Abstract
Flock house virus (FHV), a positive-strand RNA animal virus, is the only higher eukaryotic virus shown to undergo complete replication in yeast, culminating in production of infectious virions. To facilitate studies of viral and host functions in FHV replication in Saccharomyces cerevisiae, yeast DNA plasmids were constructed to inducibly express wild-type FHV RNA1 in vivo. Subsequent translation of FHV replicase protein A initiated robust RNA1 replication, amplifying RNA1 to levels approaching those of rRNA, as in FHV-infected animal cells. The RNA1-derived subgenomic mRNA, RNA3, accumulated to even higher levels of >100,000 copies per yeast cell, compared to 10 copies or less per cell for 95% of yeast mRNAs. The time course of RNA1 replication and RNA3 synthesis in induced yeast paralleled that in yeast transfected with natural FHV virion RNA. As in animal cells, RNA1 replication and RNA3 synthesis depended on FHV RNA replicase protein A and 3′-terminal RNA1 sequences but not viral protein B2. Additional plasmids were engineered to inducibly express RNA1 derivatives with insertions of the green fluorescent protein (GFP) gene in subgenomic RNA3. These RNA1 derivatives were replicated, synthesized RNA3, and expressed GFP when provided FHV polymerase in either cis or trans, providing the first demonstration of reporter gene expression from FHV subgenomic RNA. Unexpectedly, fusing GFP to the protein A C terminus selectively inhibited production of positive- and negative-strand subgenomic RNA3 but not genomic RNA1 replication. Moreover, changing the first nucleotide of the subgenomic mRNA from G to T selectively inhibited production of positive-strand but not negative-strand RNA3, suggesting that synthesis of negative-strand subgenomic RNA3 may precede synthesis of positive-strand RNA3.
Flock house virus (FHV) is a positive-strand RNA virus whose small genome, high-level replication, and other characteristics make it an attractive model for analyzing positive-strand RNA virus replication and assembly (6). FHV is the best-studied member of the Nodaviridae, the family with the smallest genomes of any animal positive-strand RNA virus. The Nodaviridae family contains viruses infecting a variety of invertebrates and vertebrates, including some viruses that cause lethal neuropathologies in fish (9, 30, 31). FHV was originally isolated from the grass grub Costelytra zealandica, near the Flock House agricultural research station in Bulls, New Zealand (13). FHV also replicates robustly in Drosophila melanogaster cells, converting 20% of total cell protein to viral protein (14, 16), and directs RNA replication in mammalian cells (5).
The FHV genome consists of two RNAs, which are both packaged in a single, nonenveloped, icosahedral virion (17, 37). RNA1 (3.1 kb) serves as mRNA for protein A (112 kDa), which contains the Gly-Asp-Asp (GDD) amino acid motif characteristic of RNA polymerases and which is essential for FHV RNA replication (4, 15). FHV RNA2 (1.4 kb) encodes the capsid protein precursor (43 kDa) (11, 15). RNA2 thus replicates only in the presence of RNA1, while RNA1 replicates efficiently in the absence of RNA2.
RNA1 (see Fig. 1) also encodes a subgenomic RNA3 (0.4 kb) containing two overlapping open reading frames (ORFs) encoding proteins B1 and B2 (20). The protein B1 ORF is the 3′ end of the protein A ORF, which extends into RNA3. Translation of this B1 fragment (10.8 kDa) of protein A is not required for any step of the FHV life cycle, and the presence of an initiation codon in the corresponding region of the protein A ORF is not conserved in other nodaviruses (22). Protein B1 production is also inefficient, perhaps because the 7-nucleotide (nt) 5′ untranslated region preceding its initiation codon is too short for efficient translation. Protein B2, translated from the second AUG in RNA3, accumulates to 20-fold-higher levels than B1 and can represent up to 5% of total cell protein (14). Protein B2 is not required for RNA1 replication or RNA3 synthesis in single-cycle replication assays but is required for maintenance of RNA1 replication upon serial RNA passaging (4). Based on these results, it was suggested that protein B2 might contribute to the fidelity of RNA replication or to regulating the balance of RNA1 translation and replication (4).
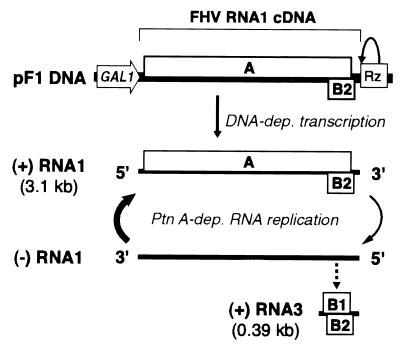
FIG. 1.
Schematic illustration of a DNA cassette to express FHV RNA1 in yeast and the use of this cassette to initiate FHV-specific, RNA-dependent RNA1 replication and subgenomic RNA3 synthesis. Within the bracketed region of FHV sequence, single lines indicate the 39-nt 5′ and 49-nt 3′ untranslated regions, while boxes A, B1, and B2 denote the corresponding ORFs. The protein B1 ORF is the 3′-terminal portion of the protein A ORF. The flanking GAL1 promoter and self-cleaving HDV Rz are indicated. (For details of plasmid structure, see Materials and Methods and Fig. 4). Initial positive-strand RNA1 synthesis is by DNA-dependent RNA polymerase II transcription. This RNA serves as mRNA for protein A, which is required for the FHV-specific, RNA-dependent RNA synthesis events indicated below positive-strand RNA1. Dashed arrow, one possible route to subgenomic RNA3 (the actual pathway to RNA3 synthesis and the negative-strand RNA template(s) for RNA3 have not been conclusively defined [see Discussion]).
We previously showed that, following transfection of FHV virion RNA into the yeast Saccharomyces cerevisiae, FHV undergoes its complete replication cycle, including the production of infectious virions (34). Furthermore, we showed that an FHV RNA2-derived replicon could express a yeast reporter gene (34). However, the free RNA replicons are readily lost from dividing yeast populations, which would greatly inhibit yeast genetic approaches to identify and study the contributions of viral and host functions in FHV infection (25).
To overcome these limitations, we describe here in vivo expression of functional FHV RNA1 from DNA plasmids in yeast, providing inducible initiation of robust FHV RNA1 replication. The level and requirements of FHV RNA1 replication in yeast were compared to those previously reported in animal cells. We also describe development of the first FHV RNA1 derivatives that harness subgenomic mRNA synthesis for reporter gene expression. This greatly facilitates reporter-based assays or screens of FHV RNA synthesis, because reporter gene expression remains dependent on FHV RNA replication and subgenomic mRNA synthesis even when the RNA1 derivative is continuously produced by DNA-dependent transcription. Green fluorescent protein (GFP) expression by this route was analyzed at the single-cell level. In the course of this work we also identified cis- and trans-acting mutations with unexpected, selective effects on subgenomic mRNA synthesis.
MATERIALS AND METHODS
Cells, growth, and transformation.
Plasmid DNAs were introduced into YPH500 (MATα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3Δ200 leu2Δ1) cells using the Frozen-EZ yeast transformation kit (Zymo Research) in accordance with the manufacturer's protocol. Transformed yeast cells were grown at 26°C in media selective for desired plasmids, with glucose or galactose as the carbon source as indicated for each experiment. Within an experiment, to allow growth of yeast strains containing only one experimental plasmid in the same medium types as those used for yeast containing two plasmids, plasmids containing the appropriate selectable markers but lacking FHV sequences were cotransformed with experimental plasmids. The WR strain of Drosophila line 1 cells (15) was propagated at 26°C.
Plasmid constructions.
All plasmids described below were based on yeast 2μm high-copy-number plasmid YEplac112 (19) or YEp351 (24), which contains the TRP1 or LEU2 selectable marker gene, respectively. For PCR mutagenesis, PCR products were first subcloned into Bluescript KS+ with T-A overhangs (3) or into pCR-XL-TOPO using the TOPO XL PCR cloning kit (Invitrogen). Site-directed mutagenesis was by the two-primer method (43), where the second primer removes a unique restriction site, thus allowing for a reduction in the frequency of nonmutant plasmids by digestion. For both PCR and site-directed mutagenesis, sequenced fragments were used to construct the desired clone. Plasmid laboratory names are indicated in parentheses.
(i) pF1ΔP (Tp71R).
pF1ΔP was described previously under the name p1R (34).
(ii) pF1 (TpG1R).
pF1 was constructed by ligating the EcoRI/SnaBI GAL1 promoter fragment of pB3MI1 (26), the PstI/PflMI fragment of p1B9SP (12) containing the 5′ end of the FHV RNA1 cDNA, the PflMI/NarI fragment of p1R (34) containing the 3′ end of the FHV RNA1 cDNA fused to the hepatitis delta virus ribozyme (HDV Rz) cDNA, and the NarI/EcoRI fragment of shuttle vector YEplac112 (19) containing the origin of replication. The sequence of the GAL1-FHV RNA1 fusion is depicted in Fig. 4.
FIG. 4.
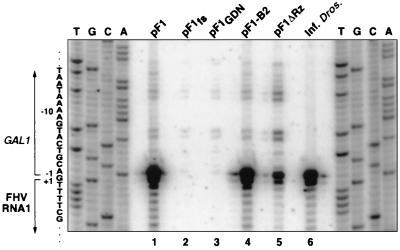
Primer extension analysis of 5′ ends of FHV RNA1 species in yeast expressing RNA1 derivatives. Yeast cells were grown, induced with galactose, and harvested for total RNA extraction 4 DPI as described for Fig. 2. A 5′ 32P-labeled oligonucleotide complementary to bases 30 to 60 of FHV RNA1 was annealed to total RNA from yeast carrying the indicated plasmids (lanes 1 to 5) or from FHV-infected Drosophila cells (lane 6) and extended with reverse transcriptase. The products were electrophoresed on a 6% polyacrylamide sequencing gel and exposed to a PhosphorImager imaging plate. The sequence ladders on each side were prepared using the same 5′-labeled primer and pF1 plasmid DNA as a template. The sequence corresponding to the sense RNA is indicated at the left, with the GAL1 promoter region and 5′ end of FHV RNA1 bracketed. Numbering is relative to the 5′ end of FHV RNA1.
(iii) pF1fs, pF1GDN, pF1-B2, and pF1ΔRz (TpG1fsR, TpG1GDNR, TpG1(8B1)R, and TpG1).
pF1fs was generated by replacing the PflMI/NarI fragment of pF1 with that from p1(fs)R (34). pF1GDN was generated by replacing the pF1 XmaI fragment with the XmaI fragment of the PCR product amplified from pF1 with primers 5′CATGGCGCCAATGGGCCATTCAAA3′ and 5′AGCCCGGGAAAGACCATTATCACCGCACTTCGGT3′. The underlined nucleotide in the second primer changes the protein A aspartic acid codon at RNA1 nt 2116 to an asparagine codon. pF1-B2 was derived from pF1 by replacing the BstXI fragment overlapping the B2 ORF with the corresponding fragment from an RNA1 plasmid containing two engineered mutations in the B2 ORF (22). The first mutation changes the B2 initiating methionine codon AUG at nt 2838 to threonine codon ACG. The second changes the B2 serine codon UCA at nt 2909 to stop codon UAA. Both changes result in silent changes at the third position of codons in the protein A and B1 ORFs. For pF1ΔRz, the SacI fragment of pF1, containing the 3′ end of the FHV RNA1 linked to the HDV Rz, was replaced with the SacI fragment of p1B9SP (12), containing the 3′ end of FHV RNA1 followed by vector sequences.
(iv) pF1-GFPC1, pF1-GFPC2, pF1fs-GFPC1, pF1fs-GFPC2, pF1fs-GFPN1, and pF1fs-GFPN2 and their derivatives (TpG1CαGR, TpG1Oβ2GR, TpG1-ECαGR, TpG1-EOβ2GR, TpG1-ENαGSR, and TpG1-ENβGSR).
The plasmids containing GFP ORF insertions were generated in three steps. First, the unique SfoI (a blunt-cutting isoschizomer of NarI) site in pF1 and pF1fs was disrupted by insertion of linker 5′CAAGCTTG3′. Second, a unique NarI site (5′GGCGCC3′) was introduced at positions N1, N2, C1, and C2 (see Fig. 5) in these pF1 derivatives. The NarI sites at N1 and C1, underlined in the primers below, were introduced in a three-way ligation between an ApaI/NarI-cut PCR fragment, a NarI/BsrGI-cut PCR fragment, and the ApaI/BsrGI-cut pF1 derivative. Primer 5′CATGGCGCCAATGGGCCATTCAAA3′, in conjunction with 5′GGCGCCCATTGGTAACGATTC3′ or 5′CATGGCGCCCTTCCGGTTGTTGGAAGGC3′, was used to generate the ApaI/NarI-cut PCR fragment containing N1 or C1, respectively. Primer 5′CATGGCGCCTCAATGATGGGTAAC3′, in conjunction with 5′GGCGCCTTAAACGATGCCAAGC3′ or 5′CATGGCGCCTGACCCCCCACCCGCA3′, was used to generate the NarI/BsrGI-cut PCR fragment containing N1 or C1, respectively. The NarI site at N2 was introduced by site-directed mutagenesis of pF1 using primers 5′CAATGTTAAACGATGGGCGCCCCAAGCAAACTCGCG3′ and 5′GGCCGGCATCACCGGTACCTGATGCGGATTT3′. The first primer introduces the NarI site, underlined, and the second primer changes 2 nt, underlined, to remove a unique SgrAI site. The NarI site at C2, underlined, was introduced into pF1 derivatives by ligation of the BlpI/BsrGI-cut pF1 derivative and the BlpI/BsrGI-cut fragment of the pF1 PCR product generated using primers 5′AGCTCAGCCACAGCCTTCCAACAACCGGAAGTGACGGCGCCCCCCCACCCGCAAAA3′ and 5′GCACAGATGCTTCGTCGACAAAGATATGCTAT3′. Third, following the introduction of the unique NarI sites in the vector, a cloned PCR product containing the GFP ORF flanked by NarI sites was ligated at each position. The GFP ORF, with or without its own stop codon (underlined), was amplified from YEGFP1 (10) using primers 5′GGCGCCTCTAAAGGTGAAGAA3′ and 5′GGCGCCTTATTTGTACAATTC3′ (for N1 or N2) or 5′GGCGCCTTTGTACAATTCATC3′ (for C1 or C2). The GFP ORFs inserted at positions N1 and N2 thus contained their own stop codons. The GFP ORFs inserted at positions C1 and C2 did not contain their own stop codons, relying instead on the stop codon in the viral ORF.
FIG. 5.
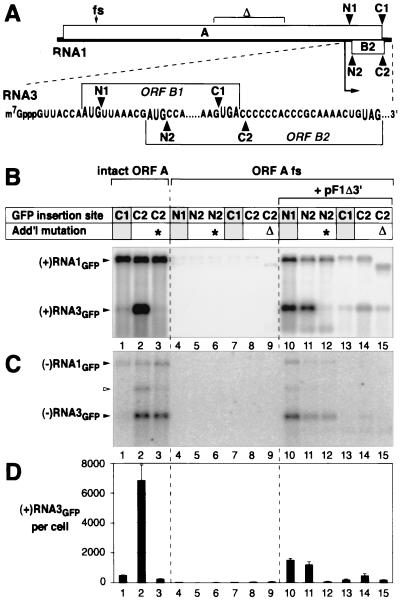
FHV-directed production of subgenomic RNA3 derivatives with GFP insertions. (A) Schematic of FHV RNA1 and its subgenomic mRNA product, RNA3. Bent arrow, start of RNA3 sequences. N1, N2, C1, and C2 denote positions at which the GFP ORF was inserted in RNA1 derivatives (see text). GFP ORF insertions at N1 and C1 were in frame with the protein A ORF and its 3′-terminal subset in RNA3, the protein B1 ORF. GFP ORF insertions at N2 and C2 were in frame with the protein B2 ORF. The positions of the protein A ORF frameshift and deletion mutations used in some RNA1/GFP derivatives are shown at the top. (B) Northern analysis of positive-strand FHV RNA1 and RNA3 accumulation for GFP insertion derivatives. As shown at the top, lanes 1 to 3 used RNA1/GFP derivatives with an intact protein A ORF. Lanes 4 to 9 and 10 to 15 used RNA1/GFP derivatives with the protein A ORF frameshifted at nt 378 (as in pF1fs; Fig. 3), without or with protein A-expressing helper plasmid pF1Δ3′, as indicated. For each lane, the table shows the site of GFP ORF insertion in RNA1 and whether the RNA1/GFP derivative contained either of two additional mutations, a G-to-T change at the first nucleotide of RNA3 (∗) and deletion of RNA1 nt 1459 to 2128 (Δ), within the middle of the protein A ORF (see panel A). Yeast cells containing the indicated plasmids were grown, induced with galactose, and harvested for total RNA extraction 4 DPI and analyzed as described for Fig. 2A using a 32P-labeled in vitro transcript probe complementary to the positive-strand GFP ORF. (C) Northern analysis of negative-strand FHV RNA1 and RNA3 accumulation, performed as for panel B except that the probe was complementary to the negative-strand GFP ORF and the image was printed at a higher intensity level. The weaker band (open arrowhead) visible above negative-strand RNA3 in some lanes contains undenatured double-stranded RNA3 (B. Lindenbach and P. Ahlquist, unpublished results). (D) Accumulation of positive-strand RNA3 per cell. Averages and standard deviations from panel B and two additional experiments are shown.
The G-to-T change at nt 2721 of pF1-GFPC2 was generated by ligating the BlpI/BamHI fragment of pF1-GFPC2, the BamHI/BbsI fragment of pF1, and the BbsI/BlpI fragment of a PCR product amplified from pF1 with primers 5′GCGATGAAGACGGCG T TGCGCCGAAC TCCGTGGACGAATCT T TACCAATGTTAAACGATG3′ and 5′GCTTCAGTAAGCCAGATG3′. The first primer changes the 5′-most nucleotide of the subgenomic RNA, underlined, from a G to a T. The G-to-T change at nt 2721 of pF1fs-GFPN2 was generated by ligating the XbaI/BamHI fragment of pF1fs-GFPN2, the BamHI/BbsI fragment of pF1, and the BbsI/XbaI fragment of a PCR product amplified from pF1fs-GFPN2 with the same primers described above. The deletion derivative of pF1fs-GFPC2, which lacks nt 1459 to 2128, was generated by digesting pF1fs-GFPC2 with BglII and SmaI, filling in with the Klenow fragment of DNA polymerase I (New England Biolabs), and religating.
(v) pF1Δ3′ (LpG13′-5R).
To generate a cDNA for the protein A mRNA, the FHV RNA1 cDNA of pF1 was mutagenized by the two-primer method with primers 5′GCCCGAAAGGGCAGGGTCGGCATGGC3′ and 5′GGCCGGCATCACCGGTACCTGATGCGGATTT3′. The first primer removes the last 5 nt of the FHV RNA1 cDNA, normally found between the underlined nucleotides, and the second primer changes 2 nt, underlined, to remove the unique NarI site. To facilitate cloning, the BlpI/BsrGI fragment of the mutagenized cDNA was ligated to the BlpI/BsrGI fragment of a pF1 derivative whose unique SfoI site was disrupted by insertion of HindIII linker 5′CAAGCTTG3′. The cDNA-containing HindIII fragment of this intermediate was then ligated to HindIII-cut YEp351 (24) to generate pF1Δ3′.
RNA analysis.
Hot-phenol extraction of total yeast RNA, formaldehyde denaturation, Northern blotting to Nytran nylon membranes (Schleicher and Schuell), and hybridization were performed as described previously (29, 32). Strand-specific 32P-labeled in vitro transcripts were generated as described previously (28). The probes for negative- and positive-strand FHV RNA1 and RNA3 corresponded to or were complementary to nt 2718 to 3064 of FHV RNA1. The probe for negative-strand GFP RNA corresponded to nt 4 to 133 of the GFP ORF. The probe for positive-strand GFP RNA was complementary to nt 572 to 714 of the GFP ORF. A Molecular Dynamics PhosphorImager digital radioactivity imaging system was used to obtain quantitative results.
Total RNA was stained with Cybergreen I (Molecular Probes, Eugene, Oreg.) and imaged with a FluorImager (Molecular Dynamics), in accordance with the manufacturer's protocol.
Primer extension was conducted essentially as described previously (3). In brief, primer 5′CAATTCAGTTCGGGTGATCTGGTGTTCTCC3′, complementary to nt 60 to 90 of RNA1, was 32P labeled at the 5′ end with polynucleotide kinase (Epicenter Technologies, Madison, Wis.) and ethanol precipitated to reduce unincorporated [γ-32P]ATP. Labeled primer (0.2 ng) was annealed to 0.5 μg of total cellular RNA at 65°C for 1.5 h in a 5-μl volume, slowly cooled to room temperature, and extended at 50°C for 45 min in a 15-μl reaction mixture with avian myeloblastosis virus reverse transcriptase (Promega). Immediately after transcription, an equal volume of 90% formamide, 20 mM EDTA, 0.1% xylene cyanol, and 0.1% bromphenol blue was added and samples were analyzed on a 6% acrylamide sequencing gel. For comparison, a DNA sequence ladder was prepared from a pF1 template and the same 32P-labeled primer using a Sequitherm kit (Epicenter Technologies).
Microscopy and flow cytometry.
GFP expression was analyzed 4 days post-galactose induction. GFP expression was imaged in cells by epifluorescence microscopy with a Zeiss Axiovert 135TV microscope equipped with a 485-nm excitation filter and a 510-nm emission filter. Images were captured using a SenSys charge-coupled device camera system (Photometrics, Tucson, Ariz.) and IPLab Spectrum, version 3.1.2, software (Scanalytics, Inc., Fairfax, Va.) and processed with Adobe Photoshop, version 5. Prior to quantitative analysis of GFP expression by fluorescence-activated cell sorting (FACS), yeast cells were diluted 1:10 in water. FACS data from 10,000 live cells per sample were collected on a Becton Dickinson (Heidelberg, Germany) FACScan equipped with a 488-nm argon-ion laser. Fluorescence emission was detected through a 530/30-nm filter. Fluorescence and forward scatter data were collected with logarithmic amplifiers and analyzed using Win MDI, version 2.8. To set the background threshold for scoring positive fluorescence, we used the observation by microscopy that 0.5% of galactose-induced yeast cells containing pF1fs-GFPN2 alone (without protein A-expressing helper plasmid pF1Δ3′) fluoresced at a weak but visible level. Accordingly, for each set of parallel FACS analyses, the threshold for positive fluorescence was set to the level that excluded 99.5% of cells containing pF1fs-GFPN2 alone.
RESULTS
High-level induction of wt FHV RNA1 replication in yeast.
Previously we described yeast plasmid pF1ΔP, which initiates FHV RNA1 replication and supports the replication of FHV RNA2 derivatives in yeast (34). pF1ΔP contains a complete, wild-type (wt) FHV RNA1 cDNA but lacks a defined yeast promoter to express this cDNA. RNA1 transcripts from pF1ΔP appear to originate from cryptic promoters upstream of the RNA1 5′ end and are terminated by the self-cleaving HDV Rz positioned to produce the authentic FHV RNA1 3′ end. Yeast cells containing pF1ΔP, when grown on glucose to a density of 107 cells per ml, display constitutive but low-level FHV RNA1 replication: such cells accumulate about 500 copies of RNA1 per cell, on average, compared to a final accumulation of almost 100,000 copies of RNA1, based on particle yield, in each yeast spheroplast successfully transfected with virion RNA from FHV-infected Drosophila cells (34).
To control and increase the DNA-directed production of transcripts with defined 5′ ends that could initiate FHV RNA1 replication and subgenomic mRNA synthesis, we incorporated an inducible yeast promoter (Fig. 1). In plasmid pF1, the major transcription initiation sites of the galactose-inducible, glucose-repressible GAL1 promoter were placed next to the authentic viral 5′ end of the FHV RNA1 cDNA, again followed by the HDV Rz cDNA (Fig. 1). When yeast cells containing this plasmid were grown in glucose medium, no positive- or negative-strand RNA1 was detected (Fig. 2A and B, lanes 8). When the yeast cells were transferred to galactose medium, this plasmid initiated FHV RNA1 replication, as demonstrated by production of negative- and positive-strand RNA1 and subgenomic RNA3 (Fig. 2, lanes 9 to 12). Maximum levels of replication products were reached between 2 and 3 days postinduction (DPI), and RNA1 levels were relatively constant from 2 to 4 DPI. This time course of RNA1 appearance was very similar to that seen after transfection of yeast cells with FHV virion RNA (34). Also as in prior experiments, the ratio of positive- to negative-strand RNA1 was approximately 70 and the ratio of positive- to negative-strand RNA3 was approximately 25.
FIG. 2.
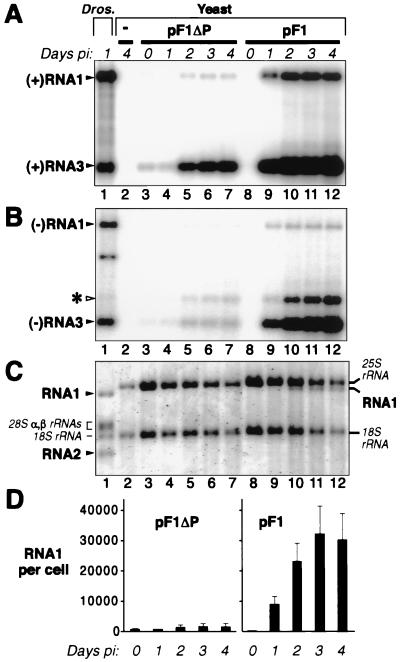
Time course of DNA-induced FHV RNA1 replication and subgenomic mRNA expression in yeast. Yeast were grown to 107 cells per ml in medium containing glucose, pelleted, and resuspended at an equal density in medium containing galactose to induce the GAL1 promoter of pF1 (Fig. 1). Cells were harvested for RNA extraction at the indicated DPI. Lane 1, positive control containing total RNA from Drosophila (Dros.) cells infected with 10 FHV PFU per cell; lane 2, negative control containing total RNA from yeast with no FHV sequences; lanes 3 to 7, total RNA from yeast bearing pF1ΔP; lanes 8 to 12, total RNA from yeast bearing pF1. (A) Northern analysis of positive-strand FHV RNA1 and RNA3 accumulation. Total RNA (0.5 μg per lane) was denatured in 50% formamide–6% formaldehyde at 65°C, electrophoresed on a 1% agarose-formaldehyde gel, transferred to a nylon membrane, hybridized to a 32P-labeled in vitro transcript probe complementary to positive-strand RNA1 and RNA3, and exposed to a PhosphorImager imaging plate. The positions of FHV RNA1 and RNA3 are indicated. RNA2, though present in the FHV-infected Drosophila cells, was not detected in lane 1 because the probe used was complementary only to RNA1 and RNA3. (B) Northern analysis of negative-strand FHV RNA1 and RNA3 accumulation, performed as for panel A except that the blot was hybridized to a 32P-labeled in vitro transcript probe complementary to negative-strand RNA1 and RNA3 and was printed at a higher intensity level. The positions of FHV RNA1 and RNA3 are indicated at the left. The band above RNA3 (asterisk), whose intensity trails that of RNA3, contains undenatured double-stranded RNA3 (B. Lindenbach and P. Ahlquist, unpublished results). The band below negative-strand RNA1 in lane 1 may represent a defective deletion derivative of RNA1 (36). (C) A 0.6% agarose-formaldehyde gel, electrophoresed longer than gels in panels A and B, was stained with Cybergreen I (Molecular Probes) and imaged on a FluorImager (Molecular Dynamics) to reveal total RNA. For lane 1, the positions of Drosophila rRNAs and the two FHV genomic RNAs, RNA1 and RNA2, are indicated at the left. For yeast lanes 2 to 12, the positions of yeast rRNAs and FHV RNA1 are indicated at the right. (D) Accumulation of positive-strand RNA1 per cell for yeast containing pF1ΔP or pF1 at the indicated times after transfer to galactose. Quantitative analysis of panel A Northern blot signals was used to calculate the number of FHV RNA1 molecules per yeast cell, given the signal from a known amount of a coelectrophoresed FHV RNA standard, the known amount of total yeast RNA loaded per lane, and an average value of 1.2 pg of total RNA per cell (38). Averages and standard deviations from panel A and two (for pF1ΔP) or three (for pF1) additional independent experiments are shown.
PhosphorImager quantitation of Northern blots from Fig. 2 and three other experiments showed that positive-strand FHV RNA1 levels in galactose-induced, pF1-containing cells (Fig. 2A, lanes 10 to 12) were nearly 50-fold higher than those in pF1ΔP-containing cells grown under standard (glucose) conditions (Fig. 2A, lane 3, and D). A fraction of this increased accumulation appeared due to the slower growth of yeast in galactose, since when pF1ΔP-containing cells were grown in galactose-containing medium, RNA1 levels increased twofold while RNA3 levels increased almost sevenfold (Fig. 2A, lanes 4 to 7, and D). Nevertheless, the majority of the increase was due to superior induction of FHV RNA replication from pF1: after 4 days in galactose, RNA1 levels in pF1 cells were over 20-fold higher than those in pF1ΔP cells, while RNA3 levels in pF1 cells were 6-fold higher.
In pF1-containing yeast, RNA1 levels per microgram of total RNA approached those in infected Drosophila cells, while RNA3 levels in pF1-containing yeast were much higher than those in FHV-infected Drosophila cells (Fig. 2A, lane 1 versus lanes 10 to 12). The lower RNA3/RNA1 ratio in Drosophila cells is due to the presence of RNA2, which inhibits positive-strand RNA3 synthesis (18, 44, 45). As shown in Fig. 2C, RNA1 was readily detectable in both cell types by total RNA staining, along with rRNAs. In pF1-containing yeast, RNA1 levels reached 32,000 molecules per cell (Fig. 2D), while RNA3 reached 120,000 molecules per cell (Fig. 2A). In contrast, over 95% of all yeast mRNAs accumulate to less than 10 copies per cell (42).
Requirements for FHV RNA1 replication and subgenomic RNA3 transcription in yeast.
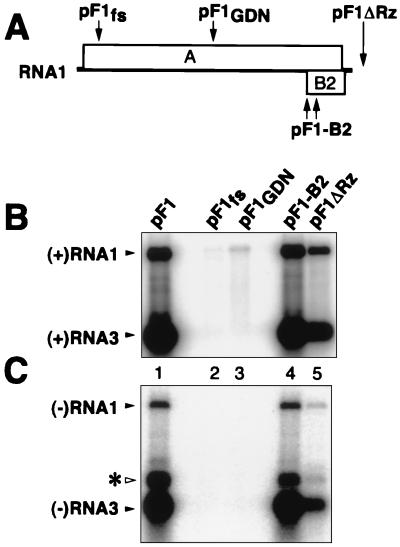
To determine the relative contributions of DNA-dependent transcription and FHV RNA-dependent RNA replication to pF1-induced RNA1 production, we tested two mutations in protein A (Fig. 3A): one due to a 4-nt frameshifting insertion at RNA1 nt 378, early in the protein A ORF (pF1fs [34]), and a single amino acid substitution changing the highly conserved RNA-dependent RNA polymerase motif GDD to GDN (pF1GDN). As expected from analysis of similar mutations (27, 34), these protein A changes abolished detectable negative-strand RNA1 and positive- or negative-strand RNA3 and reduced positive-strand RNA1 accumulation 40- to 150-fold (based on the quantitation of Fig. 3B and C and two similar experiments). Thus, in yeast bearing pF1, FHV RNA replication amplified RNA1 40- to 150-fold above the levels produced by the strong GAL1 promoter.
FIG. 3.
Requirements for FHV RNA replication and subgenomic expression in yeast. (A) Schematic of FHV RNA1 showing the positions of the indicated mutations (see text for further details). (B) Northern analysis of positive-strand FHV RNA1 and RNA3 accumulation. Yeast cells containing the indicated plasmids were grown, induced with galactose, and harvested for total RNA extraction 4 DPI, as described in the legend of Fig. 2. RNA analysis was as described for Fig. 2A. The positions of FHV RNA1 and RNA3 are indicated. To more clearly observe the weak RNA signals in lanes 2 and 3, no samples were loaded in the flanking lanes. (C) Northern analysis of negative-strand FHV RNA1 and RNA3 accumulation, performed as for panel B except that the blot was hybridized to a 32P-labeled in vitro transcript probe complementary to negative-strand RNA1 and RNA3 and was printed at a higher intensity level. The positions of FHV RNA1 and RNA3 are indicated. The weaker band above RNA3 (asterisk) contains undenatured double-stranded RNA3 (B. Lindenbach and P. Ahlquist, unpublished results).
Mutations blocking FHV protein B2 expression have no effect on an initial cycle of RNA1 replication in mammalian (BHK21) cells but dramatically inhibit subsequent RNA1 replication in second and third pools of cells infected by serial RNA passaging (4). To test for a possible B2 contribution to FHV RNA replication in yeast, B2 was inactivated by two simultaneous substitutions that were silent with respect to the overlapping protein A ORF but that changed the B2 initiation codon to ACG and B2 codon 58 to a stop codon (4, 22). This B2 inactivation had no effect on production of positive- or negative-strand RNA1 or RNA3 in the initial cycle of RNA1 replication in yeast (Fig. 3B and C, lanes 4).
In mammalian cells, 3′ extensions on FHV RNA1 transcripts inhibit replication less than do 5′ extensions, with a 12-nt 3′ extension, e.g., reducing RNA1 replication only twofold (4). To determine the contribution of a natural 3′ transcript end to RNA1 replication in yeast, we generated pF1ΔRz, which lacks the 3′ ribozyme of pF1 (Fig. 3A). Most RNA1 transcripts from pF1ΔRz likely terminate at a yeast polyadenylation signal in the 2μm origin of replication in the plasmid, 0.6 kb downstream of the RNA1 3′ end (41). Some transcripts may also terminate earlier at weaker, fortuitous polyadenylation sites, which in yeast can be generated by AU-rich sequences (40). In galactose-induced yeast containing pF1ΔRz, negative-strand RNA1 and RNA3 levels were reduced seven- and fivefold, respectively, relative to those in yeast containing pF1 (Fig. 3C, lane 5).
Selective amplification of RNA1 transcripts with natural 5′ termini.
The 5′ ends of FHV RNA1 from virions and FHV RNA1 derivatives from yeast containing pF1 and its derivatives were examined by primer extension (Fig. 4). As seen previously (4), extension of an RNA1 primer on FHV virion RNA (Fig. 4, lane 6) produced two strong bands, the lower band corresponding to the 5′ terminus of RNA1 and the upper band corresponding to cap-dependent incorporation of an additional nucleotide. Similar cap-dependent incorporation of an additional nucleotide has been described for primer extension on other RNAs (1, 2, 21). To determine the RNA1 transcript start sites in the absence of FHV RNA replication, transcripts from galactose-induced pF1fs- and pF1GDN-containing yeast were subjected to primer extension (Fig. 4, lanes 2 and 3). Both contained RNA1 transcripts with multiple 5′ ends, in keeping with corresponding multiple transcription start sites from other GAL1 promoter fusions (26). The most prominent bands corresponded to nt −1, −6, −7, −12, and −13 relative to the 5′ end of FHV RNA1 (Fig. 4, lane 6). The last four bands may represent transcription starts at −6 and −12, with a higher band from cap-dependent incorporation (see above). Close examination of this and similar primer extensions suggests that the band at −1 may be due to superposition of two weaker doublets at +1 and −1 and at −1 and −2, representing transcription starts at +1 (the 5′ end of FHV RNA1) and −1.
Primer extension of total RNA from galactose-induced pF1- and pF1-B2-containing yeast revealed dramatic, selective amplification of the bands at +1 and −1, corresponding to the natural 5′ end of FHV RNA1 (Fig. 4, lanes 1, 4, and 6). pF1ΔRz-containing yeast showed selective but weaker amplification of the same +1 and −1 bands (Fig. 4, lane 5). Thus, for each pF1 derivative from Fig. 3, the degree of amplification of natural RNA1 5′ ends (Fig. 4) agreed well with the level of protein A-dependent RNA1 replication revealed by Northern blotting (Fig. 3).
In Fig. 4, the intensity of primer extension bands for DNA-derived primary transcripts varied between some lanes, likely due to several causes. Compared to that for pF1ΔRz (lane 5), primary transcript band intensity was reduced for pF1 and pF1-B2 (lanes 1 and 4), possibly because the considerable excess of FHV-dependent RNA1 replication product (represented by bands at positions +1 and −1) competed for primer hybridization or reverse transcriptase. Primary transcript bands were weakest for pF1fs and pF1GDN (lanes 2 and 3). For pF1fs, primary transcript accumulation would be reduced by nonsense-mediated mRNA decay (23). Reasons for lower accumulation of pF1GDN transcripts are less clear, but this might suggest that RNA1 transcripts could be stabilized by wt protein A (40) or destabilized by some protein A mutants.
Synthesis of GFP-encoding RNA3 by FHV polymerase provided in cis and selective cis and trans effects on subgenomic RNA.
To link RNA-dependent RNA synthesis by FHV RNA1 to reporter gene expression in yeast, we tested insertions in subgenomic RNA3. In principle, incorporating a reporter gene in RNA3 could provide a high-copy-number mRNA whose production depends on FHV-specific negative-strand RNA and subgenomic mRNA synthesis. As a reporter, we chose the gene for a fluorescence-enhanced, yeast codon-optimized version of GFP, whose expression can be assayed in living cells (10).
Useful insertion of reporter genes in RNA3 is constrained by the tight organization of RNA1, in which the 3′ end of the protein A ORF overlaps most of RNA3 and the protein B2 ORF (Fig. 5A). To avoid disrupting essential RNA replication factor A, insertions must be made in the last 74 nt of RNA1, containing the termination codon for the proteins A and B1 ORFs, the last 19 nt of the protein B2 ORF, the B2 ORF termination codon, and an additional 49 untranslated nucleotides. In this region we tested two insertions. First, we inserted the GFP ORF immediately before the protein A ORF termination codon (position C1 in Fig. 5A), thus fusing GFP to the C terminus of protein A. While this disrupts the B2 ORF, B2 is not required for RNA1 or RNA3 synthesis in yeast (Fig. 3, lane 4). Second, we inserted the GFP ORF 1 nt after the protein A ORF termination codon (position C2 in Fig. 5A). This leaves protein A unmodified and fuses GFP near the C terminus of protein B2.
Yeast containing these pF1-GFPC1 or pF1-GFPC2 derivatives produced similar levels of positive- and negative-strand RNA1 derivatives of the expected electrophoretic mobilities for GFP insertion (Fig. 5B and C, lanes 1 and 2). Quantitative PhosphorImager analysis of these hybridization signals showed that RNA1 accumulation per cell for pF1-GFPC1 and pF1-GFPC2 was approximately 15% of that for the wt RNA1 plasmid, pF1. Yeast containing pF1-GFPC2 also produced a strong subgenomic RNA3 band with the expected electrophoretic mobility, which accumulated to even higher levels than RNA1 (Fig. 5B, lane 2). To confirm the origin of this band as a subgenomic RNA3 derivative, the 5′ RNA3 nucleotide in pF1-GFPC2 was changed from G to T. As previously found for wt RNA1 using a strand-insensitive assay (actinomycin D-resistant incorporation of [3H]uridine [4]), this substitution greatly inhibited accumulation of positive-strand RNA3 (Fig. 5B, lane 3). Unexpectedly, however, our strand-specific Northern analysis further revealed that this mutation did not reduce the level of negative-strand RNA3 (Fig. 5C, lane 3; see also Discussion). Another selective but distinct effect on subgenomic RNA was seen with pF1-GFPC1, for which accumulation of both positive- and negative-strand RNA3 was reduced almost 15-fold relative to that for pF1-GFPC2 (see Discussion).
Synthesis of GFP-encoding RNA3 by FHV polymerase provided in trans.
As an alternate strategy for linking reporter gene expression to FHV RNA3 synthesis, we also tested GFP ORF insertions in the 5′ end of RNA3. As these insertions disrupt the protein A ORF, protein A was provided in trans from helper plasmid pF1Δ3′. pF1Δ3′ contains FHV RNA1 cDNA between the GAL1 promoter and HDV Rz but differs from pF1 in two respects. First, the last 5 nt of RNA1 cDNA, which are required in cis for RNA1 replication (4), were deleted. Second, while pF1 and its other derivatives bear the TRP1 selectable marker, pF1Δ3′ bears the LEU2 selectable marker. This allows simultaneous selection for pF1Δ3′ and pF1-GFP derivatives in the same cell.
The GFP ORF was inserted in pF1 immediately after the initiation codon for protein B1 or B2 (positions N1 and N2 in Fig. 5A). To prevent possible interference effects from the large, disrupted protein A variants that would otherwise result, these plasmids also contained the protein A-truncating 4-nt frameshift at position 378 (Fig. 3). When the resulting plasmids, pF1fs-GFPN1 and pF1fs-GFPN2, were transformed into yeast individually, low-level DNA-derived RNA1 transcripts were produced but no negative-strand RNA1 or subgenomic RNA3 was detected (Fig. 5B and C, lanes 4 and 5). In contrast, yeast containing protein A-expressing helper plasmid pF1Δ3′ plus either pF1fs-GFPN1 or pF1fs-GFPN2 contained amplified positive-strand RNA1, RNA3, and their negative strands as expected for FHV RNA replication (Fig. 5B and C, lanes 10 and 11). For pF1fs-GFPN2, the G-to-T substitution at the start of RNA3 sequences was used to verify the nature of the putative subgenomic RNA3 derivative. As expected, this mutant greatly reduced the level of positive-strand but not negative-strand subgenomic RNA3 (Fig. 5B to D, lanes 12).
To compare the replication of RNA1 derivatives by protein A supplied in cis and in trans, the 4-nt protein A frameshift insertion (Fig. 3) was transferred into pF1-GFPC1 and pF1-GFPC2, yielding pF1fs-GFPC1 and pF1fs-GFPC2. As expected, no negative-strand RNA1 or subgenomic RNA3 was detected in yeast containing either plasmid alone (Fig. 5B and C, lanes 7 and 8). In yeast containing either of these plasmids plus helper plasmid pF1Δ3′, subgenomic RNA3 appeared, positive-strand RNA1 levels increased slightly, and negative-strand RNA1 was visible on long exposures (Fig. 5B and C, lanes 13 and 14). For pF1fs-GFPC2, RNA1 and RNA3 levels averaged 10- to 12-fold lower than those when protein A was provided in cis (Fig. 5B and D, lanes 2 and 14) and 2.5- to 3-fold lower than those for pF1fs-GFPN1 or pF1fs-GFPN2 (Fig. 5B and D, lanes 10 and 11). For pF1fs-GFPC1 plus pF1Δ3′, RNA1 accumulation was similarly reduced compared to that for protein A synthesis in cis (Fig. 5B, lanes 1 and 13).
In principle, in yeast containing pF1fs-GFPC2 and pF1Δ3′, DNA or RNA recombination could regenerate the RNA1 derivative encoded by pF1-GFPC2, bearing a wt protein A ORF, and able to direct its own replication in cis. Such recombinants would not be detected because they would comigrate with the parental, frameshifted RNA1 replicon. To test for such recombination, a 0.7-kb segment of the protein A ORF was deleted from pF1fs-GFPC2 to create a vector encoding an RNA1 derivative of novel size. The segment deleted (Fig. 5A) was downstream of the protein A frameshift and is not required in cis for RNA1 replication (7). Yeast bearing this deletion plasmid plus helper pF1Δ3′ produced the expected genomic and subgenomic RNA species at levels approaching those in cells with pF1fs-GFPC2 plus pF1Δ3′ (Fig. 5B, lanes 14 and 15). However, no full-sized genomic RNA1 was seen (Fig. 5B, lane 15), showing that recombination did not detectably contribute to this RNA3 production.
FHV replication-dependent GFP expression in live yeast.
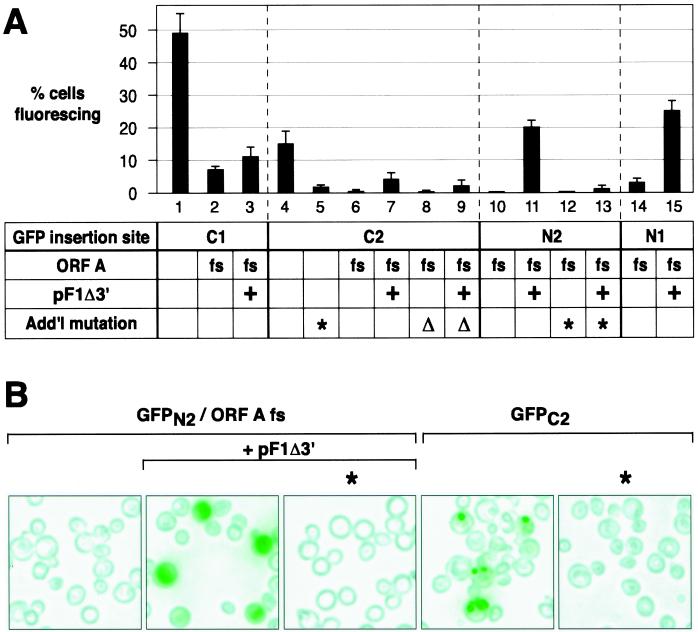
Fluorescence microscopy of live yeast containing replicating RNA1/GFP derivatives revealed green fluorescing cells, but the frequency of cells fluorescing varied with the GFP ORF insertion site and other features of the RNA1 derivative. For each FHV genotype examined in Fig. 5, FACS was used to determine the frequency of green fluorescing cells (Fig. 6A). As shown in Fig. 6B, there was generally a clear discrimination between fluorescing and nonfluorescing cells. The possible contribution of incomplete plasmid segregation (8) and other effects to the appearance of nonfluorescing cells is considered in Discussion.
FIG. 6.
Analysis of FHV-directed GFP expression at the single-cell level. Yeast cells containing the indicated plasmids, designated as in Fig. 5, were grown and induced for 4 days as described in Fig. 2 before analysis by FACS and epifluorescence microscopy. (A) For each FACS experiment (see Materials and Methods), fluorescence intensity was determined for 10,000 live cells per cell line. All cell lines were examined in one session in each of three replicate experiments. Percentages of cells fluorescing above background are shown as the averages and standard deviations from the three experiments. (B) Representative fields of living yeast cells expressing the indicated FHV derivatives. Yeast cells were excited at 485 nm, and fluorescence emission was screened with a 510-nm filter. All images were captured with a digital camera in a single session and were processed identically. Fluorescence was imaged as green and superimposed onto a brightfield image of the same field of cells.
The highest frequency of fluorescing cells, 49%, was found for the GFP ORF insertion at position C1 (Fig. 6A, lane 1). Although the GFP ORF insertion at C1 severely inhibited RNA3 production (Fig. 5B to D, lanes 1), GFP was fused to the C terminus of protein A and so was directly translatable from RNA1 (Fig. 5A). Accordingly, when the protein A ORF was disrupted by frameshifting, the frequency of fluorescence dropped sevenfold (Fig. 6A, lane 2). Providing protein A in trans from pF1Δ3′ partially restored RNA3 production (Fig. 5B and D, lanes 1, 7, and 13) but provided little or no significant increase in GFP expression (Fig. 6A, lane 3), consistent with the possibility that most GFP expression from the C1 insertion derivatives was from translation of RNA1, not RNA3.
For RNA1 with the GFP ORF inserted at C2 and N2, 15 and 20% of yeast cells, respectively, showed green fluorescence (Fig. 6A, lanes 4 and 11). As expected for these GFP ORF fusions to the protein B2 ORF (Fig. 5A), mutation of the RNA3 start site showed that this expression was dependent on production of subgenomic RNA3 (Fig. 6A, lanes 5 and 13). Similarly, GFP expression in these cases required protein A expression, supplied in either cis or trans (Fig. 6A, lanes 4, 6, 7, 10, and 11).
GFP ORF insertion at N1, which allowed significant RNA3 production (Fig. 5B to D, lanes 10), resulted in green fluorescence of 25% of yeast. Unlike insertion at C1, where little RNA3 was made, this expression occurred when the protein A ORF was frameshifted, as long as protein A was provided in trans (Fig. 6A, lanes 14 and 15).
Protein A-dependent GFP expression from RNA1 position N2, which produced GFP as an independent protein unfused to any FHV protein, resulted in green fluorescence distributed throughout the yeast cytoplasm (Fig. 6B) as found previously for free GFP in yeast (33). However, when GFP was expressed from position C2 in RNA1, green fluorescence was confined to a few punctate sites in each positive cell (Fig. 6B). This localization likely reflects an effect of B2 sequences, since the GFP ORF inserted in position C2 is expressed as a C-terminal fusion to the first 100 amino acids of protein B2. Further work is in progress to determine if these spots represent simple aggregation of the fusion protein or some more specific targeting.
DISCUSSION
The results reported here demonstrate substantial improvements in initiating FHV RNA replication from DNA in yeast and new pathways for FHV-directed gene expression and, as discussed further below, reveal unexpected aspects of FHV RNA replication and subgenomic mRNA synthesis. The ability to regulate FHV RNA1 replication and to achieve high levels of authentic replication products should prove very useful for further analysis of FHV RNA replication. In particular, induction of FHV replication and FHV-dependent reporter gene expression from DNA cassettes can greatly facilitate genetic analysis of viral and host functions in FHV replication, because such cassettes can be stably maintained by plasmid selection or chromosomal integration during genetic screens of yeast populations (25, 26).
DNA-derived induction of FHV RNA replication.
Transcription of FHV RNA1 cDNA from the GAL1 promoter allowed the inducible expression of RNAs in yeast that replicated to levels observable by staining total cellular RNA. The system was tightly repressible, with no FHV RNAs detected under noninducing conditions (Fig. 2A and B, lanes 8). However, upon induction, authentic FHV RNA1 rapidly appeared and was amplified by protein A-dependent RNA replication to levels 40- to 150-fold above those of the starting, DNA-derived transcripts from the strong GAL1 promoter (Fig. 3). The ability of FHV RNA replication in yeast to amplify RNA1 to levels approaching those of rRNA and to maintain these levels for days in culture (Fig. 2) appears particularly notable since this was done in the absence of FHV RNA2. As RNA2 encodes the capsid protein, this RNA1 was not stabilized by encapsidation. Additionally, in the absence of FHV RNA2, subgenomic RNA3 levels exceeded those of RNA1.
pF1-induced RNA1 replication and subgenomic RNA3 synthesis in yeast reflected FHV RNA synthesis in Drosophila and mammalian cells in all respects tested. Positive- and negative-strand RNA1 and RNA3 appeared in ratios similar to those seen in Drosophila cells. In the absence of RNA2, the increased accumulation of subgenomic RNA3 (18, 44, 45) was also duplicated in yeast. As in mammalian cells, RNA1 replication and RNA3 synthesis were dependent on the polymerase-like FHV protein A but independent of protein B2 (4) and RNA3 synthesis was inhibited by a G-to-T mutation at the start of RNA3 sequences in RNA1. Although GAL1-promoted DNA transcription initiated at multiple sites upstream of the RNA1 cDNA, FHV RNA replication in yeast selectively amplified RNAs with the authentic 5′ end of FHV RNA1 from Drosophila cells (Fig. 4). In addition, the kinetics of RNA1 replication and RNA3 synthesis following galactose induction paralleled those of FHV RNA replication in yeast transfected with authentic FHV virion RNA (34).
FHV-dependent reporter gene expression.
Using GFP as a convenient reporter, we have demonstrated the first expression of a foreign gene from FHV RNA1 derivatives. Experimentally useful GFP expression was obtained from each of four selected insertion sites in RNA1 (Fig. 5A), with fluorescence intensity was easily visible by microscopy or FACS (Fig. 6). For all four GFP insertions, high-level GFP expression depended on FHV RNA replication. However, as described below, differences in the insertion sites led to differences in the pathway and efficiency of GFP expression and different effects on RNA1 and RNA3 synthesis and accumulation.
GFP ORF insertion at position C1 fused GFP to the C terminus of protein A, allowing GFP expression by translation of RNA1. Accordingly, the relative level of GFP expression from RNA1 derivatives with the GFP ORF inserted at C1 correlated with the level of RNA1 accumulation rather than that of RNA3 accumulation (Fig. 5B and 6A). For the other three insertion sites, as intended, the primary mode of GFP expression appeared to be through synthesis and translation of subgenomic RNA3. GFP ORF insertions at positions C2 and N2 were not in frame with the protein A ORF and so were not translatable from RNA1. In keeping with this, GFP expression from these derivatives was largely abolished by an RNA3 start site mutation that inhibited positive-strand RNA3 production (Fig. 5B and 6). Because GFP ORF insertion at position N1 disrupted the protein A ORF, this insertion was deliberately tested in the context of an early protein A ORF frameshift mutation, blocking translation of a GFP-protein A fusion from RNA1 and making GFP expression dependent on RNA3.
Since the level of GFP expression due to all four GFP insertions was responsive to FHV RNA replication, any of the four might potentially be useful in screening for host or virus mutations affecting RNA replication. GFP ORF insertions at positions N2 and C2 may be especially useful for genetic screens because of their low background expression in the absence of FHV RNA replication (Fig. 6). The higher protein A-independent background of GFP expression for insertions at C1 and N1 appears likely to be related to in-frame fusion of the GFP ORF to the protein A ORF. Even in derivatives with an engineered early frameshift in the protein A ORF, alternate mechanisms may allow translation initiation at downstream sites along the protein A ORF, leading to expression of a fluorescing truncated protein A-GFP fusion protein. Such mechanisms may include fortuitous transcription initiation sites in the FHV cDNA, RNA splicing, and translational frameshifting or reinitiation.
The use of GFP allowed the screening of individual live cells, revealing differences not only in fluorescence per cell but also in the frequency of cells fluorescing. Nonfluorescing cells may have been due in part to the typically incomplete segregation to daughter cells of yeast 2μm plasmids such as pF1 and its derivatives, which even under selective conditions are usually absent from 20 to 30% of cells in culture (8; W.-M. Lee and P. Ahlquist, unpublished results). Such effects would be particularly acute for any of the protein A ORF frameshift derivatives, such as the N1 and N2 insertions, for which GFP expression required two plasmids, the GFP ORF-containing pF1 derivative and protein A-expressing helper plasmid pF1Δ3′. Among cells retaining the necessary plasmids, failure to initiate RNA1 replication in every cell in every generation also may have contributed to nonfluorescing cells. Consideration of these and other points discussed here should allow further improvements in FHV-directed reporter expression. Nevertheless, the reporter expression obtained in this study should be sufficient for many screening purposes.
C-terminal GFP fusion to protein A selectively inhibits RNA3 synthesis.
GFP ORF insertion at position C2 preserved expression of wt protein A, which replicated RNA1 and synthesized even larger amounts of RNA3 (Fig. 5B to D, lanes 2). GFP ORF insertion at position C1, just 4 nt upstream, fused GFP to the C terminus of protein A. The resulting protein A-GFP (GFPC1) fusion supported replication of its RNA1 to the same level as the B2-GFP (GFPC2) fusion but, remarkably, displayed a selective, 15-fold reduction in RNA3 production (Fig. 5B to D, lanes 1 and 2). In principle, this could be due to aberrant function of the protein A-GFP fusion or disruption of cis elements required for RNA3 synthesis. However, when wt protein A was provided in trans by helper plasmid pF1Δ3′, the same GFP ORF insertion at position C1 reduced average RNA3 accumulation only about twofold relative to GFP ORF insertion at position C2 (Fig. 5B and D, lanes 13 and 14). Thus, the selective defect in subgenomic RNA3 accumulation caused by GFP ORF insertion at C1 appears to be largely due to altered function in the protein A-GFP fusion. This conclusion also explains why the C1 and C2 insertions produce dramatically different subgenomic phenotypes, despite their close 4-nt spacing (Fig. 5A).
Selective inhibition of positive-strand RNA3 accumulation.
While GFP ORF insertion at C1 inhibited accumulation of both negative- and positive-strand RNA3, a more selective effect was associated with a cis-acting mutation. As previously found for wt RNA1 in animal cells (4), a G-to-T substitution at the start of RNA3 sequences in RNA1 greatly inhibited accumulation of positive-strand RNA3 (Fig. 5B, lane 3). Intriguingly, however, our strand-specific Northern analysis showed that this mutation did not reduce the level of negative-strand RNA3 (Fig. 5C, lane 3). This implies either that the mutation inhibits the stability rather than the synthesis of positive-strand RNA3 or that negative-strand RNA3 can be synthesized directly from a positive-strand RNA1 template, without prior synthesis of positive-strand RNA3. Models for subgenomic mRNA synthesis from independently generated subgenomic negative strands have been discussed for coronaviruses (35) and for red clover necrotic mottle virus (39). For FHV, as for coronaviruses, further experiments will be required to resolve the pathways and templates for negative- and positive-strand subgenomic RNA synthesis.
Cis-acting effects of GFP insertions.
GFP ORF insertion at position C1 or C2 inhibited RNA1 accumulation approximately sixfold relative to that for wt RNA1 (Fig. 5B, lanes 1 and 2). For GFP ORF insertion at C2, which preserved expression of wt protein A, this reduction must be due to cis-acting effects of the GFP ORF insertion on RNA1 replication, stability, or both. Similarly, when FHV protein A was provided in trans, RNA1 and RNA3 accumulations for GFP ORF insertions at positions N1 and N2 were two- to threefold higher than those for GFP ORF insertions at positions C1 and C2 (Fig. 5B to D, lanes 10 to 15). These results imply that some cis-acting sequences important for RNA1 replication or stability overlap or adjoin the sites of the C1 and C2 coding region insertions, which are 70 to 74 nt from the RNA1 3′ end. Positions N1 and N2, by contrast, are 367 and 377 nt from the RNA1 3′ end.
ACKNOWLEDGMENTS
We thank Brendan Cormack for providing the yeast enhanced GFP gene, Becky Hoffman and Kathy Schell for assistance with FACS analysis, Brett Lindenbach for technical assistance and helpful comments, and Cindy Luongo for helpful comments.
This research was supported by the National Institutes of Health through grant GM35072. P.A. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Ahlquist P, Janda M. cDNA cloning and in vitro transcription of the complete brome mosaic virus genome. Mol Cell Biol. 1984;4:2876–2882. doi: 10.1128/mcb.4.12.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison R F, Janda M, Ahlquist P. Infectious in vitro transcripts from cowpea chlorotic mottle virus cDNA clones and exchange of individual RNA components with brome mosaic virus. J Virol. 1988;62:3581–3588. doi: 10.1128/jvi.62.10.3581-3588.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 4.Ball L A. Requirements for the self-directed replication of flock house virus RNA 1. J Virol. 1995;69:720–727. doi: 10.1128/jvi.69.2.720-727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ball L A, Amann J M, Garrett B K. Replication of Nodamura virus after transfection of viral RNA into mammalian cells in culture. J Virol. 1992;66:2326–2334. doi: 10.1128/jvi.66.4.2326-2334.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ball L A, Johnson K L. Nodaviruses of insects. In: Miller L K, Ball L A, editors. The insect viruses. New York, N.Y: Plenum Publishing Corporation; 1998. pp. 225–267. [Google Scholar]
- 7.Ball L A, Wohlrab B, Li Y. Nodavirus RNA replication: mechanism and harnessing to vaccinia virus recombinants. Arch Virol Suppl. 1994;9:407–416. doi: 10.1007/978-3-7091-9326-6_40. [DOI] [PubMed] [Google Scholar]
- 8.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 9.Comps M, Pepin J F, Bonami J R. Purification and characterization of two fish encephalitis viruses (FEV) infecting Lates calcarifer and Dicentrarchus labrax. Aquaculture. 1994;123:1–10. [Google Scholar]
- 10.Cormack B P, Bertram G, Egerton M, Gow N A, Falkow S, Brown A J. Yeast-enhanced green fluorescent protein (yEGFP), a reporter of gene expression in Candida albicans. Microbiology. 1997;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- 11.Dasgupta R, Sgro J Y. Nucleotide sequences of three nodavirus RNA2's: the messengers for their coat protein precursors. Nucleic Acids Res. 1989;17:7525–7526. doi: 10.1093/nar/17.18.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dasmahapatra B, Dasgupta R, Saunders K, Selling B, Gallagher T, Kaesberg P. Infectious RNA derived by transcription from cloned cDNA copies of the genomic RNA of an insect virus. Proc Natl Acad Sci USA. 1986;83:63–66. doi: 10.1073/pnas.83.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dearing S C, Scotti P D, Wigley P J, Dhana S D. A small RNA virus isolate from the grass grub Costelytra zealandica (Coleoptra: Scarabaeidae) N Z J Zool. 1980;7:267–269. [Google Scholar]
- 14.Friesen P D, Rueckert R R. Black beetle virus: messenger for protein B is a subgenomic viral RNA. J Virol. 1982;42:986–995. doi: 10.1128/jvi.42.3.986-995.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friesen P D, Rueckert R R. Synthesis of black beetle virus proteins in cultured Drosophila cells: differential expression of RNAs 1 and 2. J Virol. 1981;37:876–886. doi: 10.1128/jvi.37.3.876-886.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friesen P D, Scotti P, Longworth J, Rueckert R R. Black beetle virus: propagation in Drosophila line 1 cells and an infection-resistant subline carrying endogenous black beetle virus-related particles. J Virol. 1980;35:741–747. doi: 10.1128/jvi.35.3.741-747.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallagher T M. Ph.D. thesis. Madison: University of Wisconsin-Madison; 1987. [Google Scholar]
- 18.Gallagher T M, Friesen P D, Rueckert R R. Autonomous replication and expression of RNA1 from black beetle virus. J Virol. 1983;46:481–489. doi: 10.1128/jvi.46.2.481-489.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 20.Guarino L A, Ghosh A, Dasmahapatra B, Dasgupta R, Kaesberg P. Sequence of the black beetle virus subgenomic RNA and its location in the viral genome. Virology. 1984;139:199–203. doi: 10.1016/0042-6822(84)90342-8. [DOI] [PubMed] [Google Scholar]
- 21.Gupta K C, Kingsbury D W. Complete sequences of the intergenic and mRNA start signals in the Sendai virus genome: homologies with the genome of vesicular stomatitis virus. Nucleic Acids Res. 1984;12:3829–3841. doi: 10.1093/nar/12.9.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harper T A. Ph.D. thesis. Madison: University of Wisconsin—Madison; 1994. [Google Scholar]
- 23.Hentze M W, Kulozik A E. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 24.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 25.Ishikawa M, Diez J, Restrepo-Hartwig M, Ahlquist P. Yeast mutations in multiple complementation groups inhibit brome mosaic virus RNA replication and transcription and perturb regulated expression of the viral polymerase-like gene. Proc Natl Acad Sci USA. 1997;94:13810–13815. doi: 10.1073/pnas.94.25.13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishikawa M, Janda M, Krol M A, Ahlquist P. In vivo DNA expression of functional brome mosaic virus RNA replicons in Saccharomyces cerevisiae. J Virol. 1997;71:7781–7790. doi: 10.1128/jvi.71.10.7781-7790.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jablonski S A, Morrow C D. Mutation of the aspartic acid residues of the GDD sequence motif of poliovirus RNA-dependent RNA polymerase results in enzymes with altered metal ion requirements for activity. J Virol. 1995;69:1532–1539. doi: 10.1128/jvi.69.3.1532-1539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroner P, Ahlquist P. RNA-based viruses. In: Gurr S J, McPherson M J, Bowles D J, editors. Molecular plant pathology. Vol. 1. Oxford, United Kingdom: IRL Press; 1992. pp. 23–34. [Google Scholar]
- 29.Leeds P, Peltz S W, Jacobson A, Culbertson M R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 30.Mori K, Nakai T, Muroga K, Arimoto M, Mushiake K, Furusawa I. Properties of a new virus belonging to Nodaviridae found in larval striped jack (Pseudocaranx dentex) with nervous necrosis. Virology. 1992;187:368–371. doi: 10.1016/0042-6822(92)90329-n. [DOI] [PubMed] [Google Scholar]
- 31.Munday B L, Nakai T, Nguyen H D. Antigenic relationship of the picorna-like virus of larval barramundi, Lates calcarifer Bloch, to the nodavirus of larval striped jack, Pseudocaranx dentex (Bloch & Schneider) Aust Vet J. 1994;71:384–385. doi: 10.1111/j.1751-0813.1994.tb00937.x. [DOI] [PubMed] [Google Scholar]
- 32.Newman T C, Ohme-Takagi M, Taylor C B, Green P J. DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell. 1993;5:701–714. doi: 10.1105/tpc.5.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niedenthal R K, Riles L, Johnston M, Hegemann J H. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 34.Price B D, Rueckert R R, Ahlquist P. Complete replication of an animal virus and maintenance of expression vectors derived from it in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:9465–9470. doi: 10.1073/pnas.93.18.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawicki S G, Sawicki D L. A new model for coronavirus transcription. Adv Exp Med Biol. 1998;440:215–219. doi: 10.1007/978-1-4615-5331-1_26. [DOI] [PubMed] [Google Scholar]
- 36.Selling B H. Ph.D. thesis. Madison: University of Wisconsin-Madison; 1986. [Google Scholar]
- 37.Selling B H, Rueckert R R. Plaque assay for black beetle virus. J Virol. 1984;51:251–253. doi: 10.1128/jvi.51.1.251-253.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 39.Sit T L, Vaewhongs A A, Lommel S A. RNA-mediated trans-activation of transcription from a viral RNA. Science. 1998;281:829–832. doi: 10.1126/science.281.5378.829. [DOI] [PubMed] [Google Scholar]
- 40.Sullivan M L, Ahlquist P. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J Virol. 1999;73:2622–2632. doi: 10.1128/jvi.73.4.2622-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sutton A, Broach J R. Signals for transcription initiation and termination in the Saccharomyces cerevisiae plasmid 2μm circle. Mol Cell Biol. 1985;5:2770–2780. doi: 10.1128/mcb.5.10.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wodicka L, Dong H, Mittmann M, Ho M H, Lockhart D J. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 43.Wong F, Komaromy M. Site-directed mutagenesis using thermostable enzymes. BioTechniques. 1995;18:1034–1038. [PubMed] [Google Scholar]
- 44.Zhong W. Ph.D. thesis. Madison: University of Wisconsin-Madison; 1993. [Google Scholar]
- 45.Zhong W, Rueckert R R. Flock house virus: down-regulation of subgenomic RNA3 synthesis does not involve coat protein and is targeted to synthesis of its positive strand. J Virol. 1993;67:2716–2722. doi: 10.1128/jvi.67.5.2716-2722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]