Abstract
Orofacial cleft (OFC) is a common human congenital anomaly. Epithelial-specific RNA splicing regulators ESRP1 and ESRP2 regulate craniofacial morphogenesis and their disruption result in OFC in zebrafish, mouse and humans. Using esrp1/2 mutant zebrafish and murine Py2T cell line models, we functionally tested the pathogenicity of human ESRP1/2 gene variants. We found that many variants predicted by in silico methods to be pathogenic were functionally benign. Esrp1 also regulates the alternative splicing of Ctnnd1 and these genes are co-expressed in the embryonic and oral epithelium. In fact, over-expression of ctnnd1 is sufficient to rescue morphogenesis of epithelial-derived structures in esrp1/2 zebrafish mutants. Additionally, we identified 13 CTNND1 variants from genome sequencing of OFC cohorts, confirming CTNND1 as a key gene in human OFC. This work highlights the importance of functional assessment of human gene variants and demonstrates the critical requirement of Esrp-Ctnnd1 acting in the embryonic epithelium to regulate palatogenesis.
Introduction
The study of orofacial cleft (OFC) has been foundational to genetic analysis of congenital anomalies. Craniofacial structural malformations are amenable to detailed phenotypic classification in large cohorts where genomic studies have been carried out to identify associated loci (1–8). As whole-genome sequencing (WGS) strategies and technologies advance, a growing list of genes and gene variants associated with OFC are being cataloged (1, 8–11). These approaches have uncovered the critical role of many genes regulating the embryonic oral epithelium in palate formation and OFC pathogenesis, including: TP63, IRF6, GRHL3, ESRP1/2, CTNND1 (12–24).
Because most cases of non-syndromic OFC occur sporadically, the pathogenicity of variants cannot be inferred or supported by segregation among affected family members. Therefore, determining the functional significance of gene variants remains challenging. Multiple in silico predictive algorithms such as SIFT, PolyPhen-2, MutationTaster, PROVEAN and AlphaMissense offer functional predictions for gene variants utilizing amino acid sequence information, sequence conservation, biophysical properties, or homolog alignment (25–30). However, when given the same gene variants, these predictive tools may provide null values or contradicting results (31, 32). Indeed, the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG-AMP), weights functional studies higher than in silico evidence for asserting pathogenic potential in gene variants for genes not previously established as causal for a particular disease (33–35). We and others previously showed that functional testing of human gene variants is essential, as in silico approaches alone fail to reach the necessary accuracy for clinical translation (36–40). While bioinformatics tools have greatly facilitated the functional interpretation of genetic variants (41–43), it is also important to note the essential role of functional validation of gene variants, especially for those genes where computational predictions tend to differ from experimental validation (44–50).
ESRP1 and its paralog ESRP2 are epithelial splicing regulatory proteins that co-localize with Irf6 and function in the embryonic epithelium to regulate craniofacial development and epithelial-mesenchymal transition during embryogenesis (22, 51–53). Global transcriptome analysis comparing mutant irf6 and wildtype zebrafish revealed that the epithelial-specific splicing regulator Esrp1 was differentially expressed (52). We showed that Esrp1 and Esrp2 are colocalized in the periderm and oral epithelium and are required for the formation of the anterior neurocranium (ANC), a teleost embryonic structure developmentally analogous to the mammalian primary palate in the manner that it is formed from the convergence of frontonasal derived midline prominence and paired maxillary projections (54–57). Targeted disruption of Esrp1 in the mouse resulted in bilateral cleft lip and palate (21). In the esrp1/2 double homozygote zebrafish, cleft formed in the ANC and extended to the upper edge of the mouth opening, analogous to the cleft lip and/or palate (CL/P) phenotype observed in the Esrp1/2 mutant mice (22, 52). In humans biallelic ESRP1 mutations were described to cause hearing loss (58), heterozygous ESRP2 mutations were associated with CL/P (20) and both ESRP1 and ESRP2 splicing targets were related to cancer-associated processes (59). Given the central role of ESRP1 in periderm and embryonic epithelial development, there is likely selection against deleterious ESRP1 alleles so that variants associated with hearing deficit are likely hypomorphic and homozygous or biallelic loss-of-function alleles are likely embryonic lethal and not observed clinically.
Here, we applied complementary in vivo and in vitro models to functionally interrogate human ESRP1 and ESRP2 gene variants. To increase the rigor of the functional test using another independent assay, we also examined Esrp-mediated alternative splicing in a murine Esrp1/2 double knockout Py2T cell model. The Py2T cell line has been used effectively to study epithelial mesenchymal transition and we have previously generated and characterized Esrp1 and Esrp2 double knock-out Py2T lines (23, 53). Using these independent approaches, we functionally determined the pathogenicity of the 7 ESRP1 and 12 ESRP2 human gene variants from CL/P cohorts or reported in hearing loss. We previously showed that Esrp1/2 regulated splicing of Ctnnd1 (60). Using RNAscope, we found that Ctnnd1 transcripts co-localized with Esrp1 and Esrp2 in the mouse and zebrafish embryonic oral epithelium. The esrp1/2 zebrafish model also presented a functional assay to test the function of Esrp-regulated genes such as Ctnnd1. In fact, exogenous expression of ctnnd1 mRNA in zebrafish esrp1/2 mutants partially rescued the cleft ANC, foreshortened pectoral fin and fused otolith phenotypes. Additionally, WGS of CL/P cohorts identified 13 new CTNND1 gene variants, making this one of the most frequently associated genes in OFC. Taken together, these results demonstrate the critical requirement of Esrp-Ctnnd1 operating in the embryonic epithelium to regulate palatogenesis.
Methods
Animal husbandry and breeding
Zebrafish (Danio Rerio) of the Tübingen strain were raised and bred following approved institutional protocols at Massachusetts General Hospital. Embryos were collected and raised in E3 Medium (5.0 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) containing 0.0001% Methylene blue at 28.5°C.
Gene variant identification, sequence alignment, and variant effect prediction
Three WGS datasets of 759 OFC trios from the Gabriella Miller Kids First (GMKF) Research (dbGaP; European trios, dbGaP: phs001168.v2.p2; Colombian trios, dbGaP: phs001420.v1.p1; Taiwanese trios, dbGaP: phs000094.v1.p1) were filtered for variants in ESRP1, ESRP2, and CTNND1 that were (1) heterozygous in the affected patient, (2) had a minor allele frequency no greater than 0.001 in any population in gnomAD or 1000 Genomes, and (3) had a variant consequence of missense, frameshift, stop-gain, splicing, or in-frame insertion/deletion. We further supplemented the resulting list with additional variants from ClinVar associated with an OFC or autosomal recessive deafness. In total, the ClinVar list included 12 ESRP1 and 20 ESRP2 variants. ClinVar variants were accessed in 2021, we note that new variants have been uploaded to ClinVar for ESRP1 and ESRP2, but these new variants did not include relevant clinical phenotype information so were not included in this study. OFC associated genes were based on a previously published study that curated a list of approximately 500 genes based on known clinical syndromes and association results from GWAS (61).
To further refine the variant list to identify variants for testing in mouse and zebrafish assays, we aligned the human, mouse and zebrafish Esrp1 and Esrp2 amino acid sequences using Clustal Omega (62). 7 ESRP1 and 12 ESRP2 variants at fully conserved residues were then annotated using SIFT, PolyPhen-2, and AlphaMissense to obtain the predicted change in protein function and were categorized as benign, pathogenic, or of unknown significance. We included a silent mutation from ESRP2, at threonine 475 (T475T) that served as an internal negative control. Variants were annotated to the following human transcripts: ESRP1: NM_017697.4/ENST00000433389.8; ESRP2: NM_024939.3/ENST00000473183.7; and CTNND1: NM_001085458.2/ENST00000399050.10.
All variants from this study are listed in Table 1 in the supplementary material.
Rare-variants analysis
We performed rare variant burden tests using RV-TDT (2) for protein-altering variants in ESRP1, ESRP2, and CTNND1 that had a minor allele frequency less than 0.1% in any gnomAD population. DenovolyzeR (0.2.0), an R package which compares the observed number of DNMs to the expected number of DNMs based on a mutational model developed by Samocha et al. (2014) (63), was used to determine if de novo variants were enriched in these three genes.
Plasmid generation, site-directed mutagenesis, and mRNA synthesis
mRNA from wildtype zebrafish embryos was collected at multiple time points from 6 hours post fertilization (hpf) to 4 days post fertilization (dpf), reverse transcribed, and combined to make pooled cDNA to clone the esrp1 coding sequence (CDS). esrp1 and esrp2 were each cloned into a pCS2+8 plasmid backbone using the In-Fusion HD Cloning Kit (Clontech). The resulting pCS2+8-esrp2 plasmid was mutagenized with synonymous mutations surrounding the translational start-site using the GeneArt site-directed mutagenesis (SDM) system (ThermoFisher) to generate esrp2 transcripts resistant to esrp2 morpholino binding. The 19 human ESRP1 and ESRP2 variants were each individually introduced to the pCS2+8-esrp1 or MO-resistant pCS2+8-esrp2 plasmids through the GeneArt SDM system. All generated pCS2+8 plasmids were digested with NotI at 37°C for 1hr, and capped mRNA was synthesized using the SP6 mMessage mMachine kit (ThermoFisher).
For the murine Py2T transfection experiments, we used the pIBX-C-FF(B)-mCherry-esrp1(2A)-+CKLP plasmid containing the mouse Esrp1 cDNA sequence, fused to a mCherry tag (gift from Russ Carstens, University of Pennsylvania). Mouse Esrp2 cDNA was purchased from Genomics Online. Esrp1 cDNA was cloned into the pcDNA3.1 backbone containing a CMV promoter and SV40 polyA tailing sequence for expression in mammalian cells using the In-Fusion HD Cloning Kit (Clontech) to generate the pcDNA3.1-esrp1-mCherry plasmid. An mCherry tag was fused in-frame onto the Esrp2 cDNA and introduced into the pcDNA3.1 backbone through a multi-insert in-Fusion cloning strategy, using the pIBX-C-FF(B)-mCherry-Esrp1(2A)-+CKLP as the template for the 2A-mCherry sequence to generate the pcDNA3.1-esrp2-mCherry plasmid. Selected human ESRP1 and ESRP2 gene variants were introduced using the GeneArt SDM system, as described above.
Zebrafish microinjection and esrp1/2 rescue assay
We previously generated a zebrafish line carrying homozygous loss-of-function alleles in esrp1 through CRISPR/Cas9 harboring −4 bp indels which led to a frame shift mutation and early protein truncation (52). esrp2 morpholinos (GeneTools) were reconstituted to a concentration of 8ug/uL in water and stored in single-use aliquots at RT. 2nL droplets containing (1) 8ng esrp2 morpholino, (2) 0.05% phenol red and (3) 200pg of esrp1, esrp2, or esrp gene-variant mRNA were microinjected directly into the cytoplasm of one-cell stage esrp1−/− zebrafish embryos and grown until 4dpf. (We have previously shown that the esrp2 morpholino, injected into esrp1−/− esrp2wt/wt is sufficient to phenocopy the esrp1−/−; esrp2 −/− phenotype, which is consistent with previous descriptions (22, 52). Since all the injected embryos were derived from mating of esrp1−/− males and females, all animals had the esrp1−/− genotype and did not require additional genotyping after phenotype analysis. At 4 dpf, embryos were fixed in 4% formaldehyde, stained with acid-free Alcian blue as previously described (64), and micro-dissected to inspect the anterior neurocranium (ANC). The ANC phenotype flatmount was then scored as wildtype ANC, cleft ANC or rescued ANC.
PY2T cell maintenance and transfection
Mouse Py2T cells and Esrp1/2 DKO Py2T cells were a gift from Russ Carstens from the University of Pennsylvania (23). Cells were maintained in DMEM supplemented with 10% FBS and penicillin/streptomycin and were not cultured past passage 30. 10.8ug of plasmid was transfected onto 106 cells using the 100uL Neon system (ThermoFisher) with a single, 30 second pulse at 1400V and plated onto 6-well plates. Cells were harvested for RNA after 24hr, reverse transcribed, and the cDNA was used for RT-PCR using primers spanning the splice junctions for Ctnnd1 exons 1 and 3 and Afhgef11 exons 36 and 38, Arhgef11 Forward (TCAAGCTCAGAACCAGCAGGAAGT) and Arhgef11 Reverse (TGCTCGATGGTGTGGAAGATCACA), as described (23). The gels were quantified by densitometry using Fiji/ImageJ and the results are expressed as mean ± SEM. Statistical analysis involved using GraphPad Prism 9.0 for Windows. The experiments were performed in triplicate. One-way Anova test, with each comparison standing alone was used for statistical analysis. P < 0.05 was considered statistically significant.
ctnnd1 mRNA injection into esrp1−/−; esrp2+/− intercross
To construct the mRNA in vitro transcription (IVT) template, synthetic Ctnnd1 cDNA, isoform-201 on Ensembl (ENSDART00000106048.4), was cloned into the linearized DNA template vector (Takara Bio USA). The plasmid vectors were purified by a QIAprep spin miniprep kit (QIAGEN). The plasmid was digested with Hind III HF (NEB Biolabs) at 37°C for 1hr, 80°C for 20 minutes for inactivation and mRNA was synthesized using the T7 MEGAshortscript kit (ThermoFisher).
For micro-injection, progeny of esrp1−/−; esrp2+/− inter-cross, previous described by Carroll, 2020 (52) were injected at the single cell stage with either 250 pg of ctnnd1 mRNA (along with water), or gfp mRNA, for controls. Injected embryos were raised to 4 dpf, at which time embryos were fixed in 4% formaldehyde, stained with acid-free Alcian blue, and microdissected to inspect the anterior neurocranium (ANC). The ANC was scored as wildtype ANC or cleft ANC. Additionally, the pectoral fins were also analyzed and scored as wildtype fin or curled fin. For the otolith phenotype, wildtype was scored when the otoliths were separate and the mutant phenotype when the otoliths were fused. For the paired bilateral structures, if one fin was curled or one set of otoliths were fused, the animal was scored as mutant. After the phenotypic assessments for ANC, fin and otoliths, both the mRNA injected embryos and the control injected embryos was tracked and individually genotyped. Whenever there is an animal with genotype of esrp1−/−; esrp2−/− but exhibited ANC that are not fully cleft, fins that are not fully curled and separate otoliths, these animals were scored as rescues.
RNA in situ hybridization staining (RNAScope and BaseScope)
Wildtype and esrp1−/−; esrp2+/− zebrafish were crossed and the progeny embryos raised to 4 dpf. The esrp1−/−; esrp2−/− double mutant embryos were scored at 4 dpf based on the abrogated pectoral fin phenotype. The wild type and esrp1−/−; esrp2−/− embryos were fixed in 4% formaldehyde, taken through a sucrose gradient, and then cryo embedded and sectioned. RNAScope probes were designed with assistance from ACDBio to target the region of 700–1661 base pairs of the RNA for DR Ctnnd1 XM_021476936.1, which corresponds to ENSDART00000106048.4 for ensemble 201.
Additionally, RNAScope and BaseScope probes were designed for murine Esrp1 (we have previously shown that Esrp1 and Esrp2 colocalize in the oral epithelium) (52). Hybridization and staining were performed according to the manufacturers protocol. Stained sections were imaged on a Leica SP8 confocal microscope where a Z-stack was obtained and analyzed on imageJ software to obtain optimal images. BaseScope probes were designed and purchased from ACDBio to specifically target the Ctnnd1 long and short isoforms. Staining was carried out according to the manufacturer’s protocols on both fixed, frozen, and sectioned wildtype and Esrp1/2 DKO at E15. Stained sections were imaged as above.
Statistics and Reproducibility
The results are expressed as percentage or as mean ± SEM. Statistical analysis was using GraphPad Prism 10 for Windows (GraphPad Software, San Diego, CA, www.graphpad.com). All experiments were performed at least in triplicate. Two-way analysis of variance or Student t test was used for statistical analysis. P < 0.05 was considered statistically significant.
Results
esrp1 and esrp2 are required for morphogenesis of epithelial derived tissues
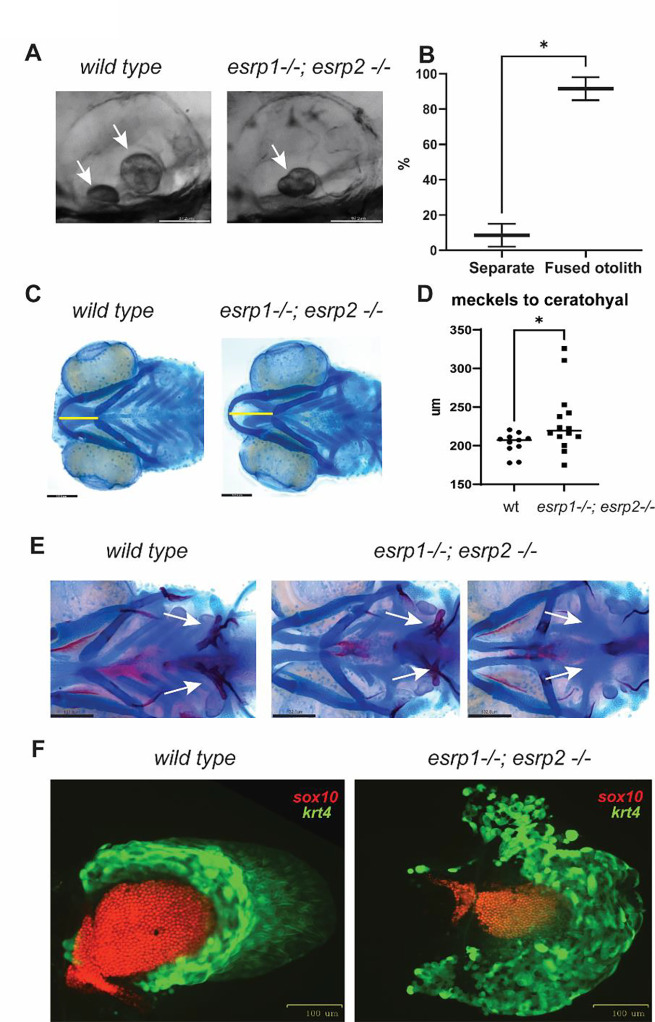
We previously described the genetic requirement of esrp1/2 in zebrafish epithelial development, disruption of which resulted in tethering of the upper mouth opening extending into a separation of the anterior neurocranium, a phenotype morphologically analogous to CL/P of amniotes (52). Given the expression of esrp1/2 and in periderm and embryonic epithelial cells broadly, we examined other structures formed by epithelial origins. It was reported that Esrp1 regulated the alternative splicing of Arhgef11, which was described to be important for proper otoliths development in zebrafish (65). When the esrp1/2 double mutants were examined at 4 dpf, we discovered that more than 90% of the mutant larvae exhibited at least one fused otolith (Figure 1A, B).
Figure 1. esrp1 and esrp2 are required for morphogenesis of epithelial derived tissue: otoliths, pharyngeal teeth and pectoral fins.
(A) zebrafish otoliths indicated by white arrows at 72 hpf. (B) Quantification and t test of zebrafish otoliths from genotyped mutants characterized as separate or fused otoliths. t-test, n=75. (C) Alcian blue representation of a 6 dpf zebrafish wildtype and esrp1−/− esrp2−/− double mutant showing cartilage stain, yellow line shows the measurement of the distance between the midline of Meckel’s and ceratohyal cartilages. (D) quantification and t-test analysis of this measurement in wildtype (n=11) and esrp1−/−; esrp2 −/− mutants (n=14). (E) Alcian blue and Alizarin red staining of larvae at 7 dpf ventral view, the pharyngeal teeth are present in wildtype (white arrows). In contrast, the esrp1−/; esrp2−/− all exhibit decreased number of teeth, and occasionally some double mutants lack all ceratobranchial cartilages and the pharyngeal teeth are absent. (F) wildtype and esrp1−/− esrp2−/− mutant pectoral fins labeled with sox10 mCherry (red) and krt4 gfp (green).
Ventral cartilages that form with epithelial-mesenchymal interactions were also dysmorphic, where the Meckel’s cartilage appeared longer in the antero-posterior axis and narrower in the coronal axis. These morphologic differences can be captured by measuring the distance between Meckel’s and ceratohyal cartilages which is extended in the esrp1/2 mutants (Figure 1C, D). We also detected partial penetrance of loss of ceratobranchial cartilages in 30% of the esrp1/2 double mutant larvae at 7 dpf, and these larvae also exhibited loss of pharyngeal teeth (Figure 1E).
Epithelial-mesenchymal interaction is also required for pectoral fin development. We observed that the esrp1/2 double mutants exhibit foreshortened and curled pectoral fins, where the sox10 labeled chondrocytes that populate the mesenchymal component and the krt4 labeled epithelial populations are both decreased in cell number in the esrp1−/−; esrp2−/− fins at 4 dpf (Figure 1F). Whereas the wildtype fins extend and fan out as they develop to 4 dpf, the fins in the esrp1−/−; esrp2−/− larvae curl proximally and are typically stuck to the torso through epithelial attachments (Figure 6A).
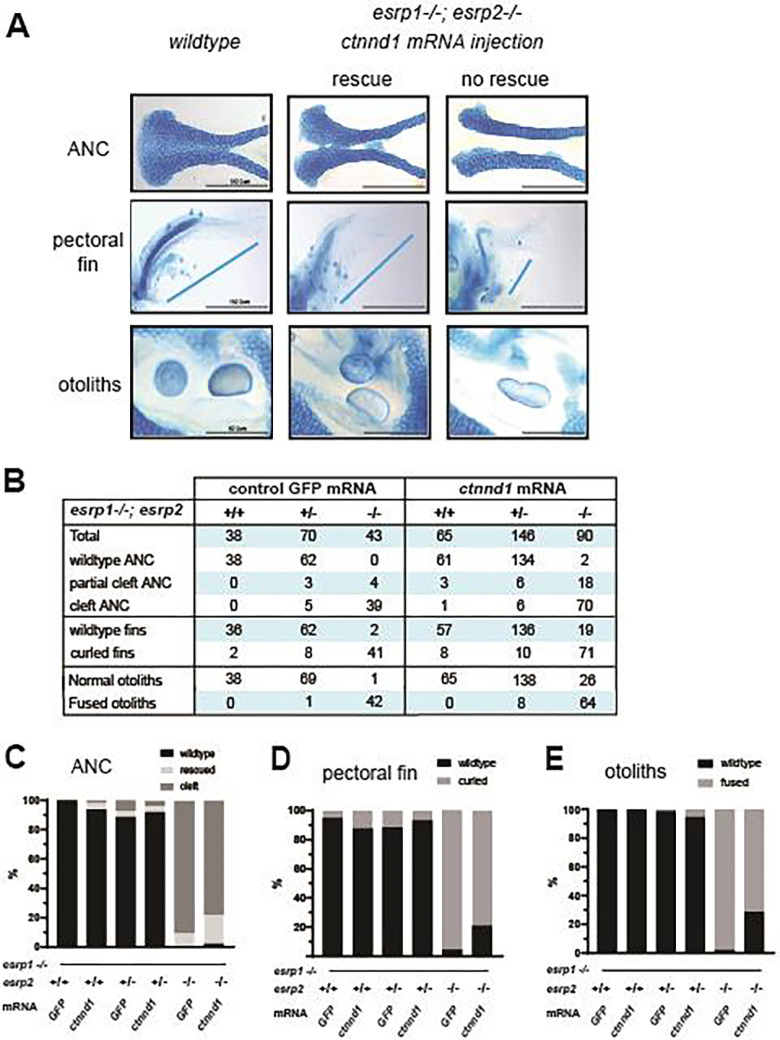
Figure 6. Over-expression of ctnnd1 rescues esrp1(−/−), esrp2(−/−) epithelial phenotypes.
(A) Image representing how wild type, intermediate and cleft ANC, pectoral fins and otoliths were sorted. (B) Representative table with the number of total fish injected and rescued by the ctnnd1 mRNA injection with GFP mRNA injection as control. Scoring of ANC phenotype (%) (C), fin phenotype (%) (D) and the otolith phenotype (%) (E) in the injected esrp1−/−; esrp2+/− inter-cross larvae confirmed by genotyping, showing 20–22% rescue of ANC, fin and otolith phenotypes in the esrp1−/−; esrp2−/− double homozygous larvae.
In vitro and in vivo assays to functionally test ESRP1 and ESRP2 human gene variants.
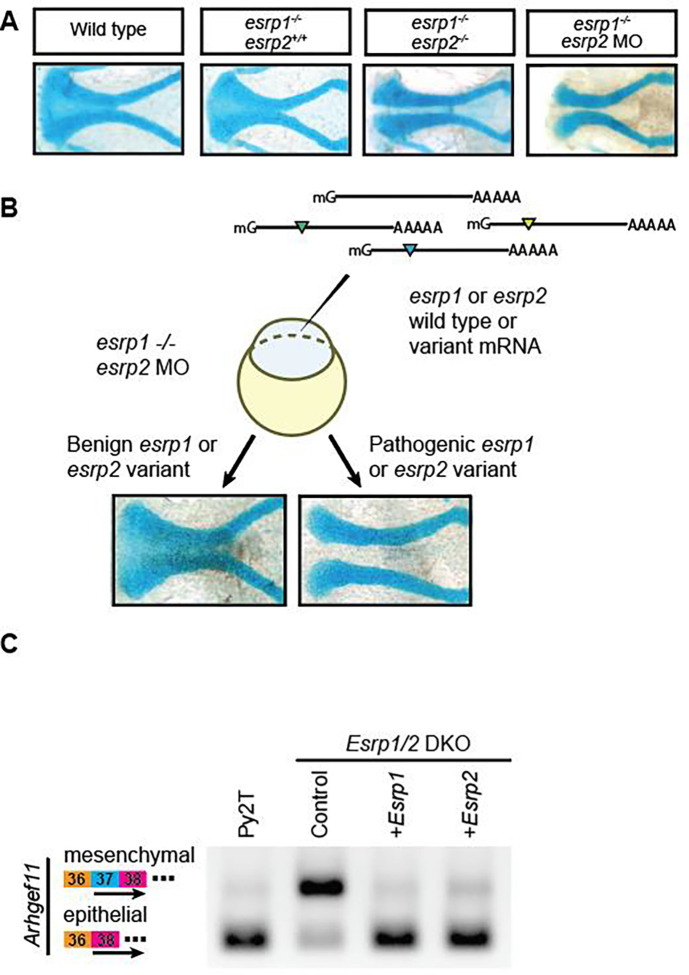
In a previous study we showed that esrp1−/−; esrp2+/− intercross yielded Mendelian ratio of 25% esrp1−/−; esrp2−/−, and that injection of morpholino against esrp2 in the esrp1−/− mutant embryos can consistently phenocopy the esrp1−/−; esrp2−/− double mutant (Figure 2A) (52). This esrp1−/−; esrp2 MO model provides significant advantages over esrp1+/−; esrp2+/− intercross, as the entire clutch of the esrp1−/− embryos injected with esrp2 MO consistently exhibited the cleft ANC phenotype greatly facilitating detection of rescue of injected ESRP1/2 mRNA to be tested.
Figure 2. Complementary in vivo and in vitro functional assays to test human ESRP1 and ESRP2 gene variants.
(A) Microdissected ANC of Alcian-blue stained embryos at 4 dpf for wild-type, esrp1−/−; esrp2+/+, esrp1−/−; esrp2−/−, and esrp1−/−; esrp2 MO embryos. (B) Schematic for the esrp morphant variant assay in zebrafish. Variants that robustly rescued the cleft ANC phenotype were scored as benign, while variants that failed to rescue the cleft ANC phenotype were scored as pathogenic. (C) RT-PCR was performed using primers spanning exons 36–38 of Arhgef11 on cDNA isolated from wild-type mouse Py2T cells, Esrp1/2 double-knockout Py2T cells, or Esrp1/2 double-knockout PyY2T cells electroporated with plasmids encoding for either Esrp1 or Esrp2 genes. Arrow markers point to the epithelial (short) isoform and mesenchymal (long) isoform retaining exon 37.
We found that over-expression of wildtype zebrafish and human ESRP1 and ESRP2 mRNA rescued the cleft ANC phenotype in esrp1−/−; esrp2 MO embryos (Figure 2B) (52). Alcian blue staining of esrp1−/−; esrp2−/− zebrafish at 4 dpf revealed a cleft ANC phenotype where a population of chondrocytes in the medial ANC is absent. A similar phenotype is observed when translation-blocking anti-esrp2 morpholinos were injected into esrp1−/− embryos (Figure 2A).
To functionally test human ESRP1 or ESRP2 gene variants, we introduced point mutations into zebrafish esrp1 or esrp2 coding sequences and subsequently co-inject 8ng of anti-esrp2 MO with either: (1) capped esrp1 mRNA, (2) capped esrp2 mRNA mutagenized with synonymous mutations at the MO binding site, or (3) either esrp1 mRNA encoding for human ESRP1 gene variants of unknown significance, or (MO-resistant) esrp2 mRNA encoding for human ESRP2 gene variants of unknown significance. We hypothesized that benign variants that preserve protein function would robustly rescue the cleft ANC phenotype like native esrp1 or esrp2 mRNA. Conversely, pathogenic human ESRP1/2 gene variants with loss-of-function would fail to rescue the cleft ANC phenotype (Figure 2B). Human ESRP1 and ESPR2 gene variants were cloned by site directed mutagenesis, and synthesized mRNA was injected with esrp2 MO into one-cell stage esrp1−/− embryos. The esrp2 cDNA was engineered to prevent hybridization of the esrp2 MO to the synthesized mRNA.
In order to gain additional functional assessment of the gene variants, we developed an independent in vitro assay using Esrp1/2 mutant Py2T cells (66). The murine Py2T epithelial cell line was developed where Esrp1 and Esrp2 were ablated using CRISPR-mediated gene editing. The Esrp1/2−/− Py2T cells exhibited splicing deficiencies in the Esrp target gene, Arhgef11 (Figure 2C) (66). RT-PCR performed on wildtype Py2T cell cDNA using primers spanning splice junctions for Arhgef11 demonstrated the presence of two major isoforms. The difference between these two isoforms is the presence or absence of exon 37, which is included in mesenchymal cells, but skipped in Py2T epithelial cells (23, 67, 68). Py2T cells carrying Esrp1 and Esrp2 loss-of-function alleles preferentially expressed the longer mesenchymal isoform of Arhgef11.
We found that over-expression of Esrp1 or Esrp2 in the Esrp1/2 DKO Py2T cells efficiently rescued RNA-splicing to generate the epithelial isoform of Arhgef11 transcript (Figure 2C).
Identifying human ESRP1 and ESRP2 gene variants
Genome sequencing efforts have deposited numerous gene variants in publicly available repositories, including the Gabriella Miller Kids First (GMKF) Pediatric Research Program and ClinVar (69–71). We filtered sequencing data from the both repositories for patients with OFC or autosomal recessive deafness (20, 58) and identified gene variants for either ESRP1 or ESRP2 to generate a list of 32 potentially disease-associated gene variants.
Because we are utilizing in vivo assay in zebrafish and in vitro assay in murine Py2T cells, we prioritized those human ESRP1 and ESRP2 gene variants residing in cross-vertebrate conserved residues. For ESRP1, the overall amino acid sequence identity was 97% and 64.68% between humans and mice, or humans and zebrafish, respectively. However, when focusing on the RNA-recognition motif (RRM) domains of ESRP1, the similarity of the sequences between humans and mice and humans and zebrafish increased to 98.82% and 94.12% for RRM1, 99.08% and 79.82% for RRM2, and 95.06% and 77.78% for RRM3. Similarly, for ESRP2, the overall amino acid sequence similarity was 98.67% between humans and mice and 85.33% between humans and zebrafish. The domain-specific amino acid sequence similarities were 98.67% and 85.33% for RRM1, 98.13% and 81.31% for RRM2, and 96.3% and 77.78% for RRM3 between humans and mice, and humans and zebrafish, respectively. Altogether, we identified 19 out of the 32 gene variants in residues fully conserved between human, mouse, and zebrafish. Gene variants were evenly spread throughout both proteins and included two variants in the RRM1 domain of ESRP1 and two variants each in the RRM1, RRM2, and RRM3 domains of ESRP2 (Figure 1 Supplementary Material).
We found that the in silico predictions from SIFT and Polyphen-2 followed one of four patterns: (1) concordant predictions from both tools annotating the variant as benign, (2) concordant predictions from both tools annotating the variant as damaging, (3) discordant predictions from both tools, (4) tools unable to predict the effect of the variant on protein function (Table 1). Altogether, two variants from ESRP1 (E194A and N643S) and two variants from ESRP2 (C372S and T475T) were predicted by both SIFT and PolyPhen-2 to have a benign effect on protein function. One variant from ESRP1 (Q90R) and four from ESRP2 (R250Q, R315H, R353Q, and R667C) were predicted by both to have a deleterious effect on protein function. SIFT and PolyPhen-2 do not offer predictions for three truncation variants (ESRP1 D222fs, ESRP2 R520*, and ESRP2 E547del). However, the remaining three ESRP1 variants (L259V, K287R and Y605F) and four ESRP2 variants (L92Q, S508L, R437H, and L665W) had discordant predictions between both algorithms. Thus, in silico predictions were not adequate to annotate roughly half of the selected gene variants and required an alternate approach to predict their effects on protein function.
Table 1:
ESRP1 and ESRP2 variants classification
| ESRP1 | Protein Domain | PolyPhen-2 | SIFT | Alpha Missense | Zf in vivo assay | Py2T in vitro assay | Interpretation |
|---|---|---|---|---|---|---|---|
| Q90R | Damaging | Damaging, LC | Benign | Rescue | n/a | Benign | |
| E194A | Benign | Benign | Benign | Rescue | n/a | Benign | |
| D222fs | n/a | n/a | n/a | Mutant | Deficient | Damaging | |
| L259V | RRM1 | Damaging | Benign | Pathogenic | Rescue | Restored | Benign |
| K287R | RRM1 | Damaging | Benign | Benign | Rescue | n/a | Benign |
| Y605F | Benign | Damaging, LC | Benign | Rescue | n/a | Benign | |
| N643S | Benign | Benign | Benign | Rescue | Restored | Benign | |
| ESRP2 | Protein Domain | PolyPhen-2 | SIFT | Alpha Missense | Zf in vivo assay | Py2T in vitro assay | Interpretation |
| L92Q | Benign | Damaging, LC | Likely pathogenic | Rescue | n/a | Benign | |
| R250Q | Damaging | Damaging | Likely pathogenic | Rescue | Restored | Benign | |
| R315H | RRM1 | Damaging | Damaging | Likely pathogenic | Mutant | Deficient | Damaging |
| R353Q | RRM1 | Damaging | Damaging | Likely pathogenic | Rescue | Restored | Benign |
| C372S | RRM2 | Benign | Benign | Ambiguous | Rescue | n/a | Benign |
| R437H | RRM2 | Damaging | Benign | Ambiguous | Rescue | n/a | Benign |
| T475T | RRM3 | Benign | Benign | n/a | Rescue | Restored | Benign |
| S508L | RRM3 | Damaging | Benign | Likely pathogenic | Rescue | Restored | Benign |
| R520STOP | RRM3 | n/a | n/a | n/a | Mutant | Deficient | Damaging |
| E547del | RRM3 | n/a | n/a | n/a | Rescue | n/a | Benign |
| L665W | Benign | Damaging, LC | Likely benign | Rescue | n/a | Benign | |
| R667C | Damaging | Damaging, LC | Ambiguous | Rescue | Restored | Benign |
Functional testing of ESRP1 and ESRP2 variants in zebrafish and murine Py2T cell assays
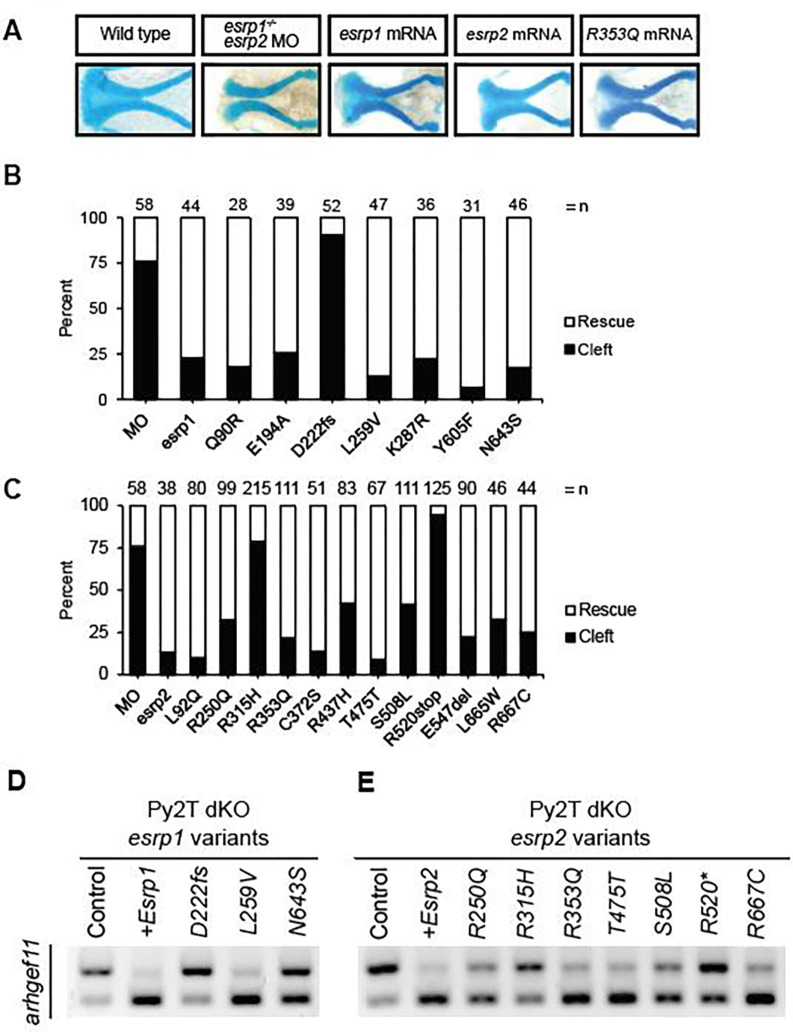
The selected 19 ESRP1 and ESRP2 gene variants were experimentally tested in zebrafish and Py2T cell assays. Site-directed mutagenesis was carried out in ESRP1 and ESRP2 cDNA sequences and cloned into the pCS2+8 vector backbone to generate capped mRNA for microinjection into zebrafish embryos. The zebrafish assay was optimized by microinjection of esrp2 translation-blocking morpholinos into esrp1−/− intercross, because the esrp2−/− females are infertile (22). However, since the esrp2 MO would also neutralize exogenous injected ESRP2 mRNA upon co-injection into zebrafish embryos, synonymous mutations were introduced in the translational start site of the pCS2+8-Esrp2 plasmid, to generate esrp2 MO-resistant ESRP2 mRNA transcripts. Co-injection of 8ng of esrp2 MO with 200pg of either ESRP1 mRNA or MO-resistant ESRP2 mRNA fully rescued the ANC phenotype in over 75% of 19 injected clutches at 4 dpf (Figure 3A).
Figure 3. Functional testing of human ESRP1 and ESRP2 gene variants.
(A) Representative images of the ANC from Alcian-blue stained larvae at 4 dpf after injection with esrp2 MO and 200pg of: esrp1 mRNA, esrp2 R353Q mRNA. ANC was scored as a rescued ANC or cleft ANC (B) ESRP1 and (C) ESRP2 gene variant rescue assay results for embryos injected with esrp2 MO and 200pg of esrp1 variant mRNA. Results presented as percentage of rescue vs. cleft as different numbers of embryos survived and were analyzed, indicated as n above each bar. (D) ESRP1 and (E) ESRP2 gene variant rescue assay by detecting alternative splicing of Arhgef11 in murine Py2T wildtype and Esrp1−/−; Esrp2−/− double knockout cells.
To test for the ability of human ESRP1/2 gene variants to rescue the cleft ANC phenotype in zebrafish, each of the 19 ESRP1 or ESRP2 gene variants was co-injected with esrp2-MO into esrp1−/− zebrafish embryos. At 4 dpf, the injected fish were fixed, stained with Alcian Blue, and analyzed. We found that for ESRP1, all 6 missense variants rescued the ANC phenotype. Only one variant, a frameshift mutation at the 222 aspartate residue (D222fs), had a large proportion of cleft ANC in the injected clutch compared to embryos injected with wildtype esrp1 mRNA, and was scored as a pathogenic variant (Figure 3B). For ESRP2, 10 out of 12 tested gene variants rescued the ANC phenotype, in a ratio like the esrp2 mRNA control and were scored as benign variants. The silent mutation T475T, served as an internal negative control and also scored as benign. The remaining 2 ESRP2 gene variants (R315H and R520*) failed to rescue the ANC phenotype and were scored as pathogenic (Figure 3C).
To independently assess the gene variant functional testing results obtained from the zebrafish model, we tested 3 ESRP1 and 8 ESRP2 human gene variants using the mouse Py2T cell assay, with epithelial-specific RNA splicing of Arhget11 as the readout (Figure 3D, 3E). We aimed to obtain an additional functional assessment for those gene variants testing results that contradicted in silico prediction. We performed site-directed mutagenesis to introduce the 11 gene variants, that were electroporated into Esrp1/2 DKO PY2T cells and performed the RT-PCR assay 24 hours post-electroporation. We found that for ESRP1, gene variant L259V restored Arhgef11 restriction to the epithelial isoform was scored as damaging for Polyphen-2 and Alpha missense and benign for SIFT (Figure 3, Table 1). The frameshift variant, D222fs, that was pathogenic in the in vivo assay was also pathogenic in this assay as it was unable to restore the epithelial isoform (Figure 3D, Table 1). Interestingly, the ESRP1 gene variant N643S partially restored some of the splicing function of Esrp1, where both epithelial and mesenchymal Arhgef11 isoforms were detected in a 1:1 ratio (Figure 3D). However, the same variant, N643S, in zebrafish rescued the phenotype. Statistical analysis for the Py2T rescue assay, can be found at Supplementary Figure 2. These results suggest that ESRP1 N643S variant may be hypomorphic, or that Arhgef11 is just one readout of Esrp1 mRNA splicing activity. Because Esrp1 shows position-dependent repression of exon splicing of Arhgef11, it is possible that some domains or regions may be required, or not, for some specific functions. It is possible that some splicing events may be differentially affected by mutations and there are other suggested functions of Esrp1 in mRNA stabilization or post-transcriptional regulation that are accounted for in the zebrafish rescue assay (60).
For ESRP2, variants R250Q, R353Q and R667C rescued the molecular splicing of Arhgef11 in the Py2T assay, (Figure 3E, Table 1). However, ESRP2 gene variants R315H, S508L, and R520* failed to rescue deficient Arhgef11 splicing in the Py2T assay and were scored as pathogenic, corroborating the pathogenic scoring from the zebrafish ANC rescue assay (Figure 3E, Table 1).
Overall, we found that the in vivo zebrafish ANC rescue assay and the in vitro Py2T splicing assays were largely concordant to determine pathogenicity of the ESRP1 and ESRP2 gene variants tested. PolyPhen-2 correctly predicted the effect of 8/18 (44.4%) tested gene variants, while SIFT correctly predicted the effect of 7/18 (38.8%) gene variants. When the predictions of both algorithms were concordant, they correctly predicted the consequence of 5 out of 7 (71.4%) gene variants on protein function (Table 1). The performance of concordant predictions was better for annotating benign variants where the algorithms correctly identified all four concordant benign variants with benign effects in both of our assays. Strikingly, the computational agreement incorrectly annotated 2 of 4 (50%) gene variants as pathogenic that had benign effects in both rescue assays. Ultimately, the algorithmic predictions were unable to determine half of the identified gene variants and greatly overestimated the prevalence of pathogenic variants (Table 1).
AlphaMissense over-interpreted pathogenic variants
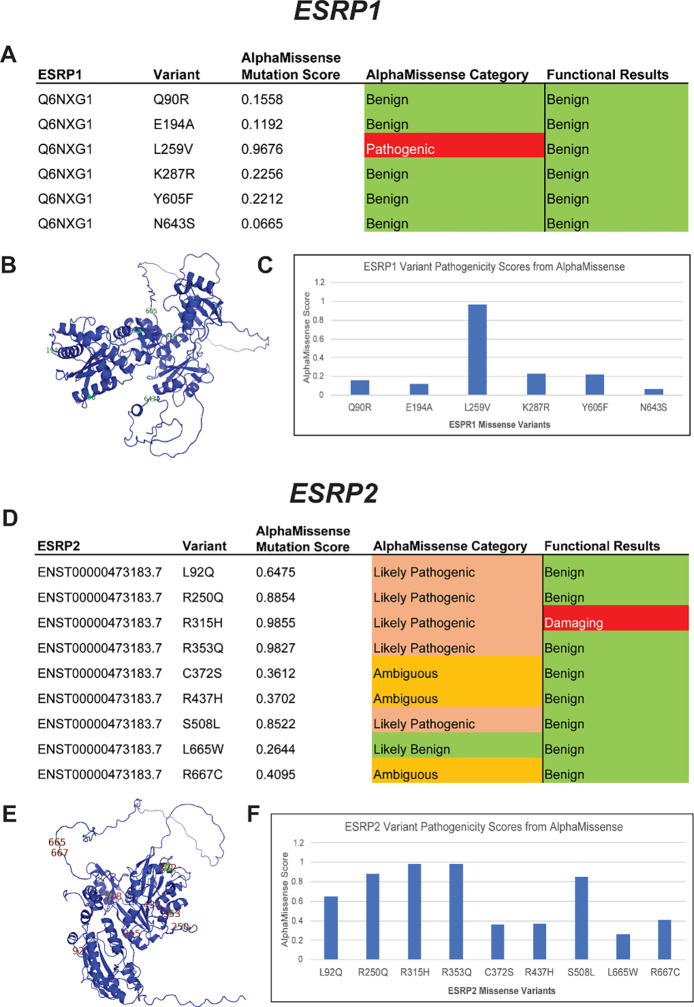
Recently a new gene variant analysis tool AlphaMissense was released and purported to improve variant calling accuracy by leveraging protein structure information predicted by machine learning algorithm AlphaFold (72). Using AlphaMissense to analyze the 6 ESRP1 and 9 ESRP2 missense variants we had functionally tested, we observed that AlphaMissense classified 5 variants as benign for ESRP1 (Q90R, E194A, K287R, Y605F, N643S) consistent with the functional tests, but called L259V as pathogenic when both the in vivo and in vitro functional tests demonstrated protein function (Figure 4).
Figure 4. AlphaMissense pathogenicity predictions for ESRP1 and ESRP2 missense variants.
ESRP1 and ESRP2 gene variants from OFC cases in the GMFK Children’s dataset and ClinVar variants associated with cleft lip and/or palate or autosomal recessive deafness were identified. 6 ESRP1 and 9 ESRP2 (A and D) missense variants were analyzed using the AlphaMissense (AM) model. The tables (C and F) show the AM-predicted pathogenicity compared to our functional test results and the AM mutation score, which is also graphed. On the left, the ESRP1 and ESRP2 AlphaFold structures (B and E), with labeled missense mutations, color-coded with the functional results.
For ESRP2, AlphaMissense and the experimental validation were only concordant on 2 variants out of 9, calling R315H as pathogenic and L665W as benign (Figure 4). AlphaMissense called 6 variants as pathogenic when they were shown to be functionally benign in both in vitro and in vivo functional tests. Therefore, our results showed that AlphaMissense may over-interpret variants as pathogenic for some genes.
Alternative splicing to generate epithelial isoform of Ctnnd1 requires Esrp1/2 function
We and others demonstrated that Esrp1 and Esrp2 regulate the alternative splicing of Ctnnd1, generating isoforms that differ between epithelial and mesenchymal cell types (20, 60, 66, 73), making Ctnnd1 an interesting Esrp1/2 target that has also been implicated in CL/P (61).
CTNND1 (p120-catenin) have been associated with Blepharocheilodontic (BCD) syndrome and non-syndromic human CL/P (19, 20, 74). Like other catenins, Ctnnd1 has dual roles: it functions as part of the adherens junction cellular scaffolding to stabilize cell adhesion molecules, as well as a transcriptional regulator (19, 75–79). Furthermore, functional differences between epithelial and mesenchymal forms of Ctnnd1 have been described (80–82). Four major isoforms for Ctnnd1 have been characterized in humans. The full-length isoform, isoform 1, has a translational start site at the first methionine in the sequence (1 Met), while isoforms 2, 3, and 4 undergo splicing events that cause a 5’ truncation of the transcript and change the translational start site to methionines 55, 102, and 324, respectively. Isoform 1 of CTNND1 is predominantly expressed in the mesenchyme, while the shorter isoform 3 is restricted to the epithelium. The remaining isoforms, 2 and 4, are less abundant and have not been thoroughly characterized (74).
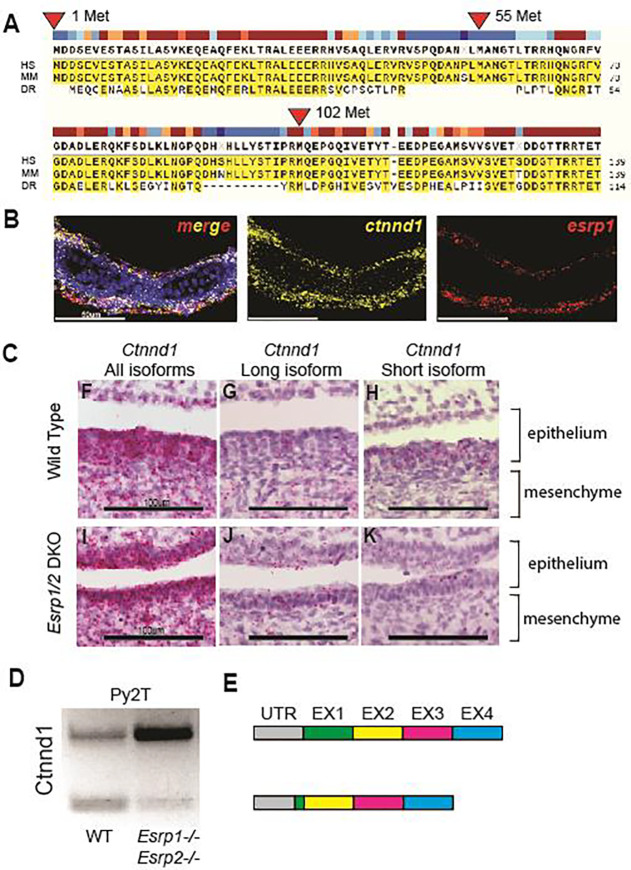
When we aligned the amino acid sequences between human, mouse, and zebrafish Ctnnd1 homologs, we found that methionine in positions 1 and 102 are conserved in all three species. Methionine 55 is part of a 14 aa stretch absent in zebrafish (Figure 5A). Given that transcripts for the long (mesenchymal) isoform shifts to the shorter (epithelial) isoform by splicing out a 5’ exon(s) and moving down to a conserved methionine, splicing pattens are well-conserved across human, mouse, and zebrafish. Cox et. al reported that ESRP2 and a short form of the full-length CTNND1 protein, identified by an antibody to the C-terminus, are colocalized in the periderm of human embryos (20). Meanwhile, RNA splicing of Ctnnd1 transcripts is deficient in the embryonic epithelium of Esrp1−/− mice (53).
Figure 5. Alternative splicing of Ctnnd1 is regulated by Esrp1/2.
(A) Amino acid sequence alignment of the first 140 residues of CTNND1 protein across human, mouse, and zebrafish. Translation for isoform 1 of CTNND1 begins at methionine 1, while isoform 3 encodes a truncated form that starts translation at methionine 102. Methionine residues at positions 55 and 324 are not conserved across all three species. (B) Detection of esrp1 and ctnnd1 gene expression in zebrafish at 4dpf, demonstrates shared localization of transcripts in the embryonic epithelium. This coronal section includes the ventral Meckel’s cartilage. (C) Detection of murine Ctnnd1 mRNA using isoform-specific base-scope probes in the oral epithelium and tongue mesenchyme. The wildtype sections show that the Ctnnd1 long isoform is present in both epithelial and mesenchymal cells. The Ctnnd1 short isoform is present preferentially in epithelial cells and not in the mesenchymal cells. In the Esrp1/2 DKO mouse, the mesenchymal Ctnnd1 long isoform is detected in epithelial and mesenchymal cells, with loss of the Ctnnd1 short isoform. (D) RT-PCR of the Ctnnd1 long and short isoforms from Py2T cells. (E) Diagrammatic representation of the ESRP-regulated CTNND1 alternative splicing to generate the shorter epithelial isoform.
We confirmed that long and shorter Ctnnd1 isoforms were found in the mouse Py2T cells by performing RT-PCR using primers spanning exon 2, which is partially skipped in the shorter isoform for Ctnnd1. In the Esrp1/2−/− Py2T cell line, the splicing pattern of Ctnnd1 shifts and is biased towards the longer mesenchymal isoform, confirming previous observations (60).
To localize Ctnnd1 and Esrp1/2 gene expression in wildtype mouse and zebrafish, we carried out RNAscope and BaseScope on wildtype and mutant mouse and zebrafish sections (Figure 5B 5C). The Ctnnd1 probe used identifies shared C-terminal exons shared in all Ctnnd1 isoforms. Only Esrp1 probe was used here as we and others have previously shown that Esrp1 and Esrp2 gene expression are co-localized in mouse and zebrafish (21, 22, 52, 66). In zebrafish, ctnnd1 and esrp1 RNAscope signals are co-localized robustly throughout the oral epithelium with sparse signals in the mesenchyme.
To assess the tissue specific distribution of the longer mesenchymal isoforms of Ctnnd1 vs. shorter epithelial isoform, BaseScope probes were used to detect the two Ctnnd1 isoforms from wildtype and Esrp1−/−; Esrp2−/− mutant mouse at E15. Similar to RNAScope result in zebrafish (Figure 5B), the murine Ctnnd1 BaseScope signals for both mesenchymal and epithelial isoforms were robust in the oral epithelium and sparsely scattered in the mesenchyme (Figure 5D and 5G). When signal is differentiated by isoform, the longer Ctnnd1 mesenchymal isoform was uniformly distributed throughout the epithelium and mesenchyme (5E and 5H). However, the shorter Ctnnd1 epithelial isoform was restricted to the epithelial cells and excluded from the muscle (5F and 5I). In the wildtype, BaseScope signals of the longer Ctnnd1 mesenchymal isoform appeared equally distributed in the mesenchyme and epithelium, and the signals of the shorter isoform was epithelial restricted. In the Esrp1−/−; Esrp2−/− mutant mouse, Ctnnd1 transcript level was significantly reduced and predominantly the longer Ctnnd1 mesenchymal isoform was detected, in both the mesenchyme and epithelium. The shorter Ctnnd1 epithelial isoform was sparsely detected via BaseScope in the Esrp1−/−; Esrp2−/− mutant, consistent with the finding where shorter isoform was significantly reduced in the Esrp1−/−; Esrp2−/− Py2T cells by qPCR. These results corroborate that Esrp1/2 is required for RNA splicing of Ctnnd1, generating the shorter isoform specifically in the epithelium but not the mesenchyme.
CTNND1 gene variants from OFC cohorts
Twenty-four CTNND1 gene variants have been reported and a growing number of new variants have been found in ongoing WGS studies of OFC cohorts (19, 26). In a recent analysis of 759 OFC trios, we identified 15 variants in CTNND1 with allele frequencies less than 0.1% in gnomAD (Figure 3 Supplementary Material). Two variants were de novo and one was inherited from an affected parent. Pathogenic variants in CTNND1 accounted for 0.8% of the cohort. Only 10% of the cohort had a pathogenic variant in 500 genes implicated in OFC that we analyzed, making CTNND1 the mostly frequently mutated variant in this cohort (61). In the gene-based burden test, rare variants were nominally over-transmitted to affected children (p=0.06); de novo variants are enriched in CTNND1 (p=0.005 for loss-of-function de novo variants; 0.001 for protein-altering de novo variants). Nearly all the missense variants were classified as variants of unknown significance, indicating that functional testing is critical. In fact, we estimate that CTNND1 mutations account for at least 1.5% of CL/P cases. By comparison, IRF6 mutations are estimated to be the most common cause of CL/P, accounting for 2% of cases. Taken together, CTNND1 stands to be as important as IRF6 in contributing to the genetic risk of syndromic and non-syndromic CL/P.
Ctnnd1 over-expression rescue esrp 1−/−; esrp 2−/− cleft ANC, curled fin and fused otolith phenotypes
To functionally assess the relationship between Esrp and Ctnnd1, we injected the zebrafish ctnnd1 isoform-201 (ENSDART00000106048.4) mRNA into esrp1−/−; esrp2+/− offspring at the 1-cell stage. Mutants and control embryos were analyzed at 4 dpf, assessing the ANC, the pectoral fin, and otoliths phenotypes, followed by genotyping (figure 6A).
Control gfp mRNA injected esrp1−/−; esrp2−/− larvae, exhibited cleft ANC, the pectoral fins were hypoplastic and stuck to the thorax, and fused otoliths, the mutant phenotypes were fully penetrant and reliably scored (Figure 6B–E). In the ctnnd1 mRNA injected esrp1−/−; esrp2−/− larvae, 22% (n = 20 of 90, p<0.01) demonstrated a full or partial rescue of the ANC (Figure 6B–E). Correspondingly, the injected esrp1−/−; esrp2−/− larvae exhibited significant rescue of the abrogated fin phenotype, with 21% (n = 19 of 90, p<0.01) exhibiting extension of the pectoral fin and angling away from the thorax. The fused otolith phenotype was scored as either separate or fused, and demonstrated 26% (n = 26 of 90, p<0.01) rescue (Figure 6A). The morphogenesis of the ANC, pectoral fin and the otoliths all reflect different aspects of embryonic epithelium development and interaction with the associated mesenchyme of the esrp 1−/−; esrp 2−/− embryos. The ctnnd1 mRNA over-expression rescuing the epithelial defects in the esrp 1−/−; esrp 2−/− suggests that a key function of esrp1/2 in epithelial biology is to regulate ctnnd1 function.
Discussion
Several independent lines of evidence corroborate that the ESRP1 and ESRP2 genes are important OFC loci in humans. ESRP1 was proposed to be the most likely candidate CL/P risk gene in the 8q22.1 locus (83, 84). Ectopic expression of p63 converted human fibroblasts to keratinocyte-like cells and ESRP1 was transcriptionally induced together with activation of an epithelial enhancer within a topologically associated domains (TADs) containing a non-syndromic CL/P risk locus (85). This is consistent with the biological observation and p63, Irf6 and Esrp1/2 co-localize in the embryonic epithelium, and that mutations of these 3 genes result in OFC phenotypes. Further, a whole exome sequencing study of non-syndromic CL/P in multi-affected families identified pathogenic variants in ESRP2 with an autosomal dominant inheritance pattern (20).
Several studies showed in mouse and zebrafish models that Esrp1 and Esrp2 are important in craniofacial development. We showed that Esrp1 and Esrp2 are co-localized with Irf6 in the embryonic oral epithelium, and when Ersp1/2 are disrupted, cleft of the lip and palate formed, validating that mouse and zebrafish are robust animal models of human OFC (21, 52, 53).
There is growing recognition that RNA binding proteins that regulate alternative splicing play vital roles in craniofacial morphogenesis. Clinically, spliceosomopathies are often associated with syndromic craniofacial abnormalities due to disruption of splicing factors such as PUF60, ETUD2, SF3B4, RBM10, and ESRP2 (86). Animal models defective in RNA splicing that exhibit craniofacial phenotypes include: Esrp1/2, Rbfox2, Srsf3, and Sf3b2 (21, 22, 52, 87, 88). The ESRP proteins are uniquely expressed in epithelial structures and direct post-transcriptional modifications that distinguish protein isoforms between epithelium and mesenchyme. We applied complementary phenotypic and molecular assays to interrogate the functional consequence of identified ESRP1/2 gene variants in cohorts of autosomal recessive deafness and CL/P.
As the magnitude of available WGS data increases, the need for assigning clinically actionable information continues to grow. The sequence variant interpretation (SVI) working group from ACMG-AMP frequently reconvenes to update, revise, and refine the ACMG criteria to provide the clearest guidance possible (33, 34). Most recently, the working group provided further guidance regarding functional assays and experimental model systems. Among these, they highlighted the need to ascertain the gene variants’ physiologic context and molecular consequence. Here, we applied complementary phenotypic assays in the zebrafish ANC rescue, in addition to the Py2T splicing assay, to assess the physiologic and molecular consequences of ESRP1/2 gene variants observed in clinical cohorts. These functional tests identified 7 pathogenic variants out of 18 ESRP1/2 variants examined. Moreover, these functional readouts of orthologous systems across species attest to the strongly conserved nature of epithelial splicing by the ESRPs in craniofacial morphogenesis. These results highlight the need for experimental models to enhance the validity of in silico predictions of protein function. We found that while the SIFT and PolyPhen-2 algorithms have a positive predictive value when they align in predicting benign variants, they tend to overestimate the prevalence of pathogenic variants.
While AlphaMissense provided slightly better predictions for ESRP1 than SIFT and PolyPhen-2, in the case of ESRP2, AlphaMissense over-interpreted benign variants as pathogenic. A similar high false positive rate was seen in a different disease, cystic fibrosis transmembrane conductance regulator (89), and for epithelial master regulator IRF6 (90). This work highlights that protein structure and machine learning approaches today are still insufficient to accurately predict pathogenicity, where functional tests are indispensable to validate the pathogenicity of variants.
These functional assays revealed novel insights into ESRP1/2 protein function and downstream targets spliced by the ESRPs. We found that the gene variants with the largest effect size for the zebrafish ANC rescue assay lie in RRM1 and RRM3 of ESRP2. Variants R250Q and R353Q were predicted by PolyPhen-2, SIFT and AlphaMissense to be damaging or likely pathogenic, but in both independent functional tests corroborated to be benign variants. In contrast, R315H was functionally tested by both assays to be a deleterious variant, consistent with prior work demonstrating R315 to impact RNA binding based on protein structure analysis (91). Furthermore, we provide molecular evidence that Esrp transcripts rescue molecular splicing patterns of putative Esrp-target genes Arhgef11 and Ctnnd1. Moreover, gene variants with pathogenic potential do not restore splicing patterns of Arhgef11, providing evidence that the gene variants impair Esrp function and likely contribute to disease pathogenicity. These functional assays provide key data to satisfy the ACMG-AMP standards, where molecular assays are used to contribute to our understanding of mechanisms for disease.
Mutations in CTNND1 and CDH1 (E-cadherin) are the known cause of BCD, which includes abnormal eyelids, upper lip, palate, and teeth development (20, 74, 92). The precise pathological mechanism remains to be elucidated, but in healthy epithelial cells CTNND1 binds to E-cadherin to stabilize adherens junctions and desmosomes, and therefore displacement of CTNND1 causes endocytosis of CDH1 and loss of the junction. Another possibility is disruption of the canonical WNT pathway signaling, as CTNND1 is known to modulate transcription by binding to transcription factors such as Kaiso in the Wnt pathway (93, 94). It is known, and further supported by the evidence in this work, that alternatively spliced isoforms of CTNND1 are differentially expressed in the epithelium and mesenchyme, and here we show that those distinct splicing patterns are dependent on Esrp1/2 activity. However, it is not known how the alternatively spliced isoforms differ in function, alter embryonic and craniofacial morphogenesis, or contribute to disease. Thus, further studies into the functional differences between CTNND1 isoforms are warranted and would provide insight into the disease etiology of BCD or the mechanism of the cleft palate from ESRP loss-of-function.
Supplementary Material
Acknowledgements
We thank the Aquatics Core at Massachusetts General Hospital and Children’s Hospital of Philadelphia (CHOP) for their dedication to fish health and maintenance of our colonies. We thank the CHOP IDDRC Biostatistics and Data Science core (HD105354) for consultation. We are grateful for the funding support from the National Institutes of Health (HG013031) to K.W. and (DE032332, DE027983) to E.C.L., research and fellowship grants from Shriners Hospitals for Children and institutional support from Children’s Hospital of Philadelphia.
References
- 1.Bishop MR, Diaz Perez KK, Sun M, Ho S, Chopra P, Mukhopadhyay N, et al. Genome-wide Enrichment of De Novo Coding Mutations in Orofacial Cleft Trios. Am J Hum Genet. 2020;107(1):124–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bureau A, Parker MM, Ruczinski I, Taub MA, Marazita ML, Murray JC, et al. Whole exome sequencing of distant relatives in multiplex families implicates rare variants in candidate genes for oral clefts. Genetics. 2014;197(3):1039–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlson JC, Taub MA, Feingold E, Beaty TH, Murray JC, Marazita ML, et al. Identifying Genetic Sources of Phenotypic Heterogeneity in Orofacial Clefts by Targeted Sequencing. Birth Defects Res. 2017;109(13):1030–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leslie EJ. Genetic models and approaches to study orofacial clefts. Oral Dis. 2021. [DOI] [PubMed] [Google Scholar]
- 5.Mukhopadhyay N, Feingold E, Moreno-Uribe L, Wehby G, Valencia-Ramirez LC, Muneton CPR, et al. Genome-Wide Association Study of Non-syndromic Orofacial Clefts in a Multiethnic Sample of Families and Controls Identifies Novel Regions. Front Cell Dev Biol. 2021;9:621482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beaty TH, Marazita ML, Leslie EJ. Genetic factors influencing risk to orofacial clefts: today’s challenges and tomorrow’s opportunities. F1000Res. 2016;5:2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011;12(3):167–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welzenbach J, Hammond NL, Nikolic M, Thieme F, Ishorst N, Leslie EJ, et al. Integrative approaches generate insights into the architecture of non-syndromic cleft lip with or without cleft palate. HGG Adv. 2021;2(3):100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson JC, Anand D, Butali A, Buxo CJ, Christensen K, Deleyiannis F, et al. A systematic genetic analysis and visualization of phenotypic heterogeneity among orofacial cleft GWAS signals. Genet Epidemiol. 2019;43(6):704–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtis SW, Chang D, Lee MK, Shaffer JR, Indencleef K, Epstein MP, et al. The PAX1 locus at 20p11 is a potential genetic modifier for bilateral cleft lip. HGG Adv. 2021;2(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi M, Hosomichi K, Yamaguchi T, Nagahama R, Yoshida H, Maki K, et al. Whole-genome sequencing in a pair of monozygotic twins with discordant cleft lip and palate subtypes. Oral Dis. 2018;24(7):1303–9. [DOI] [PubMed] [Google Scholar]
- 12.Richardson R, Mitchell K, Hammond NL, Mollo MR, Kouwenhoven EN, Wyatt ND, et al. p63 exerts spatio-temporal control of palatal epithelial cell fate to prevent cleft palate. PLoS Genet. 2017;13(6):e1006828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferretti E, Li B, Zewdu R, Wells V, Hebert JM, Karner C, et al. A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Developmental cell. 2011;21(4):627–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, et al. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet. 2002;32(2):285–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingraham CR, Kinoshita A, Kondo S, Yang B, Sajan S, Trout KJ, et al. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6). Nat Genet. 2006;38(11):1335–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richardson RJ, Dixon J, Malhotra S, Hardman MJ, Knowles L, Boot-Handford RP, et al. Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nat Genet. 2006;38(11):1329–34. [DOI] [PubMed] [Google Scholar]
- 17.de la Garza G, Schleiffarth JR, Dunnwald M, Mankad A, Weirather JL, Bonde G, et al. Interferon regulatory factor 6 promotes differentiation of the periderm by activating expression of Grainyhead-like 3. J Invest Dermatol. 2013;133(1):68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peyrard-Janvid M, Leslie EJ, Kousa YA, Smith TL, Dunnwald M, Magnusson M, et al. Dominant mutations in GRHL3 cause Van der Woude Syndrome and disrupt oral periderm development. Am J Hum Genet. 2014;94(1):23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alharatani R, Ververi A, Beleza-Meireles A, Ji W, Mis E, Patterson QT, et al. Novel truncating mutations in CTNND1 cause a dominant craniofacial and cardiac syndrome. Hum Mol Genet. 2020;29(11):1900–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cox LL, Cox TC, Moreno Uribe LM, Zhu Y, Richter CT, Nidey N, et al. Mutations in the Epithelial Cadherin-p120-Catenin Complex Cause Mendelian Non-Syndromic Cleft Lip with or without Cleft Palate. Am J Hum Genet. 2018;102(6):1143–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bebee TW, Park JW, Sheridan KI, Warzecha CC, Cieply BW, Rohacek AM, et al. The splicing regulators Esrp1 and Esrp2 direct an epithelial splicing program essential for mammalian development. Elife. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burguera D, Marquez Y, Racioppi C, Permanyer J, Torres-Mendez A, Esposito R, et al. Evolutionary recruitment of flexible Esrp-dependent splicing programs into diverse embryonic morphogenetic processes. Nat Commun. 2017;8(1):1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S, Cieply B, Yang Y, Peart N, Glaser C, Chan P, et al. Esrp1-Regulated Splicing of Arhgef11 Isoforms Is Required for Epithelial Tight Junction Integrity. Cell Rep. 2018;25(9):2417–30 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y, Carstens RP. Alternative splicing regulates distinct subcellular localization of Epithelial splicing regulatory protein 1 (Esrp1) isoforms. Sci Rep. 2017;7(1):3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7(10):e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–81. [DOI] [PubMed] [Google Scholar]
- 28.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang H, Thomas PD. Tools for Predicting the Functional Impact of Nonsynonymous Genetic Variation. Genetics. 2016;203(2):635–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X, Li C, Mou C, Dong Y, Tu Y. dbNSFP v4: a comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 2020;12(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frazer J, Notin P, Dias M, Gomez A, Min JK, Brock K, et al. Disease variant prediction with deep generative models of evolutionary data. Nature. 2021;599(7883):91–5. [DOI] [PubMed] [Google Scholar]
- 32.Oliver JD, Turner EC, Halpern LR, Jia S, Schneider P, D’Souza RN. Molecular Diagnostics and In Utero Therapeutics for Orofacial Clefts. J Dent Res. 2020;99(11):1221–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bean LJH, Funke B, Carlston CM, Gannon JL, Kantarci S, Krock BL, et al. Diagnostic gene sequencing panels: from design to report-a technical standard of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2020;22(3):453–61. [DOI] [PubMed] [Google Scholar]
- 35.Jaravine V, Balmford J, Metzger P, Boerries M, Binder H, Boeker M. Annotation of Human Exome Gene Variants with Consensus Pathogenicity. Genes (Basel). 2020;11(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harnish JM, Deal SL, Chao HT, Wangler MF, Yamamoto S. In Vivo Functional Study of Disease-associated Rare Human Variants Using Drosophila. J Vis Exp. 2019(150). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Liu Z, Bellen HJ, Yamamoto S. Navigating MARRVEL, a Web-Based Tool that Integrates Human Genomics and Model Organism Genetics Information. J Vis Exp. 2019(150). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raud L, Ka C, Gourlaouen I, Callebaut I, Ferec C, Le Gac G, et al. Functional analysis of novel RHD variants: splicing disruption is likely to be a common mechanism of variant D phenotype. Transfusion. 2019;59(4):1367–75. [DOI] [PubMed] [Google Scholar]
- 39.Schaid DJ, Chen W, Larson NB. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat Rev Genet. 2018;19(8):491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li EB, Truong D, Hallett SA, Mukherjee K, Schutte BC, Liao EC. Rapid functional analysis of computationally complex rare human IRF6 gene variants using a novel zebrafish model. PLoS Genet. 2017;13(9):e1007009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim SS, Dey KK, Weissbrod O, Marquez-Luna C, Gazal S, Price AL. Improving the informativeness of Mendelian disease-derived pathogenicity scores for common disease. Nat Commun. 2020;11(1):6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Itan Y, Shang L, Boisson B, Ciancanelli MJ, Markle JG, Martinez-Barricarte R, et al. The mutation significance cutoff: gene-level thresholds for variant predictions. Nat Methods. 2016;13(2):109–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y, Yeung WSB, Chiu PCN, Cao D. Computational approaches for predicting variant impact: An overview from resources, principles to applications. Front Genet. 2022;13:981005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang M, Zhang J, Zhao H, Ievlev V, Zhong W, Huang W, et al. Functional Characterization of a Novel IRF6 Frameshift Mutation From a Van Der Woude Syndrome Family. Front Genet. 2020;11:562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Griesemer D, Xue JR, Reilly SK, Ulirsch JC, Kukreja K, Davis JR, et al. Genome-wide functional screen of 3’UTR variants uncovers causal variants for human disease and evolution. Cell. 2021;184(20):5247–60 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Findlay GM, Daza RM, Martin B, Zhang MD, Leith AP, Gasperini M, et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562(7726):217–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glazer AM, Wada Y, Li B, Muhammad A, Kalash OR, O’Neill MJ, et al. High-Throughput Reclassification of SCN5A Variants. Am J Hum Genet. 2020;107(1):111–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giacomelli AO, Yang X, Lintner RE, McFarland JM, Duby M, Kim J, et al. Mutational processes shape the landscape of TP53 mutations in human cancer. Nat Genet. 2018;50(10):1381–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mighell TL, Evans-Dutson S, O’Roak BJ. A Saturation Mutagenesis Approach to Understanding PTEN Lipid Phosphatase Activity and Genotype-Phenotype Relationships. Am J Hum Genet. 2018;102(5):943–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jia X, Burugula BB, Chen V, Lemons RM, Jayakody S, Maksutova M, et al. Massively parallel functional testing of MSH2 missense variants conferring Lynch syndrome risk. Am J Hum Genet. 2021;108(1):163–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33(5):591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carroll SH, Macias Trevino C, Li EB, Kawasaki K, Myers N, Hallett SA, et al. An Irf6-Esrp1/2 regulatory axis controls midface morphogenesis in vertebrates. Development. 2020;147(24). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee S, Sears MJ, Zhang Z, Li H, Salhab I, Krebs P, et al. Cleft lip and cleft palate in Esrp1 knockout mice is associated with alterations in epithelial-mesenchymal crosstalk. Development. 2020;147(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schilling TF, Kimmel CB. Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development. 1997;124(15):2945–60. [DOI] [PubMed] [Google Scholar]
- 55.Wada N, Javidan Y, Nelson S, Carney TJ, Kelsh RN, Schilling TF. Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development. 2005;132(17):3977–88. [DOI] [PubMed] [Google Scholar]
- 56.Dougherty M, Kamel G, Grimaldi M, Gfrerer L, Shubinets V, Ethier R, et al. Distinct requirements for wnt9a and irf6 in extension and integration mechanisms during zebrafish palate morphogenesis. Development. 2013;140(1):76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swartz ME, Sheehan-Rooney K, Dixon MJ, Eberhart JK. Examination of a palatogenic gene program in zebrafish. Dev Dyn. 2011;240(9):2204–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rohacek AM, Bebee TW, Tilton RK, Radens CM, McDermott-Roe C, Peart N, et al. ESRP1 Mutations Cause Hearing Loss due to Defects in Alternative Splicing that Disrupt Cochlear Development. Dev Cell. 2017;43(3):318–31 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Freytag M, Kluth M, Bady E, Hube-Magg C, Makrypidi-Fraune G, Heinzer H, et al. Epithelial splicing regulatory protein 1 and 2 (ESRP1 and ESRP2) upregulation predicts poor prognosis in prostate cancer. BMC Cancer. 2020;20(1):1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peart NJ, Hwang JY, Quesnel-Vallieres M, Sears MJ, Yang Y, Stoilov P, et al. The global Protein-RNA interaction map of ESRP1 defines a post-transcriptional program that is essential for epithelial cell function. iScience. 2022;25(10):105205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Diaz Perez KK, Curtis SW, Sanchis-Juan A, Zhao X, Head T, Ho S, et al. Rare variants found in clinical gene panels illuminate the genetic and allelic architecture of orofacial clefting. Genet Med. 2023;25(10):100918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sievers F, Higgins DG. The Clustal Omega Multiple Alignment Package. Methods Mol Biol. 2021;2231:3–16. [DOI] [PubMed] [Google Scholar]
- 63.Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46(9):944–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walker MB, Kimmel CB. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech Histochem. 2007;82(1):23–8. [DOI] [PubMed] [Google Scholar]
- 65.Panizzi JR, Jessen JR, Drummond IA, Solnica-Krezel L. New functions for a vertebrate Rho guanine nucleotide exchange factor in ciliated epithelia. Development. 2007;134(5):921–31. [DOI] [PubMed] [Google Scholar]
- 66.Warzecha CC, Shen S, Xing Y, Carstens RP. The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol. 2009;6(5):546–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Itoh M, Radisky DC, Hashiguchi M, Sugimoto H. The exon 38-containing ARHGEF11 splice isoform is differentially expressed and is required for migration and growth in invasive breast cancer cells. Oncotarget. 2017;8(54):92157–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 2011;7(8):e1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Landrum MJ, Chitipiralla S, Brown GR, Chen C, Gu B, Hart J, et al. ClinVar: improvements to accessing data. Nucleic Acids Res. 2020;48(D1):D835–D44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sayers EW, Beck J, Bolton EE, Bourexis D, Brister JR, Canese K, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2021;49(D1):D10–D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mukhopadhyay N, Bishop M, Mortillo M, Chopra P, Hetmanski JB, Taub MA, et al. Whole genome sequencing of orofacial cleft trios from the Gabriella Miller Kids First Pediatric Research Consortium identifies a new locus on chromosome 21. Hum Genet. 2020;139(2):215–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheng J, Novati G, Pan J, Bycroft C, Zemgulyte A, Applebaum T, et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science. 2023;381(6664):eadg7492. [DOI] [PubMed] [Google Scholar]
- 73.Faux MC, King LE, Kane SR, Love C, Sieber OM, Burgess AW. APC regulation of ESRP1 and p120-catenin isoforms in colorectal cancer cells. Mol Biol Cell. 2021;32(2):120–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kievit A, Tessadori F, Douben H, Jordens I, Maurice M, Hoogeboom J, et al. Variants in members of the cadherin-catenin complex, CDH1 and CTNND1, cause blepharocheilodontic syndrome. Eur J Hum Genet. 2018;26(2):210–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Davis MA, Ireton RC, Reynolds AB. A core function for p120-catenin in cadherin turnover. J Cell Biol. 2003;163(3):525–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fukumoto Y, Shintani Y, Reynolds AB, Johnson KR, Wheelock MJ. The regulatory or phosphorylation domain of p120 catenin controls E-cadherin dynamics at the plasma membrane. Exp Cell Res. 2008;314(1):52–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ireton RC, Davis MA, van Hengel J, Mariner DJ, Barnes K, Thoreson MA, et al. A novel role for p120 catenin in E-cadherin function. J Cell Biol. 2002;159(3):465–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ishiyama N, Lee SH, Liu S, Li GY, Smith MJ, Reichardt LF, et al. Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion. Cell. 2010;141(1):117–28. [DOI] [PubMed] [Google Scholar]
- 79.Reynolds AB, Daniel J, McCrea PD, Wheelock MJ, Wu J, Zhang Z. Identification of a new catenin: the tyrosine kinase substrate p120cas associates with E-cadherin complexes. Mol Cell Biol. 1994;14(12):8333–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reynolds AB, Daniel JM, Mo YY, Wu J, Zhang Z. The novel catenin p120cas binds classical cadherins and induces an unusual morphological phenotype in NIH3T3 fibroblasts. Exp Cell Res. 1996;225(2):328–37. [DOI] [PubMed] [Google Scholar]
- 81.Schackmann RC, Tenhagen M, van de Ven RA, Derksen PW. p120-catenin in cancer - mechanisms, models and opportunities for intervention. J Cell Sci. 2013;126(Pt 16):3515–25. [DOI] [PubMed] [Google Scholar]
- 82.Stairs DB, Bayne LJ, Rhoades B, Vega ME, Waldron TJ, Kalabis J, et al. Deletion of p120-catenin results in a tumor microenvironment with inflammation and cancer that establishes it as a tumor suppressor gene. Cancer Cell. 2011;19(4):470–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Overhoff J, Rabideau MM, Bird LM, Schweitzer DN, Haynes K, Schultz RA, et al. Refinement of the 8q22.1 microdeletion critical region associated with Nablus mask-like facial syndrome. Am J Med Genet A. 2014;164A(1):259–63. [DOI] [PubMed] [Google Scholar]
- 84.Yu Y, Zuo X, He M, Gao J, Fu Y, Qin C, et al. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nat Commun. 2017;8:14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lin-Shiao E, Lan Y, Welzenbach J, Alexander KA, Zhang Z, Knapp M, et al. p63 establishes epithelial enhancers at critical craniofacial development genes. Sci Adv. 2019;5(5):eaaw0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Griffin C, Saint-Jeannet JP. Spliceosomopathies: Diseases and mechanisms. Dev Dyn. 2020;249(9):1038–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dennison BJC, Larson ED, Fu R, Mo J, Fantauzzo KA. Srsf3 mediates alternative RNA splicing downstream of PDGFRalpha signaling in the facial mesenchyme. Development. 2021;148(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Timberlake AT, Griffin C, Heike CL, Hing AV, Cunningham ML, Chitayat D, et al. Haploinsufficiency of SF3B2 causes craniofacial microsomia. Nat Commun. 2021;12(1):4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eli Fritz McDonald KEO, Jonathan P. Schlebach, Jens Meiler, Lars Plate. Pre-print Benchmarking AlphaMissense Pathogenicity Predictions Against Cystic Fibrosis Variants BioRxiv. 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Murali H, Wang P, Liao EC, Wang K. Genetic variant classification by predicted protein structure: A case study on IRF6. Comput Struct Biotechnol J. 2024;23:892–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dominguez C, Fisette JF, Chabot B, Allain FH. Structural basis of G-tract recognition and encaging by hnRNP F quasi-RRMs. Nat Struct Mol Biol. 2010;17(7):853–61. [DOI] [PubMed] [Google Scholar]
- 92.Ghoumid J, Stichelbout M, Jourdain AS, Frenois F, Lejeune-Dumoulin S, Alex-Cordier MP, et al. Blepharocheilodontic syndrome is a CDH1 pathway-related disorder due to mutations in CDH1 and CTNND1. Genet Med. 2017;19(9):1013–21. [DOI] [PubMed] [Google Scholar]
- 93.Del Valle-Perez B, Casagolda D, Lugilde E, Valls G, Codina M, Dave N, et al. Wnt controls the transcriptional activity of Kaiso through CK1epsilon-dependent phosphorylation of p120-catenin. J Cell Sci. 2011;124(Pt 13):2298–309. [DOI] [PubMed] [Google Scholar]
- 94.Park JI, Kim SW, Lyons JP, Ji H, Nguyen TT, Cho K, et al. Kaiso/p120-catenin and TCF/beta-catenin complexes coordinately regulate canonical Wnt gene targets. Developmental cell. 2005;8(6):843–54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.