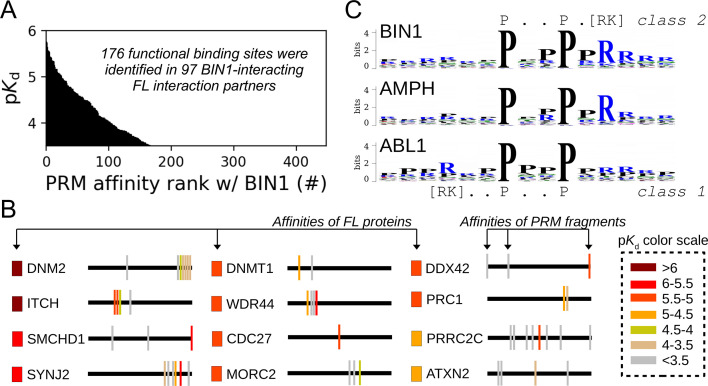
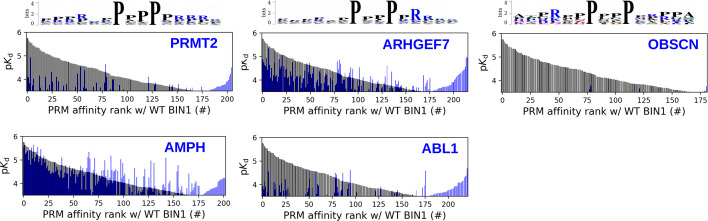
Figure 5. Affinity measurements between the SH3 domain of BIN1 and isolated PRM fragments.
(A) Affinity profile of BIN1_SH3 measured using fragmentomic holdup against 448 synthetic PRMs found in FL interaction partners previously identified by nHU-MS. 176 PRMs were found to bind to BIN1 displaying affinities ranging from low micromolar to a few hundreds of micromolar dissociation constants. These motifs were found in 97 proteins, matching at least a single functional binding site for half of the identified FL interaction partners. (B) The combination of native and fragmentomic holdup reveals biophysical properties of FL proteins and elementary binding sites. The measured affinities of intact proteins are indicated with colored boxes and site-specific affinities of individual PRMs are indicated with colored spikes, where colors were adjusted to measured steady-state affinities of FL proteins and PRM sites, respectively. Note that the protein schemes are not to scale to the actual protein length, but are the approximate relative positions of indicated PRMs. (C) Affinity-weighted specificity logo of the SH3 domains of BIN1, AMPH, and ABL1. BIN1 and AMPH nearly uniquely interacts with class 2 PxxP motifs, while ABL1 prefers to bind class 1 PxxP motifs. See Figure 5—figure supplement 1 and Supplementary file 2 for further details.