Abstract
The transmembrane (TM) glycoprotein gp41 of human immunodeficiency virus type 1 possesses an unusually long (∼150 amino acids) and highly conserved cytoplasmic region. Previous studies in which this cytoplasmic tail had been deleted partially or entirely have suggested that it is important for virus infectivity and incorporation of the gp120-gp41 glycoprotein complex into virions. To determine which regions of the conserved C-terminal domains are important for glycoprotein incorporation and infectivity, several small deletions and amino acid substitutions which modify highly conserved motifs were constructed in the infectious proviral background of NL4.3. The effects of these mutations on infectivity and glycoprotein incorporation into virions produced from transfected 293-T cells and infected H9 and CEM×174 cells were determined. With the exception of a mutation deleting amino acids QGL, all of the constructs resulted in decreased infectivity of the progeny virus both in a single-round infectivity assay and in a multiple-infection assay in H9 and CEM×174 cells. For most mutations, the decreased infectivity was correlated with a decreased incorporation of glycoprotein into virions. Substitution of the arginines (residues 839 and 846) with glutamates also reduced infectivity, but without a noticeable decrease in the amount of glycoprotein incorporated into virus produced from infected T cells. These results demonstrate that minor alterations in the conserved C-terminal region of the gp41 cytoplasmic tail can result in reductions in infectivity that correlate for most but not all constructs with a decrease in glycoprotein incorporation. Observed cell-dependent differences suggest the involvement of cellular factors in regulating glycoprotein incorporation and infectivity.
For human immunodeficiency virus type 1 (HIV-1), the presence of the envelope glycoproteins in virus particles is essential for infectivity. The env gene encodes the envelope glycoproteins, which are synthesized, as in other retroviruses, as a polypeptide precursor (gp160). The precursor is enzymatically cleaved into the surface subunit (SU, gp120) and the transmembrane subunit (TM, gp41), which remain noncovalently associated (36). The main function of SU is to facilitate the initial steps of virus attachment by interacting with the major cellular receptor molecule (CD4) and a coreceptor molecule. These interactions trigger conformational changes within the ectodomain of TM which allow fusion to occur between the viral envelope and the host cell membrane (2, 4, 17, 19).
The TM proteins of HIV and other lentiviruses possess, in contrast to all other retrovirus genera, an unusually long and highly conserved cytoplasmic region (tail) of ∼150 amino acids. The exact function of this long cytoplasmic tail is not clearly understood, although it is believed to have some important function in vivo, as only one infectious HIV clone with a truncated cytoplasmic tail has been isolated to date (28, 29). Mutational studies from several laboratories (6, 12, 15, 22, 23, 38) in which truncations and deletions of various lengths were introduced into the cytoplasmic tail of gp41 have indicated, with one exception (35), that this region is important for infectivity. The decreased infectivity of mutants with truncated cytoplasmic tail regions was postulated to be due in some cases to reduced glycoprotein incorporation into virus particles (12, 38).
Several studies demonstrated that the HIV glycoproteins are incorporated into virus particles via an interaction with the Gag matrix (MA) domain (3, 9, 10, 13, 14, 24, 37). Residues within the N-terminal region of MA, which are important for this interaction, were identified by Freed and Martin (14), but the exact region of the gp41 cytoplasmic tail involved is still controversial. While Freed and Martin (13) implicated the region of gp41 between amino acids 761 and 791, also referred to as helix 2, Cosson (9) demonstrated, using a direct binding assay, that truncation of the most C-terminal 20 amino acids of gp41 (834 to 854) was sufficient to abrogate the interaction with MA.
The only structural information available for the cytoplasmic tail of gp41 to date are computer predictions which indicate the presence of two amphipathic regions between amino acids 772 and 790 and amino acids 828 and 848 (34). These two predicted amphipathic helices are also referred to as helices 2 and 1 or lentiviral lytic peptides 2 and 1 (LLP-2 and -1), respectively. Extensive work using synthetic peptides comprising these regions demonstrated that they are able to bind (20, 31, 33) and perturb (1, 7, 8, 25) membranes and also interact with calmodulin (30, 32). The role of these regions in the context of the entire glycoprotein is not yet fully understood. Studies have, however, reported results similar to those observed with the synthetic peptides, including association with cell membranes (16) and interaction with calmodulin (18, 27).
Here we used a mutational analysis to determine whether highly conserved domains within the cytoplasmic tail of HIV-1 gp41 play critical roles in infectivity and glycoprotein incorporation. Several highly conserved amino acids within the LLP-1 region of the gp41 cytoplasmic tail were deleted or replaced, and the effects of these mutations on glycoprotein incorporation into virus and virus infectivity in the lymphoid cell lines H9 and CEM×174 were assessed. Our results indicate that some of the conserved C-terminal amino acids within the LLP-1 region play an important role in glycoprotein incorporation and affect infectivity. Further, we suggest an additional role for this region of the gp41 cytoplasmic tail during infection since the reduced infectivity of some constructs could not be explained solely by reduced glycoprotein incorporation.
MATERIALS AND METHODS
Cell culture.
293-T cells were obtained from the American Type Culture Collection (Manassas, Va.) and maintained in Dulbecco's modified Eagle medium (GIBCO, Grand Island, N.Y.) containing 10% fetal bovine serum, l-glutamine (2 mM), penicillin G (100 U/ml), and streptomycin sulfate (0.1 mg/ml). MAGI-X4 cells (HeLa-CD4-LTR-β-galactosidase and CXCR4 coreceptor) were maintained in the same medium as 293-T cells, with the addition of G418 sulfate (0.2 mg/ml) and hygromycin B (0.1 mg/ml). The lymphoid cell lines CEM×174 and H9 were maintained in RPMI 1640 (GIBCO) containing 10% fetal bovine serum, l-glutamine (2 mM), penicillin G (100 U/ml), and streptomycin sulfate (0.1 mg/ml). The T-cell lines and MAGI-X4 cells were obtained through the AIDS Reference and Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases.
Mutagenesis.
Mutations in the cytoplasmic tail region of gp41 were generated in the HXB2 background using PCR mutagenesis or the Altered Sites mutagenesis system (Promega, Madison, Wis.). The 422-bp BamHI-XhoI fragment of HXB2 was subcloned into the pNL4.3 infectious provirus. As all mutations were generated in the HXB2 background, the WT (wild-type) construct was generated by subcloning the HXB2 BamHI-XhoI fragment into the NL4.3 background, which differs only by four amino acids (Fig. 1), and all results were normalized to WT. All mutations were verified by DNA sequencing and tested for normal expression of viral proteins in 293-T cells.
FIG. 1.
Amino acid sequences of constructs used in this study. Mutations in the highly conserved C-terminal region of gp41 of HIV-1 were engineered in the HXB2 background, and the BamHI-XhoI fragment was then subcloned into the proviral pNL4.3 clone (see Materials and Methods for details). The top depicts the consensus sequence of clade B viruses starting at the BamHI site, where uppercase letters indicate 100% conserved residues, lowercase letters depict 50% conserved residues, and ? denotes variable amino acids (17). The C-terminal region starting with amino acid 811 (NL4.3 numbering, equivalent to amino acid 813 in HXB2) is depicted for NL4.3 as well as for all mutants. An asterisk represents the conversion of that amino acid to a stop codon, a dash represents a deletion of the corresponding amino acid, and bold letters indicate amino acid substitutions. Note that there are only four amino acid differences in the entire BamHI-XhoI fragment between the NL4.3 sequence and the HXB2 sequence (highlighted in bold in WT).
Transfections.
Purified proviral DNA (10 μg) was used to transfect 293-T cells at 50 to 70% confluency grown in 100-mm-diameter culture plates with a modified calcium phosphate method as described previously (5).
Metabolic labeling and immunoprecipitation.
293-T cells were metabolically labeled 36 to 48 h posttransfection. After starvation in cysteine-methionine-deficient medium for 30 min at 37°C, the cells were labeled for 30 min at 37°C using 1 mCi of [35S]methionine-cysteine protein labeling mix (Dupont, NEN). After a chase period in 20% complete medium for 12 to 16 h, cells were washed in phosphate-buffered saline (PBS) and lysed in Berman lysis buffer (1% Nonidet P-40, 0.1% sodium dodecyl sulfate [SDS], 0.5% deoxycholate, phenylmethylsulfonyl fluoride, and aprotinin in PBS). After pelleting of nuclei at 14,000 rpm, cleared supernatants were immunoprecipitated in two steps, using first HIV-negative human serum and then HIV-positive patient sera. Staphylococcus aureus fixed cells were used for the precipitation. Immunoprecipitated proteins bound to the cells were washed three times in wash buffer (1% Nonidet P-40–0.1% SDS in PBS) and once in 20 mM Tris (pH 6.8) before denaturation in protein loading buffer (50 mM Tris [pH 6.8], 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, 10% glycerol) and separation by SDS-polyacrylamide gel electrophoresis (PAGE) on an 8% gel.
Glycoprotein incorporation analysis.
The amount of glycoprotein incorporated into virus particles obtained from transfected 293-T cells or from H9 cells infected with NL4.3-based constructs was analyzed. Transfected or infected cells were metabolically labeled as described above. After a 12- to 16-h chase in 20% complete medium, supernatants were collected, filtered (0.45-μm-pore-size filter), and pelleted through a 20% (wt/wt) sucrose cushion at 100,000 × g (Ti 70.1 rotor; Beckman) for 2 h. The virus pellet was resuspended in 0.5 ml of Berman lysis buffer and immunoprecipitated with AIDS patient sera (1:1,000) as described above. After separation by SDS-PAGE, gels (8%) were enhanced (Enhance, Dupont, NEN) and dried. Dried gels were exposed to film at −80°C and to phosphor screens at room temperature. A PhosphorImager (Molecular Dynamics) and the software ImageQuant were used to quantitate the ratio of incorporated gp120 or gp41 to p24. Results for mutants are expressed as percentage of WT incorporation.
Infectivity assay.
To determine the ability of the NL4.3-based mutant constructs to infect lymphoid cell lines (H9 and CEM×174), supernatants of transiently transfected 293-T cells were collected and filtered (0.45-μm-pore-size filter) 36 to 48 h after transfection. Virus content in filtered supernatants was quantitated using a reverse transcriptase (RT) assay as described previously (11). Virus-containing supernatants were normalized for RT activity, and 10,000 RT units were used to infect 2 × 106 to 3 × 106 cells for 4 h. Experiments were performed in triplicate for each construct. Cells were cultured and every 3 to 4 days were counted with a hemocytometer and either split or fed to maintain the same number of cells for all constructs. Supernatant samples (1 ml) were collected and frozen at −80°C on days when cells were split. All collected supernatants were analyzed for RT activity at the end of the experiment. The infection rate index was determined as (day of peak RT activity of WT)/(day of peak RT activity of mutant).
One-step infectivity assay in MAGI-X4 cells.
Supernatants from either transiently transfected 293-T cells or infected H9 cells were normalized for RT activity as above and used to infect 0.8 × 105 MAGI-X4 cells/well in 24-well plates for 12 h. Cells were washed after 12 h, and new medium was added. After 48 to 60 h, cells were fixed with 1% formaldehyde–0.2% glutaraldehyde in PBS and stained for β-galactosidase. The number of blue foci was determined either for the whole well or for at least three fields of view per well. Results for all constructs are expressed as percentage of the number of blue foci counted for WT.
RESULTS
Construction of mutants.
The alterations introduced in the highly conserved cytoplasmic tail of gp41 from HIV-1 are depicted in Fig. 1. These mutations were designed to determine the role of conserved domains in the LLP-1 region of the cytoplasmic tail in infectivity and glycoprotein incorporation into virus particles. WT was constructed to control for the four amino acid changes in the BamHI-XhoI fragment between NL4.3 and HXB2. Constructs Δ6 and Δ19 have been described and studied previously by this laboratory in the HXB2.BH10 background (12), which, in contrast to the NL4.3 provirus used here, lacks Vpr and Nef. The other constructs either deleted (ΔQGL, ΔPRRIR, and ΔSD1) or altered (RR/KK and RR/EE) highly conserved motifs within the LLP-1 region without altering the rev open reading frame. The RR/KK mutation replaced the two adjacent positively charged arginines of the PRRIR motif with positively charged lysine residues. The RR/EE mutation, which replaces the conserved positively charged arginine residues at positions 841 and 848 (HXB2 numbering, equivalent to residues 839 and 846, respectively, in NL4.3) with negatively charged glutamates, has so far been described only in peptide studies, where it is referred to as analog 3 (25).
Mutations in conserved regions of the cytoplasmic domain of gp41 affect the incorporation of glycoprotein into virus particles in transfected 293-T cells.
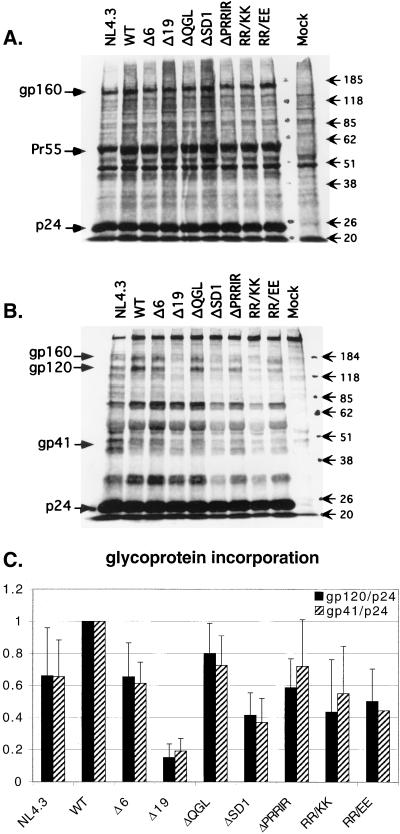
The mutations in the proviral NL4.3 constructs were evaluated for their effects on glycoprotein incorporation. Transiently transfected 293-T cells were metabolically labeled, and cell lysates as well as viral pellets were separated by SDS-PAGE (8% gel) after immunoprecipitation. Typical examples of the separated cell lysate and viral pellet proteins are shown in Fig. 2A and B, respectively. Similar levels of glycoprotein expression were observed in cell lysates of all constructs (Fig. 2A). Some gp160 was observed in the viral pellets of all constructs (Fig. 2B). This appears to be a property of transfected 293-T cells, since we generally observe less of this protein in virions released from COS-1 or T-cell lines. The absence of p25 in the virus pellets argues against cellular contamination of these samples. Glycoprotein incorporation into virions was quantitated as gp120/p24 or gp41/p24 by phosphorimage analysis and normalized to WT levels for each experiment (Fig. 2C). Importantly, both calculations of glycoprotein incorporation, gp120/p24 and gp41/p24, showed very similar results for each mutant, suggesting that shedding of gp120 from the surface was not altered by the mutations. This was also confirmed by equivalent amounts of gp120 observed in the supernatants (data not shown). It is worth noting that even conservative changes such as RR/KK as well as deletions of some conserved regions within the C-terminal cytoplasmic tail are able to significantly decrease the glycoprotein incorporation. The largest decrease of incorporated glycoprotein, observed for Δ19, is most likely due to the instability of this protein as described previously (12). The deletion of the conserved motif QGL had the least effect on glycoprotein incorporation, while the other constructs exhibited reduced glycoprotein incorporation at 40 to 60% of the WT level, with ΔSD1 consistently having the largest reduction in glycoprotein incorporation, followed by RR/KK and RR/EE. It is worth noting that NL4.3 had consistently lower glycoprotein incorporation compared to WT, although these constructs differ by only four amino acids in the gp41 cytoplasmic tail (Fig. 1). With the exception of Δ19, where decreased surface expression of Env was observed by flow cytometry, glycoprotein incorporation did not correlate with decreased surface expression of the proteins (data not shown).
FIG. 2.
Effects of mutations in the gp41 cytoplasmic tail on glycoprotein incorporation in transfected 293-T cells. Immunoprecipitated proteins from cell lysates (A) and from viral pellets from the same experiment (B) are shown on an SDS–8% polyacrylamide gel. The gp120/p24 and gp41/p24 ratios were calculated after phosphorimaging. (C) Quantitation of glycoprotein incorporation, normalized to WT in each experiment. Data represent averages of 8 to 13 experiments for gp120/p24 and 6 to 9 experiments for gp41/p24. Error bars show standard deviations. Sizes are indicated in kilodaltons.
Mutations in conserved regions of the cytoplasmic domain of gp41 affect infectivity in H9 and CEM×174 cell lines.
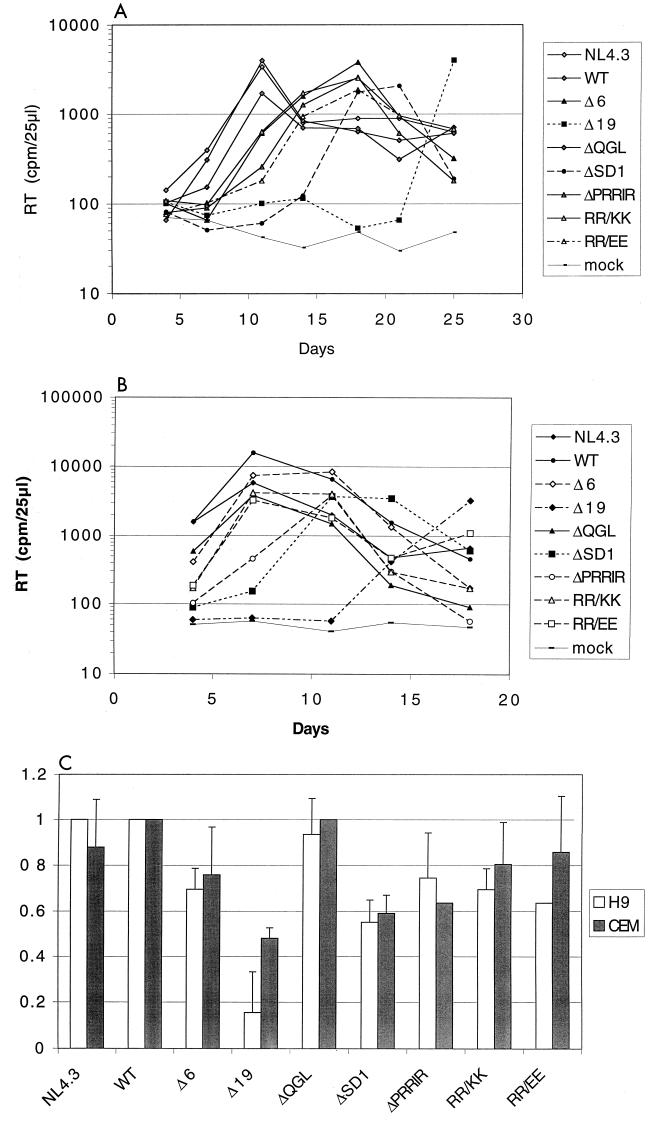
Supernatants from transfected 293-T cells, normalized for RT activity, were used to infect the T-cell lines H9 and CEM×174 to determine the ability of the mutants to establish a productive infection. RT activity was determined when cells were split every 3 to 4 days; typical results for H9 and CEM×174 cells are shown in Fig. 3A and B.
FIG. 3.
Effects of mutations in the gp41 cytoplasmic tail on infectivity of T-cell lines. Supernatants from transfected 293-T cells were normalized for RT activity and used to infect 3 × 106 cells. Cells were split on the days indicated, and RT activity of supernatants was determined. Experiments were performed twice in triplicate; typical results are shown for H9 (A) and CEM×174 (B). (C) Infection rate index, determined as (day of peak RT activity of WT)/(day of peak RT activity of mutant). Averages and standard deviations are shown. Missing standard deviations indicate that all cultures peaked at the same day, resulting in a standard deviation of zero.
In H9 cells, a delayed peak of infection was observed for all mutants except ΔQGL. Mutant ΔSD1 consistently exhibited the longest delay (10 days) in reaching peak RT levels, whereas Δ6, ΔPRRIR, RR/KK, and RR/EE had intermediate delays (∼6 days). The Δ19 mutant established a productive infection in only 5 out of 10 cultures.
It was noted in an initial experiment that the CEM×174 cell line was by far more susceptible to infection with HIV (NL4.3) and that the infection spread more rapidly. When the same amount of virus input (10,000 cpm) was used as for H9 cells, all mutants reached a peak RT on the same day (day 3 [not shown]). In subsequent experiments, a 1:20 dilution of input virus (RT of 500 cpm) was used for infection of CEM×174 cells. Under these conditions, a delayed-infectivity phenotype similar (albeit compressed) to that observed in the H9 cell line was observed for the mutants (compare Fig. 3A and B). Again the RT activity of ΔQGL peaked at the same day as those of WT and NL4.3; activities of Δ6, ΔPRRIR, RR/KK, and RR/EE were slightly delayed, and that of ΔSD1 was most delayed, in reaching peak values. In contrast to the H9 cells, Δ19 was able to cause a productive infection in all infected CEM×174 cultures, but RT levels similar to the WT level were observed only after more than 18 days of culture. To correct for differences between experiments, an infection rate index was calculated and normalized to WT (Fig. 3C). Despite the faster replication kinetics of CEM×174 cells, the infection rate indices were comparable for most constructs in both H9 and CEM×174 cells. For both cell types, minor sequence changes or small deletions in the cytoplasmic tail of gp41 caused a decrease in infectivity which, for most constructs, appeared to correlate to the amount of glycoprotein incorporated into virus particles produced from transfected 293-T cells (compare Fig. 3C and 2C). These effects were not limited to lymphoid cell lines, since similar delays in infectivity for ΔSD1 were also observed in peripheral blood mononuclear cells (not shown).
Mutations in conserved regions of the cytoplasmic domain of gp41 affect infectivity in a single-round assay in MAGI-X4 cells.
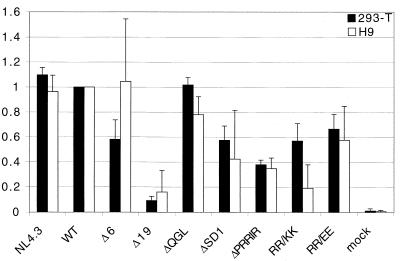
To determine the infectivity of mutants in a single-round assay, supernatants from transfected 293-T cells and from infected H9 cells, normalized for RT activity, were used to infect MAGI-X4 cells. The number of infected cells as indicated by staining for β-galactosidase was determined and normalized to WT (Fig. 4). In general, the infectivities of each of the different mutants in this single-round assay were consistent irrespective of the source of the infectious supernatants (from transfected 293-T cells or infected H9 cells). The mutations had effects on single-round infectivity similar to those observed for multiple-round infectivity assays in H9 cells, with the exceptions of ΔPRRIR and RR/KK, which were both more defective in the single-round assay (compare Fig. 4 and 3C). The results obtained with supernatants from 293-T cells further correlated with the glycoprotein incorporation determined for virus released from transfected 293-T cells, with two exceptions. Construct ΔPRRIR had a decreased ability to replicate in the single-round assay despite near-normal glycoprotein incorporation, while NL4.3 had decreased glycoprotein incorporation but replicated like WT in the single-round assay (Fig. 2C). The results shown for virus derived from H9 cells were obtained using supernatants at the time of peak RT levels for all mutants including Δ19 and do not include supernatants of cells that had not been productively infected. Despite this fact, the Δ19 virus from supernatants at late time points remained defective for infection in a single-round assay. It is also worth noting that mutants ΔPRRIR, RR/KK, and RR/EE were more infectious than ΔSD1 in the multiple-infection assay in both H9 and CEM×174 cells, whereas this was not the case in the single-round MAGI assay.
FIG. 4.
Effects of mutations in the gp41 cytoplasmic tail on infectivity in a single-round assay using MAGI-X4 cells. Supernatants from transfected 293-T or infected H9 cells at the peak of RT production were normalized for RT activity and used to infect MAGI-X4 cells. Cells were stained for β-galactosidase 48 h after infection. Blue foci were counted either as the total number per well or in three fields of view per well, and results are compared to WT. Data represent results from at least two independent transfections/infections, and triplicates for each mutant were analyzed. Error bars show standard deviations.
Mutations in conserved regions of the cytoplasmic domain of gp41 affect the incorporation of glycoprotein into virus particles in infected H9 cells.
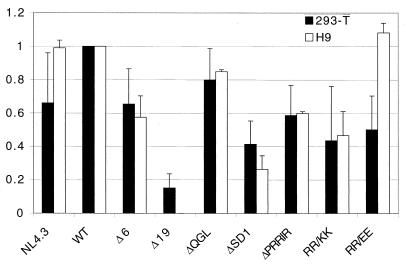
In the multiple-infection assay, only the first round of infection was caused by virus particles produced in transfected 293-T cells (for which the glycoprotein incorporation had been determined [Fig. 2C]), while the following rounds of infections were initiated by virus produced in H9 or CEM×174 cells (not shown). Although there was general agreement between the results from the multiple-infection assay and the single-round assay for most constructs, we wanted to rule out that the minor differences observed for ΔPRRIR, RR/KK, and RR/EE were a reflection of differences in glycoprotein incorporation into virus particles depending on the cell line in which the virus was produced. We therefore metabolically labeled infected H9 cells and quantitated the amount of glycoprotein incorporated into virus particles produced at peak of infection for each mutant in the same way as described for transfected 293-T cells (see Materials and Methods). Comparison of the amounts of glycoprotein incorporated into virus particles originating from 293-T and H9 cells quantitated as gp120/p24 is shown in Fig. 5. With two exceptions, levels of glycoprotein incorporation were similar irrespective of the source of virus. NL4.3 had consistently lower glycoprotein incorporation than WT in transfected 293-T cells, while there was no distinguishable difference in infected H9 cells. Glycoprotein from RR/EE, on the other hand, was incorporated at WT levels into virus from infected cells but at only about 50% of the WT level in transfected 293-T cells. Interestingly, the other two of the three mutations that exhibited the largest difference between results in the long-term and single-round infectivity assays (ΔPRRIR and RR/KK) showed very little variation of glycoprotein incorporation into particles when produced in different cell lines, suggesting that those differences in infection were not due to variations in glycoprotein incorporation.
FIG. 5.
Amount of glycoprotein incorporated into virus particles for each construct (quantitated as gp120/p24 as described in Materials and Methods) derived from transfected 293-T cells or infected H9 cells. Data for 293-T cells are the same as in Fig. 2C; data for H9 cells are averages from two independent experiments, with error bars indicating ranges.
DISCUSSION
We have previously determined the effects of larger truncations of the gp41 cytoplasmic tail on infectivity and suggested that the observed decreased infectivity may have been due in part to inefficient glycoprotein incorporation into virus particles (12). In the present study we focused on several highly conserved, small regions within the C-terminal 37 amino acids of gp41 to determine their role in glycoprotein incorporation and infectivity. The small deletions and amino acid substitutions introduced in this region of the cytoplasmic tail had measurable effects on both glycoprotein incorporation and infectivity.
No significant effect on either glycoprotein incorporation or infectivity was observed for the deletion of the invariant three amino acids, QGL, indicating that these residues are dispensable for the investigated functions of gp41.
The majority of mutations displayed a decreased infectivity which correlated somewhat with reduced glycoprotein incorporation. These included Δ6, Δ19, ΔSD1, ΔPRRIR, RR/KK, and RR/EE if one compares glycoprotein incorporation into virus from transfected 293-T cells and the infection rate index in the multiple- round assay. Of these mutations, however, ΔPRRIR and RR/KK exhibited variable effects in the infectivity assay that differed between the single-round MAGI assay and the multiple-round assay. Both mutations were less infectious in the single-round MAGI assay than in the multiple-infection assay. This did not appear to be due to altered glycoprotein incorporation in H9 cells compared to that of 293-T cells. The lack of efficient infection in the MAGI assay was neither correlated with glycoprotein incorporation nor reflected in decreased infectivity in the multiple-round assay, suggesting that this effect may have been specific to the MAGI-X4 cells.
The only constructs where the effect on glycoprotein incorporation differed from the effect on infectivity were NL4.3 and RR/EE. Despite the fact that the NL4.3 gp41 fragment differs from that of WT by only four amino acids (Fig. 1), glycoprotein incorporation for NL4.3 in transfected 293-T cells was consistently lower than that of WT and was at a level equivalent to that observed for some of the replication-defective mutants (Δ6 and ΔPRRIR). Nevertheless, no reduction in infectivity was observed in either infectivity assay. Moreover, the kinetics of infectivity were similar for NL4.3 and WT in both H9 and CEM×174 cells even when 50-fold dilutions of virus input were used to infect CEM×174 cells (data not shown). The unaltered infectivity kinetics of NL4.3 in comparison to WT that we have observed here is in agreement with observations by Wilk and colleagues (35) that subcloning of the equivalent env fragment (BamHI-XhoI) from BH10 to NL4.3 had no effect on infection kinetics.
For the mutant RR/EE, glycoprotein incorporation in H9 cells was equivalent to WT, yet the infectivity of the virus was delayed in both assays. It is interesting that the equivalent mutation to RR/EE in synthetic peptide studies (referred to as analog 3) had been shown to block calmodulin binding as well as the pore-forming ability of LLP-1. Taken together the results from these two constructs (NL4.3 and RR/EE) clearly demonstrate that glycoprotein incorporation is not the only factor contributing to differences in infectivity. This further suggests that mediating glycoprotein incorporation and infectivity may be two separate functions of the cytoplasmic tail governed by different regions within the tail. Despite the general correlation between the effects of most mutants on glycoprotein incorporation and infectivity, the fact that in two constructs infectivity was not explained by glycoprotein incorporation strongly suggests that the cytoplasmic tail may have other important functions influencing infectivity. A similar discrepancy between glycoprotein incorporation and infectivity had previously been observed for a C-terminal 12-amino-acid deletion of the gp41 cytoplasmic tail (38).
While other studies have previously implicated the importance of the cytoplasmic tail of gp41 in infectivity (6, 12, 38) and cytopathicity (23), this is the first report investigating the role of highly conserved domains of the C terminus of the cytoplasmic tail. Taken together, our results indicate that the C-terminal region of gp41 plays an important modulatory role in both glycoprotein incorporation and infectivity. However, these two events (glycoprotein incorporation and infectivity) may not be linked, at least for some of our constructs. Recently, Freed and Martin (13) suggested that helix 2 of the cytoplasmic tail is involved in the interaction of TM with MA which facilitates glycoprotein incorporation, at least in the presence of a long (not truncated) cytoplasmic tail. Data presented here support a role for the more C-terminal region of gp41 in glycoprotein incorporation, which is further strengthened by direct binding studies with purified glutathione S-transferase–gp41 and glutathione S-transferase–MA (9), as well as by similar observations for simian immunodeficiency virus (S. A. González, personal communication; unpublished data).
With two notable exceptions, the results obtained with virus from 293-T and H9 cells were similar for all assays. The exceptions were for glycoprotein incorporation into the NL4.3 and RR/EE virions. For both constructs, less glycoprotein was consistently incorporated into virus from transfected 293-T cells than into virus from infected H9 cells. This result suggests that cell-specific factors are involved in modulating glycoprotein incorporation. Although we can not exclude the possibility that second-site mutations that occurred during the infection of H9 cells may have altered the glycoprotein incorporation, this is unlikely given that the results are averages from two independent transfections and infections. Moreover, for RR/EE the increased Env incorporation was not accompanied by an increase in virus infectivity. A recent study by Murakami and Freed (26) also revealed cell-specific differences in glycoprotein incorporation of molecules in which the cytoplasmic tail of gp41 was truncated. This is presumably due to host cell factors which need to be identified in future studies.
A pronounced reduction of glycoprotein incorporation upon deletion of the conserved region of PRRIR or replacement of the central RR with KK was observed. The fact that the deletion of the adjacent amino acids QGL had no effect suggests that the PRRIR sequence may be directly involved in the process facilitating the glycoprotein incorporation and/or that the three-dimensional structure rather than the linear sequence is important for the function of the cytoplasmic tail. These findings hence support earlier reports suggesting that specific sequences or conformations may be more important than the length of the cytoplasmic tail (23). Based on early computer predictions, Venable and colleagues (34) proposed that the two amphipathic helices of the cytoplasmic tail may be traversing the bilayer. Recent studies with synthetic peptides of LLP-1 suggest that the amphipathic helix is inserted in the lipid phase parallel to the lipid/water interface (20, 21). In such a conformation the charged arginines of the PRRIR motif would be exposed on the hydrophilic side, either interacting with the lipid headgroups or being available for interaction with other proteins. This could explain why the mutation of the PRRIR region had a greater effect than the mutation of the adjacent QGL region which may be embedded in the lipid membrane.
The interpretation of these and other mutational studies of the gp41 cytoplasmic tail will become more apparent once a detailed structure for this region of TM becomes available. We are currently assessing the involvement of these C-terminal regions in functions that have been attributed to the corresponding synthetic LLP-1 peptide, such as calmodulin binding and membrane perturbation, in order to shed light on the controversial role of this conserved region of gp41.
ACKNOWLEDGMENTS
This work was supported by research grant AI33319 from the National Institute of Health to E.H.
MAGI-X4, H9, and CEM×174 cells were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases. We thank Silvia González for critical review of the manuscript, and we thank Susan Dubay and Tshana Thomas for excellent technical assistance.
REFERENCES
- 1.Arroyo J, Boceta M, Gonzalez M E, Michel M, Carrasco L. Membrane permeabilization by different regions of the human immunodeficiency virus type 1 transmembrane glycoprotein gp41. J Virol. 1995;69:4095–4102. doi: 10.1128/jvi.69.7.4095-4102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Binley J, Moore J P. HIV-cell fusion. The viral mousetrap. Nature. 1997;387:346–348. doi: 10.1038/387346a0. [DOI] [PubMed] [Google Scholar]
- 3.Bugelski P J, Maleeff B E, Klinkner A M, Ventre J, Hart T K. Ultrastructural evidence of an interaction between Env and Gag proteins during assembly of HIV type 1. AIDS Res Hum Retroviruses. 1995;11:55–64. doi: 10.1089/aid.1995.11.55. [DOI] [PubMed] [Google Scholar]
- 4.Chan D C, Kim P S. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 5.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen S S, Ferrante A A, Terwilliger E F. Characterization of an envelope mutant of HIV-1 that interferes with viral infectivity. Virology. 1996;226:260–268. doi: 10.1006/viro.1996.0654. [DOI] [PubMed] [Google Scholar]
- 7.Chernomordik L, Chanturiya A N, Suss-Toby E, Nora E, Zimmerberg J. An amphipathic peptide from the C-terminal region of the human immunodeficiency virus envelope glycoprotein causes pore formation in membranes. J Virol. 1994;68:7115–7123. doi: 10.1128/jvi.68.11.7115-7123.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comardelle A M, Norris C H, Plymale D R, Gatti P J, Choi B, Fermin C D, Haislip A M, Tencza S B, Mietzner T A, Montelaro R C, Garry R F. A synthetic peptide corresponding to the carboxy terminus of human immunodeficiency virus type 1 transmembrane glycoprotein induces alterations in the ionic permeability of Xenopus laevis oocytes. AIDS Res Hum Retroviruses. 1997;13:1525–1532. doi: 10.1089/aid.1997.13.1525. [DOI] [PubMed] [Google Scholar]
- 9.Cosson P. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 1996;15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 10.Dorfman T, Mammano F, Haseltine W A, Gottlinger H G. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1994;68:1689–1696. doi: 10.1128/jvi.68.3.1689-1696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubay J W, Roberts S J, Brody B, Hunter E. Mutations in the leucine zipper of the human immunodeficiency virus type 1 transmembrane glycoprotein affect fusion and infectivity. J Virol. 1992;66:4748–4756. doi: 10.1128/jvi.66.8.4748-4756.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubay J W, Roberts S J, Hahn B H, Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J Virol. 1992;66:6616–6625. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freed E O, Martin M A. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freed E O, Martin M A. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J Virol. 1995;69:1984–1989. doi: 10.1128/jvi.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabuzda D H, Lever A, Terwilliger E, Sodroski J. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 1992;66:3306–3315. doi: 10.1128/jvi.66.6.3306-3315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haffar O K, Dowbenko D J, Berman P W. The cytoplasmic tail of HIV-1 gp160 contains regions that associate with cellular membranes. Virology. 1991;180:439–441. doi: 10.1016/0042-6822(91)90054-f. [DOI] [PubMed] [Google Scholar]
- 17.Hunter E. gp41, a multifunctional protein involved in HIV entry and pathogenesis, p. iii-55–iii-73. In: Korber B, Hahn B, Foley B, Mellors J W, Leitner T, Myers G, McCutchan F, Kuiken C L, editors. Human retroviruses and AIDS 1997: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1997. [Google Scholar]
- 18.Ishikawa H, Sasaki M, Noda S, Koga Y. Apoptosis induction by the binding of the carboxyl terminus of human immunodeficiency virus type 1 gp160 to calmodulin. J Virol. 1998;72:6574–6580. doi: 10.1128/jvi.72.8.6574-6580.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kilby J M, Hopkins S, Venetta T M, DiMassimo B, Cloud G A, Lee J Y, Alldredge L, Hunter E, Lambert D, Bolognesi D, Matthews T, Johnson M R, Nowak M A, Shaw G M, Saag M S. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 20.Koenig B W, Bergelson L D, Gawrisch K, Ward J, Ferretti J A. Effect of the conformation of a peptide from gp41 on binding and domain formation in model membranes. Mol Membr Biol. 1995;12:77–82. doi: 10.3109/09687689509038499. [DOI] [PubMed] [Google Scholar]
- 21.Koenig B W, Ferretti J A, Gawrisch K. Site-specific deuterium order parameters and membrane-bound behavior of a peptide fragment from the intracellular domain of HIV-1 gp41. Biochemistry. 1999;38:6327–6334. doi: 10.1021/bi982800g. [DOI] [PubMed] [Google Scholar]
- 22.Kowalski M, Potz J, Basiripour L, Dorfman T, Goh W C, Terwilliger E, Dayton A, Rosen C, Haseltine W, Sodroski J. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science. 1987;237:1351–1355. doi: 10.1126/science.3629244. [DOI] [PubMed] [Google Scholar]
- 23.Lee S J, Hu W, Fisher A G, Looney D J, Kao V F, Mitsuya H, Ratner L, Wong-Staal F. Role of the carboxy-terminal portion of the HIV-1 transmembrane protein in viral transmission and cytopathogenicity. AIDS Res Hum Retroviruses. 1989;5:441–449. doi: 10.1089/aid.1989.5.441. [DOI] [PubMed] [Google Scholar]
- 24.Mammano F, Kondo E, Sodroski J, Bukovsky A, Gottlinger H G. Rescue of human immunodeficiency virus type 1 matrix protein mutants by envelope glycoproteins with short cytoplasmic domains. J Virol. 1995;69:3824–3830. doi: 10.1128/jvi.69.6.3824-3830.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller M A, Cloyd M W, Liebmann J, Rinaldo C R, Jr, Islam K R, Wang S Z, Mietzner T A, Montelaro R C. Alterations in cell membrane permeability by the lentivirus lytic peptide (LLP-1) of HIV-1 transmembrane protein. Virology. 1993;196:89–100. doi: 10.1006/viro.1993.1457. [DOI] [PubMed] [Google Scholar]
- 26.Murakami T, Freed E O. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc Natl Acad Sci USA. 2000;97:343–348. doi: 10.1073/pnas.97.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radding W, Pan Z Q, Hunter E, Johnston P, Williams J P, McDonald J M. Expression of HIV-1 envelope glycoprotein alters cellular calmodulin. Biochem Biophys Res Commun. 1996;218:192–197. doi: 10.1006/bbrc.1996.0034. [DOI] [PubMed] [Google Scholar]
- 28.Shimizu H, Hasebe F, Tsuchie H, Morikawa S, Ushijima H, Kitamura T. Analysis of a human immunodeficiency virus type 1 isolate carrying a truncated transmembrane glycoprotein. Virology. 1992;189:534–546. doi: 10.1016/0042-6822(92)90577-c. [DOI] [PubMed] [Google Scholar]
- 29.Shimizu H, Morikawa S, Yamaguchi K, Tsuchie H, Hachimori K, Ushijima H, Kitamura T. Shorter size of transmembrane glycoprotein of an HIV-1 isolate. AIDS. 1990;4:575–576. doi: 10.1097/00002030-199006000-00013. [DOI] [PubMed] [Google Scholar]
- 30.Srinivas S K, Srinivas R V, Anantharamaiah G M, Compans R W, Segrest J P. Cytosolic domain of the human immunodeficiency virus envelope glycoproteins binds to calmodulin and inhibits calmodulin-regulated proteins. J Biol Chem. 1993;268:22895–22899. [PubMed] [Google Scholar]
- 31.Srinivas S K, Srinivas R V, Anantharamaiah G M, Segrest J P, Compans R W. Membrane interactions of synthetic peptides corresponding to amphipathic helical segments of the human immunodeficiency virus type-1 envelope glycoprotein. J Biol Chem. 1992;267:7121–7127. [PubMed] [Google Scholar]
- 32.Tencza S B, Mietzner T A, Montelaro R C. Calmodulin-binding function of LLP segments from the HIV type 1 transmembrane protein is conserved among natural sequence variants. AIDS Res Hum Retroviruses. 1997;13:263–269. doi: 10.1089/aid.1997.13.263. [DOI] [PubMed] [Google Scholar]
- 33.Trommeshauser D, Galla H J. Interaction of a basic amphipathic peptide from the carboxyterminal part of the HIV envelope protein gp41 with negatively charged lipid surfaces. Chem Phys Lipids. 1998;94:81–96. doi: 10.1016/s0009-3084(98)00047-4. [DOI] [PubMed] [Google Scholar]
- 34.Venable R M, Pastor R W, Brooks B R, Carson F W. Theoretically determined three-dimensional structures for amphipathic segments of the HIV-1 gp41 envelope protein. AIDS Res Hum Retroviruses. 1989;5:7–22. doi: 10.1089/aid.1989.5.7. [DOI] [PubMed] [Google Scholar]
- 35.Wilk T, Pfeiffer T, Bosch V. Retained in vitro infectivity and cytopathogenicity of HIV-1 despite truncation of the C-terminal tail of the env gene product. Virology. 1992;189:167–177. doi: 10.1016/0042-6822(92)90692-i. [DOI] [PubMed] [Google Scholar]
- 36.Willey R L, Bonifacino J S, Potts B J, Martin M A, Klausner R D. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc Natl Acad Sci USA. 1988;85:9580–9584. doi: 10.1073/pnas.85.24.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu X, Yuan X, Matsuda Z, Lee T H, Essex M. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol. 1992;66:4966–4971. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu X, Yuan X, McLane M F, Lee T H, Essex M. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of Env proteins into mature virions. J Virol. 1993;67:213–221. doi: 10.1128/jvi.67.1.213-221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]