Abstract
The distribution and levels of simian immunodeficiency virus (SIV) in tissues and plasma were assessed in naturally infected African green monkeys (AGM) of the vervet subspecies (Chlorocebus pygerythrus) by limiting-dilution coculture, quantitative PCR for viral DNA and RNA, and in situ hybridization for SIV expression in tissues. A wide range of SIV RNA levels in plasma was observed among these animals (<1,000 to 800,000 copies per ml), and the levels appeared to be stable over long periods of time. The relative numbers of SIV-expressing cells in tissues of two monkeys correlated with the extent of plasma viremia. SIV expression was observed in lymphoid tissues and was not associated with immunopathology. Virus-expressing cells were observed in the lamina propria and lymphoid tissue of the gastrointestinal tract, as well as within alveolar macrophages in the lung tissue of one AGM. The range of plasma viremia in naturally infected AGM was greater than that reported in naturally infected sooty mangabeys. However, the degree of viremia in some AGM was similar to that observed during progression to AIDS in human immunodeficiency virus-infected individuals. Therefore, containment of viremia is an unlikely explanation for the lack of pathogenicity of SIVagm in its natural host species, AGM.
Human immunodeficiency virus type 1 (HIV-1) is a member of a large family of nonhuman primate lentiviruses which have been designated simian immunodeficiency viruses (SIVs). At the present time, SIVs from at least seven African monkey species have been identified and molecularly characterized. The genetic relationships among five of these SIV strains isolated from sooty mangabey monkeys (Cercocebus atys; SIVsm), mandrills (Mandrillus sphinx; SIVmnd), Sykes monkeys (Cercopithecus albogularis mitis; SIVsyk), African green monkeys, (AGM; Chlorocebus spp.; SIVagm), and chimpanzees (Pan troglodytes; SIVcpz) have been reviewed previously (23, 25, 44). Briefly, SIV isolates segregate phylogenetically, based upon their species of origin, into at least five lineages represented by SIVsm, SIVagm, SIVsyk, SIVlhoest (9, 26), and SIVcpz with a number of additional novel strains from other primates recently characterized (12, 18). The SIVagm lineage consists of four distinct subtypes that cluster depending upon the species of AGM from which they were isolated, i.e., grivets (Chlorocebus aethiops; SIVagm/gri [14, 29]), vervets (Chlorocebus pygerythrus; SIVagm/ver [1, 4, 5, 15, 20, 29]), sabaeus monkeys (Chlorocebus sabaeus; SIVagm/sab [1–3, 28]), and tantalus monkeys (Chlorocebus tantalus; SIVagm/tan [22, 33]).
The lack of virulence of SIV isolates for their particular natural host species is intruiging (reviewed in references 3 and 21). SIV-infected African monkeys such as sooty mangabeys infected with SIVsm or AGM infected with SIVagm exhibit no evidence of immunodeficiency (3, 7, 8, 13, 16, 20, 21, 34, 36, 37, 42). However, experimental infection of Asian species of monkeys (Macaca sp.) with SIVsm or the highly related SIVmac strain (6, 11, 31, 36, 45, 48) or SIVagm (20) can induce an AIDS-like syndrome. SIVlhoest isolated from L'Hoest monkeys also appears to induce AIDS in macaques while apparently resulting in asymptomatic infection in its natural host (26). The comparative study of the pathogenesis of such viruses in their natural host and a disease-susceptible host such as macaques thus may shed light on the pathogenesis of AIDS in humans. The SIV strains which have been the best characterized in terms of virulence for macaques are SIVsm (6, 21, 31, 48) and the related SIVmac strains (11, 41, 45).
In the susceptible macaque host, the semi-steady-state level at which plasma viremia plateaus after the primary phase of infection is an excellent predictor of the subsequent disease course in SIVsm- and SIVmac-infected macaques (24, 47). Macaques with persistently high plasma viremia succumb more rapidly to disease than those with lower levels of plasma viremia. This correlation suggests that the extent of viral replication is a major determinant of disease pathogenesis. A similar association has been observed in HIV-1-infected humans (32).
However, the paradigm that pathogenesis is associated with the extent of viral replication does not appear to be maintained in some of the natural models of SIV infection. Recent viral load studies of naturally infected sooty mangabey monkeys demonstrated active ongoing viral replication as measured by plasma viral RNA assays (42). As observed in SIVsm-infected macaques (24) and HIV-infected humans (38), the degree of plasma viremia was reflected in the extent of virus expression in lymphoid tissues. Since the viral load in asymptomatic mangabeys is comparable to that observed in macaques inoculated experimentally with pathogenic SIV (24, 47), it appears unlikely that strict containment of viremia is a likely explanation for the lack of pathogenicity of SIVsm in its natural host species.
Similar to the species-specific virulence of SIVsm in sooty mangabeys and macaques, AGM infected with SIVagm remain asymptomatic (3, 7, 8, 13, 19, 20, 21, 34, 35, 37) whereas experimental transmission of virus to pigtailed (PT) macaques (Macaca nemestrina) can result in an AIDS-like syndrome (20). As confirmation that this virus has not changed in pathogenesis by passage in macaques, experimental infection of SIV-seronegative AGM with this pathogenic isolate does not result in disease (20). Quantitative studies of viral loads in naturally infected AGM revealed that the proviral load in peripheral blood mononuclear cells (PBMC) and lymph nodes of such animals is similar to that observed during asymptomatic infection of humans with HIV-1 (8, 19), and viral populations in infected AGM appear to undergo significant evolution (34). The purpose of the present study was to characterize the virus load and distribution in tissues and blood of healthy AGM of the vervet species (C. pygerythrus) that were naturally infected with SIVagm.
MATERIALS AND METHODS
Animals and tissues.
Two SIVagm-infected, healthy, wild-caught AGM of the vervet species (AGM90 and AGM155) that were imported from Kenya in 1987 (29) were euthanatized, and a complete necropsy was performed. Fresh lymphoid tissues collected at autopsy were disrupted by gentle douncing, and the resulting mononuclear cells were washed and viably frozen in 90% fetal calf serum–10% dimethyl sulfoxide in liquid nitrogen. Representative samples were also collected from a variety of gastrointestinal sites and parenchymal organs, including the kidneys, liver, salivary glands, and brain. Tissues samples were collected in formalin for routine histopathology and in situ hybridization (ISH) and aliquots were frozen on dry ice in phosphate-buffered saline (PBS) and stored in liquid nitrogen. Sequential plasma samples (collected with EDTA as an anticoagulant) and formalin-fixed lymph node biopsies were obtained from an additional AGM (AGM219) from the same cohort. Plasma samples were assayed for SIV RNA by real-time reverse transcriptase (RT)-PCR, and lymph node biopsies were assessed for SIV expression by ISH. Plasma samples and PBMC were also obtained from five naturally infected vervets, and paraffin-embedded tissue blocks were obtained from another six naturally infected vervet monkeys from the Paul Ehrlich Institute, Langen, Germany. Finally, sequential plasma samples were collected from two vervets and two sabaeus monkeys inoculated with SIVagm9063 (20). Animals were housed in accordance with the guidelines of the National Institutes of Health Animal Care and Use Committee.
Tissue culture and cells.
Virus was isolated from viably frozen, disrupted lymph node mononuclear cells (LNMC) from lymphoid tissues of AGM155 and AGM90 by cocultivation with CEMss cells. The minimum number of infected cells was determined by limiting 10-fold dilution coculture of LNMC with CEMss cells; each dilution was cocultured in quadruplicate, and a 50% tissue culture-infective dose (TCID50) was determined based on RT-expressing wells. Virus was isolated from cryopreserved tissues such as those of the gastrointestinal tract, brain, lungs, and liver following thawing on ice by douncing tissues in Hanks balanced saline solution. The resulting homogenate was centrifuged, and the pelleted material was used to isolate genomic DNA. The supernatant was then filtered (0.45-μm-pore-size filter), and the filtrate was used to isolate SIV by incubation with CEMss cells. These cultures were sequentially monitored for supernatant RT activity. Plasma samples were evaluated for the presence of SIV p27gag antigen using a commercial antigen capture assay (Coulter Corp., Hialeah, Fla.), and also the infectious titer of SIV within these samples was evaluated by incubation of limiting 10-fold dilutions with CEMss cells.
To evaluate cell populations in the peripheral blood that harbored SIV, PBMC were separated by centrifugation through lymphocyte separation medium (Organon Teknica). The monocyte population was allowed to adhere to plastic tissue culture wells for 5 days in RPMI 1640 medium containing 10% fetal calf serum and 10% normal rhesus macaque serum, washed extensively to remove nonadherent cells, and cocultivated with CEMss cells. Purification of CD4 and CD8 subsets was performed using positive selection with CD4 (Leu3A) or CD8 (Leu2), followed by Dynabeads conjugated with anti-mouse immunoglobulin G. Genomic DNA was isolated from a portion of these CD4+ or CD8+ cells for PCR analysis, and another portion was cocultivated with CEMss cells after blasting for 4 days in RPMI 1640 with 10% fetal calf serum–5 μg of phytohemagglutinin per ml–10% interleukin-2. Cell cultures were monitored for supernatant RT activity by standard methods.
SIV-specific ISH.
Nonradioactive ISH for SIV expression was performed as previously described on formalin-fixed, paraffin-embedded tissues utilizing sense or antisense digoxigenin-labeled riboprobes that spanned the entire SIVagm9063-2 genome (20). SIVagm9063-2 is an infectious, pathogenic, molecularly cloned provirus derived from a virus isolate from a PT macaque (PT63) inoculated a year previously with SIVagm90 isolated directly from naturally infected vervet AGM90. Briefly, slides were heated for 5 min and washed sequentially in xylene, 100% ethanol, 95% ethanol, and diethylpyrocarbonate-treated water. Slides were then incubated with 5 mM levamisole–1× SSC (0.15 M NaCl plus 0.015 M sodium citrate)–25 μg of proteinase K per ml of 10 mM Tris (pH 7.4)–0.2 mM CaCl2 at 37°C, followed by 0.1 M glycine in PBS, 1× PBS, and 0.1 M triethanolamine–0.25% acetic anhydride. Prehybridization under a coverslip in a preheated humidity chamber was performed for 15 min, followed by hybridization with either the sense or the antisense probe at a final concentration of 2.5 ng/ml overnight at 52°C. Following hybridization, the sections were washed in 2× SSC–50% formamide solution at 50°C, followed by a wash in 2× SSC, and treated with RNase solution. Slides were blocked with a buffer containing 2% horse serum, 0.3% Tween 20, 150 mM NaCl, and 100 mM Tris (pH 7.4) for 1 h. Slides were then incubated for 1 h with a 1:5,000 dilution of sheep antidigoxigenin alkaline phosphatase conjugate in 2% horse serum, washed twice, and incubated with the nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate substrate in the dark, at room temperature, overnight. This technique is as sensitive as radioactive methods (unpublished data). For example, SIVsm riboprobes are sensitive enough to detect the initial infected cells following rectal inoculation of macaques with SIVsmPBj (27).
QC-PCR for viral RNA in plasma.
To quantitate viral RNA levels in plasma, a PCR assay was developed based upon an internally controlled quantitative competitive (QC)-PCR-based assay (40) adapted for SIVagm from vervets. A 270-bp region of gag was generated by PCR amplification from pSIVagm9063-2 using primers 2548 and 2549 (see below), which incorporated BamHI and EcoRI sites to facilitate cloning. The PCR product was cloned into the BamHI and EcoRI sites of pGEM-7Zf (Promega, La Jolla, Calif.) to generate wild-type plasmid pGagWT. A 93-bp deletion was introduced by inverse PCR with primers gagF and gagR (see below) to generate the competitive template (pGagCT), where the underlined bases highlight an introduced EcoRV site used to facilitate religation. Primers RTgagF and RTgagR for the RT-PCR were designed in a region of gag that is highly conserved among SIVagm clones from vervet monkeys (see Fig. 2A). The following primers amplify 277- and 184-bp fragments, respectively, from the wild-type and competitive templates: 2548-F (5′-gttgaattctaacagggagacaacagcgccacctgg-3′) and 2549-R (5′-tacggatccggtggtgggtgtgttacatcccactgg-3′), gagF-2565 (5′-gttgatatctccttgttgttgcgctgggaa-3′) and gagR-2566 (5′-tacgatatccaagcactctcagaagggtgt-3′), and RTgagF (5′-ctggtggcagtcaaatttcccagcgcaac-3′) and RTgagR (5′-cagtgggatgtaacacacccaccacc-3′).
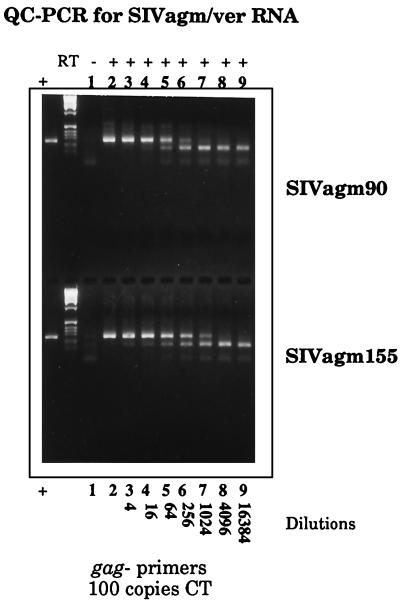
FIG. 2.
Analysis of SIVagm isolates by QC-PCR. PCR amplifications of reactions containing 100 copies of the competitive template (CT) and of serial fourfold dilution series of viral RNAs extracted from SIVagm90- and SIVagm155-infected-cell culture supernatants are shown. The plus and minus signs on the top line indicate whether RT was included in the reaction mixture. Virus was used undiluted (lane 2) or diluted 1:4 (lane 3), 1:16 (lane 4); 1:64 (lane 5), 1:256 (lane 6), 1:1,024 (lane 7), 1:4,096 (lane 8), or 1:16,384 (lane 9).
RT-PCR was employed for quantitation of viral RNA in plasma using a purified in vitro T7 runoff transcript from pGagCT as the competitive template, a GeneAmp RNA PCR Core kit (Perkin-Elmer AB, Foster City, Calif.), and 2548-F, RTgagF, and RTgagR as primers. Plasma samples for analysis were collected using acid citrate dextrose as an anticoagulant and stored in a freezer at −70°C. Virions were pelleted by ultracentrifugation, and viral RNA was isolated using a QIAmp-Viral RNA kit (Qiagen Inc., Valencia, Calif.). Fourfold dilutions of the test RNA were mixed with various known copy numbers of the competitive template and subjected to reverse transcription at 42°C for 30 min. The resulting cDNA was then amplified (45 cycles of 94°C for 1 min, 55°C for 2 min, and 72°C for 1 min), and the products were separated by electrophoresis on 3% agarose HT gels (ISS Corp.) containing ethidium bromide at 0.25 μg/ml. Viral RNA levels were calculated using the dilution factor for the sample dilution which gave a wild-type visual signal equivalent to that of the competitive template, normalized to the volume of plasma extracted, and expressed as the number of SIV RNA copies per milliliter of plasma. As controls for the accuracy of RNA determinations of the competitive template, the number of copies was quantified by measurements of A260 based on the extinction coefficient calculated for the transcript sequence. The copy number of the standard was confirmed by RT-PCR of serial 10-fold dilutions with detection of a percentage of the replicates predicted to contain one copy. The efficiencies of amplification of the competitive and wild-type RNA templates were similar when assessed separately in modeling experiments. Finally, SIVagm155-4 and SIVagm9063-2 virus stocks were prepared as described above for plasma samples and subjected to QC-PCR analysis.
Semiquantitative PCR assay for viral DNA in tissues and blood.
The number of viral DNA copies in total cellular DNA samples extracted from tissues was estimated using a semiquantitative limiting-dilution PCR assay. A 250-bp region of gag was amplified by nested PCR (60 cycles of 94°C for 1 min, 55°C for 2 min, and 72°C for 1 min with outer primers 1807 and 1808 and nested inner primers 1809 and 1810 [see below]). Serial 10-fold dilutions of cellular DNA ranging from 50 pg to 500 ng were subjected to PCR amplification. PCR products were separated on a 0.9% agarose gel and transferred to nitrocellulose, and Southern blot hybridization was performed using a probe synthesized by PCR from pSIVagm155 cloned DNA using primers 1923 and 1924 (see below). Assuming the average DNA content of a eukaryotic cell to be 5 pg, these samples were equivalent to 10 (50 pg) to 100,000 (500 ng) cells. Based upon detection of a PCR signal in a dilution, the minimum number of cells required to produce a PCR signal was extrapolated. This assay was validated using 10 μg of total cellular DNA extracted from uninfected CEMss cells to which 14 pg of full-length SIVagm9063-2 plasmid had been added, which is roughly equivalent to infected cells containing one viral DNA copy per cell. Dilutions of this DNA were assayed by limiting-dilution PCR analysis of 500 ng to 0.05 pg, with a specific amplification product observed in samples containing 5 pg of DNA or more (data not shown). The primer sequences were as follows: 1807, 5′-cctcagagctgcataaaagcagat-3′; 1808, 5′-tcactcaagtccctgttcgggcgc-3′; 1809, 5′-tactaggagaccagcttgagcctg-3′; 1810, 5′-tgctggagtttctctcgcctgggt-3′; 1923, 5′-tgttcgctggttagcctaacc-3′; 1924, 5′-gagagagaacccagtaaggaa-3′; 2013, 5′-tgccactaagagaaataacataca-3′; 2014, 5′-cagtgctgaggtagacgcccccat-3′; 2015, 5′-atttttcgcactttttaaaagaaa-3′; 2016, 5′-actcaagtccctgttcgggcgcca-3′.
Real-time RT-PCR assay.
A real-time RT-PCR assay for levels of viral RNA in plasma was developed using methodology based on the Prism 7700 Sequence Detection System (Applied Biosystems, Foster City, Calif.) that was used previously for SIVsm/mac-specific real-time PCR (46). The real-time PCR assay required three adjacent areas with a high degree of nucleotide conservation, and the fragment amplified by the gag primers derived for QC-PCR was not sufficiently conserved to allow the derivation of a conserved probe sequence. Therefore, an alignment of known SIVagm full-length sequences (SIVagm155-4, SIVagmTYO-1, SIVagm 3, and SIVagm9063-2) from vervets was used to identify a highly conserved region within the transmembrane glycoprotein-encoding region of the envelope (see Fig. 1). The forward and reverse primers used to amplify a 122-nucleotide fragment and an internal fluorogenic probe primer were generated using inosine (i) residues at positions that were not conserved between SIVagm sequences. The sequences were as follows: AgmF, 5′-GTC CAG TCT CiG CAi TTi CTT G-3′; AgmR, 5′-CGG GCA TTG AGG TTT TTi AC-3′; probe, 5′-R-CAG iTG TTG AAG CTi ACC ATT TGG G-Q-3′ (where R indicates a FAM group and Q indicates a TAMRA group conjugated through linker arm nucleotide linkage). A high-performance liquid chromatography-purified probe was obtained from Applied Biosystems or Perkin-Elmer.
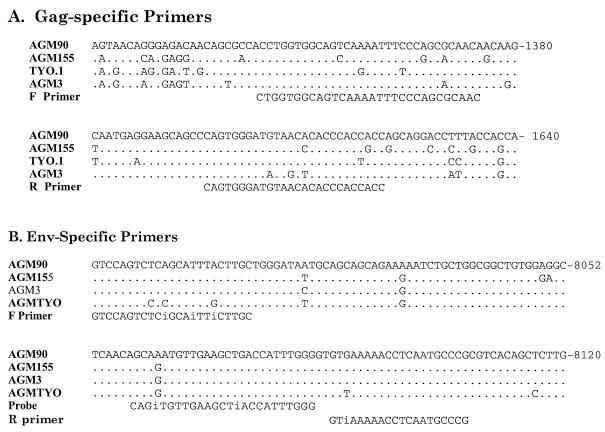
FIG. 1.
Oligonucleotide primers used for quantitative PCR for viral RNA. (A) Primers used for QC-PCR assay. Shown is an alignment of portions of the gag gene of SIVagm clones from vervet monkeys compared with SIVagm9063-2, where identity is shown as a dot and nucleotide substitutions are indicated. Nucleotide positions within the genome of SIVagm9063-2 are indicated at the right. The forward (F) and reverse (R) primers used for the QC-PCR assay are shown below the sequence alignment. (B) Primers used for real-time PCR assay. Shown is an alignment of portions of the envelope gene of SIVagm clones compared with that of SIVagm9063-2, where identity is shown as a dot and nucleotide substitutions are indicated. The primers used for real-time PCR analysis are shown below the alignment, with the letter i indicating insertion of an inosine at a variable amino acid.
A 1.9-kb fragment (HindIII/HincII) of the envelope of pSIVagm9063-2 was cloned into the pTRI-10 [poly(A)] plasmid vector, and sense RNA was transcribed from an EcoRI-linearized plasmid using T7 polymerase (46). After DNase treatment, the full-length RNA transcript was purified using oligo(dT) beads and quantitated by determining the A260. The copy number of the standard was confirmed by RT-PCR of serial 10-fold dilutions with detection of a percentage of the replicates predicted to contain one copy. Four serial 10-fold dilutions, followed by two 5-fold dilutions, of 109 copies of the RNA transcript were used as a standard for the assay (1:5,000 to 1:125,000,000 dilutions) to develop a standard curve that exhibited minimal interrun variability. Plasma samples were isolated as described above and assayed in triplicate. RT reactions were performed with 96-well plates, and reaction mixtures contained identical concentrations of the following components in RNase-free water: random hexamers (2.5 μM; Promega, Madison, Wis.), 5.0 mM MgCl2, 1.0 mM deoxynucleoside triphosphates, 5.0 mM dithiothreitol, and 20 U of Superscript RT.
RESULTS
In the present study, viral loads were assessed by using a combination of ISH, semiquantitative virus isolation, and measurement of viral RNA levels in plasma samples from naturally infected AGM with a focus on monkeys of the vervet species (C. pygerythyrus). We focused our efforts on one subtype of SIVagm because of the tremendous diversity within SIVagm isolates from different AGM species (as much as 30% in conserved genes such as gag and pol). To gain insight into the tissue distribution and viral loads in such monkeys, lymphoid tissues from eight naturally infected vervet monkeys, plasma samples from six naturally infected vervets, and plasma samples from two vervets and two sabaeus monkeys that had been experimentally inoculated with SIVagm9063 were evaluated. A wide range of tissues were available from two naturally infected vervets, allowing the evaluation of tissue-associated viral loads and distribution. These two vervets (AGM90 and AGM155) are the sources of infectious molecular clones SIVagm9063-2 and SIVagm155-4, respectively. Sequential plasma samples from one naturally infected vervet and two experimentally infected vervets provided us the opportunity to evaluate viral-load stability over time.
QC-PCR assay for viral RNA in plasma.
To evaluate viral RNA levels in the plasma of infected animals, a QC-RT-PCR assay specific for SIVagm was developed using primers based on a region of gag conserved among SIVagm155-4, SIVagmTYO-1, SIVagm3, and SIVagm9063-2 (Fig. 1A). The effectiveness of these primers was evaluated on viral RNAs extracted from culture supernatants of SIV isolates from AGM90 and AGM155. As shown in Fig. 2, both of these viral genomes were amplified efficiently. The efficiencies of amplification of the wild-type and competitive RNA templates were similar. Serial dilutions of viral RNA samples were amplified with a constant amount of a competitive template that contained a distinguishing 93-bp deletion. An equivalence point was achieved for each sample that allowed semiquantiative evaluation of the number of RNA genomes present in these two samples. As summarized in Table 1, the viral stocks contained 1.5 × 107 and 4 × 106 copies of viral RNA per ml, respectively.
TABLE 1.
Quantitation of SIVagm155 and SIVagm90 in culture supernatants of SIV isolates and plasma of naturally infected monkeys
| Samplea | SIV p27 level (ng/ml) | No. of viral RNA copies/mlb | TCID50/ml | No. of EMc particles/ml |
|---|---|---|---|---|
| SIVagm155 | 5.05 | 1.5 × 107 | 3.2 × 103 | 2.4 × 106 |
| SIVagm90 | 1.20 | 4.0 × 106 | 2.0 × 103 | 1.9 × 106 |
| AGM155 plasma | <0.05 | <1,000 | 0 | NDd |
| AGM90 plasma, 1991 | <0.05 | 20,000 | 1 | ND |
| AGM90 plasma, 1993 | <0.05 | 10,000 | ND | ND |
Viral stocks to be used for quantitation were generated by infecting CEMss cells with cell-free virus isolated directly from naturally infected animals AGM90 and AGM155.
The data used to derive the numbers of viral RNA copies per milliliter are shown in Fig. 2.
EM, electron microscopic.
ND, not done.
To evaluate whether this represented a valid estimate of the number of viral particles in the samples, parallel analysis of particle numbers by electron microscopy, p27 antigen content determination (Coulter Corp.), and TCID50 measurement was also performed. The ratio of the p27 antigen level to the number of viral RNA copies was similar to that which we have observed for SIVsm isolates and plasma samples (∼4,000 copies of viral RNA per pg of p27). As expected, a similar rank order of these various assays was observed. In all of the assays used to quantitate virus (antigen, particles), the number of viral particles quantitated was always slightly higher in the SIVagm155 virus stock. The TCID50 was approximately 5,000-fold lower than the levels of viral RNA. Thus, we concluded that this assay system efficiently detected both SIVagm155 and SIVagm90 in these viral isolates.
As shown in the bottom portion of Table 1, we then applied this assay to heparinized plasma samples collected from AGM90 and AGM155. Viral RNA in the plasma of AGM155 was below the limit of detection of the assay (<1,000 copies per ml). In contrast, two sequential plasma samples from AGM90 contained 10,000 and 20,000 viral RNA copies per ml, respectively. Virus was isolated from undiluted plasma from AGM90 but could not be rescued from AGM155 plasma samples, consistent with a higher level of plasma viremia for AGM90. Since these initial samples contained heparin, which could inhibit a PCR, these estimates of viral RNA represented a minimum estimate. To obtain a more accurate picture of the viral load in such monkeys, plasma samples from eight additional SIVagm-infected vervets were assayed using a real-time assay.
Measurement of viral RNA in plasma using a real-time PCR assay.
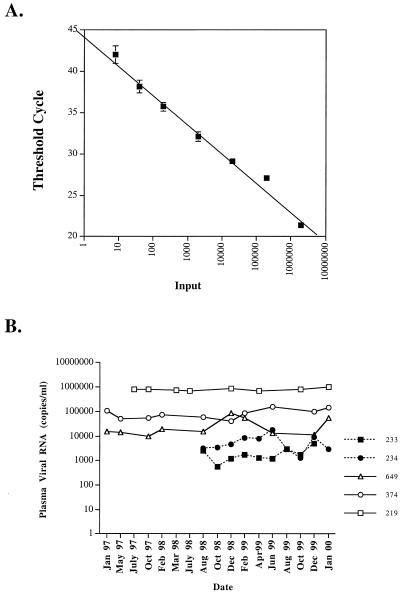
Since the two samples were widely divergent in terms of viral RNA levels, we obtained fresh plasma samples from six additional naturally infected vervets and two vervets inoculated experimentally with SIVagm9063 (Table 2). Virus was isolated by coculture of CEMss cells with PBMC of two of these animals (386 and 5319), and new isolates of SIVagm155 and SIVagm90 were generated. A real-time RT-PCR assay specific for the vervet subtype of SIVagm was developed using primers specific for a highly conserved region of the envelope (Fig. 1B) which readily amplified SIVagm90, SIVagm155, SIVagm386, and SIVagm5319. Sequence analysis of the 120-bp product of the real-time PCR primers demonstrated that the SIVs infecting these five vervets were relatively well conserved in this region of the genome (96 to 97% nucleotide identity). A dilution series of an RNA transcript of the SIVagm9063-2 envelope was used to develop a standard curve (Fig. 3A) from which viral RNA levels were extrapolated.
TABLE 2.
Quantitation of SIVagm in culture supernatants of SIV isolates and plasma of naturally infected monkeys using real-time RT-PCR
| Animala | Virus isolate
|
No. of viral RNA copies/ml of plasma | |
|---|---|---|---|
| SIV p27 level (ng/ml) | No. of viral RNA copies/ml | ||
| Natural infection | |||
| AGM155 | 99 | 3.3 × 108 | NDc |
| AGM90 | 98 | 5.5 × 108 | ND |
| AGMF8 | ND | ND | 3,200 |
| AGM386 | 136 | 3.0 × 107 | 4,700 |
| AGM5319 | 115 | 1.0 × 107 | <1,000 |
| AGM5332 | ND | ND | 13,000 |
| AGM5387 | ND | ND | 41,000 |
| AGM219 | ND | ND | 800,000b |
| Experimental SIVagm9063 infection | |||
| AGM374 | ND | ND | 86,000b |
| AGM649 | ND | ND | 28,000b |
| AGM233 | ND | ND | 2,000b |
| AGM234 | ND | ND | 6,100b |
| PT9532 | ND | ND | 60,000,000 |
| PT9323 | ND | ND | 210,000,000 |
Viral stocks to be used for quantitation were generated by infecting CEMss cells with cell-free virus isolated directly from naturally infected animals. Plasma samples from PT macaques PT638 and PT9323 were collected at 9 days after intravenous inoculation with SIVagm9063-2.
A mean of 8 to 10 independent plasma samples were collected sequentially over a period of 2 to 3 years.
ND, not determined.
FIG. 3.
Analysis of SIVagm RNA levels in plasma of naturally and experimentally infected AGM using a real-time RT-PCR assay. This plot of the input control template (in vitro envelope transcript) copy number versus the threshold cycle demonstrates the assay's precision and broad dynamic range. The plotted values represent mean values ± 1 standard deviation for triplicate determinations. (B) SIV RNA levels in plasma through 3 years of chronic infection in a naturally infected vervet (AGM219). Sequential SIV RNA levels in plasma are also shown for two vervet monkeys (374 and 649) and two sabaeus monkeys (233 and 234) that were experimentally inoculated with SIVagm9063 in 1994.
Quantitation was not performed on the plasma samples from AGM90 and AGM155 due to the confounding presence of heparin. Viral RNA levels in plasma samples from six additional naturally infected AGM ranged from <1,000 to 800,000 copies (Table 2), consistent with the range observed in samples from AGM90 and previously. The viral RNA levels in plasma samples from the experimentally infected AGM were within the range observed in the naturally infected vervets, with mean levels of 28,000 and 86,000 copies/ml (Table 2). These moderate levels contrasted with the high SIV RNA levels in the plasma of two PT macaques that had been experimentally inoculated with SIVagm9063-2. As shown in Fig. 3B, longitudinal analysis of plasma samples collected from the two experimentally inoculated vervet monkeys (AGM374 and AGM649) during the period from January 1997 to January 2000 revealed that the viral RNA levels in plasma were remarkably stable over time. Similarly, sequential viral RNA levels in the plasma of the one naturally infected vervet for which multiple samples were available were also stable over time (Fig. 3). Interestingly, viral RNA levels in plasma were consistently lower in two sabaeus monkeys (AGM233 and AGM234) that had been experimentally inoculated with SIVagm9063-2.
Distribution of virus in naturally infected AGM.
As detailed in Table 3, virus isolation, PCR and ISH were used to determine the distribution and loads of SIV in tissues of AGM90 and AGM155. Virus isolation from a range of tissues was attempted utilizing CEMss to rescue infectious virus. Virus was isolated from all lymphoid tissues (lymph nodes, spleen, and thymus) and samples from the gastrointestinal tract (esophagus, stomach, duodenum, jejunum, ileum, cecum, and colon). Virus was not isolated from other tissues, such as the kidneys, liver, salivary glands, skin, muscle, or brain, with the exception of the kidneys and liver of AGM90.
TABLE 3.
Detection of infectious virus by virus isolation, viral DNA by PCR, and viral RNA by ISH in tissues from naturally infected AGM
| Tissue | AGM155
|
AGM90
|
||||
|---|---|---|---|---|---|---|
| VIa | PCRb | ISHc | VI | PCR | ISH | |
| Lymphoid | ||||||
| Spleen | + (1) | + (10) | 0 | + (10) | + (1) | 2 |
| Inguinal lymph node | + (10) | + (10) | 0 | + (10) | + (10) | 2 |
| Axillary lymph node | + (1) | + (10) | 1 | + (10) | + (10) | 2 |
| Mesenteric lymph node | + (1) | + | 1 | + (10) | + | 2 |
| Thymus | + | + (10) | NTd | + | + (1) | 2 |
| Bone marrow | − | − | NT | + | + (1) | NT |
| PBMC | + (1) | + (1) | NT | + (10) | + | NT |
| Gastrointestinal | ||||||
| Salivary gland | + | + | NT | + | + | NT |
| Esophagus | + | + | NT | + | + | NT |
| Stomach | + | + | 0 | + | − | 1 |
| Duodenum | + | + | 0 | + | + | 1 |
| Jejunum | + | + | 1 | + | + | 1 |
| Ileum | + | + | 1 | + | + | 2 |
| Ileocecal valve | + | + | 1 | + | + | 2 |
| Cecum | + | + | 1 | + | + | 2 |
| Other | ||||||
| Kidney | − | − | 0 | + | + | 0 |
| Liver | − | − | 0 | + | − | 0 |
| Heart | − | − | 0 | − | − | 0 |
| Brain | − | − | 0 | − | + | 0 |
| Lung | − | − | 0 | + | + | 3 |
| Skin | − | − | NT | − | − | NT |
| Muscle | − | − | NT | NT | − | NT |
| Adrenal gland | + | + | NT | − | + | NT |
| Ovary | − | + | NT | NT | + | NT |
| Urinary bladder | − | + | NT | NT | − | NT |
VI, virus isolation. In parentheses are the TCID50/105 cells required for virus isolation.
Amplification of viral DNA by PCR. +, positive amplification; −, no amplification. In parentheses are the numbers of provirus-positive cells per 105 cells.
Numbers of SIV-expressing cells were assessed semiquantitatively. Values: 0, no positive cells detected; 1, one or two cells per section; 2, zero to two positive cells per high-power field; 3, one to five positive cells per high-power field.
NT, not tested.
To evaluate the cell types in the blood that harbored SIV, virus isolations were performed on activated PBMC, monocyte-derived macrophages, and CD4+ lymphocytes (selected with CD4 Dynabeads [Dynal]). Virus was isolated from PBMC of both animals. In addition, SIVagm was isolated from the monocyte-derived macrophage populations of both animals and from CD4-selected and CD8-selected cells of AGM90. A minimum of 104 cells was required for isolation of SIV from PBMC from AGM90, whereas isolation of virus from AGM155 required 106 cells. The isolation of virus from CD8+ lymphocytes is not entirely unexpected, since the majority of CD4+ lymphocytes of AGM coexpress CD8 (35).
Analysis of tissues by PCR for viral DNA yielded results that were generally concordant with virus isolation; 32 of 38 virus isolation-PCR pairs were concordant. Discordant pairs were the AGM90 stomach, which was positive for virus isolation but negative for proviral DNA and AGM155 urinary bladder, ovary, liver, kidney, and pancreas, for which proviral DNA was detected but virus was not isolated. The latter virus isolations were performed on frozen-tissue homogenates that could potentially reduce the sensitivity of virus isolation.
In order to provide a more accurate quantitation of the virus load in tissues, limiting-dilution coculture was performed on viable single-cell suspensions from various lymphoid tissues, as summarized in Table 3. Virus isolation from lymphoid tissues required a minimum of 104 to 105 cells, consistent with a viral load of 10 to 100 infected cells per 106 total lymphocytes. This estimate of infected cell numbers was compared with the approximate number of cells harboring provirus as evaluated by a semiquantitative PCR assay for viral DNA in tissues. Based on reconstruction experiments using genomic DNA spiked with full-length pSIVagm155-4 plasmid DNA to estimate 1 copy per cell, the semiquantitative PCR assay was capable of detecting a minimum of 1 to 10 copies. Viral-load estimates based on the PCR assay were similar to those obtained for virus isolation, although in some individual tissues the estimates differed by 10-fold. For example, PCR analysis estimated 103 cells per 106 in the mesenteric lymph node tissue of AGM90 harbored viral DNA, whereas the estimate based on recovery of infectious virus was 100 cells per 106. This could be due to the relative sensitivities of the two assays or the presence of a minor population of latently infected cells. Overall, the combined PCR and virus isolation data suggest that these naturally infected AGM had moderate viral burdens and the general agreement in the estimates of cells harboring infectious virus and those containing provirus is consistent with the lack of a significant reservoir of latently infected cells.
Variable SIV expression in tissues revealed by ISH analysis.
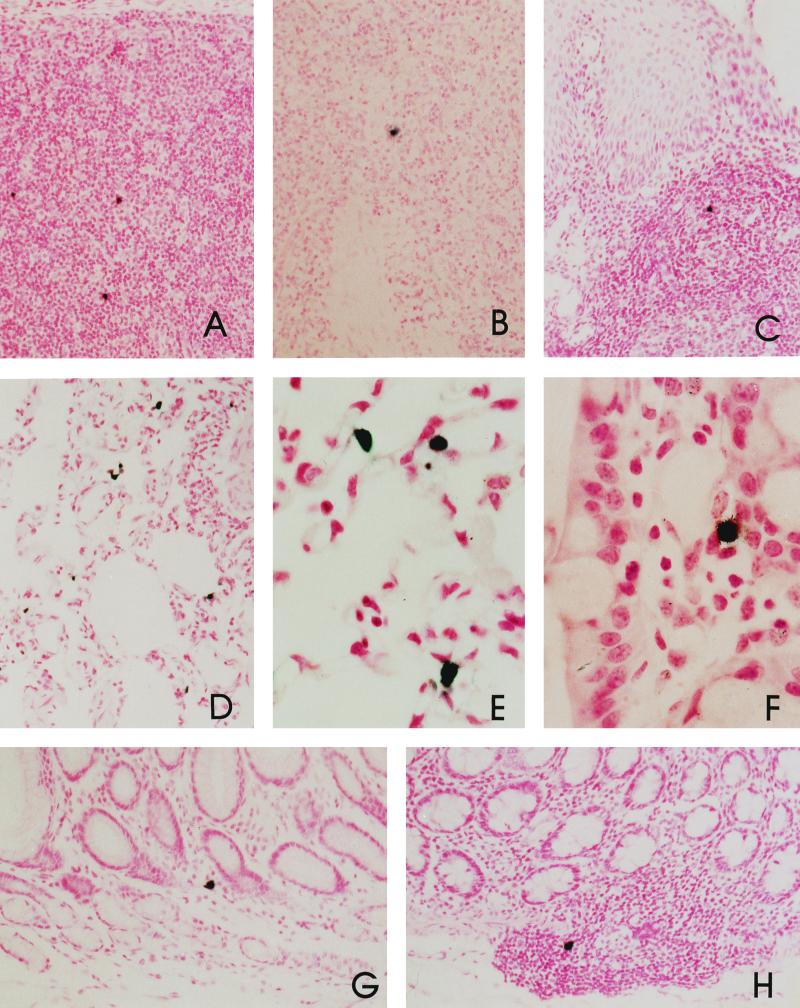
We next analyzed expression of SIV in tissues of AGM90 and AGM155. ISH for SIV expression was performed on a variety of tissues to map the distribution of infected cells in tissues. As shown in Fig. 4, tissue sections were hybridized with sense or antisense probes in parallel to confirm the specificity of hybridization and in all cases hybridizing cells were observed only in sections incubated with the antisense probe. The expression of viral RNA correlated well with the presence of viral DNA as detected by PCR or virus isolation and with the assessment of the relative viral load. SIV-expressing cells were observed only rarely in lymphoid tissues and gastrointestinal sections from AGM155 and were not observed in other tissues (brain, lung, kidney, and liver). In contrast, SIV-expressing cells were more frequently detected in tissues of AGM90 (Table 3). SIV-expressing cells were observed in the spleen, the lymph nodes (axillary, bronchial, inguinal, and mesenteric), the gut-associated lymphoid tissues, and the lamina propria and submucosa of the gastrointestinal tract (Fig. 5). SIV expression was observed in cells within bronchus-associated lymphoid tissues and in alveolar macrophages of the lung tissue of AGM90 (a tissue positive by PCR analysis). As predicted from the virus load assessment, only occasional cells (zero to two per high-power field) expressing SIV RNA were observed in tissues of AGM155. Within lymphoid tissues, these SIV-expressing cells were generally within the paracortical regions of the lymph node. Riboprobe deposition in germinal centers, as observed in SIVmac and SIVsm infection of macaques, was not observed. Indeed, the lymph nodes of these infected AGM were quiescent, with no evidence of either follicular or paracortical hyperplasia.
FIG. 4.
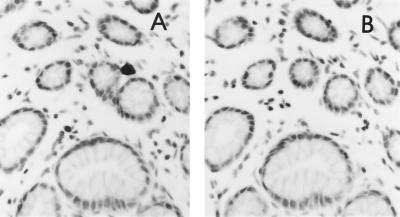
Specificity of detection of SIV expression by ISH. SIV expression in the gastrointestinal tract of AGM90 showing the specificity of hybridization. (A) Section hybridized with the antisense SIV probe showing an SIV-expressing cell near an intestinal crypt. (B) Same field as in panel A on a serial section hybridized with the sense SIV probe demonstrating no specific hybridization.
FIG. 5.
Examples of SIV-expressing cells in lymphoid and nonlymphoid tissues. SIV expression in the gastrointestinal tracts of naturally infected AGM90 and AGM155 was detected by ISH. Panels: A, mesenteric lymph node of AGM90; B, spleen of AGM90; C, tonsil of AGM90; D and E, macrophages within the lung tissue of AGM90; F, ileum of AGM155; G, stomach of AGM90; H, colon of AGM90.
Variable expression of SIV in six additional naturally infected AGM.
To determine whether these two vervets were representative of the spectrum of viral expression in tissues of naturally infected AGM, we examined lymph node biopsies from a selection of seven additional naturally infected AGM (Table 4). One of these monkeys was the original source for the SIVagm3 molecular clone (AGM37; reference 5). Some of these animals had been evaluated previously for viral loads in plasma and PBMC (8, 19), and these data, which were similar to those measured in the present study, are summarized in Table 4. Plasma samples from these vervets were negative for SIV antigen and contained antibodies that reacted solely with the envelope glycoproteins. Of the four plasma samples evaluated, only two contained low levels of infectious virus (2 and 0.2 TCID50/ml for AGM37 and AGM38). A variety of tissues from these animals were evaluated for SIV expression by ISH as detailed in Table 4. SIV-expressing cells were detectable only in the lymph nodes of three of these AGM (Z4, F3, and AGM37). SIV-expressing cells were infrequent in lymph node biopsies of two of the animals (Z4 and F3). AGM37 tissues were particularly interesting, since high virus expression was restricted to peripheral lymph node biopsies and, to a lesser extent, within the spleen. Sections of lung, intestine, mesenteric lymph node, and brain tissues from AGM37 were negative by ISH. Interestingly, virus expression was not observed in lymph node biopsies from AGM219, despite moderate to high plasma viremia (Table 2 and Fig. 3). Thus, AGM90 and AGM155 appeared to fit within the spectrum of viral expression levels of this larger subset of SIV-infected AGM. While the majority of naturally infected AGM had restricted virus expression, higher expression levels were observed in two animals (AGM90 and AGM37).
TABLE 4.
Summary of viral-load measurements and ISH of naturally infected AGM
| Monkey | STLV-1a | Malariab | TCID50/105 LNMC | LNMC source | ISHd |
|---|---|---|---|---|---|
| AGM90 | + | − | 10.0 | Table 3 | ++ |
| AGM155 | + | − | 1.0 | Table 3 | + |
| AGM37 | + | + | NDc | Peripheral lymph nodes | ++ |
| Mesenteric lymph node | − | ||||
| Spleen | + | ||||
| Ileum, lung, brain | − | ||||
| AGM38 | + | ND | 1.0 | Lymph node | − |
| AGMF3 | − | ND | ND | Lymph node (n = 3) | + |
| AGMF8 | + | ND | 0.2 | Lymph node | − |
| AGMZ4 | + | + | 1.0 | Lymph node, spleen | + |
| Lung | − | ||||
| AGMZ8 | + | + | 10 | Lymph node | − |
| Spleen, kidney | − |
STLV-1 infection was determined by Western blot analysis with HTLV-1 antigen.
Malarial infection was inferred from pigment within spleen.
ND, not determined due to lack of appropriate samples or pathology report.
++, one to five virus-positive cells per high-power field; +, less than one virus-positive cell per high-power field; −, no detectable virus-positive cells.
DISCUSSION
The lack of immunodeficiency in SIV-infected African monkeys provides a valuable opportunity to examine the host and viral factors responsible for the virulence of primate lentiviruses. Many potential mechanisms have been proposed to explain the lack of disease in African monkeys that are naturally infected with their own unique SIV isolates. Some of these mechanisms include reduced in vivo viral replication due to intrinsic target cell resistance or CD8 suppressor factors, lack of cytopathology of SIV for CD4+ lymphocytes of these monkeys, selective infection of macrophages rather than lymphocytes, immune tolerance, and lack of immune activation as a consequence of SIV infection. AGM and sooty mangabeys are the only two monkey species naturally infected with SIV that have been studied to any extent. Unfortunately, the assays performed to assess viral loads in naturally infected AGM and sooty mangabeys have not allowed comparisons of these two animal models. Recent studies of sooty mangabeys demonstrated high levels of viral RNA in plasma and extensive expression of SIVsm in lymphoid tissues (42), whereas viral RNA levels in plasma and distribution of SIV-expressing cells in tissue have not been reported for naturally infected AGM. Recent studies of primary infection in AGM experimentally inoculated with a primary SIVagm isolate revealed extensive primary viremia that was rapidly and strongly controlled (13).
In the present report, AGM naturally infected with SIVagm were evaluated for viral expression in tissue and plasma. Viral RNA levels in the plasma of naturally infected AGM ranged from undetectable to 800,000 copies per ml of plasma, a considerably wider range than that observed in sooty mangabeys. The viral DNA loads in lymph node mononuclear cells from AGM (2 to 100 copies per 106 cells) were also 100-fold lower than the viral DNA loads observed in naturally infected sooty mangabey monkeys (103 to 104 copies per 106 cells). Suppressed plasma viremia in two AGM in the present study fell within the range of plasma viremia observed in long-term nonprogression (LTNP) after SIVsm or HIV infection (10, 39). The moderate viremia observed in the other five AGM was comparable to the spectrum of viremia observed during the chronic stage of primate lentivirus infection (24, 40, 47). This study suggests that the viral load in SIVagm-infected AGM is generally lower than that observed in SIVsm-infected sooty mangabeys. In addition, this study suggests that viral loads may vary widely between individual infected AGM without apparent adverse pathologic consequences.
In an attempt to understand the basis for the lack of disease in natural models of SIV infection, it is useful to compare them with pathogenic infections such as SIVsm or SIVmac infection of macaques and HIV-1 infection of humans. The most obvious comparison is with LTNP in SIV and HIV infections. A hallmark of LTNP in SIV-infected macaques and HIV-infected humans is marked suppression of viral replication to almost undetectable levels (10, 39). Clearly, SIVsm-infected sooty mangabeys do not fit this profile and the present study suggests that ongoing viral replication in many naturally infected AGM is considerably higher than in HIV-infected human LTNP. Comparative virologic studies with a common SIVagm isolate will be important to elucidate the differences in pathogenesis between AGM and macaques.
Most studies compare the virologic characteristics of naturally infected AGM to the asymptomatic phase of HIV infection in humans (8, 13, 19). However, many characteristics of the immune responses in natural SIVagm infection are considerably different from those observed in asymptomatic HIV infection (10, 39). Naturally infected AGM frequently lack a Gag-specific antibody response (3, 37); such reduced Gag-specific antibody responses are never observed in the asymptomatic phase of HIV infection but are associated with advanced disease progression and development of AIDS. In addition, although moderate levels of envelope-specific antibody are observed, this antibody does not appear to neutralize their own SIV isolates (37). SIV-infected AGM also maintain normal lymphoid morphology (8). This differentiates it from the lymphadenopathy, characterized by pronounced follicular and paracortical lymphoid hyperplasia and trapping of viral RNA complexes on follicular dendritic cells in germinal centers, that is evident even in asymptomatic HIV infection (38). Finally, CD4 lymphocytes of AGM, whether SIV infected or naive, are highly underrepresented in the peripheral blood and coexpress the CD8 antigen (35). Less than 10% of the lymphocytes of the AGM in the present study expressed CD4, resulting in absolute CD4 lymphocyte counts of 100 to 200 per μl of blood. Such a CD4 lymphocyte count in an HIV-infected human would be consistent with AIDS and would likely be associated with the onset of opportunistic infections. Clearly, neither of these animals exhibited any evidence of immunodeficiency.
One of the most intriguing findings was the viral-load range in naturally infected AGM. The range of normal viremia in SIVagm-infected AGM overlapped the lower range of viremia observed in pathogenic models of SIV infection, such as SIVsm or SIVmac infection of macaques (24, 47). The range of plasma viremia was similar to that observed recently in experimentally inoculated AGM of the sabaeus species (13). Longitudinal assessment of SIV RNA in the plasma of five different AGM revealed a semi-steady-state level that was characteristic for the individual monkey, similar to that observed in HIV infection of humans and SIV infection of macaques (24, 32, 47). The lack of AIDS in AGM suggests that the consequences of persistent viremia are quite different from those in humans infected with HIV-1 or macaques infected with either SIVagm or SIVsm/mac. Our data suggest that at least some naturally infected AGM appear to strictly contain viral replication whereas other individuals exhibit persistent viral replication. There did not appear to be any obvious trivial explanations for the difference in virus expression between AGM155 and AGM90. Both were geriatric, and both were coinfected with simian T-cell lymphotropic virus type 1 (STLV-1). Virus expression in tissues of AGM90, however, was increased almost uniformly throughout the lymphoid tissues and gastrointestinal tract relative to that observed in AGM155. However, comparison of semiquantitative limiting-dilution coculture and PCR (Table 3) revealed only minor differences, suggesting that the higher viremia in AGM90 may be the result of enhanced viral expression. The virus distribution in AGM90 was also broadened to include expression within the lung. Based upon morphology, SIV-expressing cells appeared to be macrophages, suggesting an altered spectrum of cell types infected in AGM90. In addition, pathologic evaluation of the lungs of AGM90 revealed interstitial infiltration of lymphocytes and eosinophils, as well as evidence of mite debris consistent with active lung mite (Pneumonyssis sp.) infestation. Since there was no evidence of lung mite infestation in AGM155, increased expression of SIV in the lungs of AGM90 suggests a role of this parasitic infection in enhancing virus replication.
In summary, the present study suggests that suppression of in vivo viral replication is an unlikely explanation for the lack of virulence of SIVagm in its natural host. This conclusion is consistent with that reached in the study of SIVsm infection of sooty mangabeys. However, the range of plasma viremia observed among naturally infected AGM was wider than that reported for sooty mangabeys (42). Despite viral-load differences between these two animal models, it is unlikely that suppression of viral replication in vivo will explain the lack of AIDS-like disease in either of these two host species. The mechanisms responsible for attenuation of disease in AGM are likely to be multifactorial and complex, including decreased destruction of CD4+ lymphocytes. Continued comparative studies of the virologic and immune responses in these two models will be important to delineate the mechanistic basis for the avirulence of SIV in natural host species.
ACKNOWLEDGMENTS
The first two authors contributed equally.
We thank R. Byrum at Bioqual, Inc., for assistance with the SIV-infected monkeys; R. Elkins for performing necropsies; C. Erb, S. Whitted, and R. Goeken for technical assistance; and S. Norley of the Paul Ehrlich Institute and A. Schmeel from Chiron-Behring, Marburg, Germany, for providing samples from naturally infected AGM.
REFERENCES
- 1.Allan J S, Short M, Taylor M E, Su S, Hirsch V M, Johnson P R, Shaw G M, Hahn B H. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J Virol. 1991;65:2816–2828. doi: 10.1128/jvi.65.6.2816-2828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allan J S, Kanda P, Kennedy R C, Cobb E K, Anthony M, Eichberg J W. Isolation and characterization of simian immunodeficiency viruses from two subspecies of African Green monkeys. AIDS Res Hum Retrovir. 1990;6:275–285. doi: 10.1089/aid.1990.6.275. [DOI] [PubMed] [Google Scholar]
- 3.Allan J S. Pathogenic properties of simian immunodeficiency viruses in nonhuman primates. Annu Rev AIDS Res. 1991;1:191–206. [Google Scholar]
- 4.Baier M, Garber C, Cichutek K, Kurth R. Complete nucleotide sequence of a simian immunodeficiency virus from African green monkeys: a novel type of intragroup divergence. Virology. 1990;176:216–221. doi: 10.1016/0042-6822(90)90246-n. [DOI] [PubMed] [Google Scholar]
- 5.Baier M, Werner A, Cichutek K, Garber C, Müller C, Kraus G, Ferdinand F J, Hartung S, Papas T S, Kurth R. Molecularly cloned simian immunodeficiency virus SIVagm3 is highly divergent from other SIVagm isolates and is biologically active in vitro and in vivo. J Virol. 1989;63:5119–5123. doi: 10.1128/jvi.63.12.5119-5123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baskin G B, Murphey-Corb M, Watson E A, Martin L N. Necropsy findings in rhesus monkeys experimentally infected with cultured simian immunodeficiency virus (SIV/Delta) Vet Pathol. 1988;25:456–467. doi: 10.1177/030098588802500609. [DOI] [PubMed] [Google Scholar]
- 7.Beer B, Denner J, Brown C R, Norley S, zur Megede J, Coulibaly C, Plesker R, Holzammer S, Baier M, Hirsch V M, Kurth R. Simian immunodeficiency virus of African green monkeys is apathogenic in the newborn natural host. J Acquir Immune Defic Syndr. 1998;18:210–220. doi: 10.1097/00042560-199807010-00003. [DOI] [PubMed] [Google Scholar]
- 8.Beer B, Scherer J, zur Megede J, Norley S, Baier M, Kurth R. Lack of dichotomy between virus load of peripheral blood and lymph nodes during long-term simian immunodeficiency virus infection of African green monkeys. Virology. 1996;219:367–375. doi: 10.1006/viro.1996.0262. [DOI] [PubMed] [Google Scholar]
- 9.Beer B E, Bailes E, Goeken R, Dapolito G, Coulibaly C, Norley S G, Kurth R, Gautier J-P, Gautier-Hion A, Vallet D, Sharp P M, Hirsch V M. Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): evidence for host-dependent evolution of SIV within the C. lhoesti superspecies. J Virol. 1999;73:7734–7744. doi: 10.1128/jvi.73.9.7734-7744.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao Y, Qin L, Zhang L, Safrit J, Ho D D. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N Engl J Med. 1995;332:201–208. doi: 10.1056/NEJM199501263320401. [DOI] [PubMed] [Google Scholar]
- 11.Chalifoux L V, Ringler D, King N W, Seghal P K, Desroisiers R C, Daniel M D, Letvin N L. Lymphadenopathy in macaques experimentally infected with simian immunodeficiency virus (SIV) Am. J Pathol. 1987;128:104–110. [PMC free article] [PubMed] [Google Scholar]
- 12.Clewley J P, Lewis J C M, Brown D W G, Gadsby E L. A novel simian immunodeficiency virus (SIVdrl) pol sequence from the drill monkey, Mandrillus leucophaeus. J Virol. 1998;72:10305–10309. doi: 10.1128/jvi.72.12.10305-10309.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diop O M, Gueye A, Dias-Tavares M, Kornfeld C, Faye A, Ave P, Huerre M, Corbet S, Barre-Sinoussi F, Müller-Trutwin M C. High levels of viral replication during primary simian immunodeficiency virus SIVagm infection are rapidly and strongly controlled in african green monkeys. J Virol. 2000;74:7538–7547. doi: 10.1128/jvi.74.16.7538-7547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fomsgaard A, Hirsch V M, Allan J S, Johnson P R. A highly divergent proviral DNA clone of SIV from a distinct species of African green monkey. Virology. 1991;182:397–402. doi: 10.1016/0042-6822(91)90689-9. [DOI] [PubMed] [Google Scholar]
- 15.Fukasawa M, Miura T, Hasegawa A, Morikawa S, Tsujimoto H, Miki K, Kitamura T, Hayami M. Sequence of simian immunodeficiency virus from African green monkey, a new member of the HIV/SIV group. Nature. 1988;333:457–461. doi: 10.1038/333457a0. [DOI] [PubMed] [Google Scholar]
- 16.Fultz P, McClure H M, Anderson D C, Swenson R B, Anand R, Srinivasan A. Isolation of a T-lymphotropic retrovirus from naturally-infected sooty mangabey monkeys (Cercocebus atys) Proc Natl Acad Sci USA. 1986;83:5286–5290. doi: 10.1073/pnas.83.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao F, Bailes E, Robertson D L, Chen Y, Rodenburg C M, Michael S F, Cummins L B, Arthur L O, Peeters M, Shaw G M, Sharp P M, Hahn B H. Origin of HIV-1 in chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 18.Georges-Corbot M C, Lu C Y, Makuwa M, Telfer P, Onanga R, Dubreuil G, Chen Z, Smith S M, Georges A, Gao F, Hahn B H, Marx P A. Natural infection of a household pet red-capped mangabey (Cercocebus torquatus torquatus) with a new simian immunodefieincy virus. J Virol. 1998;72:600–608. doi: 10.1128/jvi.72.1.600-608.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartung S, Boller K, Cichutek K, Norley S G, Kurth R. Quantitation of a lentivirus in its natural host: simian immunodeficiency virus in African green monkeys. J Virol. 1992;66:2143–2149. doi: 10.1128/jvi.66.4.2143-2149.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirsch V M, Dapolito G, Johnson P R, Elkins W R, London W T, Montali R, Goldstein S, Brown C. Induction of AIDS by simian immunodeficiency virus from an African green monkey: species-specific variation in pathogenicity correlates with the extent of in vivo replication. J Virol. 1995;69:955–967. doi: 10.1128/jvi.69.2.955-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirsch V M, Johnson P R. Pathogenic diversity of simian immunodeficiency viruses. Virus Res. 1994;32:183–203. doi: 10.1016/0168-1702(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 22.Hirsch V M, McGann C, Dapolito G, Goldstein S, Ogen-Odoi A, Biryawaho B, Lakwo T, Johnson P R. Identification of a new subgroup of SIVagm in tantalus monkeys. Virology. 1993;197:426–430. doi: 10.1006/viro.1993.1606. [DOI] [PubMed] [Google Scholar]
- 23.Hirsch V M, Johnson P R. Genetic diversity and phylogeny of primate lentiviruses. In: Morrow J, Haigwood N, editors. HIV molecular organization pathogenicity and treatment. Amsterdam, The Netherlands: Elsevier Science Publishers; 1993. pp. 221–240. [Google Scholar]
- 24.Hirsch V M, Fuerst T R, Sutter G, Carroll M W, Yang L C, Goldstein S, Piatak M, Jr, Elkins W R, Alvord W G, Montefiori D C, Moss B, Lifson J D. Patterns of viral replication correlate with outcome in simian immunodeficiency virus (SIV)-infected macaques: effect of prior immunization with a trivalent SIV Vaccine in modified vaccinia virus Ankara. J Virol. 1996;70:3741–3752. doi: 10.1128/jvi.70.6.3741-3752.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirsch V M, Dapolito G, Goeken R, Campbell B. Phylogeny and natural history of the primate lentiviruses, SIV and HIV. Curr Opin Genet Dev. 1997;5:798–806. doi: 10.1016/0959-437x(95)80014-v. [DOI] [PubMed] [Google Scholar]
- 26.Hirsch V M, Campbell B J, Bailes E, Goeken R, Brwon C, Elkins W R, Axthelm M, Murphey-Corb M, Sharp P M. Characterization of a novel simian immunodeficiency virus (SIV) from l'hoest monkeys (Cercopithecus l'hoesti): Implication for the origins of SIVmnd and other primate lentiviruses. J Virol. 1999;73:1036–1045. doi: 10.1128/jvi.73.2.1036-1045.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirsch V M, Sharkey M E, Brown C R, Brichacek B, Goldstein S, Wakefield J, Byrum R, Elkins W R, Hahn B H, Lifson J D, Stevenson M. Vpx is required for dissemination and pathogenesis of SIVSM PBj: evidence of macrophage-dependent viral amplification. Nat Med. 1998;4:1401–1408. doi: 10.1038/3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin M J, Hui H, Robertson D L, Muller M C, Barre-Sinoussi F, Hirsch V M, Allan J S, Shaw G M, Sharp P M, Hahn B H. Mosaic genome structure of simian immunodeficiency virus from West African green monkeys. EMBO J. 1994;13:2935–2947. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson P R, Fomsgaard A, Allen J S, Gravell M, London W T, Olmsted R A, Hirsch V M. Simian immunodeficiency viruses from African green monkeys display unusual genetic diversity. J Virol. 1990;64:1086–1092. doi: 10.1128/jvi.64.3.1086-1092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koup R A, Safrit J T, Cao Y, Andrews C A, McLeod G, Borkowsky W, Farthing C, Ho D D. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McClure H M, Anderson D C, Fultz P N, Ansari A A, Lockwood E, Brodie A. Spectrum of disease in macaque monkeys chronically infected with SIV/SMM. Vet Immunol Immunopathol. 1989;21:13–24. doi: 10.1016/0165-2427(89)90126-8. [DOI] [PubMed] [Google Scholar]
- 32.Mellors J W, Munoz A, Giorgi J V, Margolick J B, Tassoni C J, Gupta P, Kingsley L A, Todd J A, Saah A J, Detels R, Phair J P, Rinaldo C J. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997;126:945–954. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 33.Müller M C, Saksena N K, Nerrienet E, Chappey C, Hervé V M A, Durand J-P, Legal-Campodonico P, Lang M-C, Digoutte J-P, Georges A J, Georges-Courbot M-C, Sonigo P, Barré-Sinoussi F. Simian immunodeficiency viruses from central and western Africa: evidence for a new species-specific lentivirus in tantalus monkeys. J Virol. 1993;67:1227–1235. doi: 10.1128/jvi.67.3.1227-1235.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller-Trutwin M C, Corbet S, Tavares M D, Herve V M A, Nerrienet E, Georges-Corbot M-C, Saurin W, Sonigo P, Barre-Sinoussi F. The evolutionary rate of nonpathogenic simian immunodeficiency virus (SIVagm) is in agreement with a rapid and continuous replication in vivo. Virology. 1996;223:89–102. doi: 10.1006/viro.1996.0458. [DOI] [PubMed] [Google Scholar]
- 35.Murayama Y, Amano A, Mukai R, Shibata H, Matsunaga S, Takahashi H, Yoshikawa Y, Hayami M, Noguchi A. CD4 and CD8 expressions in African green monkey T lymphocytes: implication for resistance to SIV infection. Int Immunol. 1997;9:843–851. doi: 10.1093/intimm/9.6.843. [DOI] [PubMed] [Google Scholar]
- 36.Murphey-Corb M, Martin L N, Rangan S R, Baskin G B, Gormus B J, Wolf R H, Andes W A, West M, Montelero R C. Isolation of an HTLV-III-related retrovirus from macaques with simian AIDS and it origin in asymptomatic mangabeys. Nature. 1986;321:435–437. doi: 10.1038/321435a0. [DOI] [PubMed] [Google Scholar]
- 37.Norley S G, Kraus G, Ennen J, Bonilla J, Konig H, Kurth R. Immunological studies of the basis for the apathogenicity of simian immunodeficiency virus from African green monkeys. Proc Natl Acad Sci USA. 1990;87:9067–9071. doi: 10.1073/pnas.87.22.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pantaleo G, Graziosi C, Demarest J F, Butini L, Montroni M, Fox C H, Orenstein J M, Kotler D P, Fauci A S. HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease. Nature. 1993;362:355–358. doi: 10.1038/362355a0. [DOI] [PubMed] [Google Scholar]
- 39.Pantaleo G, Menzo S, Vaccarezza M, Graziosi C, Cohen O J, Demarest J F, Montefiori D, Orenstein J M, Fox C, Schrager L K, Fauci A S. Studies in subjects with long-term nonprogressive human immunodeficiency virus infection. N Engl J Med. 1995;332:209–216. doi: 10.1056/NEJM199501263320402. [DOI] [PubMed] [Google Scholar]
- 40.Piatak M, Jr, Saag M S, Yang L C, Clark S J, Kappes J C, Luk K C, Hahn B H, Shaw G M, Lifson J D. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science. 1993;259:1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- 41.Reimann K A, Tenner-Racz K, Racz P, Montefiori D C, Yasutomi Y, Lin W, Ransil B J, Letvin N L. Immunopathogenic events in acute infection of rhesus monkeys with simian immunodeficiency virus of macaques. J Virol. 1994;68:2362–2370. doi: 10.1128/jvi.68.4.2362-2370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rey-Cuille M A, Berthier J L, Bomsel-Demontoy M C, Chaduc Y, Montagnier L, Hovanessian A G, Chakrabarti L A. Simain immunodeficiency virus replicates to high levels in sooty mangabeys without inducing disease. J Virol. 1998;72:3872–3886. doi: 10.1128/jvi.72.5.3872-3886.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmitz J E, Kuroda M J, Santra S, Sasseville V G, Simon M A, Lifton M A, Racz P, Tenner-Racz K, Dalesandro M, Scallon B J, Ghrayeb J, Forman M A, Monefiori D C, Rieber E P, Letvin N L, Reimann K A. Control of viremia in simian immunodeficiency virus infection by CD8(+) lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 44.Sharp P M, Robertson D L, Hahn B H. Cross-species transmission and recombination of “AIDS” viruses. Philos Trans R Soc Lond Ser B. 1995;349:4122–4149. doi: 10.1098/rstb.1995.0089. [DOI] [PubMed] [Google Scholar]
- 45.Simon M A, Brodie S J, Sassevile V G, Chalifoux L V, Desrosiers R C, Ringler D J. Immunopathogenesis of SIVmac. Virus Res. 1994;32:227–251. doi: 10.1016/0168-1702(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 46.Suryanarayana K, Wiltrout T A, Vasques G M, Hirsch V M, Lifson J D. Plasma SIV RNA viral load determination by real-time quantification of product generation in reverse transcriptase-polymerase chain reaction. AIDS Res Hum Retrovir. 1998;14:183–189. doi: 10.1089/aid.1998.14.183. [DOI] [PubMed] [Google Scholar]
- 47.Watson A, Ranchalis J, Travis B, McClure J, Sutton W, Johnson P R, Hu S-L, Haigwood N L. Plasma viremia in macaques infected with simian immunodeficiency virus: plasma viral load early in infection predicts survival. J Virol. 1997;71:284–290. doi: 10.1128/jvi.71.1.284-290.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J, Martin L N, Watson E A, Montelaro R C, West M, Epstein L, Murphey-Corb M. Simian immunodeficiency virus/delta-induced immunodeficiency disease in rhesus monkeys: relation of antibody response and antigenemia. J Infect Dis. 1988;158:1277–1286. doi: 10.1093/infdis/158.6.1277. [DOI] [PubMed] [Google Scholar]