SUMMARY
The CA3 region is central to hippocampal function during learning and memory and has a unique connectivity. CA3 pyramidal neurons are the targets of huge, excitatory mossy fiber synapses from DG axons and have a high degree of excitatory recurrent connectivity. Thus, inhibition likely plays an outsized importance in constraining excitation and shaping CA3 ensembles during learning and memory. Here, we investigate the function of a never-before studied set of dendrite-targeting, GABAergic neurons defined by expression of the synaptic adhesion molecule, Kirrel3. We discovered that activating Kirrel3-expressing GABAergic neurons specifically impairs memory discrimination and inhibits CA3 pyramidal neurons in novel contexts. Kirrel3 is required for DG-to-GABA synapse formation and variants in Kirrel3 are strong risk factors for neurodevelopmental disorders. Thus, our work suggests that Kirrel3-GABA neurons are a critical source of feed-forward inhibition from DG to CA3 during the encoding and retrieval of contextual memories, a function which may be specifically disrupted in some brain disorders.
Keywords: interneuron, inhibition, memory discrimination, memory generalization, CA3, hippocampus, contextual fear conditioning, Kirrel3, mossy fiber synapse
INTRODUCTION
The hippocampus is essential for learning and memory and, in particular, the CA3 region is critical for encoding, recalling and integrating contextual information1–7. CA3 pyramidal neurons, the principal excitatory cells of the region, receive excitatory inputs from the DG. These DG inputs are large mossy fiber boutons that synapse onto multi-headed CA3 spines. These powerful, excitatory synapses have up to 30 active zones and are known as “detonator” synapses because it is possible that release from a single bouton activates a CA3 neuron8,9. In addition, CA3 pyramidal neurons are interconnected via numerous excitatory recurrent collateral synapses. This combination of mossy fiber detonator synapses and extensive recurrent connectivity means that inhibition likely has an outsized importance in shaping CA3 activity and preventing runaway excitation10–13. In fact, DG mossy fiber synapses sprout filopodia directly from the presynaptic bouton to synapse with local GABAergic (GABA) inhibitory neurons that provide feed-forward inhibition to CA311,14. Despite the importance of inhibition in CA3, most hippocampal studies focus on inhibition in the DG and CA1; regions that are easier to access and have more traditional synaptic structures than CA3. Recent work has begun to describe CA3 GABA neuron dynamics15 and appreciate the role of feed-forward inhibition in memory16–18, but there remain large gaps in knowledge about how different types of GABA neurons modulate contextual learning and memory in the CA3 region.
GABA neurons are far fewer in number than excitatory neurons, but they are incredibly diverse and essential for proper brain function11. Studies estimate the hippocampus houses up to 30 transcriptionally distinct types of GABA neurons19–23. It is thought that different transcriptional profiles impart distinct morphological and functional properties to GABA neurons. Despite this diversity, the genetic toolset to identify and manipulate subpopulations of GABA neurons in mouse models is limited and, consequently, only a few cardinal GABA subpopulations are well studied. In contrast, we investigated the function of a previously unstudied subset of GABA neurons identified by the synaptic cell adhesion molecule Kirrel3. We previously discovered Kirrel3 is necessary to properly form DG-to-GABA mossy fiber filopodia synapses that provide feed-forward inhibition to CA39,24 and, consequently, loss of Kirrel3 increases CA3 neuron activity24. These observations suggest that Kirrel3 regulates a critical source of feed-forward inhibition between DG and CA3, and that this inhibition is mediated by Kirrel3-expressing GABA (K3-GABA) neurons. Interestingly, K3-GABA neurons are not identified by any one cardinal GABA neuron marker24. Thus, we generated a new transgenic mouse line that expresses Flp recombinase in Kirrel3-expressing cells and used intersectional genetics to specifically study K3-GABA neurons.
Here, we demonstrate that activating K3-GABA neurons, but not parvalbumin neurons, reduces memory precision by increasing generalization in a contextual fear conditioning task. We further show that K3-GABA neurons in the CA3 and hilus are a transcriptionally heterogeneous population of dendrite-targeting GABA neurons that selectively inhibit CA3 pyramidal activity in novel contexts. Thus, our work supports studying GABA neurons based on synapse-specific molecules, which have potential to drive a shared connectivity pattern among otherwise transcriptionally heterogenous groups of neurons25. Moreover, because Kirrel3 variants are associated with neurodevelopmental disorders26–40, our findings and new tools lay the foundation for studying the specific circuits and behaviors that may be disrupted by genetic changes to Kirrel3.
RESULTS
Kirrel3-expression marks a subpopulation of GABA neurons.
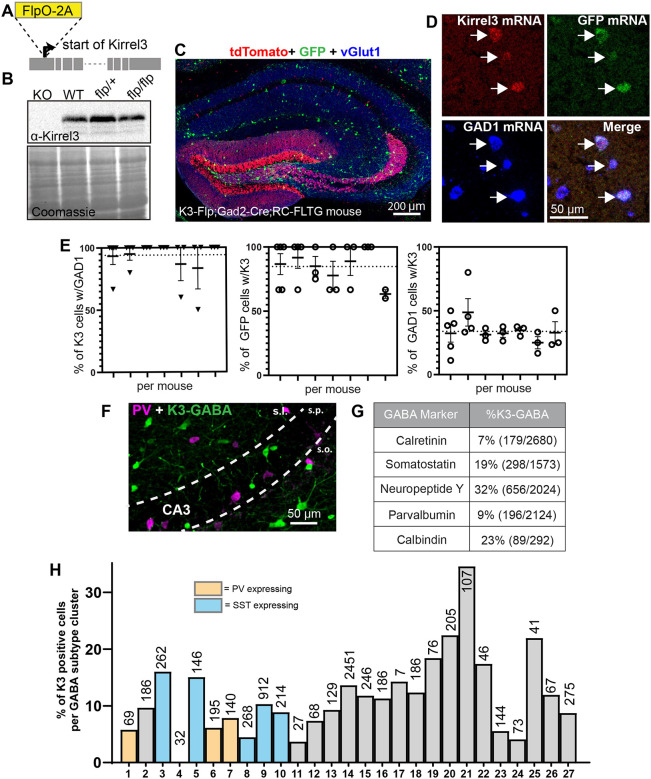
To study K3-GABA neurons, we generated a mouse line that expresses Flp recombinase under control of the endogenous Kirrel3 gene. We inserted the Flp sequence just after the start codon of Kirrel3 (Figure 1A) along with a viral 2A sequence to allow for normal expression of Kirrel3 protein (Figure 1B). To test the specificity of our new mouse line and identify K3-GABA neurons but not DG neurons, which also express Kirrel3, we crossed the Kirrel3-Flp line with a Gad2-Cre line expressing Cre in all GABA neurons41 to a reporter line known as RC:FLTG42. This reporter labels cells co-expressing both Flp and Cre with GFP (K3-GABA neurons) and cells expressing Flp alone with tdTomato (DG neurons) (Figure 1C). There is no suitable antibody to label Kirrel3 protein in tissue, so we used fluorescent in situ hybridization (FISH) with RNA probes against Kirrel3, GAD1, and GFP to test if the GFP positive cells in the triple transgenic line are GABA neurons that express Kirrel3 (Figure 1D). We imaged cells in area CA3 and found that the transgenic line functions as expected. 94% ± 14 of Kirrel3 cells in CA3 are GABA neurons (mean ± sd, Figure 1E) and 86% ± 17 of GFP cells express Kirrel3 (mean ± sd, Figure 1E). This experiment also indicates that 34% ± 14 of all GABA neurons express Kirrel3 mRNA (mean ± sd, Figure 1E). This is slightly more than we previously reported (~19%) using immunohistochemistry on Kirrel3 knockout mice, which express GFP instead of Kirrel324 and is likely due to different sensitivities of each method.
Figure 1. Kirrel3 expression marks a heterogenous group of GABA neurons.
A) Schematic of the FlpO-2A insertion site in the mouse Kirrel3 gene. B) Western blot of brain lysates from 2 months old mice show that the Flp transgenic line expresses normal levels of Kirrel3 protein. Lysates from Kirrel3 knockout (KO) and wildtype (WT) are used as controls. Coomassie stained membrane indicates equal protein loading. C) A hippocampal section from an adult Kirrel3-Flp;Gad2-Cre;RC-FLTG triple transgenic mouse (heterozygous for all transgenes indicated) stained with antibodies against TdTomato (red), GFP (green), and vGlut1 (blue). D) A magnified image of a hippocampal section labeled by fluorescent in situ hybridization (FISH). Arrows indicate K3-GABA neurons co-expressing Kirrel3, GFP, and GAD1 mRNA. E) Quantification of FISH as indicated. Each column represents a mouse and each dot represents one hippocampal section from that mouse. Average percentages from all data points are shown as a dotted line. n = 7 adult mice (3 male and 4 female) Error bars represent SEM. F) An adult triple transgenic mouse stained for GFP to label K3-GABA neurons (green) and parvalbumin (magenta). Note the lack of overlap. G) Table showing the percent of K3-GABA neurons that express the indicated marker as assessed by immunostaining triple transgenic mice similar to F. n = 3 female adult mice. Total number of neurons counted for each marker is indicated. H) Graph showing the percent of cells with at least one Kirrel3 RNA read in each transcriptomic class of GABA neuron from the DropViz data set. Classes that express parvalbumin (PV) and somatostatin (SST) are indicated. The total number of cells in each class is indicated on or above each bar. No Kirrel3 transcripts were found in any cells in group 4.
We previously tested if K3-GABA neurons could be described by a common GABA subtype marker but did not find a match24. We revisited this idea using our new reporter mouse. We stained K3-Flp;Gad2-Cre;RC:FLTG mice for GFP to mark K3-GABA neurons and a selection of GABA markers, including parvalbumin as shown in Figure 1F. We again found that K3-GABA neurons are not identified by any one GABA marker (Figure 1G). Instead of continuing to examine individual marker proteins one at a time, we next used publicly available single cell sequencing data from mouse hippocampus to determine if K3-GABA neurons fall into a single transcriptomic GABA class. Data from the DropViz database uses unsupervised clustering to group hippocampal GABA neurons into 27 distinct transcriptomic classes22. We determined the percent of cells in each group that had one or more reads of a Kirrel3 transcript. All classes except one, had some cells that express Kirrel3, no class had more than 30% of its cells express Kirrel3, and most classes had between 5–15% of their cells with at least a single Kirrel3 transcript (Figure 1H, Table S1). Similar results were obtained using data from the Allen Institute23 (not shown). Taken together, we conclude that Kirrel3 mRNA is expressed by up to 1/3 of all hippocampal GABA neurons and theyz are a heterogenous group that is not easily categorized by transcriptomics.
Activation of K3-GABA neurons using intersectional viral vectors.
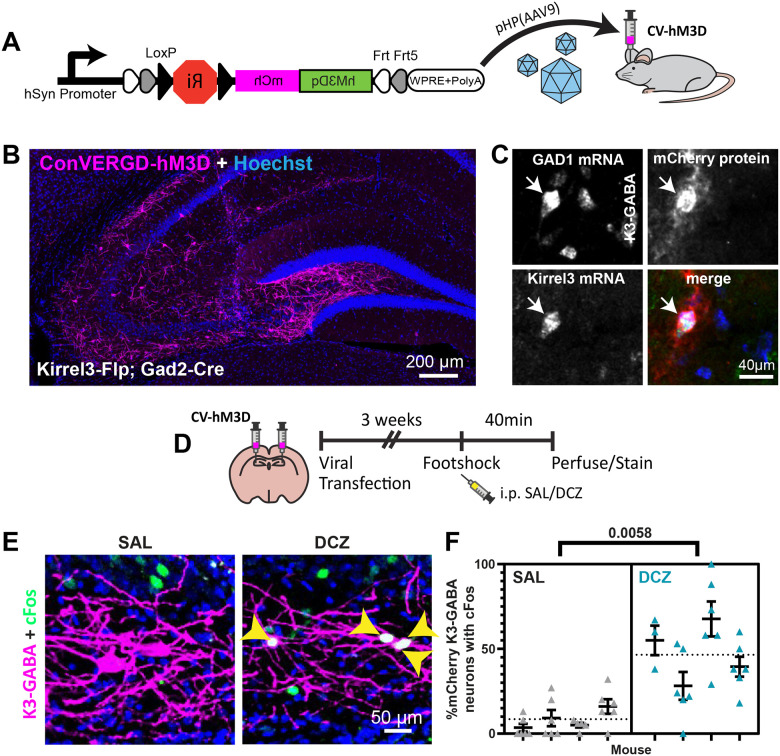
To study the function of K3-GABA neurons in area CA3, we needed a tool to selectively activate them in vivo. We used a newly developed ConVERGD intersectional expression vector that is Cre and Flp-dependent43. It contains a LoxP-flanked ribozyme, which needs to be removed by Cre to prevent mRNA degradation, and an inverted, Frt-flanked payload driven by the human synapsin promoter43. We tested the functionality of an AAV-ConVERGED vector with the activating DREADD payload hM3D-mCherry (Figure 2A). We first confirmed that hM3D-mCherry expression depended on both Flp and Cre in vitro (Figure S2A). Next, we injected the hM3D-mCherry ConVERGD virus into hippocampal area CA3 of K3-Flp;Gad2-Cre double heterozygous mice to specifically express hM3D-mCherry in K3-GABA neurons. We observed cell labeling in a pattern consistent with the known location of K3-GABA neurons with no off-target expression in K3-Flp or wildtype mice (Figure 2B and S2B). We used a dual FISH/immunostaining protocol to confirm that the hM3D-mCherry protein is selectively expressed in cells that express Kirrel3 and Gad1 mRNA (K3-GABA neurons) (Figure 2C). Taken together, our data indicates that we specifically target K3-GABA neurons.
Figure 2. Selective activation of K3-GABA neurons.
A) Schematic showing the ConVERGD(CV)-hM3D-mCherry AAV construct, AAV packaging, and brain injections. B) Representative hippocampal image from a Kirrel3-Flp;Gad2-Cre mouse with CV-hM3D-mCherry targeted to area CA3. C) Magnified image from a hippocampal section processed for dual FISH/immunohistochemistry from a Kirrel3-Flp;Gad2-Cre mouse infected with CV-hM3D-mCherry. D) Schematic of the experimental design for cFos analysis in E-F. E) Representative images showing cFos (green) in K3-GABA neurons expressing hM3D-mCherry (magenta) specifically after DCZ injection (right). Yellow arrowheads indicate K3-GABA neurons that express cFos (merge is white). E) Quantification of the % of infected K3-GABA neurons that express cFos for saline and DCZ treated mice after foot shock. n = 4 males each. Each column represents one mouse, up to 3 sections and 6 hippocampi counted per mouse. Average from all data points is shown as a dotted line. Error bars represent SEM, nested t-test.
We next tested if the ConVERGD-hM3D-mCherry activates K3-GABA neurons after injection with the agonist, deschloroclozapine (DCZ). K3-Flp; Gad2-Cre mice were injected with the ConVERGD virus. Three weeks later, mice were intraperitoneally injected with saline or DCZ 10 minutes before placing them in a behavior box and subjecting them to 4-foot shocks as used for fear conditioning (Figure 2D). 40 minutes later, mice were perfused, and sections were stained for the immediate early gene cFos, a common measure of recent neuronal activity. We found that DCZ administration significantly increased the percent of K3-GABA neurons expressing cFos compared to saline (Figure 2E, F), indicating that the ConVERGD-hM3D expression activates K3-GABA neurons in the presence of DCZ as expected.
Activating K3-GABA neurons impairs memory discrimination.
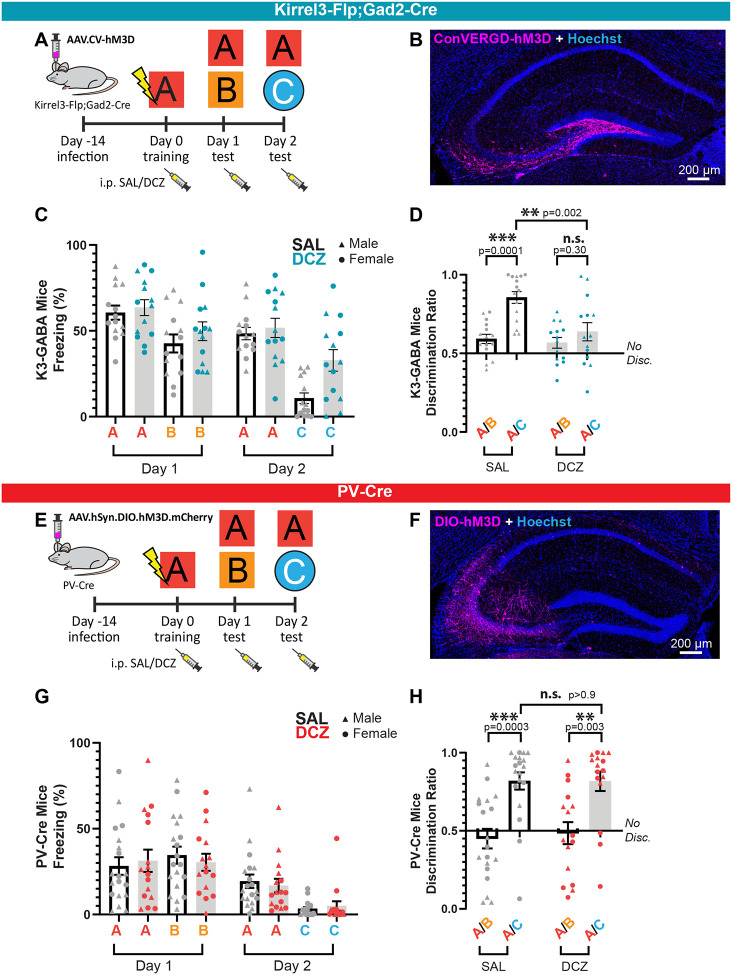
We next sought to determine if activating K3-GABA neurons alters mouse behavior in a hippocampal-dependent learning and memory task. To do this, we adapted a contextual fear conditioning paradigm capable of probing context fear acquisition, generalization, and discrimination across multiple contexts (Figures 3A and S3A)44,45. We conditioned mice in context A where they received 4 unsignaled foot shocks (1.0mA, 2 sec duration, 60 sec inter-stimulus-interval). 24 hours later, we tested their memory by measuring their freezing behavior when placing them back in context A without foot shock. In addition, we tested the degree to which they were able to discriminate this context from a similar but slightly different context, B. The next day, we tested whether animals were able to discriminate context A from an entirely novel and very distinct context, C. For each day, the context test order was counterbalanced. The time spent freezing in each context was quantified and the difference in freezing relative to context A was used to generate a discrimination ratio (A/(A+B)) between the two contexts on each day46. Wildtype mice are generally poor at distinguishing between similar contexts A and B, but reliably discriminate between distinct contexts A and C47–49, an effect we found to be present in our hands as well (Figure S3B, C). This paradigm allows us to evaluate several aspects of learning and memory upon manipulation of K3-GABA neuron activity including, if the mice have either impaired or enhanced memory formation or discrimination in similar or distinct contexts.
Figure 3. K3-GABA activation impairs memory discrimination.
A) Schematic of viral transfection and behavior paradigm for Kirrel3-Flp;Gad2-Cre mice. B) Example of a Kirrel3-Flp;Gad2-Cre mouse with a CA3 injection of the Flp and Cre-dependent CV-hM3D-mCherry AAV. C) Time spent freezing (%) when placed in indicated contexts after saline (gray) or DCZ (blue) injection. All mice were adult K3-Flp; Gad2-Cre heterozygotes injected with CV-hM3D-mCherry. n = 15 saline, 14 DCZ. D) Data from C plotted as a discrimination ratio relative to context A, where 0.5 marks no discrimination and is calculated by the percent freezing time for A/A+X, where X is either context B or C depending on the day. E) Schematic of viral transfection and behavior paradigm for PV-Cre homozygous mice. F) Example of a PV-Cre mouse with a CA3 injection of the Cre-dependent DIO-hM3D-mCherry AAV. G) Time spent freezing (%) when placed in indicated contexts after saline (gray) or DCZ (red) injection. All mice were adult PV-Cre heterozygous mice injected with Cre-dependent hM3D-mCherry. F) Discrimination ratios for PV-Cre mice showed normal discrimination for saline and DCZ injected mice, n = 19 saline, 16 DCZ. Error bars represent SEM, ANOVA multiple comparisons. Males and females are indicated and each dot represents a mouse.
Next, we tested if activating K3-GABA neurons alters the learning and memory of mice using this paradigm. Male and female K3-Flp;Gad2-Cre heterozygous mice were bilaterally injected in the CA3 region with ConVERGD-hM3D-mCherry AAVs and randomly divided into a control group that received saline injections or an experimental group that received DCZ injections. Because the function and timing of K3-GABA neurons is completely unknown, we opted to give saline or DCZ injections prior to all contexts (Figure 3A). All mice conditioned normally to foot shock on training day 0 (Figure S3D–F) and showed normal fear conditioning in context A and B on day 1 (Figure 3C, D). However, on day 2 we observed a significant increase in the fear response when K3-GABA neurons were activated by DCZ in the neutral context C (Figure 3C). This resulted in a significant decrease in the A/C discrimination ratio compared to saline controls (Figure 3D), and the loss of a difference between the A/B and A/C discrimination ratios in the DCZ condition (Figure 3D). Importantly, mice that did not express the DREADD but were injected with DCZ had normal behavior (Figure S3A, B). Thus, our data indicates that exogenous activation of K3-GABA neurons impairs memory discrimination by increasing fear generalization to a neutral context.
To test if this phenotype is specific to K3-GABA neurons or if it would result from activating any similar sized subpopulation of GABA neurons, we expressed hM3D-mCherry in parvalbumin (PV) neurons (Figure 3E, F), which have little overlap with K3-GABA neurons (Figure 1F–H). PV-Cre mice were bilaterally injected with a Cre-dependent hM3D-mCherry and tested in the same contextual fear conditioning paradigm. We found that, as expected, Cre-driven hM3D-mCherry expression was high in PV immunostained neurons (Figure S3G). However, we found that activating PV-GABA neurons does not alter learning or memory discrimination on either day compared to saline controls (Figure 3G, H). PV-Cre mice conditioned normally to foot shock on Day 0 (Figure S3H–J) and although, PV-Cre mice have a lower baseline fear response compared to wildtype and K3-Flp;Gad2-Cre mice, their discrimination ratios and relative fear response are similar. We confirmed proper hM3D-mCherry expression and targeting in all mice used for experiments (Figure S4A, B. Together, our data indicate that K3-GABA neurons in area CA3 have a specific function in memory discrimination that is not shared with PV neurons.
Activating K3-GABA neurons decreases CA3 activity in novel contexts.
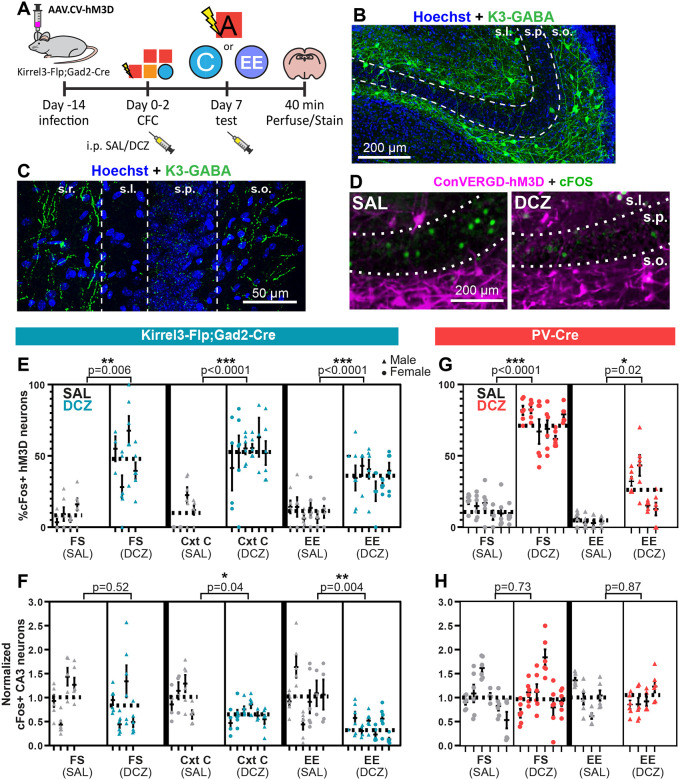
To begin to understand how activating K3-GABA neurons impairs memory discrimination at the circuit level, we sought to determine if activating K3-GABA neurons affects CA3 pyramidal neuron activity. We imaged K3-GABA projections to determine generally where they contact CA3 neurons by sparsely labeling K3-GABA neurons with a fluorescent reporter. We could clearly see axonal labeling in the CA3 SR and SO layers and exclusion from the pyramidal cell body layer (Figure 4A, B), suggesting K3-GABA neurons target CA3 pyramidal cell dendrites.
Figure 4. K3-GABA neurons inhibit CA3 in novel contexts.
A, B) Kirrel3-Flp;Gad2-Cre heterozygous mouse infected with an AAV that expresses GFP in a Flp and Cre-dependent manner to label K3-GABA neurons. B shows a magnified image of CA3 from a section that is sparsely labeled. s.o.; stratum oriens, s.p.; stratum pyramidale, s.l.; stratum lucidum, s.r.; stratum radiatum. C) Schematic of experimental design for cFos analysis. D) Example images of cFos staining in the CA3 pyramidal layer for Kirrel3-Flp;Gad2-Cre mice infected with CVD-hM3D-mCherry AAV and injected with saline (left) or DCZ (right) before exposure to an enriched environment. E) % of hM3D-expressing K3-GABA neurons that are cFos positive after saline (grey) and DCZ injection (blue) in indicated environments. FS = foot shock, CxtC = context C, EE = enriched environment. F) Number of cFos-positive CA3 neurons per mm2 of the pyramidal layer normalized to saline controls for saline (grey) and DCZ treated (blue) mice in indicated contexts. G) % of hM3D-expressing PV neurons that are cFos positive after saline (grey) and DCZ injection (red) in indicated environments. H) Number of cFos-positive CA3 neurons per mm2 normalized to saline controls for saline (grey) and DCZ treated (red) mice in indicated contexts. For all graphs in the figure (E-H), each dot represents a hippocampus section, each column represents a mouse, the average for all data points in a condition is shown as a dotted line. n = 4–8 mice per group. Nested t-test p-value comparisons are indicated and error bars show s.e.m.
Next, we tested if activating K3-GABA neurons with DREADDs inhibits CA3 neuron activity using cFos staining. One week after mice were run through the full contextual fear conditioning paradigm, we injected mice with either saline or DCZ and placed them back in the familiar context A with foot shock (FS) (Figure 4C, D). Following cFos immunostaining, we found that DCZ injection activates K3-GABA neurons compared to saline as reported in Figure 2 (Figure 4E, left panels), but surprisingly, we observed no corresponding change in CA3 neuron activity as measured by the density of cFos positive cells in the pyramidal cell layer in the same mice (Figure 4F, left panels). Because the DCZ injected mice behaved normally in every exposure to context A in the behavioral tests, we next analyzed cFos activity 40 minutes after mice were exposed to the neutral context C (CxtC), in which K3-GABA DCZinjected mice had abnormal behavior. In this condition, we again observed an increase in cFos-positive K3-GABA neurons, indicating that the DREADD is working (Figure 4E, middle panels). But this time, we also observed a significant decrease in the number of cFos-positive CA3 neurons (Figure 4F, middle panels).
We reasoned that the mice had been placed in context A, with and without foot shock, several times before the cFos analysis but they had only been exposed to context C one time before the cFos analysis. This suggests that activating K3-GABA neurons selectively decreases net CA3 neuron activity in response to novel, but not familiar, contexts. To further test this hypothesis, we placed another cohort of mice in a completely novel and neutral enriched environment (EE), which is entirely different to any of the behavior boxes that mice were exposed to during fear conditioning (see Materials and Methods). This time we observed an even greater depression of CA3 activity, despite having a similar number of active K3-GABA neurons in all contexts (Figure 4D, E, F, right panels). Interestingly, DCZ alone was not enough to activate K3-GABA neurons in the home cage condition, nor were any changes observed in CA3 activity (Figure S4C–E), suggesting that K3-GABA neurons can only be activated under specific environmental conditions. For comparison, we also analyzed CA3 activity in PV-Cre mice expressing hM3D-mCherry in the same experiment. We found that though PV neurons were activated following DCZ administration, they did not inhibit CA3 pyramidal neurons in either the foot shock context A or the neutral enriched environment (Figure 4G, H). This is consistent with the fact that activating PV neurons did not affect the contextual fear conditioning behavior. Together, our results strongly suggest that activating K3-GABA neurons selectively inhibits CA3 pyramidal neurons in novel environments and thereby reduces memory precision by increasing memory generalization.
DISCUSSION
Here, we demonstrated that activating K3-GABA neurons decreases the number of active CA3 neurons and causes mice to generalize fear to a novel, neutral environment. These results are consistent with what is broadly known about memory and ensemble size, which is that decreasing ensemble size (i.e. the number of active CA3 neurons) reduces memory discrimination, while increasing ensemble size improves it50–54.
Because discrimination is calculated as a ratio, reduced memory discrimination can be caused if mice show less of a conditioned response in the conditioned context (context A) or more in the neutral context (context C). In our experiments, K3-GABA neuron activation caused mice to generalize conditioned fear to the neutral context. Why might this happen at the cellular level? We speculate that is because at any given time active CA3 neurons come from two pools; some CA3 neurons are activated directly from the DG by the huge mossy fiber detonator synapses, while others are activated by subsequent recurrent connectivity and entorhinal inputs. It is unlikely that dendrite-targeting K3-GABA neurons would prevent powerful mossy fiber synapse transmission. However, K3-GABA neuron activity may constrain CA3 activity by limiting the amount of downstream recurrent CA3 activation and entorhinal inputs. If the strong DG inputs encode core features of the memory, such as a mouse being taken out of their home cage and put in a behavior box, while recurrent CA3 activity contributes to encoding or retrieving contextual details, then activating K3-GABA neurons would retain the core fear memory but lose contextual detail. If so, it would be predicted that mice in which K3-GABA neurons are over-activated would fear a neutral context as we observed. There are certainly other ways that K3-GABA neurons could affect CA3 activity, including via feed-back inhibition to DG and by modulating other types of GABA neurons or entorhinal inputs7,48,55–58. Moreover, because this is the first study investigating K3-GABA neuron function, we activated K3-GABA neurons with the DREADD ligand prior to each context presentation. Therefore, our results cannot determine if K3-GABA neurons primarily function during the contextual encoding, retrieval, or both. Future work will need to map the timing of K3-GABA activity as well as their precise input and output connectivity of K3-GABA neurons in more detail, but our work clearly implicates K3-GABA neurons as an important modulator of CA3 activity and lays the foundation for future work.
Our results presented here also reveal several intriguing features about the specificity of K3-GABA neuron activity. First, we showed that activation of K3-GABA neurons, and not PV neurons, decreases memory discrimination. This indicates that K3-GABA neurons have a distinct function from PV neurons and behavioral changes we observed here are not due to simply modulating any similar sized population of GABA neurons. Second, we observed that activating K3-GABA neurons significantly decreased CA3 activity in novel contexts but not in a familiar context. This suggests that CA3 neuron activity is tightly controlled by different kinds of neurons, and they are uniquely vulnerable to K3-GABA neuron activity only under certain conditions like novel contexts. Third, we observed that K3-GABA neurons expressing the activating DREADD are not responsive to DCZ in the home cage condition, but they are upon contextual stimulation. This suggests that the DREADD works as expected but its activity is masked in K3-GABA neurons in the home cage condition. This could occur if K3-GABA neurons are strongly inhibited in the familiar home environment. In addition, using cFos as a readout for K3-GABA and CA3 activity cannot reveal subtle changes in activity dynamics but is an important foundation for understanding their net activity in vivo before and after behavior. Moreover, the highly specific activity of K3-GABA neurons could relate to the synapse-specific nature of the Kirrel3 protein. DG neurons make synapses with many types of GABA neurons via two types of synapses; typical en passant synapses along the entire length of the axon and mossy fiber filopodia synapses that are only found protruding from large mossy fiber boutons9,14,59. We previously found that loss of Kirrel3 protein specifically reduces the density of DG-to-GABA mossy fiber filopodia synapses but not DG-to-GABA en passant synapses9. Thus K3-GABA neurons in area CA3 may be highly specialized to receive mossy fiber filopodia input and could be an important avenue to investigate this poorly understood aspect of the hippocampal circuit.
Finally, our work is one of the first to study a group of GABA neurons based on the expression of a molecule that is a driver of synaptic connectivity. Thus, K3-GABA neurons, though transcriptionally heterogenous, may be functionally linked in the circuit by sharing common input and output neurons driven, at least in part, by Kirrel3. Cell type-specific studies of GABA neurons using cardinal markers like PV, SST, and VIP enabled groundbreaking discoveries in circuit motifs, but it is becoming clear that even these tools label heterogenous populations of GABA neurons with diverse circuit functions60,61. The generation of new tools to study distinct cell types, combined with intersectional approaches like those used here, will propel our understanding of circuit function in the future. Moreover, missense variants and copy number changes in Kirrel3 are strongly associated with increased risk for autism spectrum disorders, intellectual disabilities, and other neurodevelopmental disorders26–40. Thus, understanding how changing K3-GABA neuron activity affects brain circuits could have significant clinical relevance and taken together, bridges molecular, cellular, and systems neuroscience.
MATERIALS AND METHODS
Mouse lines:
The Kirrel3-Flp line FlpO-P2A was inserted in frame and immediately upstream of the Kirrel3 start codon so that Kirrel3 protein expression is unperturbed. We inserted FlpO at the start because the Kirrel3 gene undergoes alternative splicing at its C-terminus. Transgenic mice were generated at the University of Nebraska School of Medicine Transgenic core using the Easi-CRISPR method of homologous recombination62. Briefly, single cell C57Bl6 mouse zygotes were injected with preassembled Cas9 ribonucleotide complexes containing Cas9 protein, a Kirrel3 crRNA (AGGAATGAGACCTTTCCAGC), and tracrRNA along with a 1,536 bp single strand DNA (ssDNA) repair template containing the FlpO-P2A sequence and ~100bp homology arms. A founder mouse was fully sequenced to ensure there were no mutations at insertion sites and the mouse was bred to a wildtype C57Bl6 mouse to propagate the strain. The donor ssDNA sequence is as follows: GCCGCGCTTGAAGAGAAACTAACTGCACACCCAAGTTGCCCGCCGGCTGCCCGCGCGCTGAGGAATGAGACCTTTCGCTAGCATGGCTCCTAAGAAGAAGAGGAAGGTGATGAGCCAGTTCGACATCCTGTGCAAGACCCCCCCCAAGGTGCTGGTGCGGCAGTTCGTGGAGAGATTCGAGAGGCCCAGCGGCGAGAAGATCGCCAGCTGTGCCGCCGAGCTGACCTACCTGTGCTGGATGATCACCCACAACGGCACCGCCATCAAGAGGGCCACCTTCATGAGCTACAACACCATCATCAGCAACAGCCTGAGCTTCGACATCGTGAACAAGAGCCTGCAGTTCAAGTACAAGACCCAGAAGGCCACCATCCTGGAGGCCAGCCTGAAGAAGCTGATCCCCGCCTGGGAGTTCACCATCATCCCTTACAACGGCCAGAAGCACCAGAGCGACATCACCGACATCGTGTCCAGCCTGCAGCTGCAGTTCGAGAGCAGCGAGGAGGCCGACAAGGGCAACAGCCACAGCAAGAAGATGCTGAAGGCCCTGCTGTCCGAGGGCGAGAGCATCTGGGAGATCACCGAGAAGATCCTGAACAGCTTCGAGTACACCAGCAGGTTCACCAAGACCAAGACCCTGTACCAGTTCCTGTTCCTGGCCACATTCATCAACTGCGGCAGGTTCAGCGACATCAAGAACGTGGACCCCAAGAGCTTCAAGCTGGTGCAGAACAAGTACCTGGGCGTGATCATTCAGTGCCTGGTGACCGAGACCAAGACAAGCGTGTCCAGGCACATCTACTTTTTCAGCGCCAGAGGCAGGATCGACCCCCTGGTGTACCTGGACGAGTTCCTGAGGAACAGCGAGCCCGTGCTGAAGAGAGTGAACAGGACCGGCAACAGCAGCAGCAACAAGCAGGAGTACCAGCTGCTGAAGGACAACCTGGTGCGCAGCTACAACAAGGCCCTGAAGAAGAACGCCCCCTACCCCATCTTCGCTATCAAGAACGGCCCTAAGAGCCACATCGGCAGGCACCTGATGACCAGCTTTCTGAGCATGAAGGGCCTGACCGAGCTGACAAACGTGGTGGGCAACTGGAGCGACAAGAGGGCCTCCGCCGTGGCCAGGACCACCTACACCCACCAGATCACCGCCATCCCCGACCACTACTTCGCCCTGGTGTCCAGGTACTACGCCTACGACCCCATCAGCAAGGAGATGATCGCCCTGAAGGACGAGACCAACCCCATCGAGGAGTGGCAGCACATCGAGCAGCTGAAGGGCAGCGCCGAGGGCAGCATCAGATACCCCGCCTGGAACGGCATCATCAGCCAGGAGGTGCTGGACTACCTGAGCAGCTACATCAACAGGCGGATCGGAAGCGGAGCTACTAACTTCAGCCTGCTGAAGCAGGCTGGAGACGTGGAGGAGAACCCTGGACCTATGAGACCTTTCCAGCTGGATTTGCTCTTCCTCTGCTTCTTCCTCTTCAGTCAAGgtaggaacccggcgcgccggagttggagcaaagtcgg.
Other mice used are: Gad2-IRES-Cre (JAX stock #010802)41, RC::FLTG (JAX stock #026932)42, and PV-Cre (JAX stock #017320), and C57BL/6J wildtype mice (JAX stock #000664). All mice were maintained and conducted in accordance with NIH guidelines on the care and use of animals approved by the University of Utah IACUC committee.
Immunohistochemistry:
Mice were transcardially perfused with 1X phosphate-buffered saline (PBS) solution prepared from 10X stock consisting of 25.6g of Na2HPO4.7H2O, 80g of NaCl, 2g KCL, 2g KH2PO4, and pH’d to 7.4. P Mice are then perfused with 4% paraformaldehyde (PFA) prior to brain extraction. Brains were then stored in 4% PFA overnight and cut in 50μm sections around the hippocampal formation. Sections were incubated in blocking solution (PBS with 3% bovine serum albumin (BSA) and 0.2% Triton-X100) for 1 hour. Sections were then left overnight with gentle shaking in primary antibody solution, which was prepared with the antibody diluted in PBS with 3% BSA. After 3 washes with PBS, sections were then incubated with secondary antibody solution for 2 hours at room temperature. A final 3 washes were performed with the third wash adding 2.5ug of Hoechst in PBS prior to mounting.
Antibodies:
Primary antibodies were used as follows: goat anti-GFP 1:5000 (Abcam, ab6673), rabbit anti-cFos 1:500 (Santa Cruz Biotech, sc-253; Synaptic Systems, 226008), rabbit anti-calretinin 1:2000 (Swant, 7699/3), mouse anti-parvalbumin 1:5000 (Swant, PV235), rat anti-somatostatin 1:500 (Chemicon, MAB354), rabbit anti-calbindin d28k 1:2000 (Swant, CB38a), rabbit anti-VIP 1:500 (Immunostar), rabbit anti-PV 1:250 (Swant, PV27a), rabbit anti-RFP 1:1000 (Fisher, NBP267102), goat anti-mCherry (Novus, NBP3–05558). All secondary antibodies were made in donkey and are from Jackson ImmunoResearch Laboratories Inc. and used at 1:1000.
Stereotaxic surgeries:
Adult mice over the age of 8-weeks were injected with Buprenorphine (0.1mg/kg) intraperitoneally an hour prior to operation. Mice were anesthetized in an airtight container ventilated with oxygen and isoflurane and then readied in stereotaxic surgery area. An injection of lidocaine (2mg/kg) is administered subcutaneously near the intended incision area which is shaved and sterilized. Small holes were drilled in the skull to expose the region of interest for viral intracranial injection. 0.5ml of AAV was delivered via picospritzer to the desired region with a 0.03 ms pulse at 60 psi. Upon completion, incisions were sealed with VetBond (Amazon) and mice were injected subcutaneously with Rimadyl (5mg/kg Carprofen). Mice were allowed to recover on heating pad and observed for signs of distress before returning to their home cage. Follow-ups continued for three days and consisted of weighing mice and administering Rimadyl to ensure healthy recovery.
Viruses:
Adeno-associated viruses (AAVs) were made using standard iodixanol gradient purification and the pHP.eb capsid63. Viral titers for all viruses used were 1011 to 1013. Viruses used are as follows: ConVERGD construct (pHP.eb)hSyn.hM3Dm.Cherry and (AAV8)hSyn.hM3D.HA43 provided by Lindsay Schwarz lab; (AAV9)hSyn.DIO.hM3D.mCherry (Addgene 44361); INTRSCT construct (pHP.eb)hSyn.Cre-on/Flp-on.YFP (Addgene 55650)64.
Enriched environment:
Mice are removed from their home cage and injected with either saline or 1.0mg/kg DCZ then immediately placed into a 40cm × 40cm plastic container with 5 novel objects evenly spaced apart. Mice are allowed to explore freely for 40-minutes prior to perfusion.
Contextual Fear Conditioning:
Mice underwent a 3-day contextual fear conditioning protocol. Mice were habituated up for 3 days prior to conditioning day (Day 0) for 1 hour. On conditioning Day 0 and after habituation, mice are then injected i.p. with either saline or 1.0mg/kg DCZ 10-minutes prior to being placed into behavior boxes (MedAssociates NIR). Behavior boxes are furnished with distinguishing features such as floor grating, light, scent, roofing, and sound that they will associate with conditioned “Context A”. Mice 4 shocks (1mA, 2-second duration, 1-minute inter-shock interval) are delivered after an initial 3-minute acclimation period. Mice then rest for an additional minute, for a total 8-minute duration conditioning protocol. On Test Day 1, mice are habituated and injected as described earlier. Half of conditioned mice are exposed to conditioned Context A for 8-minutes or “intermediate” Context B (see Figure 3B). Mice are returned to their home cage and mouse room for a minimum of 5 hours before their next context exposure. Mice initially exposed to Context A are now exposed to Context B, and vice versa. This process is repeated for Test Day 1, with “neutral” Context C replacing intermediate Context B
cFos analysis:
Confocal images (Zeiss 980) were taken at 10X and 20X objective as Z-stacks and tiles, exported as OME-TIFF files, and analyzed in FIJI blind to condition. For image analysis in FIJI, all images were z-projected for maximum intensity, despeckled, and split channels. ROIs are drawn for the entire CA3 region in the virus mCherry channel, and only the CA3 pyramidal region in the cFos channel to measure area. The cFos channel background is subtracted by a rolling ball radius value of 15, then thresholded to three times the inflection point of the signal histogram values for a consistent signal-to-noise ratio. Particles are analyzed with a circularity range of 0–1.0 and size of 30-infinity. The counts are then reviewed and saved as ROI to overlap with the virus mCherry channel and count co-expressing cells. The percent signal area within and outside of the ROI of the mCherry channel is recorded, while cell bodies are individually counted by hand-drawn ROIs. Then the number of overlapping ROIs for mCherry cell bodies and cFos counts are recorded. Counts are verified by merging channels and examining merged fluorescence. Analysis was done blind to genotype and condition.
Fluorescent in situ hybridization (FISH):
Fluorescent in situ hybridization chain reaction (HCR) HCR was performed as previously described65. Mouse brains were cryo-sectioned to 30 mm slices, mounted on slides, fixed [4% paraformaldehyde (PFA)] and washed in PBS. Before processing samples according to protocol HCR v3.0 (Invitrogen), slices were treated with 1mg/ml proteinase K-treated (TE buffer) and equilibrated in SSC buffer. HCR probes were designed and generated by Molecular Instruments for Gad1, Kirrel3, and GFP. After nuclear staining with Hoechst in PBS, coverslips were mounted in Fluoromount-G (Southern Biotech catalog #0100–01) and imaged on a confocal microscope. For protein amplification, samples are incubated in freshly prepared 4% PFA made with DEPC water and PBS for 10 minutes, then washed in 1X DEPC treated PBS (pH 7.4) for 5 minutes at room temperature three times. Samples are then blocked for 1 hour at room temperature (100 mL/slide applied over top of sections) inside humidified chamber (does not need to be RNAse free) and 75μL primary antibody solution (anti-mCherry 1:1000) is added per slide and covered with an RNAse free coverslip. Samples are incubated for 1 hour at room temperature and then overnight at 4°C in humidified chamber. The next day, samples are washed in 1X DEPC treated PBST (pH 7.4) for 5 minutes at room temperature for a total of three washes before adding the 2° antibody at 1:1000 then incubating for 1 hour at room temperature inside humidified chamber. Samples are washed in 1X DEPC treated PBST (pH 7.4) for 5 minutes at room temperature another three times and mounted by adding 250μL Fluoromount-G mounting reagent on top of sample, then cover-slipped for microscopy.
Supplementary Material
ACKNOWLEDGMENTS
We thank past and present members of the Williams lab for advice and technical help. We thank Drs Lindsey Schwarz and Alex Hughes for sharing ConVERGD reagents prior to publication. This work was supported by the following funding sources: NIH T32HD007491 (A.T-O), seed grants from the University of Utah (M.E.W.) and Brain Research Foundation (M.E.W.), and NIH grants MH125943 (M.E.W.) and MH134515 (M.E.W.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Cherubini E. & Miles R. The CA3 region of the hippocampus: how is it? what is it for? how does it do it? Front. Cell. Neurosci. 9, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Florian C. & Roullet P. Hippocampal CA3-region is crucial for acquisition and memory consolidation in Morris water maze task in mice. Behav. Brain Res. 154, 365–374 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Hunsaker M. R., Rosenberg J. S. & Kesner R. P. The role of the dentate gyrus, CA3a,b, and CA3c for detecting spatial and environmental novelty. Hippocampus 18, 1064–1073 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Gold A. E. & Kesner R. P. The role of the CA3 subregion of the dorsal hippocampus in spatial pattern completion in the rat. Hippocampus 15, 808–814 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Kesner R. P. Behavioral functions of the CA3 subregion of the hippocampus. Learn. Mem. 14, 771–781 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Rolls E. T. A quantitative theory of the functions of the hippocampal CA3 network in memory. Front. Cell. Neurosci. 7, 98 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leutgeb J. K., Leutgeb S., Moser M.-B. & Moser E. I. Pattern Separation in the Dentate Gyrus and CA3 of the Hippocampus. Science 315, 961–966 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Rollenhagen A. & Lübke J. H. R. The Mossy Fiber Bouton: the “Common” or the “Unique” Synapse? Front. Synaptic Neurosci. 2, 2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin E. A., Woodruff D., Rawson R. L. & Williams M. E. Examining Hippocampal Mossy Fiber Synapses by 3D Electron Microscopy in Wildtype and Kirrel3 Knockout Mice. eNeuro 4, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miles R., Le Duigou C., Simonnet J., Telenczuk M. & Fricker D. Recurrent synapses and circuits in the CA3 region of the hippocampus: an associative network. Front. Cell. Neurosci. 7, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrence J. J. & McBain C. J. Interneuron Diversity series: Containing the detonation – feedforward inhibition in the CA3 hippocampus. Trends Neurosci. 26, 631–640 (2003). [DOI] [PubMed] [Google Scholar]
- 12.McKenzie S. Inhibition shapes the organization of hippocampal representations. Hippocampus 28, 659–671 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vancura B., Geiller T. & Losonczy A. Organization and Plasticity of Inhibition in Hippocampal Recurrent Circuits. 2023.03.13.532296 Preprint at 10.1101/2023.03.13.532296 (2023). [DOI]
- 14.Acsády L., Kamondi A., Sík A., Freund T. & Buzsáki G. GABAergic Cells Are the Major Postsynaptic Targets of Mossy Fibers in the Rat Hippocampus. J. Neurosci. 18, 3386–3403 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vancura B., Geiller T., Grosmark A., Zhao V. & Losonczy A. Inhibitory control of sharp-wave ripple duration during learning in hippocampal recurrent networks. Nat. Neurosci. 26, 788–797 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo N. et al. Dentate granule cell recruitment of feedforward inhibition governs engram maintenance and remote memory generalization. Nat. Med. 24, 438–449 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shih Y.-T., Alipio J. B. & Sahay A. An inhibitory circuit-based enhancer of DYRK1A function reverses Dyrk1a-associated impairment in social recognition. Neuron 111, 3084–3101.e5 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neubrandt M., Oláh V. J., Brunner J. & Szabadics J. Feedforward inhibition is randomly wired from individual granule cells onto CA3 pyramidal cells. Hippocampus 27, 1034–1039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu J. et al. Intersectional mapping of multi-transmitter neurons and other cell types in the brain. Cell Rep. 40, 111036 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paul A. et al. Transcriptional Architecture of Synaptic Communication Delineates GABAergic Neuron Identity. Cell 171, 522–539.e20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudy B., Fishell G., Lee S. & Hjerling-Leffler J. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev. Neurobiol. 71, 45–61 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saunders A. et al. Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174, 1015–1030.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao Z. et al. A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222–3241.e26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin E. A. et al. The intellectual disability gene Kirrel3 regulates target-specific mossy fiber synapse development in the hippocampus. eLife 4, e09395 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Z. J. & Paul A. The diversity of GABAergic neurons and neural communication elements. Nat. Rev. Neurosci. 20, 563–572 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ben-David E. & Shifman S. Networks of Neuronal Genes Affected by Common and Rare Variants in Autism Spectrum Disorders. PLoS Genet. 8, e1002556 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhalla K. et al. Alterations in CDH15 and KIRREL3 in Patients with Mild to Severe Intellectual Disability. Am. J. Hum. Genet. 83, 703–713 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Rubeis S. et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Interstitial deletion of 11q-implicating the KIRREL3 gene in the neurocognitive delay associated with Jacobsen syndrome - Guerin - 2012 - American Journal of Medical Genetics Part A - Wiley Online Library. https://onlinelibrary.wiley.com/doi/10.1002/ajmg.a.35621. [DOI] [PubMed]
- 30.Guo H. et al. Genome sequencing identifies multiple deleterious variants in autism patients with more severe phenotypes. Genet. Med. Off. J. Am. Coll. Med. Genet. 21, 1611–1620 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iossifov I. et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaminsky E. B. et al. An evidence-based approach to establish the functional and clinical significance of CNVs in intellectual and developmental disabilities. Genet. Med. Off. J. Am. Coll. Med. Genet. 13, 777–784 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leblond C. S. et al. Both rare and common genetic variants contribute to autism in the Faroe Islands. NPJ Genomic Med. 4, 1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J. et al. Targeted sequencing and functional analysis reveal brain-size-related genes and their networks in autism spectrum disorders. Mol. Psychiatry 22, 1282–1290 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Michaelson J. J. et al. Whole Genome Sequencing in Autism Identifies Hotspots for De Novo Germline Mutation. Cell 151, 1431–1442 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neale B. M. et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485, 242–245 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talkowski M. E. et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell 149, 525–537 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang T. et al. De novo genic mutations among a Chinese autism spectrum disorder cohort. Nat. Commun. 7, 13316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuen R. K. et al. Genome-wide characteristics of de novo mutations in autism. NPJ Genomic Med. 1, 16027 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor M. R. et al. Kirrel3-Mediated Synapse Formation Is Attenuated by Disease-Associated Missense Variants. J. Neurosci. 40, 5376–5388 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.A Resource of Cre Driver Lines for Genetic Targeting of GABAergic Neurons in Cerebral Cortex - PMC. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3779648/. [DOI] [PMC free article] [PubMed]
- 42.Plummer N. W. et al. Expanding the power of recombinase-based labeling to uncover cellular diversity. Dev. Camb. Engl. 142, 4385–4393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hughes A. C. et al. A single-vector intersectional AAV strategy for interrogating cellular diversity and brain function. Nat. Neurosci. 1–11 (2024) doi: 10.1038/s41593-024-01659-7. [DOI] [PubMed] [Google Scholar]
- 44.Nakashiba T. et al. Young Dentate Granule Cells Mediate Pattern Separation, whereas Old Granule Cells Facilitate Pattern Completion. Cell 149, 188–201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun X. et al. Functionally Distinct Neuronal Ensembles within the Memory Engram. Cell 181, 410–423.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poulos A. M. et al. Conditioning- and time-dependent increases in context fear and generalization. Learn. Mem. 23, 379–385 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Besnard A. & Sahay A. Adult Hippocampal Neurogenesis, Fear Generalization, and Stress. Neuropsychopharmacology 41, 24–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bernstein E. E., van der Does F., Orr S. P. & McNally R. J. Poor Mnemonic Discrimination Predicts Overgeneralization of Fear. J. Psychopathol. Behav. Assess. 43, 152–161 (2021). [Google Scholar]
- 49.Lovett-Barron M. et al. Dendritic Inhibition in the Hippocampus Supports Fear Learning. Science 343, 857–863 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Josselyn S. A. & Tonegawa S. Memory engrams: Recalling the past and imagining the future. Science 367, eaaw4325 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pettit N. L., Yap E.-L., Greenberg M. E. & Harvey C. D. Fos ensembles encode and shape stable spatial maps in the hippocampus. Nature 609, 327–334 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sweis B. M., Mau W., Rabinowitz S. & Cai D. J. Dynamic and heterogeneous neural ensembles contribute to a memory engram. Curr. Opin. Neurobiol. 67, 199–206 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leake J., Zinn R., Corbit L. H., Fanselow M. S. & Vissel B. Engram Size Varies with Learning and Reflects Memory Content and Precision. J. Neurosci. 41, 4120–4130 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stefanelli T., Bertollini C., Lüscher C., Muller D. & Mendez P. Hippocampal Somatostatin Interneurons Control the Size of Neuronal Memory Ensembles. Neuron 89, 1074–1085 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Rolotti S. V. et al. Local feedback inhibition tightly controls rapid formation of hippocampal place fields. Neuron 110, 783–794.e6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amer T. & Davachi L. Extra-hippocampal contributions to pattern separation. eLife 12, e82250 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raza S. A. et al. Fear engrams and NPYergic circuit in the dorsal dentate gyrus determine remote fear memory generalization. 2022.04.26.489543 Preprint at 10.1101/2022.04.26.489543 (2022). [DOI]
- 58.Xu W. & Südhof T. C. A Neural Circuit for Memory Specificity and Generalization. Science 339, 1290–1295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szabadics J. & Soltesz I. Functional Specificity of Mossy Fiber Innervation of GABAergic Cells in the Hippocampus. J. Neurosci. 29, 4239–4251 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brzdąk P., Lebida K., Wyroślak M. & Mozrzymas J. W. GABAergic synapses onto SST and PV interneurons in the CA1 hippocampal region show cell-specific and integrin-dependent plasticity. Sci. Rep. 13, 5079 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu S. J. et al. Cortical somatostatin interneuron subtypes form cell-type-specific circuits. Neuron 111, 2675–2692.e9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quadros R. M. et al. Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol. 18, 92 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chatterjee D. et al. Enhanced CNS transduction from AAV.PHP.eB infusion into the cisterna magna of older adult rats compared to AAV9. Gene Ther. 29, 390–397 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fenno L. E. et al. INTRSECT: single-component targeting of cells using multiple-feature Boolean logic. Nat. Methods 11, 763–772 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trivedi V., Choi H. M. T., Fraser S. E. & Pierce N. A. Multidimensional quantitative analysis of mRNA expression within intact vertebrate embryos. Development 145, dev156869 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.