Abstract
All retrovirus glycoproteins have a cytoplasmic domain that plays several roles in virus replication. We have determined whether and how the cytoplasmic domains of oncoretrovirus glycoproteins modulate their intracellular trafficking, by using chimeric proteins that combined the α-chain of the interleukin-2 receptor with the glycoprotein cytoplasmic domains of five oncoretroviruses: human T-cell leukemia virus type 1 (HTLV-1), Rous sarcoma virus (RSV), bovine leukemia virus (BLV), murine leukemia virus (MuLV), and Mason-Pfizer monkey virus (MPMV). All of these proteins were synthesized and matured in the same way as a control protein with no retrovirus cytoplasmic domain. However, the amounts of all chimeric proteins at the cell surface were smaller than that of the control protein. The protein appearing at and leaving the cell surface and endocytosis were measured in stable transfectants expressing the chimera. We identified two groups of proteins which followed distinct intracellular pathways. Group 1 included chimeric proteins that reached the cell surface normally but were rapidly endocytosed afterwards. This group included the chimeric proteins with HTLV-1, RSV, and BLV cytoplasmic domains. Group 2 included chimeric proteins that were not detected at the cell surface, despite normal intracellular concentrations, and were accumulated in the Golgi complex. This group included the chimeric proteins with MuLV and MPMV cytoplasmic domains. Finally, we verified that the MuLV envelope glycoproteins behaved in the same way as the corresponding chimeras. These results indicate that retroviruses have evolved two distinct mechanisms to ensure a similar biological feature: low concentrations of their glycoproteins at the cell surface.
Retrovirus envelope glycoproteins are heterodimers consisting of surface and transmembrane (TM) subunits. All retrovirus TM subunits have an intracellular cytoplasmic domain that is generally less than 50 amino acids long, but it is 150 amino acids long in lentiviruses. A number of functions have been assigned to the cytoplasmic domains of retrovirus glycoproteins. They modulate the cell-to-cell fusion ability of glycoproteins (4, 19, 21, 28, 31, 32, 43, 47) and the incorporation of envelope glycoproteins into viral particles, at least for lentiviruses such as the human immunodeficiency virus (HIV) (9, 40, 44). The glycoprotein cytoplasmic domains of the Moloney murine leukemia virus (Mo-MuLV) and the human T-cell leukemia virus type 1 (HTLV-1) are also involved in steps following incorporation and are required for infectivity (7, 14).
The cytoplasmic domains of cell membrane proteins contain sorting signals that specify their intracellular trafficking and allow the transport of newly synthesized proteins to a variety of destinations on the cell surface and inside the cell (reviewed in reference 13). There are also reasons to believe that retroviral glycoproteins are no exception to this rule. Retrovirus envelope glycoproteins are addressed only to the basolateral membrane in polarized epithelial cells (2, 6, 15–17), a property assigned to their cytoplasmic domains. The glycoprotein cytoplasmic domains of lentiviruses also interact with adaptor proteins of clathrin-coated vesicles (3, 24) and harbor motifs that drive sorting of the glycoproteins to the endocytic pathway (34, 35). This probably explains the augmented fusion phenotype produced by deletions of cytoplasmic domains, a consequence of increased protein at the cell surface due to reduced endocytosis of the truncated glycoproteins. However, little is known about the intracellular routing determined by the glycoprotein cytoplasmic domains in retroviruses that do not belong to the Lentivirus genus.
We have generated chimeric proteins composed of the cytoplasmic domains of several oncovirus glycoproteins and the α-chain of the interleukin-2 (IL-2) receptor and used them to determine whether and how the cytoplasmic domain of oncovirus glycoproteins modulates their intracellular trafficking. The cytoplasmic domains were those of HTLV-1, Rous sarcoma virus (RSV), bovine leukemia virus (BLV), Mo-MuLV, and Mason-Pfizer monkey virus (MPMV). We find that all these domains reduced the amount of the chimera at the cell surface. They did so through one of two systems: one involved endocytosis of protein at the cell surface (HTLV-1, RSV, and BLV), and the other involved intracellular retention (Mo-MuLV and MPMV). The MuLV envelope glycoproteins had the same Golgi intracellular localization as the corresponding chimeric proteins. These results suggest that the cytoplasmic domains of retroviral envelope glycoproteins contain sufficient information to limit their amount at the cell surface.
MATERIALS AND METHODS
Cells, MAbs, and reagents.
HeLa cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% decomplemented fetal calf serum (FCS) and 2 mM l-glutamine (complete medium). Stably transfected HeLa cells were grown in the same medium supplemented with 200 μg of hygromycin per ml (Calbiochem, La Jolla, Calif.).
The 2A3A1H and 7G7B6 monoclonal antibodies (MAbs) are directed against the α-chain of the IL-2 receptor (CD25). The H48 MAb raised against the MuLV SU glycoproteins was a gift from M. Sitbon (IGM, Montpellier, France). The anti-Rab6 antibody was from a rabbit polyclonal antiserum (gift of B. Goud, Institut Curie, Paris, France). The transferrin receptor (TFR) was revealed by using a human transferrin-cyanine 3 conjugate.
Plasmids.
The TX-O plasmid is a mammalian expression vector containing the complete cDNA of the α-chain of the IL-2 receptor (CD25) modified at the 3′ end of the CD25 cDNA to create a HindIII-XbaI cloning cassette. The CD25-TFR construct is a TX-O derivative, which encodes the CD25 sequence with a C-terminal insertion of the endocytic motif of the TFR, YTRF (37).
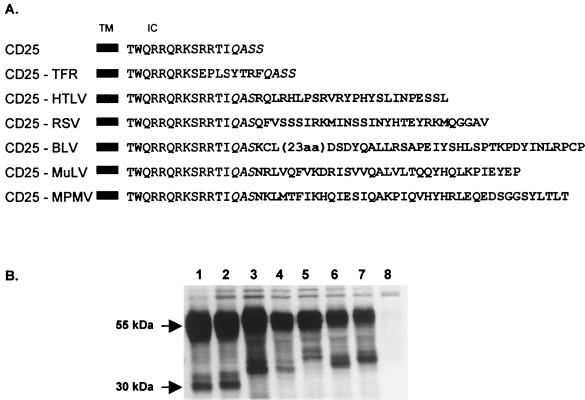
The CD25-HTLV and CD25-BLV constructs were obtained by PCR amplification with the CMV-ENV plasmid (8) or the pSG-env-BLV plasmid (gift from A. Burny, Faculté d'Agronomie, Gembloux, Belgium) to generate a HindIII-XbaI insert encoding the cytoplasmic domains of the HTLV-1 or BLV envelope glycoproteins, respectively. These PCR fragments were then inserted into the TX-O vector, previously cut with HindIII and XbaI. The other three CD25 constructs (CD25-RSV, CD25-MuLV, and CD25-MPMV) were obtained by subcloning synthetic oligonucleotides encoding the different retrovirus glycoprotein cytoplasmic domains (Life Technologies, Cergy Pontoise, France; Genaxis Biotechnology, Montigneux le Bretonneux, France) into the HindIII-XbaI cloning cassette of the TX-O plasmid. All constructs were verified by automatic sequencing. The amino acid sequences of the resulting chimeric proteins are shown in Fig. 1A.
FIG. 1.
Chimeric proteins used in this study and their immunoprecipitation after transient transfection of HeLa cells. (A) All chimeric proteins were obtained by inserting the corresponding glycoprotein cytoplasmic domain at the C terminus of the IL-2 receptor α-chain (CD25) (boldface letters). The CD25-TFR chimeric protein consists of the insertion of the endocytic motif of the TFR into the CD25 cytosolic tail. The chimeric proteins are named CD25-retrovirus. TM, transmembrane domain; IC, intracytoplasmic domain. (B) Immunoprecipitation of chimeric proteins after transient transfection of HeLa cells. Cells were radiolabeled 48 h after transfection and lysed. Lanes: 1, CD25; 2, CD25-TFR; 3, CD25-HTLV; 4, CD25-RSV; 5, CD25-BLV; 6, CD25-MuLV; 7, CD25-MPMV; 8, negative control vector (pcDNA3).
The pCEL/F Friend MuLV envelope glycoprotein expressor was a gift from M. Sitbon (IGM).
Cell transfections.
Stable transfectants were generated after transfection of 3.105 HeLa cells per well of six-well plates with 2.7 μg of the CD25 construct of interest and 0.3 μg of pHyg plasmid expressing the hygromycin resistance gene (provided by T. Issad, ICGM, Paris, France), by the calcium phosphate method. Forty-eight hours posttransfection, the cells were split into 10 plates. Selection was applied 24 h later by supplementing the complete medium with hygromycin (200 μg/ml). Hygromycin-resistant clones were then assayed for chimeric protein expression with an indirect immunofluorescence assay with the anti-CD25 7G7B6 MAb (ascitic fluid [1/500]) and a cyanine 3-coupled goat anti-mouse immunoglobulin (Ig) (Jackson Immunoresearch Laboratories, Inc. [1/300]).
Transient transfections were performed by the calcium phosphate procedure. For flow cytometry analysis, 7 × 105 HeLa cells plated in 10-mm-diameter dishes were cotransfected with 2.5 or 8 μg of the plasmids of interest and 2 μg of pEGFP1 vector (provided by M. Alizon, ICGM), which allowed the detection of transfected cells by the synthesis of green fluorescent protein (GFP). The total quantity of DNA was normalized to 10 μg by adding empty vector. For immunoprecipitation assays, 3 × 105 HeLa cells plated per well of six-well plates were transfected with 3 μg of plasmid DNA. For immunofluorescence assays, 2 × 104 cells plated per well of 24-well plates were transfected with 300 ng of plasmid. The total quantity of DNA was normalized to 1 μg by adding empty vector.
Flow cytometry.
Cells were collected 18 h after transfection by incubation with PBS containing 5 mM EDTA (37°C for 10 min), pelleted, and suspended in ice-cold PBS. They were then incubated for 1 h with the anti-CD25 2A3A1H MAb (ascitic fluid [1/2,000]) in 100 μl of PBS at 4°C, washed twice with PBS, and stained with phycoerythrin-conjugated goat anti-mouse Ig (Caltag, South San Francisco, Calif.) for 1 h at 4°C. The cells were washed again and fixed in PBS containing 2% formaldehyde (FAD) and analyzed by flow cytometry. GFP-negative cells were excluded from the analysis.
Endocytosis of chimeric proteins.
Stably transfected cells (107) were collected as described above, incubated with the anti-CD25 2A3A1H MAb (ascitic fluid [1/2,000]) for 1 h on ice, and washed in chilled PBS. They were then warmed to 37°C for the indicated times, rapidly cooled to 4°C, and washed once. MAbs bound to the cell surface were revealed by incubating cells with phycoerythrin-coupled goat anti-mouse Ig for 1 h at 4°C. Cells were washed twice with chilled PBS, fixed in PBS–2% FAD solution, and analyzed by flow cytometry. The internalization rate was calculated as follows:
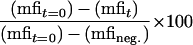 |
where mfit is the mean fluorescence intensity of cells harvested after incubation for t min at 37°C and mfineg. is the background staining without primary antibody.
Immunoprecipitation.
Thirty-six hours posttransfection, cells were washed and incubated overnight with 200 μCi of Promix 35S protein-labeling mixture (Amersham, Courtaboeuf, France) diluted in methionine- and cysteine-free DMEM containing 10% dialyzed fetal calf serum, l-glutamine, and antibiotics. Radiolabeled cells were lysed in 500 μl of MacDougal buffer (20 mM Tris-HCl [pH 8.0], 120 mM NaCl, 200 μM EGTA, 0.2 μM NaF, 0.2% sodium deoxycholate, 0.5% Nonidet P-40) supplemented with a protease inhibitor cocktail (Complete; Roche Diagnostic, France) and centrifuged at 20,000 × g for 30 min. Radiolabeled supernatants were collected and cleared by incubation for 2 h with anti-actin rabbit serum immobilized on protein G-Sepharose beads. In parallel, for immunoprecipitation of CD25 chimeric proteins, 100 μl of protein G-Sepharose was washed twice with 1 ml of MacDougal buffer and incubated with 3 μl of the anti-CD25 7G7B6 MAb (ascitic fluid) for 1 h on ice. Normal unlabeled HeLa cell lysate (500 μl) was then added to the antibody for 2 h at 4°C. Finally, the radiolabeled cleared supernatants were added, and the mixture was incubated overnight at 4°C. The beads were washed 15 times with MacDougal buffer, and the immune complexes were released from the beads by boiling the samples for 5 min in 40 μl of 1× sample buffer (125 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 9% glycerol, 0.7 M β-mercaptoethanol, 0.005% bromophenol blue). Radiolabeled proteins were separated by SDS–10% polyacrylamide gel electrophoresis.
Cell surface appearance of chimeric proteins.
The presence of the chimeric proteins was monitored by giving a radioactive pulse followed by a chase and biotinylation of the cell surface proteins followed by precipitation with streptavidin-agarose beads (surface fraction). The total cell content of chimeric proteins was measured in parallel by immunoprecipitation (total fraction). Briefly, confluent cells grown in six-well plates were rinsed twice with PBS and incubated for 1 h in methionine- and cysteine-free DMEM containing 10% dialyzed FCS. They were then incubated for 15 min with 200 μCi of Promix 35S protein-labeling mixture in 1 ml of incubation medium, followed by a chase in complete medium for 0, 30, 120, 240, or 360 min. The cells were chilled on ice, washed twice with ice-cold PBS (pH 8.0) containing 0.7 mM CaCl2 and 0.25 mM MgSO4 (PBS++), and incubated with 1 ml of sulfo-NHS-LC-LC-biotin (Pierce) solution (0.5 mg/ml in PBS++) for 30 min on ice. Biotinylation was stopped by adding 100 μl of PBS++ and 1 M glycine, followed by incubation for 5 min on ice. Cells were washed with PBS–0.1 M glycine (pH 7.4) and lysed with 500 μl of MacDougal buffer containing a protease inhibitor cocktail and 0.1 M glycine (MacDougal/glycine buffer). The lysates were immunoprecipitated overnight as described above with the anti-CD25 7G7B6 MAb. The immune complexes were released from the beads by boiling the samples for 5 min in 100 μl of SDS buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 2% SDS). Aliquots (20 μl) of supernatant were diluted in 2× sample buffer and frozen at 80°C. This represented the “total” chimeric protein fraction. The other 80 μl was placed in 1 ml of MacDougal/glycine buffer containing 40 μl of streptavidin-Sepharose beads (Pierce) and incubated overnight at 4°C. The beads were then washed five times in MacDougal/glycine buffer, and the biotinylated proteins were eluted from the streptavidin-Sepharose beads by boiling the sample for 5 min in 40 μl of 1× sample buffer. This represented the “surface” chimeric protein fraction. Radiolabeled proteins were separated by SDS–12% polyacrylamide gel electrophoresis.
Immunofluorescence.
Cells grown on glass coverslips (Polylabo, Strasbourg, France) were fixed in PBS–4% paraformaldehyde for 15 min at room temperature, quenched for 15 min in PBS–0.1 M glycine, and permeabilized for 40 min with PBS containing 0.05% saponin and 0.2% bovine serum albumin. They were then incubated for 1 h with the first antibody (anti-CD25 7G7B6 MAb, ascitic fluid  ) diluted in the permeabilizing buffer, washed, and incubated for 1 h with cyanine 3-coupled goat anti-mouse Ig (Jackson Immunoresearch Laboratories, Inc. [1/300 in permeabilizing buffer]). The cells were mounted in a solution containing 100 mg of mowiol per ml (Calbiochem), 30% (wt/vol) glycerol, and 100 mM Tris-HCl (pH 8.5) and examined under a confocal microscope (model MRC-1024; Bio-Rad).
) diluted in the permeabilizing buffer, washed, and incubated for 1 h with cyanine 3-coupled goat anti-mouse Ig (Jackson Immunoresearch Laboratories, Inc. [1/300 in permeabilizing buffer]). The cells were mounted in a solution containing 100 mg of mowiol per ml (Calbiochem), 30% (wt/vol) glycerol, and 100 mM Tris-HCl (pH 8.5) and examined under a confocal microscope (model MRC-1024; Bio-Rad).
Colocalization experiments were performed with the same protocol. Cells were incubated with rabbit polyclonal anti-Rab6 antibodies (1/200) and the anti-CD25 7G7B6 MAb or the anti-MuLV SU H48 MAb and then with cyanine 3-coupled goat anti-rabbit Ig (Jackson Immunoresearch Laboratories, Inc. [1/300]) and fluorescein isothiocyanate (FITC)-coupled goat anti-mouse Ig (Jackson Immunoresearch Laboratories, Inc. [1/300]). The endocytosis pathway was revealed by incubating cells in serum-free medium for 30 min and then with transferrin-cyanine 3 conjugate (100 nM) for 20 min. Cells were then fixed and treated as described above to reveal CD25 chimeric proteins.
RESULTS
Influence of the cytoplasmic domains of oncovirus envelope glycoproteins on the amounts of chimeric proteins at the cell surface.
We compared the properties of the cytoplasmic domains of several oncoviral envelope glycoproteins by using five constructs expressing chimeric proteins. The chimeric proteins were the full-length sequences of the cytoplasmic domains of HTLV-1, RSV, BLV, MuLV, or MPMV glycoproteins inserted at the carboxy terminus of the IL-2 receptor α-chain (CD25). The resulting constructs were named CD25-HTLV, CD25-RSV, CD25-BLV, CD25-MuLV, and CD25-MPMV (Fig. 1A).
The cell contents of all chimeric proteins were similar to that of the wild-type CD25, and they underwent normal maturation in transiently transfected HeLa cells (Fig. 1B, lanes 3 to 7). The mature glycosylated form of the IL-2 receptor α-chain migrated at 55 kDa, whereas the immature forms migrated at around 30 kDa (41). The differences in the migration of the immature forms of the chimera were due to the different lengths of the engrafted cytoplasmic domains.
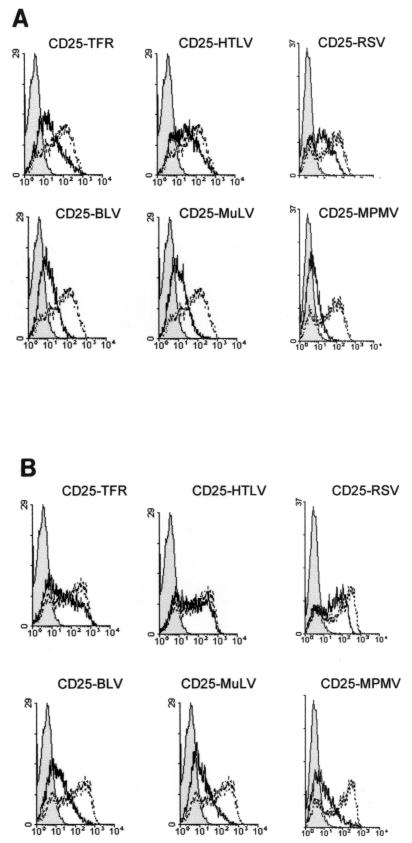
The profiles of the various chimeric proteins at the cell surface were obtained by transient transfection of HeLa cells followed by flow cytometry analysis (Fig. 2). There was considerable CD25 wild-type protein at the cell surface, even when small amounts of DNA were transfected (2.5 μg) (Fig. 2A). In contrast, cells transfected under the same conditions with DNA encoding the CD25 chimeric proteins had less protein at the cell surface, whatever the sequence of the engrafted virus cytoplasmic domain (Fig. 2A). This effect was most drastic for the CD25-BLV, CD25-MuLV, and CD25-MPMV chimeras.
FIG. 2.
Chimeric proteins at the cell surface after transient transfection of HeLa cells. HeLa cells were cotransfected with vectors expressing GFP and 2.5 μg (A) or 8 μg (B) of vector expressing the chimeric proteins. Chimeric proteins at the cell surface were measured 18 h posttransfection by flow cytometry on GFP-positive cells. One representative experiment out of at least three performed is shown. Black line profiles represent the background reactivity of the anti-CD25 MAb to HeLa cells transfected with a negative control vector (pcDNA3). Bold line profiles represent chimeric proteins, and broken line profiles represent CD25 wild-type antigen, at the cell surface, both detected with the anti-CD25 2A3A1H MAb.
We then tested whether increasing the amount of transfected DNA to 8 μg could compensate for the small amounts of chimeric proteins at the cell surface (Fig. 2B). There was almost as much of the CD25-HTLV and CD25-RSV chimeras at the cell surface as with the CD25 wild-type protein under these conditions (Fig. 2B). This effect was similar to that observed with the CD25-TFR reference chimeric protein (Fig. 2B), which contained the endocytic motif responsible for the down-regulation of the TFR (37). In contrast, the amounts of CD25-BLV, CD25-MuLV, and CD25-MPMV chimeras at the cell surface were less influenced when larger amounts of DNA were transfected (Fig. 2B). This was not due to poor intracellular protein production, because the synthesis of these chimeric proteins was comparable to that of the other proteins (Fig. 1B [compare lanes 5 to 7 to the other lanes]).
We showed that the 10-amino-acid spacing of the retroviral tails away from the membrane in CD25 chimeric proteins had no effect, by using CD8 chimeric proteins containing the HTLV-1 or MuLV cytoplasmic domains engrafted at the carboxy terminus of the CD8 protein, with no additional amino acid. Again, the amounts of the chimeric proteins at the cell surface level were smaller than those of the wild type (data not shown).
Thus, the cytoplasmic domains of HTLV-1, RSV, BLV, MuLV, and MPMV envelope proteins reduced the amounts of CD25 chimeric proteins at the cell surface. This reduction could be completely (HTLV and RSV) or partially (BLV, MuLV, and MPMV) offset by overproduction of the chimeric proteins, suggesting that the mechanisms involved in this down-regulation are saturable.
Influence of the cytoplasmic domain of the chimeric proteins on their appearance and stability at the cell surface.
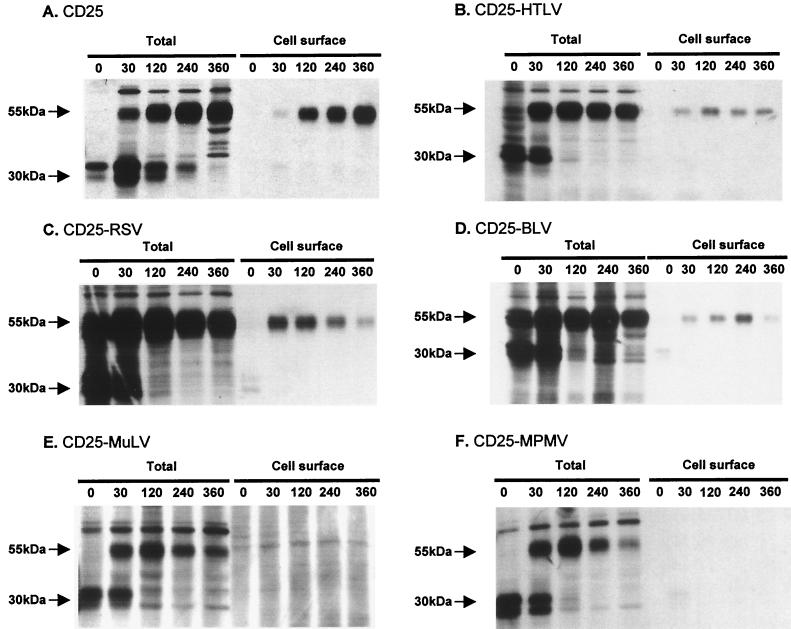
We further examined the way in which the cytoplasmic domains modulated the amounts of chimeric proteins at the cell surface by using cell lines stably expressing each of the chimeras or the CD25 wild-type construct. This was done to avoid overproduction of the proteins, which could disturb protein trafficking in saturable pathways (18). We then determined whether the various cytoplasmic domains modified the kinetics of appearance of the chimeric proteins at the cell surface (Fig. 3).
FIG. 3.
Kinetics of chimeric protein maturation and appearance at the cell surface in stably transfected HeLa cells. Stably transfected HeLa cells were radiolabeled with a 15-min pulse, followed by a chase for different times. Cell surface proteins were biotinylated, and then cells were lysed, and proteins were immunoprecipitated with the 7G7B6 MAb. “Total” indicates immunoprecipitation of chimeric proteins in whole-cell lysates. “Cell surface” indicates streptavidin precipitation of 7G7B6-immunoprecipitated proteins. (A) CD25-expressing clone. (B) CD25-HTLV-expressing clone. (C) CD25-RSV-expressing clone. (D) CD25-BLV-expressing clone. (E) CD25-MuLV-expressing clone. (F) CD25-MPMV-expressing clone. The experiments were performed for at least two independent clones in each case with similar results.
The CD25 wild-type protein was first synthesized as a 30-kDa polypeptide, which was then glycosylated at two potential glycosylation sites, giving a final 55-kDa product (Fig. 3A [total]) (41). This process was completed in 2 to 4 h, as for all chimeric proteins (Fig. 3B, C, D, E, and F [total]).
Thirty minutes was sufficient for the CD25 molecule to reach the cell surface, where it accumulated for the next 6 h (Fig. 3A [cell surface]). The CD25-HTLV, CD25-RSV, and CD25-BLV chimeric proteins reached the cell surface with kinetics similar to that of CD25, also appearing at the cell surface after 30 min (Fig. 3 B, C, and D [cell surface]). However, they did not accumulate afterwards. The highest concentration of the CD25-HTLV proteins at the cell surface was at 2 h, and the concentration then decreased (Fig. 3B [cell surface]). The chimeric protein CD25-RSV did not accumulate after 30 min but quickly decreased (Fig. 3C [cell surface]). The CD25-BLV chimeric protein was accumulated a little later than the previous ones (until 4 h after the pulse) and then rapidly decreased (Fig. 3D [cell surface]).
In contrast, the CD25-MuLV and CD25-MPMV chimeric proteins were not detected at the cell surface (Fig. 3E and F [cell surface]), despite their correct intracellular maturation (Fig. 3E and F [total]). There was also accelerated intracellular degradation of the mature CD25-MPMV chimeric protein (Fig. 3E [total]).
Thus, there were three profiles: (i) the CD25 profile, with proteins efficiently transported to and accumulated at the cell surface; (ii) the CD25-HTLV, CD25-RSV, and CD25-BLV profiles, with proteins efficiently transported to the cell surface, but which did not accumulate there; and (iii) the CD25-MuLV and CD25-MPMV profile, in which the mature proteins remained in the cell and were not detected at the cell surface.
Influence of the cytoplasmic domains of HTLV, RSV, and BLV envelope glycoproteins on internalization of the chimeric proteins.
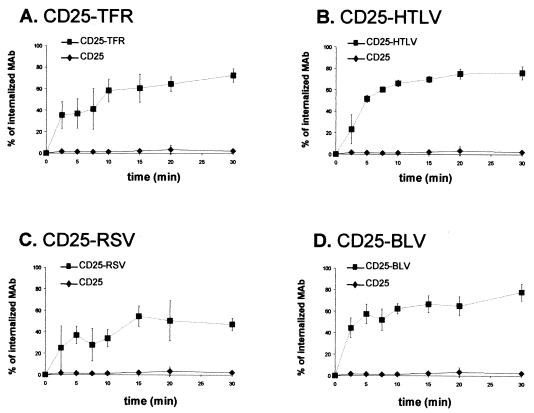
The phenotype of CD25-HTLV, CD25-RSV, and CD25-BLV chimeric proteins was examined by determining whether they were internalized once they had reached the cell surface. The endocytosis of these proteins was measured and compared to that of the CD25 or CD25-TFR chimera, with an anti-CD25 MAb as a ligand.
The CD25 wild-type protein was not internalized, whereas the CD25-TFR control chimeric protein, which contained a functional endocytic signal, was rapidly internalized (Fig. 4A). The CD25-HTLV, CD25-RSV, and CD25-BLV chimeric proteins were internalized (Fig. 4B, C, and D, respectively). The CD25-HTLV and CD25-BLV chimeras were internalized to a slightly greater extent than the CD25-RSV chimeric protein, with 70% of the bound ligand being internalized after 30 min for CD25-HTLV and CD25-BLV, compared to 50% for CD25-RSV.
FIG. 4.
Endocytosis of chimeric proteins at the cell surface. Values are percentage of anti-CD25 MAb internalized compared to MAb bound at time 0. Each curve represents the mean of three independent experiments performed in two distinct stable clones. Error bars show the standard deviation. At time 0 min, the medium fluorescence intensities were 4.93 ± 1.88, 0.83 ± 0.14, 0.73 ± 0.19, 1.18 ± 0.95, and 0.33 ± 0.10 for the CD25-, CD25-TFR (A)-, CD25-HTLV (B)-, CD25-RSV (C)-, and CD25-BLV (D)-expressing clones, respectively (mean between medium fluorescence intensity obtained at time 0 min in endocytosis experiments after deduction of the background value ± standard error).
The disappearance of the CD25-HTLV, CD25-RSV, and CD25-BLV chimeric proteins from the cell surface (Fig. 3) could thus be due to their rapid internalization, probably followed by degradation of a fraction of the proteins.
Targeting of the chimeric proteins to the TFR endocytosis pathway by the cytoplasmic domains of HTLV, RSV, and BLV envelope glycoproteins.
We first analyzed the intracellular distribution of the CD25-HTLV, CD25-RSV, and CD25-BLV chimeric proteins in stably transfected HeLa cell lines at steady state. The intracellular distribution of the retrovirus chimera differed from that of the two control proteins, CD25 and CD25-TFR. The CD25-HTLV, CD25-RSV, and CD25-BLV chimeric proteins were found in a perinuclear area and in peripheral dots (Fig. 5 [CD25-HTLV, CD25-RSV, and CD25-BLV panels]). These observations were similar in all of the clones tested. The CD25 control protein was detected mainly at the cell surface (Fig. 5 [CD25 panel]), whereas the CD25-TFR control protein was present in intracellular vesicles with a typical early and recycling endosome staining (Fig. 5 [TFR panel]).
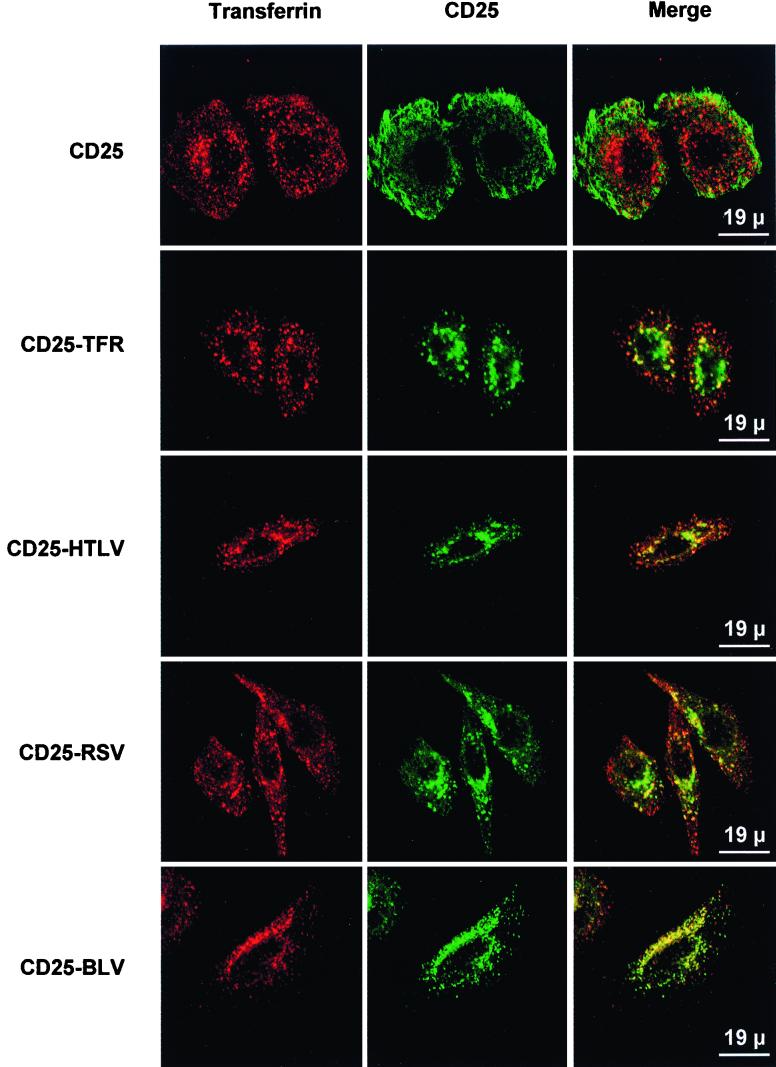
FIG. 5.
Colocalization of CD25-HTLV, CD25-RSV, and CD25-BLV chimeric proteins with transferrin, as observed by confocal microscopy. Stably transfected HeLa cells were incubated with cyanine 3-coupled transferrin prior to fixation and permeabilization. CD25 chimeric proteins were detected by using the 7G7B6 MAb. One representative experiment of at least three performed is shown.
Colocalization experiments used markers of different cellular compartments, including Rab6 GTPase (medial and trans-Golgi) and transferrin (early and recycling endosomes). The CD25 protein did not colocalize with any of these intracellular markers at steady state, and the CD25-TFR protein partly colocalized with transferrin (Fig. 5 [CD25-TFR panel] and data not shown). The CD25-HTLV, CD25-RSV, and CD25-BLV chimeric proteins also partly colocalized with the transferrin marker (Fig. 5 [CD25-HTLV, CD25-RSV, and CD25-BLV panels, respectively]). Thus, the cytoplasmic domains of HTLV, RSV, and BLV envelope proteins can target a chimeric protein to the endocytic pathway.
Localization of CD25-MuLV and CD25-MPMV chimeric proteins in the Golgi apparatus at steady state.
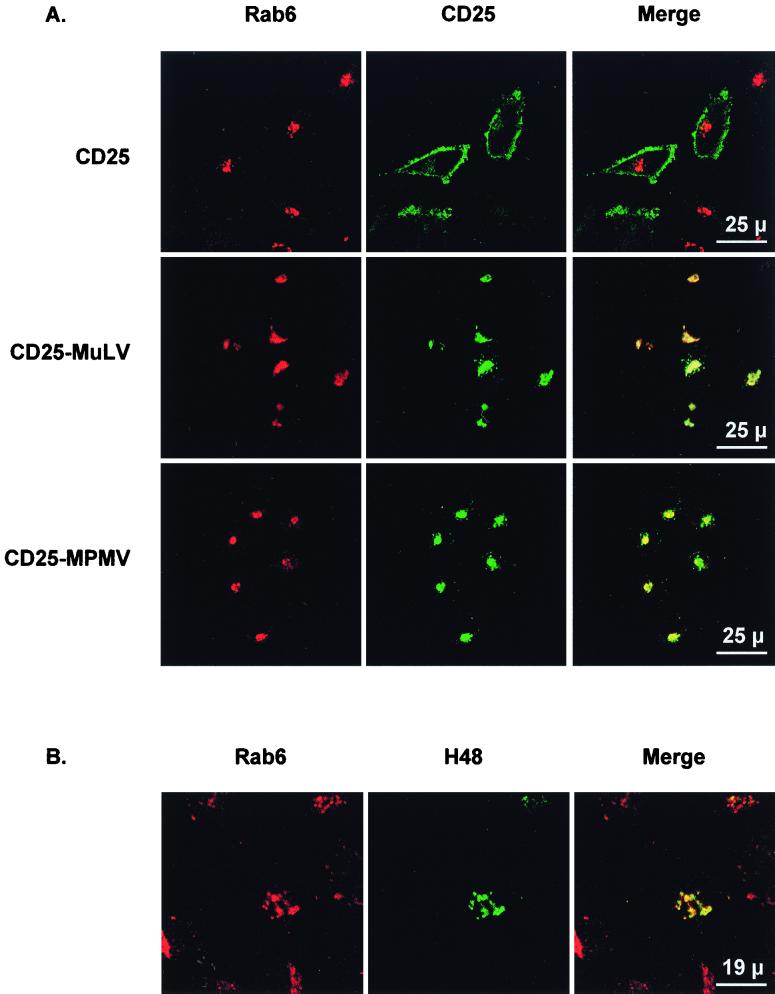
We investigated the intracellular distribution of the CD25-MuLV and the CD25-MPMV chimeric proteins in stably transfected HeLa cells at steady state. The CD25-MuLV and CD25-MPMV chimeric proteins were restricted to a perinuclear region (Fig. 6A [CD25-MuLV and CD25-MPMV panels]). These proteins were present in neither the sorting endosomes, revealed by the transferrin marker, nor the trans-Golgi network, revealed by the furin convertase marker (data not shown). However, there was a significant colocalization with the Rab6 marker, indicating large amounts of the chimeric proteins in the Golgi apparatus (Fig. 6A). These results suggest that these proteins can accumulate in the Golgi and that exit from the Golgi apparatus could be a limiting step in the intracellular transport of the CD25-MuLV and CD25-MPMV chimeric proteins.
FIG. 6.
(A) Colocalization of CD25-MuLV and CD25-MPMV chimeric proteins with the Rab6 Golgi marker, as observed by confocal microscopy. Stably transfected HeLa cells were fixed and permeabilized. The Golgi complex was revealed with an anti-Rab6 rabbit polyclonal antiserum, and the CD25 chimeric proteins were revealed with the 7G7B6 MAb. One representative experiment of at least three performed is shown. (B) Intracellular distribution of MuLV envelope glycoproteins. HeLa cells were transiently transfected with the pCEL/F MuLV envelope protein expression vector (300 ng). Forty-eight hours posttransfection, cells were fixed and permeabilized. The Golgi complex was revealed with an anti-Rab6 rabbit polyclonal antiserum, and the MuLV envelope proteins were revealed with the H48 MAb.
Similar intracellular distribution of MuLV envelope glycoproteins and the corresponding CD25-MuLV chimeric protein.
We checked the retention of CD25-MuLV and MPMV chimeras by examining the intracellular distribution of the complete MuLV envelope glycoproteins in transiently transfected HeLa cells (Fig. 6B). As for the CD25-MuLV chimeric proteins, MuLV envelope proteins were restricted to a perinuclear compartment, where they mainly colocalized with the Rab6 Golgi marker (Fig. 6B). Thus, the MuLV envelope proteins were mainly in the Golgi complex at steady state, as was the corresponding CD25-MuLV chimeric protein.
DISCUSSION
We investigated the effects of the glycoprotein cytoplasmic domains from five retroviruses, HTLV-1, RSV, BLV, MuLV, and MPMV, on the intracellular trafficking and appearance at the cell surface of chimeric proteins. The cytoplasmic domains were grafted onto the α-chain of the IL-2 receptor (CD25), so that we could examine the intrinsic properties of each of these domains and compare them in a common context.
All of the oncoviral cytoplasmic domains tested reduced the amounts of chimeric proteins at the cell surface, compared to the wild-type CD25 protein. This reduction did not result from reduced intracellular contents of the chimeric proteins compared to the wild-type protein, because the amounts of proteins inside the transfected cells were comparable. Adding the cytoplasmic domains of MuLV and HTLV-1 to the C terminus of the external and TM domains of the CD8 resulted in a similar reduction in proteins at the cell surface. This showed that the 10 amino acids of CD25 separating the retrovirus cytoplasmic domains from the plasma membrane play no role in the phenotype, because the CD8 chimeras had no additional amino acids. Earlier studies also showed that the behavior of such CD25 chimeric proteins in the cell is strictly dependent upon the presence of specific grafted motifs (18, 20, 24, 37). Thus, engrafting any amino acid sequence is not sufficient to reduce the amount of CD25 chimeric protein at the cell surface.
The reduced amount of proteins at the cell surface was due to one of two intracellular pathways. The first pathway was addressed by the cytoplasmic domains of HTLV-1, RSV, and BLV glycoproteins, which permitted the chimeric proteins to reach the cell surface, followed by their rapid endocytosis. This was confirmed by three sets of data, including the kinetics with which they appeared at the cell surface and decreased thereafter, their rate of endocytosis, and colocalization with transferrin, a marker of the endosomal recycling compartment.
Our results corroborate a recent study showing that the RSV glycoprotein cytoplasmic domain may provide it with an internalization phenotype (23). Results obtained for HTLV-1 also substantiate our previous data showing the in vitro interaction of the glycoprotein cytoplasmic domain with the adaptor complex, which recruits integral membrane proteins to clathrin-coated pits (3). Lentivirus envelope glycoproteins also undergo rapid constitutive endocytosis (34, 35), which is blocked by the presence of Gag proteins for HIV-1 (10). Taken together, these results demonstrate that many retrovirus glycoproteins tend to be eliminated from the cell surface by endocytosis.
The cytoplasmic domains of MuLV and MPMV resulted in a second phenotype. They could prevent the chimeric proteins from reaching the cell surface, by greatly restricting their transport out of the Golgi apparatus. The chimeric proteins were not detected at the plasma membrane in stable transfectants by flow cytometric analysis or by biotinylation of cell surface proteins, and much of the chimeric proteins colocalized with the Rab6 Golgi marker. Moreover, the kinetics of intracellular maturation of both chimeric proteins were similar to that of the CD25 protein, which indicated that the MuLV and MPMV cytoplasmic domains did not influence their transport from the endoplasmic reticulum to the Golgi complex. Thus, the Golgi retention appears to be different from the “quality control” occurring in the endoplasmic reticulum (12).
We also examined the intracellular distribution of the MuLV envelope proteins to confirm the biological significance of the retention. These glycoproteins were not detected at the cell surface at steady state but were in a perinuclear compartment and mainly colocalized with the Rab6 Golgi complex marker. Thus, the MuLV envelope glycoproteins behaved like the corresponding chimera. These results suggest that the MuLV and MPMV retrovirus have evolved a common retention mechanism to limit the amounts of their envelope proteins at the cell surface.
At least two models have been proposed for Golgi retention of resident enzymes such as glycosyltransferases. According to one, the length of the transmembrane domain would result in their segregation in the Golgi complex and their inefficient sorting from this compartment. The other model proposes that the hetero-oligomerization of these Golgi resident enzymes results in hetero-oligomers too large to be incorporated into transport vesicles (22). The mechanism by which CD25-MuLV and CD25-MPMV chimeric proteins are retained in the Golgi is probably different, because the TM domain of CD25 does not allow retention and because the retention we observed depends upon the grafted cytoplasmic domain. Other virus spike proteins also have Golgi retention signals residing in their cytosolic tail (1). This domain might interact with Golgi resident proteins (36) or contain a retrieval signal for keeping proteins in the Golgi. Alternatively, the exit from the Golgi could be a very limiting step for proteins addressed from the Golgi to specific compartments, such as the mannose 6-phosphate receptors or lysosomal membrane proteins (33).
Cytoplasmic domains of MuLV and MPMV glycoproteins possess similarities: an R peptide that is cleaved in the virus particle by the viral protease (5, 11) and 10 conserved amino acids (43). These elements could be essential for the trafficking of the envelope glycoproteins. Moreover, deletion of the R peptide is always correlated with increased cell fusion (4), which might be due to the missorting of truncated glycoproteins to the cell surface (31, 43).
Our results may appear surprising, because envelope glycoproteins need to be present at the cell surface, where these retroviruses bud. We cannot exclude that a very small, undetectable amount of proteins may have passed to the cell surface and was later internalized (45). However, CD25-MuLV and CD25-MPMV proteins were commonly detected at the cell surface in transient transfection experiments when proteins were overproduced in the cell. This suggests that the cellular pool of components involved in the retention of CD25-MuLV and CD25-MPMV chimeric proteins in the Golgi can be saturated, allowing their relocalization to the plasma membrane. Such an accumulation of proteins at the cell surface due to overproduction has been described for other TM proteins (18). Studies showing MuLV or MPMV glycoproteins at the cell surface could be interpreted in the same way. They were all performed with transient transfections or infections with recombinant vaccinia viruses (4, 30, 42, 43), which allowed overproduction of proteins in the cells. Thus, the proteins detected at the cell surface could result from saturation of the intracellular trafficking.
The propensity of the MuLV or MPMV glycoproteins to remain intracellular during the virus cycle may be overcome during the late steps of virus replication, when virus particles are produced, in two ways. First, infected cells could produce or accumulate large amounts of the glycoproteins, thus overcoming their intracellular retention, letting them reach the cell surface. Second, Gag proteins, which were absent from our assays, could help MuLV and MPMV glycoproteins to reach the cell surface. It will be interesting to determine whether homologous Gag proteins can influence the egress of glycoproteins from the intracellular compartment, where they tend to be retained.
All of the retrovirus cytoplasmic domains tested in this study were able to direct glycoproteins to intracellular compartments and reduce the amounts of glycoproteins at the cell surface. The transit of retrovirus glycoproteins into intracellular compartments could be required for interactions between the cytosolic Gag and membrane spike components required for virus assembly, as in several other families of viruses (29, 39). This would generalize the findings in polarized epithelial cells, where the site of retrovirus particle budding is determined by the basolateral addressing of the glycoproteins, suggesting that intracellular interactions precede the interactions of the viral components at the cell membrane and the egress of the virus.
It is also important for retroviruses in particular, and probably for viruses in general (25–27, 38, 39, 46), to minimize the amounts of envelope glycoproteins at the cell surface. Viral envelope proteins are naturally exposed at the surface of infected cells and constitute a major target of the immune response. Endocytosis can be viewed as a way of eliminating virus glycoproteins not incorporated into virions during budding. This would both limit cytopathogenic effects due to fusion effects in vivo and avoid the elimination of infected cells, which is essential for the survival of viruses causing chronic infection. The intracellular retention produced by MuLV and MPMV cytoplasmic domains can also be understood by this logic as a way of minimizing the amount of glycoproteins at the cell surface.
Thus, either of two pathways can reduce the amount of oncovirus glycoproteins at the cell surface. It will be important to define the protein motifs involved in these processes and to determine whether other virus components influence these phenomena.
ACKNOWLEDGMENTS
We thank Franck Letourneur and Nicolas Lebrun for help in plasmid sequencing, Bruno Goud for the gift of antibodies, M. Sitbon for the gifts of pCEL/F plasmid and H48 MAb, and C. Berlioz-Torrent for the gift of CD8 chimeric constructs. The English text was edited by Owen Parkes.
This work was supported by a grant from the Association Nationale pour la Recherche sur le SIDA (ANRS, Paris, France) and by the Association pour la Recherche sur le Cancer (ARC, Villejuif, France).
REFERENCES
- 1.Andersson A M, Melin L, Bean A, Pettersson R F. A retention signal necessary and sufficient for Golgi localization maps to the cytoplasmic tail of a Bunyaviridae (Uukuniemi virus) membrane glycoprotein. J Virol. 1997;71:4717–4727. doi: 10.1128/jvi.71.6.4717-4727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball J M, Mulligan M J, Compans R W. Basolateral sorting of the HIV type 2 and SIV envelope glycoproteins in polarized epithelial cells: role of the cytoplasmic domain. AIDS Res Hum Retrovir. 1997;13:665–675. doi: 10.1089/aid.1997.13.665. [DOI] [PubMed] [Google Scholar]
- 3.Berlioz-Torrent C, Shacklett B L, Erdtmann L, Delamarre L, Bouchaert I, Sonigo P, Dokhelar M C, Benarous R. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J Virol. 1999;73:1350–1361. doi: 10.1128/jvi.73.2.1350-1361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brody B A, Rhee S S, Hunter E. Postassembly cleavage of a retroviral glycoprotein cytoplasmic domain removes a necessary incorporation signal and activates fusion activity. J Virol. 1994;68:4620–4627. doi: 10.1128/jvi.68.7.4620-4627.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brody B A, Rhee S S, Sommerfelt M A, Hunter E. A viral protease-mediated cleavage of the transmembrane glycoprotein of Mason-Pfizer monkey virus can be suppressed by mutations within the matrix protein. Proc Natl Acad Sci USA. 1992;89:3443–3447. doi: 10.1073/pnas.89.8.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Compans R W. Virus entry and release in polarized epithelial cells. Curr Top Microbiol Immunol. 1995;202:209–219. doi: 10.1007/978-3-642-79657-9_14. [DOI] [PubMed] [Google Scholar]
- 7.Delamarre L, Pique C, Rosenberg A R, Blot V, Grange M-P, Le Blanc I, Dokhélar M-C. The Y-S-L-I tyrosine-based motif in the cytoplasmic domain of the human T-cell leukemia virus type 1 envelope is essential for cell-to-cell transmission. J Virol. 1999;73:9659–9663. doi: 10.1128/jvi.73.11.9659-9663.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delamarre L, Rosenberg A R, Pique C, Pham D, Dokhélar M-C. A novel human T-leukemia virus type 1 cell-to-cell transmission assay permits definition of SU glycoprotein amino acids important for infectivity. J Virol. 1997;71:259–266. doi: 10.1128/jvi.71.1.259-266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubay J W, Roberts S J, Hahn B H, Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J Virol. 1992;66:6616–6625. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egan M A, Carruth L M, Rowell J F, Yu X, Siliciano R F. Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of the Pr55gag precursor protein. J Virol. 1996;70:6547–6556. doi: 10.1128/jvi.70.10.6547-6556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green N, Shinnick T M, Witte O, Ponticelli A, Sutcliffe J G, Lerner R A. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc Natl Acad Sci USA. 1981;78:6023–6027. doi: 10.1073/pnas.78.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammond C, Helenius A. Quality control in the secretory pathway. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 13.Heilker R, Spiess M, Crottet P. Recognition of sorting signals by clathrin adaptors. Bioessays. 1999;21:558–567. doi: 10.1002/(SICI)1521-1878(199907)21:7<558::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 14.Januszeski M M, Cannon P M, Chen D, Rozenberg Y, Anderson W F. Functional analysis of the cytoplasmic tail of Moloney murine leukemia virus envelope protein. J Virol. 1997;71:3613–3619. doi: 10.1128/jvi.71.5.3613-3619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lodge R, Delamarre L, Lalonde J-P, Alvarado J, Sanders D A, Dokhélar M-C, Cohen E A, Lemay G. Two distinct oncornaviruses harbor an intracytoplasmic tyrosine-based basolateral targeting signal in their viral envelope glycoprotein. J Virol. 1997;71:5696–5702. doi: 10.1128/jvi.71.7.5696-5702.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lodge R, Göttlinger H, Gabuzda D, Cohen E A, Lemay G. The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J Virol. 1994;68:4857–4861. doi: 10.1128/jvi.68.8.4857-4861.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lodge R, Lalonde J-P, Lemay G, Cohen E A. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 1997;16:695–705. doi: 10.1093/emboj/16.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marks M S, Woodruff L, Ohno H, Bonifacino J S. Protein targeting by tyrosine- and di-leucine-based signals: evidence for distinct saturable components. J Cell Biol. 1996;135:341–354. doi: 10.1083/jcb.135.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melikyan G B, Markosyan R M, Brener S A, Rozenberg Y, Cohen F S. Role of the cytoplasmic tail of ecotropic Moloney murine leukemia virus Env protein in fusion pore formation. J Virol. 2000;74:447–455. doi: 10.1128/jvi.74.1.447-455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morelon E, Dautry-Varsat A. Endocytosis of the common cytokine receptor γc chain. Identification of sequences involved in internalization and degradation. J Biol Chem. 1998;273:22044–22051. doi: 10.1074/jbc.273.34.22044. [DOI] [PubMed] [Google Scholar]
- 21.Mulligan M J, Yamshchikov G V, Ritter G D, Jr, Gao F, Jin M J, Nail C D, Spies C P, Hahn B H, Compans R W. Cytoplasmic domain truncation enhances fusion activity by the exterior glycoprotein complex of human immunodeficiency virus type 2 in selected cell types. J Virol. 1992;66:3971–3975. doi: 10.1128/jvi.66.6.3971-3975.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munro S. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 1995;14:4695–4704. doi: 10.1002/j.1460-2075.1995.tb00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ochsenbauer C, Dubay S R, Hunter E. The Rous sarcoma virus Env glycoprotein contains a highly conserved motif homologous to tyrosine-based endocytosis signals and displays an unusual internalization phenotype. Mol Cell Biol. 2000;20:249–260. doi: 10.1128/mcb.20.1.249-260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohno H, Aguilar R C, Fournier M C, Hennecke S, Cosson P, Bonifacino J S. Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology. 1997;238:305–315. doi: 10.1006/viro.1997.8839. [DOI] [PubMed] [Google Scholar]
- 25.Olson J K, Grose C. Endocytosis and recycling of varicella-zoster virus Fc receptor glycoprotein gE: internalization mediated by a YXXL motif in the cytoplasmic tail. J Virol. 1997;71:4042–4054. doi: 10.1128/jvi.71.5.4042-4054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olson J K, Grose C. Complex formation facilitates endocytosis of the varicella-zoster virus gE:gI Fc receptor. J Virol. 1998;72:1542–1551. doi: 10.1128/jvi.72.2.1542-1551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olson J K, Santos R A, Grose C. Varicella-zoster virus glycoprotein gE: endocytosis and trafficking of the Fc receptor. J Infect Dis. 1998;178(Suppl. 1):S2–S6. doi: 10.1086/514255. [DOI] [PubMed] [Google Scholar]
- 28.Pique C, Pham D, Tursz T, Dokhélar M-C. The cytoplasmic domain of the human T-cell leukemia virus type I envelope can modulate envelope functions in a cell type-dependent manner. J Virol. 1993;67:557–561. doi: 10.1128/jvi.67.1.557-561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radsak K, Eickmann M, Mockenhaupt T, Bogner E, Kern H, Eis-Hubinger A, Reschke M. Retrieval of human cytomegalovirus glycoprotein B from the infected cell surface for virus envelopment. Arch Virol. 1996;141:557–572. doi: 10.1007/BF01718317. [DOI] [PubMed] [Google Scholar]
- 30.Ragheb J A, Anderson W F. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J Virol. 1994;68:3220–3231. doi: 10.1128/jvi.68.5.3220-3231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rein A, Mirro J, Haynes J G, Ernst S M, Nagashima K. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J Virol. 1994;68:1773–1781. doi: 10.1128/jvi.68.3.1773-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritter G D, Jr, Mulligan M J, Lydy S L, Compans R W. Cell fusion activity of the simian immunodeficiency virus envelope protein is modulated by the intracytoplasmic domain. Virology. 1993;197:255–264. doi: 10.1006/viro.1993.1586. [DOI] [PubMed] [Google Scholar]
- 33.Rohn W M, Rouille Y, Waguri S, Hoflack B. Bi-directional trafficking between the trans-Golgi network and the endosomal/lysosomal system. J Cell Sci. 2000;113:2093–2101. doi: 10.1242/jcs.113.12.2093. [DOI] [PubMed] [Google Scholar]
- 34.Rowell J F, Stanhope P E, Siliciano R F. Endocytosis of endogenously synthesized HIV-1 envelope protein. Mechanism and role in processing with class II MHC. J Immunol. 1995;155:473–488. [PubMed] [Google Scholar]
- 35.Sauter M M, Pelchen-Matthews A, Bron R, Marsh M, LaBranche C C, Vance P J, Romano J, Haggarty B S, Hart T K, Lee W M, Hoxie J A. An internalization signal in the simian immunodeficiency virus transmembrane protein cytoplasmic domain modulates expression of envelope glycoproteins on the cell surface. J Cell Biol. 1996;132:795–811. doi: 10.1083/jcb.132.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slusarewicz P, Nilsson T, Hui N, Watson R, Warren G. Isolation of a matrix that binds medial Golgi enzymes. J Cell Biol. 1994;124:405–413. doi: 10.1083/jcb.124.4.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subtil A, Delepierre M, Dautry-Varsat A. An α-helical signal in the cytosolic domain of the interleukin 2 receptor β chain mediates sorting towards degradation after endocytosis. J Cell Biol. 1997;136:583–595. doi: 10.1083/jcb.136.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tirabassi R S, Enquist L W. Role of envelope protein gE endocytosis in the pseudorabies virus life cycle. J Virol. 1998;72:4571–4579. doi: 10.1128/jvi.72.6.4571-4579.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tirabassi R S, Enquist L W. Mutation of the YXXL endocytosis motif in the cytoplasmic tail of pseudorabies virus gE. J Virol. 1999;73:2717–2728. doi: 10.1128/jvi.73.4.2717-2728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vzorov A N, Compans R W. Assembly and release of SIV env proteins with full-length or truncated cytoplasmic domains. Virology. 1996;221:22–33. doi: 10.1006/viro.1996.0349. [DOI] [PubMed] [Google Scholar]
- 41.Waldmann T A. The structure, function, and expression of interleukin-2 receptors on normal and malignant lymphocytes. Science. 1986;232:727–732. doi: 10.1126/science.3008337. [DOI] [PubMed] [Google Scholar]
- 42.Yang C, Compans R W. Analysis of the cell fusion activities of chimeric simian immunodeficiency virus-murine leukemia virus envelope proteins: inhibitory effects of the R peptide. J Virol. 1996;70:248–254. doi: 10.1128/jvi.70.1.248-254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang C, Compans R W. Analysis of the murine leukemia virus R peptide: delineation of the molecular determinants which are important for its fusion inhibition activity. J Virol. 1997;71:8490–8496. doi: 10.1128/jvi.71.11.8490-8496.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu X, Yuan X, McLane M F, Lee T-H, Essex M. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of Env proteins into mature virions. J Virol. 1993;67:213–221. doi: 10.1128/jvi.67.1.213-221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu Y, Wong P K. Studies on compartmentation and turnover of murine retrovirus envelope proteins. Virology. 1992;188:477–485. doi: 10.1016/0042-6822(92)90501-f. [DOI] [PubMed] [Google Scholar]
- 46.Zhu Z, Gershon M D, Hao Y, Ambron R T, Gabel C A, Gershon A A. Envelopment of varicella-zoster virus: targeting of viral glycoproteins to the trans-Golgi network. J Virol. 1995;69:7951–7959. doi: 10.1128/jvi.69.12.7951-7959.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zingler K, Littman D R. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein increases Env incorporation into particles and fusogenicity and infectivity. J Virol. 1993;67:2824–2831. doi: 10.1128/jvi.67.5.2824-2831.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]