991;19:3243–3249. doi: 10.1093/nar/19.12.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Truniger V, Blanco L, Salas M. Role of the “YxGG/A” motif of Phi29 DNA polymerase in protein-primed replication. J Mol Biol. 1999;286:57–69. doi: 10.1006/jmbi.1998.2477. [DOI] [PubMed] [Google Scholar]
- 61.van der Vliet P C. The role of transcription factors in enhancement of adenovirus DNA replication. Semin Virol. 1991;2:271–281. [Google Scholar]
- 62.van der Vliet P C. Roles of transcription factors in DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 87–118. [Google Scholar]
- 63.Wang E, Henner D, Furth J J. Duplication of single stranded DNA catalyzed by calf thymus DNA polymerase alpha. Nucleic Acids Res. 1976;3:129–147. doi: 10.1093/nar/3.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J, Sattar A K, Wang C C, Karam J D, Konigsberg W H, Steitz T A. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 65.Webster A, Leith I R, Hay R T. Domain organization of the adenovirus preterminal protein. J Virol. 1997;71:539–547. doi: 10.1128/jvi.71.1.539-547.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Webster A, Leith I R, Nicholson J, Hounsell J, Hay R T. Role of preterminal protein processing in adenovirus replication. J Virol. 1997;71:6381–6389. doi: 10.1128/jvi.71.9.6381-6389.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wong S W, Wahl A F, Yuan P M, Arai N, Pearson B E, Arai K, Korn D, Hunkapiller M W, Wang T S. Human DNA polymerase alpha gene expression is cell proliferation dependent and its primary structure is similar to both prokaryotic and eukaryotic replicative DNA polymerases. EMBO J. 1988;7:37–47. doi: 10.1002/j.1460-2075.1988.tb02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang G, Lin T, Karam J, Konigsberg W H. Steady-state kinetic characterization of RB69 DNA polymerase mutants that affect dNTP incorporation. Biochemistry. 1999;38:8094–8101. doi: 10.1021/bi990653w. [DOI] [PubMed] [Google Scholar]