Abstract
GB virus-B (GBV-B) causes an acute hepatitis in tamarins characterized by increased alanine transaminase levels that quickly return to normal as the virus is cleared. Phylogenetically, GBV-B is the closest relative to hepatitis C virus (HCV), and thus GBV-B infection of tamarins represents a powerful surrogate model system for the study of HCV. In this study, the course of infection of GBV-B in tamarins was followed using a real-time 5′ exonuclease (TaqMan) reverse transcription-PCR assay to determine the level of GBV-B in the serum. Peak viremia levels exceeded 109 genome equivalents/ml, followed by viral clearance within 14 to 16 weeks. Rechallenge of animals that had cleared infection resulted in viremia that was limited to 1 week, suggestive of a strong protective immune response. A robust tissue culture system for GBV-B was developed using primary cultures of tamarin hepatocytes. Hepatocytes obtained from a GBV-B-infected animal maintained high levels of cell-associated viral RNA and virion secretion for 42 days of culture. In vitro infection of normal hepatocytes resulted in rapid amplification of cell-associated viral RNA and secretion of up to 107 genome equivalents/ml of culture supernatant. In addition, infection could be monitored by immunofluorescence staining for GBV-B nonstructural NS3 protein. This model system overcomes many of the current obstacles to HCV research, including low levels of viral replication, lack of a small primate animal model, and lack of a reproducible tissue culture system.
Hepatitis C virus (HCV) is a major worldwide health problem, with an estimated 2% of the population chronically infected with this virus. Chronic HCV infection can cause significant liver disease and cirrhosis of the liver and, in some patients, lead eventually to liver cancer. The current animal model for the study of HCV is the chimpanzee. However, this model system suffers from the limited availability of chimpanzees and the high cost associated with conducting studies on large nonhuman primates. A smaller, less expensive model system would be desirable.
One alternative model is the hepatitis GB virus-B (GBV-B) in Saguinus species (tamarins). The GB agents, GBV-A, -B, and -C, are members of Flaviviridae (21); GBV-B is the virus most closely related to HCV (22). GBV-A and GBV-B were isolated from tamarins inoculated with a blood sample from a surgeon (GB) suffering from acute hepatitis (10). Although GBV-A and GBV-B were isolated from a tamarin inoculated with human serum believed to contain a human hepatitis virus, GBV-A and GBV-B are considered tamarin viruses. While GBV-A has been isolated from a number of tamarins (5), GBV-B has been isolated only once, and its origins are unclear. A third GB agent, GBV-C (27), also known as hepatitis G virus (19), was isolated from human serum samples in attempts to isolate new hepatitis viruses; however its association with hepatitis is tenuous (1).
GBV-A causes no recognized disease in tamarins, while GBV-B causes an acute, self-limited hepatitis, as evidenced by a reproducible rise in the serum level of alanine transaminase (ALT), an indication of liver damage. HCV infection in humans frequently results in persistent infection with associated disease sequelae, while GBV-B-infected tamarins appear to clear the viral infection with no long-lasting ill effects. However, relatively few animals have been followed long term by reverse transcription-PCR (RT-PCR) since the virus was identified in 1995 (28), and so the potential for a low percentage of chronic infections exists. The acute nature of GBV-B infections in tamarins distinguishes this hepatitis from HCV infections in humans, although the viruses share many characteristics. GBV-B is the virus most closely related to HCV genetically (22). The polyproteins possess approximately 25 to 30% homology at the amino acid level (21), while the 5′ and 3′ untranslated regions are more distinct (6, 21, 24). The functional similarities between the viruses were demonstrated by the correct processing of the HCV polyprotein by the GBV-B NS3 protease and the creation of functional chimeric NS3 proteins between HCV and GBV-B (7, 25). The studies support the premise that antiviral compounds with activity against GBV-B might show similar activities against HCV.
In this report, we describe the development of a tissue culture system for GBV-B that utilizes primary cultures of tamarin hepatocytes. Our studies were initiated by the development of a quantitative, real-time 5′ exonuclease PCR (TaqMan) assay for GBV-B and the characterization of the infection profile of GBV-B in the absence of GBV-A in two tamarins. GBV-B replication in culture was documented for 42 days using hepatocytes from an infected tamarin. Replication could also be demonstrated by detection of the NS3 protein in infected cells by immunofluorescence. In vitro infection of normal hepatocytes was highly efficient and was accompanied by high levels of replication and secretion of infectious virions. This system should provide a much-needed surrogate tissue culture system for HCV.
MATERIALS AND METHODS
Animals.
Tamarins were housed at the Southwest Regional Primate Research Center at the Southwest Foundation for Biomedical Research. Animals were cared for by members of the Department of Laboratory Animal Medicine in accordance with Guide for the Care and Use of Laboratory Animals (9), and all protocols were approved by the Institutional Animal Care and Use Committee. Two species of tamarins were used in this study. Characterization of GBV-B replication in vivo was performed with two cottontop tamarins (Saguinus oedipus); in vitro tissue culture studies used hepatocytes obtained from two moustached tamarins (S. mystax). Tamarins 12024 and 12026 were inoculated with GBV-B, followed through the course of the infection, and subsequently rechallenged with a GBV-B inoculum. Infected and uninfected primary tamarin hepatocytes were obtained from tamarins 12036 and 12035, respectively.
Hepatitis GB inoculum.
Hepatitis GB inoculum was obtained from the American Type Culture Collection (ATCC) (VR-806). A 100-μl aliquot was used as inoculum to infect tamarin 12024 by intravenous injection. Following inoculation, the animal was bled biweekly and serum was collected to assay for GBV-B and GBV-A RNA and for antibodies to GBV-B. Two additional tamarins, 12026 and 12036, were injected with 100 μl of a 1:100 dilution of the 16-day postinjection plasma from tamarin 12024 (referred to as the challenge inoculum). A portion of this 16-day postinjection plasma has been deposited with the ATCC.
Cloning of GBV-A and GBV-B genomic fragments.
Fragments of the GBV-A and GBV-B genomes were amplified using RNA templates isolated from 25 μl of the original ATCC GB serum using a High Pure RNA isolation kit (Boehringer Mannheim). DNA fragments of 337 and 217 bp, representing GBV-A and GBV-B sequences, respectively, were amplified using a single-tube Access RT-PCR kit (Promega). A 45-min cDNA reaction at 48°C preceded 55 rounds of amplification using denaturation for 30 s at 94°C, annealing for 30 s at 50°C, and extension for 30 s at 60°C. The primer sets used were those cited by Schlauder et al. (26): GBV-A primers 3011F (5′GAAAGCTTGGTTGGTTGTGG) and 3348R (5′CAATAGCACAATCTTCCTTGG), and GBV-B primers 5539F (5′CAAAATGTTCCTGTCATTATTTG) and 5756R (5′GATCCATAGTGAGCCACTCAC). Both fragments were cloned into the T-tailed vector pGEM-T (Promega) for the production of hybridization probes and synthetic RNA copy number standards.
Cloned GBV-B DNA (a gift from Stanley Lemon) was used to amplify a 924-bp fragment of the putative NS3 protein, using primers 4064F (5′CCGGATCCTTTACAGCACATATGGCATGTACC) and 4987R (5′GGGAATTCGGGTCATTGGGAGCAGCATAGCC). The NS3 fragment was released from pGEM-T by restriction at the 5′ BamHI and 3′ EcoRI sites within the primers. The fragment was cloned into pGEX-3X (Amersham Pharmacia) for expression as glutathione S-transferase (GST) fusion proteins in Escherichia coli. The identity of the cloned fragment was confirmed by sequencing.
Expression of the GST-NS3 fusion protein and generation of polyclonal antisera.
The GST-NS3 fusion protein, representing a portion of NS3 identified as being immunogenic in infected animals (23), was expressed in E. coli to serve as antigen to generate polyclonal rabbit antisera. Initial attempts to express the GST-NS3 protein resulted in an insoluble pellet of GST-NS3 protein upon lysis of the bacteria by sonication, precluding affinity purification on glutathione beads. The protein pellet was solubilized in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, and the GST-NS3 polypeptide was purified by SDS-PAGE. The GST-NS3 protein, visualized by staining with Coomassie blue in water, was excised and eluted from macerated acrylamide by shaking in water. A single rabbit was immunized with the gel-purified material to generate the anti-GST-NS3 antiserum used for immunofluorescence.
Anti-NS3 ELISA.
Antibodies to GST-NS3 in GBV-B-infected animals were monitored by enzyme-linked immunosorbent assay (ELISA) using soluble GST-NS3 protein isolated from E. coli by sonication in the presence of 4% Tween 20. The soluble GST-NS3 protein was purified on glutathione beads using established methods. Purified GST-NS3 protein (10 ng per well) was bound to Immulon 2 96-well plates (Dynatech Laboratories, Chantilly, Va.) in borate-buffered saline (145 mM NaCl, 6 mM NaOH, 48 mM H3BO3, 50 mM KCl [pH 8.2]). All ELISA incubations were for 1 h at 37°C except for the final substrate incubation; between incubation steps, wells were washed four times with phosphate-buffered saline (PBS)–0.05% Tween 20. Unoccupied protein binding sites were blocked with 5% nonfat dry milk in PBS. Serial tamarin serum samples were diluted 1:40 in antibody diluent, 0.5% nonfat dry milk–PBS–0.05% Tween 20, before incubation. Bound antibody was detected with goat anti-human immunoglobulin G (IgG)-horseradish peroxidase conjugate diluted 1:1,000 in antibody diluent. The substrate [100 μl of 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS; 1 mg/ml; Sigma] was incubated at room temperature until color development was stopped by the addition of 50 μl of 1% SDS. Plates were read at 405 nm.
TaqMan quantification of GBV-B RNA.
GBV-B RNA was isolated from serum, cells, or medium either by using a High Pure RNA isolation kit or by extraction with RNAzol (Biotecx Laboratories). GBV-B RNA was quantified by a real-time, 5′ exonuclease PCR (TaqMan) assay using a primer-probe combination that recognized a portion of the GBV-B capsid gene. The primers (558F [5′AACGAGCAAAGCGCAAAGTC] and 626R [5′CATCATGGATACCAGCAATTTTGT]) and probe (579P [5′6FAM-AGCGCGATGCTCGGCCTCGTA-TAMRA]) (6FAM = 6-carboxyfluorescein; TAMRA = N,N,N′,N′-tetramethyl-6-carboxyrhodamine) were selected using the Primer Express software designed for this purpose (PE Biosystems, Foster City, Calif.) and were obtained from PE Biosystems. The primers were used at 15 pmol/50-μl reaction, and the probe was used at 10 pmol/50-μl reaction. The reactions were performed using a TaqMan Gold RT-PCR kit (PE Biosystems) and included a 30-min 48°C reverse transcription step, followed by 10 min at 95°C and then 45 cycles of amplification using the universal TaqMan standardized conditions, a 15-s 95°C denaturation step followed by a 1-min 60°C annealing/extension step. Standards to establish genome equivalents were synthetic RNAs transcribed from the cloned capsid gene. Synthetic RNA was prepared using a SP6 Megascript kit (Ambion) and was purified by DNase treatment, RNAzol extraction, and ethanol precipitation. RNA was quantified by optical density, and 10-fold serial dilutions were prepared from 10 to 1 million copies, using tRNA as a carrier. These standards were run in duplicate in all TaqMan assays in order to calculate genome equivalents in the experimental samples. The calibration curves from one preparation of synthetic RNA to the next are essentially identical. Although no international standard for GBV-B RNA is available, similar assays developed for HCV yield values comparable to those for commercially available assays.
Hepatocyte cultures.
Tamarin 12036 was inoculated with 100 μl of a 1:100 dilution of 16-day postinjection plasma from tamarin 12024. Hepatocytes were isolated 31 days postinjection using collagenase perfusion as described elsewhere (17). Uninfected hepatocytes were isolated from naive tamarin 12035 and used for in vitro infections. Hepatocytes were seeded into 12- or 6-well collagen-coated tissue culture dishes or into 60-mm-diameter dishes containing glass coverslips for immunofluorescence. Hepatocytes were maintained in serum-free medium containing hormones and growth factor supplements as previously described (17). Culture medium was changed three times per week, and samples were harvested periodically by addition of 500 μl of Trizol (Life Technologies) to the wells. Coverslips were harvested at 4 days postseeding and used for immunofluorescence studies as previously described (15). Coverslips were stained with rabbit antiserum directed against E. coli-expressed GST-NS3 protein at 1:1,000 and then detected using a goat anti-rabbit IgG-fluorescein conjugate at 1:20. Aliquots of primary hepatocytes were frozen by established methods (17), and most studies were performed with frozen cells that had been thawed and plated.
RESULTS
Detection of GBV-A and GBV-B viruses.
The original description of the molecular cloning of the GB agents reported that the GB inoculum contained both GBV-B and GBV-A (28). To analyze the GB inoculum and serum from infected animals, RT-PCR assays were developed to detect GBV-B and GBV-A. Analysis of the GB agent present in the original deposit at the ATCC revealed that it contained both GBV-B and GBV-A. In contrast, analysis of serial bleeds from tamarin 12024 inoculated with the ATCC GB material (see below) failed to detect GBV-A (data not shown), while GBV-B could be readily detected. The failure to induce a GBV-A infection could have been due to a number of factors, including the age of the inoculum or preexisting immunity from previous GBV-A exposure. Nonetheless, the animals described in this study were infected with GBV-B alone, and the serum from these animals represents a source of GBV-B inoculum not contaminated with GBV-A.
The initial RT-PCR assay for GBV-B was a gel-based assay combined with Southern hybridization. To obtain a quantitative measure of GBV-B RNA for our studies, we developed a real-time, 5′ exonuclease RT-PCR (TaqMan) assay. The capsid region was chosen as the target site for the TaqMan primers and probe. The assay readily detected synthetic GBV-B RNA down to 10 copies and was linear over a 6-log range from 10 to 1 million copies, based on genome equivalents calculated using synthetic RNA transcribed from the GBV-B capsid clone (see Materials and Methods).
Course of GBV-B infection in tamarins.
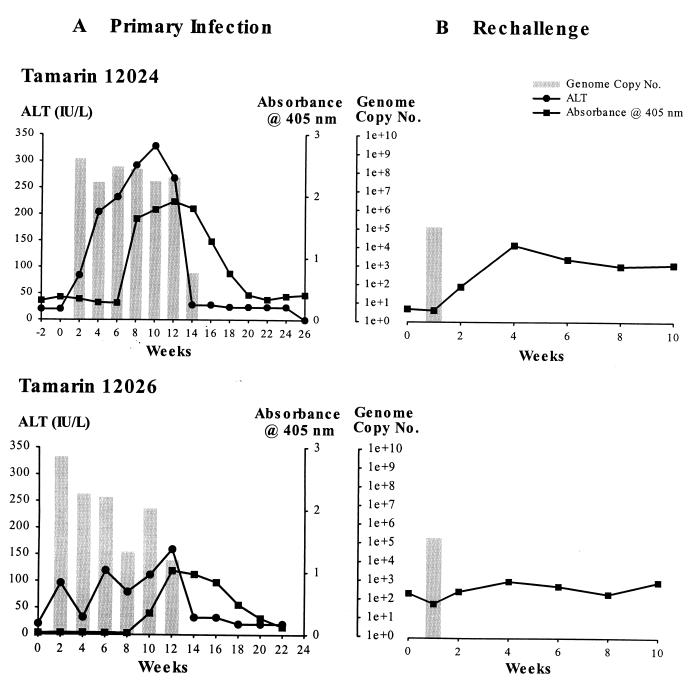
Tamarin 12024 was injected with 100 μl of the ATCC GB inoculum. The animal was bled every 2 weeks, and the serum was tested for ALT as an indication of liver damage, for viral RNA by TaqMan RT-PCR, and for antibodies to NS3 by ELISA (Fig. 1A, top). A robust infection was initiated with the ATCC GB inoculum in tamarin 12024. By week 2 postinoculation (p.i.), the first postinoculation bleed, the serum viral RNA titer was 4.8 × 108 genome equivalents (ge)/ml and remained elevated through week 12 p.i. The serum viral RNA levels rapidly declined to 3.4 × 102 ge/ml at week 14 p.i. Viral RNA was no longer detectable by week 16 p.i. The ALT levels also increased rapidly and closely paralleled the pattern for serum viral RNA. The ALT level was elevated to 84 by week 2 p.i., peaked at 328 by week 10 p.i., and had dropped to normal levels by week 14 p.i. The tamarin seroconverted for anti-NS3 antibody on week 8 p.i., and peak ELISA values were observed on week 14 p.i.; however, the ELISA values rapidly declined in the absence of viral replication and were near background levels by week 20 p.i.
FIG. 1.
Infection profiles of GBV-B infected tamarins. (A) Progress of initial GBV-B infection in tamarins 12024 (top) and 12026 (bottom). GBV-B levels are shown in comparison to the liver damage, as measured by ALT, and rise in serum antibody titers to the GBV-B NS3 protein over the 24- to 26-week course of the experiment. (B) GBV-B levels and serum antibody titers to the NS3 protein during the course of rechallenge infection of 12024 (top) and 12026 (bottom) with a standard inoculum of GBV-B-containing 12024 tamarin serum. GBV-B TaqMan data, measured in genome equivalents per milliliter of serum, are shown as bar graphs, with line graphs representing ALT and NS3 ELISA absorbance superimposed.
A second tamarin was infected with the serum derived from 12024. The day 16 plasma (7.4 × 107 ge/ml) was chosen to be a standardized inoculum for future experiments. The use of 100 μl of a 1:100 dilution of the serum provided a dose of 7.4 × 104 ge. Tamarin 12026 was challenged with this inoculum. The course of infection closely resembled the infection pattern observed in 12024 (Fig. 1A, bottom). Serum viral RNA levels peaked at 3.1 × 109 ge/ml at 2 weeks p.i. The virus titer declined to 9.1 × 103 by 12 weeks p.i. and was below the level of detection by week 14 p.i. The higher initial virus titer in tamarin 12026 than in 12024 probably represents interanimal variation rather than the difference in the inoculum. ALT levels were elevated by 2 weeks p.i. but fluctuated considerably, reaching a peak value of 160 on week 12 p.i. The animal seroconverted for antibody to NS3 by week 10 p.i. These data indicate that a standardized GBV-B inoculum lacking GBV-A has been produced for future in vivo and in vitro studies.
GBV-B rechallenge of previously infected tamarins.
To determine whether animals that had previously cleared GBV-B infection would be protected from a rechallenge with homologous inoculum, tamarins 12024 and 12026 were injected with 100 μl of a 1:100 dilution of 12024, the challenge inoculum. Very little evidence of reinfection was observed by TaqMan analysis of serial serum samples. Both animals were PCR positive 1 week p.i., with titers of 1.2 × 105 and 5.3 × 105 for tamarins 12024 and 12026, respectively (Fig. 1B). All subsequent bleeds taken at 1-week intervals were negative. The viral RNA detected at week 1 p.i. could not represent carry-forward of the inoculum, since the viral RNA levels per milliliter of serum were equivalent to the entire inoculum. Thus, the rapid decrease in viral RNA values seen at week 1 p.i. represents an enhanced clearance of an attenuated infection. Serum from each animal was tested by ELISA to determine whether an increase in antibody titer followed the second exposure to the virus. Tamarin 12024 showed a marked increase in serum antibody titers to the NS3 protein by week 4 postrechallenge (Fig. 1B, top); in contrast, tamarin 12026 showed an essentially flat response to the GBV-B rechallenge (Fig. 1B, bottom).
GBV-B replication in in vivo-infected hepatocytes.
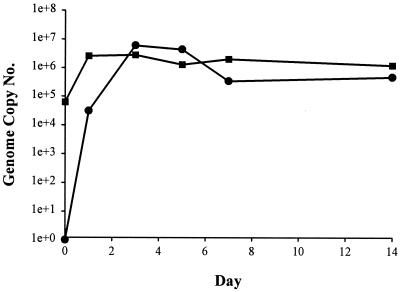
One of our major priorities with the GBV-B system was the creation of a tissue culture system that would permit studies that have been difficult or impossible to conduct with HCV. The high level of replication of GBV-B observed in tamarins suggested that many of the types of studies impossible with HCV might be feasible with this system. To initiate the culture studies, we first determined whether tamarin hepatocytes were permissive for maintaining GBV-B replication in culture when the hepatocytes were obtained from a GBV-B-infected animal. Hepatocytes were isolated from liver tissue by collagenase perfusion from tamarin 12036 at 31 days p.i. with the standardized inoculum. Hepatocytes were maintained using the serum-free medium and culture conditions that we had established for HCV replication in chimpanzee hepatocytes (17). Cultures were maintained for up to 6 weeks, and individual wells were harvested at intervals for analysis of viral RNA by TaqMan assay. Analysis of an aliquot of the cells prior to plating (day 0) determined that the cells contained 8 × 107 ge/μg of total cell RNA (Fig. 2). On day 2 postplating, the cells contained 2.6 × 107 ge/μg of RNA, indicating that the culture system was maintaining viral RNA levels similar to that observed in vivo. Some decline in viral RNA levels was observed over time, with a decrease of approximately 2 logs in titer by day 42 postisolation (Fig. 2). Medium collected at day 42 postplating contained 9.2 × 105 ge/ml after 18 medium changes, indicating that the cells continued to secrete virus for prolonged periods postplating (data not shown).
FIG. 2.
Persistence of GBV-B replication in primary tamarin hepatocytes. GBV-B levels were monitored for 42 days in primary hepatocytes isolated from GBV-B-infected tamarin 12036. Day 0 represents RNA isolated from hepatocytes harvested on the day of isolation from the liver.
Susceptibility of normal tamarin hepatocytes for in vitro GBV-B infection.
To determine the level of susceptibility of tamarin hepatocytes to in vitro infection with GBV-B, normal hepatocytes were isolated from an uninfected animal (tamarin 12035). The hepatocytes were plated in collagen-coated dishes and exposed to GBV-B-containing tamarin serum under various conditions. A number of parameters were examined to determine whether GBV-B could efficiently infect hepatocytes in vitro. A crucial step in the analysis was to determine whether the level of GBV-B increases over time following inoculation of primary tamarin hepatocytes. An in vitro growth curve was conducted by inoculation of hepatocytes on day 3 postplating with 10 μl of undiluted serum from 12024 day 16 plasma (7.4 × 105 ge; multiplicity of infection of 0.75 ge/cell). Following a 6-h exposure to inoculum, the cells were washed extensively, and wells were harvested for RNA on the days indicated (Fig. 3). Cells harvested following the 6-h exposure period (day 0) contained 6.3 × 104 ge/μg of RNA, representing virus that was bound to or had been internalized by the cells. By day 1, the level of cell-associated GBV-B RNA increased 41-fold, to 2.6 × 106 ge/μg of RNA. The level of cell-associated GBV-B RNA remained approximately the same through the 14 day experiment. Although peak cell-associated RNA levels were attained by day 1, secreted virus did not peak until day 3. The level of viral RNA in the medium on day 1, 3.1 × 104 ge/ml, increased 193-fold, to 6.0 × 106 ge/ml, on day 3. Viral RNA levels declined by approximately 1 log by day 7 and remained constant to day 14 p.i. (Fig. 3).
FIG. 3.
Growth curve of GBV-B in primary tamarin hepatocytes. Tamarin hepatocytes were inoculated with GBV-B-containing plasma, using an adsorption period of 6 h. Cultures were washed extensively to remove unadsorbed virus and then harvested in duplicate at various times over a 14-day period. Time zero (immediately after washing away the inoculum) represents the level of virus attached or internalized by the cells. Cell-associated (squares) or secreted (circles) GBV-B RNA was quantified by TaqMan RT-PCR and expressed as genome equivalents per microgram of cellular RNA or milliliter of culture medium, respectively.
TaqMan values in these assays were expressed in genome equivalents per microgram of cell RNA as a means of normalizing independent cultures irrespective of variation in cell number between cultures and efficiency in recovery of cell RNA. The values can be extrapolated to a per-cell basis by assuming that 1 μg of cell RNA is derived from 5 × 104 cells. In addition, the percentage of cells infected as determined by immunofluorescence (20% [see below]) can be used to refine the estimation of virions produced per infected cell. Given these assumptions, these values indicate that each infected cell contained 260 genomes and that 60 virions per infected cell were secreted every 2 days (medium was changed every other day). This ratio suggests that a significant fraction of the newly synthesized virions remain cell associated.
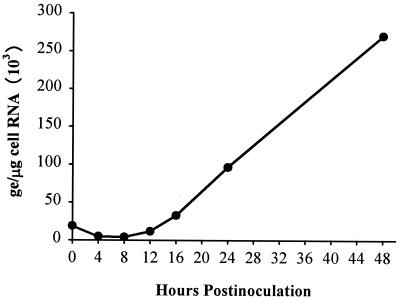
Estimation of doubling time for GBV-B.
Since the level of cell-associated GBV-B RNA had reached a maximum level within 24 h p.i., the first time point examined, a compressed growth curve study was performed using duplicate cultures. To examine earlier time points, the inoculum was adsorbed to the cells for only 1 h. Following the adsorption period, cells were extensively washed, and cultures were harvested to determine the level of viral RNA adsorbed to the cells at the 0-h time point. Cultures were then harvested at multiple times during the first 24 h p.i. and at 48 h p.i. (Fig. 4). At time zero, 18.7 × 103 ge was adsorbed to the cultures; this decreased to 4.8 × 103 and 4.0 × 103 ge at the 4- and 8-h time points, respectively. The first increase in viral RNA levels was observed at 12 h p.i. with 11.5 × 103 ge. The RNA levels increased threefold at each 4-h time interval between 8, 12, and 16 h, from 4.0 × 103 to 11.5 × 103 to 32.5 × 103 ge, respectively. The increase in viral RNA levels from 8 to 12 h and 12 to 16 h provides an estimated doubling time of 2.6 h. The level of viral RNA increased a further 3- and 2.8-fold at 24 and 48 h p.i., to 96.5 × 103 and 270 × 103 ge/μg of cell RNA, respectively. The maximum level of cell-associated viral RNA was lower in this experiment than in the previous growth curve, presumably due to the reduced exposure time to the inoculum, since results of subsequent studies were nearly identical to results of the initial growth curve (see below and compare Fig. 3 to Table 1).
FIG. 4.
Estimation of doubling time for GBV-B in primary tamarin hepatocytes. Tamarin hepatocytes were inoculated with GBV-B-containing plasma using a shortened adsorption period (1 h). Cultures were washed extensively to remove unadsorbed virus and harvested in duplicate at various times over a 48-h period. Time (immediately after washing away the inoculum) represents the level of virus attached or internalized by the cells. Cell-associated GBV-B RNA was quantified by TaqMan RT-PCR.
TABLE 1.
Time of maximum susceptibility
| Day of infectiona | RT-PCRb
|
|
|---|---|---|
| Well A | Well B | |
| 1 | 1.2 × 107 | 2.1 × 107 |
| 3 | 3.3 × 107 | 1.4 × 107 |
| 7 | 7.0 × 105 | 7.1 × 105 |
| 14 | 3.2 × 105 | 3.8 × 105 |
Cultures were infected on various days after plating and were harvested 7 days after infection.
TaqMan value, expressed in genome equivalents of GBV-B RNA per infected culture.
Time of maximum susceptibility for in vitro infection.
Since cultured hepatocytes are known to lose differentiated status to various degrees depending on the culture conditions, it was necessary to determine the optimum time postplating for in vitro infection or the window of maximum susceptibility. Hepatocytes from tamarin 12035 were inoculated for 6 h in duplicate on days 1, 3, 7, and 14 postisolation. Each well was harvested 7 days postinfection, and total cell RNA was analyzed by TaqMan RT-PCR. The duplicate wells showed nearly identical levels of GBV-B RNA, with less than twofold variation (Table 1). Considering that the culture system uses primary primate hepatocytes and measurements are based on RT-PCR, this level of reproducibility is remarkable. Cultures infected on days 1 and 3 postplating yielded comparable values of 2 × 106 to 3 × 106 ge/μg of RNA (Table 1), which were in turn nearly identical to the values obtained in the in vitro growth curve where the cultures were inoculated under similar conditions (6-h adsorption on day 3 postplating) and were harvested 7 days p.i. (Fig. 3). However, the levels of GBV-B RNA were decreased significantly in wells inoculated on days 7 and 14 postplating. Although the results from the latter time points could still be considered an efficient culture system for GBV-B, it is clear that some required function was diminished at latter times.
Titration of inoculum.
For the initial in vitro infection experiments, 10 μl of undiluted 16-day postinfection plasma from tamarin 12024 was arbitrarily selected. Higher levels of plasma can often form fibrin clots on primary hepatocytes and occasionally induce toxicity. TaqMan analysis of this plasma indicated that it contains 7.4 × 107 ge/ml. This would provide a multiplicity of infection of approximately 0.74 ge per cell at 10 μl per well (106 cells). To determine the minimum inoculum that provides efficient infection, a titration of the inoculum was performed (Table 2). Primary hepatocytes were inoculated 3 days postplating with dilutions of GBV-B plasma containing from 5 × 102 to 1 × 106 ge, and cultures were harvested 1 and 3 days p.i. Early harvests were used such that the initial level of infection could be determined prior to cell-to-cell spread of the infection. In this experiment, TaqMan values were extrapolated to genome equivalents per culture by multiplying the total cellular RNA harvested from each culture by the genome equivalents per microgram of cell RNA determined in TaqMan assays. Although expression of values in genome equivalents-per-microgram yields normalized values for all cultures, it does not present an accurate representation of the total yield per culture. The results demonstrate a 100-fold amplification of the input RNA by 24 h p.i. for the lowest inoculum used. This value further increased to 240-fold above the input RNA by day 3. Increasing the inoculum resulted in dramatic increases in the yield of viral RNA, with the highest dose having genome equivalent-per-culture values 480-fold above the lowest dose. However, the increase was not linear. This probably reflects the inefficiency of adsorption of high doses of inoculum. At all doses of inoculum considerable, amplification of the input RNA level occurred by 24 h, and further increase was observed at 3 days for doses below 5 × 104 ge. The relative increases in cell-associated RNA levels in comparison to the total input RNA values represent an underestimation of replication, since the data from the 0-h time point in Fig. 3 can be used to estimate that less than 10% of the inoculum adsorbs to the cells, and it is unlikely that all adsorbed virions enter a productive replication cycle.
TABLE 2.
Titration of inoculum
| Inoculum (ge)a | Cell-associated viral RNA (ge of GBV-B RNA/infected culture)
|
|
|---|---|---|
| Day 1 | Day 3 | |
| 5.0 × 102 | 5.0 × 104 | 1.2 × 105 |
| 2.0 × 103 | 8.0 × 104 | 3.5 × 105 |
| 1.0 × 104 | 7.1 × 105 | 1.5 × 106 |
| 5.0 × 104 | 2.1 × 106 | 8.0 × 106 |
| 2.0 × 105 | 1.1 × 107 | 2.4 × 107 |
| 1.0 × 106 | 2.4 × 107 | 2.5 × 107 |
Cultures were inoculated 3 days after plating with various doses of GBV-B and were harvested 1 and 3 days p.i.
Infectivity of tissue culture derived GBV-B.
To determine whether GBV-B from 12036 in vivo-infected hepatocytes was infectious upon passage to new cultures, uninfected hepatocytes from tamarin 12035 were inoculated with the medium derived from day 42 cultures of the in vivo-infected cultures of 12036 (Fig. 2). Cultures were inoculated 3 days postplating with 0.5 ml of medium (4.6 × 105 ge) or 0.5 ml of a 10-fold concentrate of the medium, and cultures were harvested 7 days p.i. Analysis of the cells for cell-associated viral RNA or the medium for secreted virus indicated that both inocula induced infection, with approximately a 10-fold increase in the average level of cell-associated viral RNA in cultures inoculated with the 10-fold-concentrated medium (Table 3). Although the TaqMan values were lower for the cell-associated viral RNA in comparison to cultures inoculated with tamarin serum, the level of secreted virus was comparable to that obtained for the day 7 harvest in the in vitro growth curve (Fig. 3). The data demonstrate that GBV-B released into the medium from infected primary tamarin hepatocytes is infectious in vitro. Subsequent studies have confirmed the infectivity of secreted virions, as well as demonstrated multiple log reductions in specific infectivity for particles produced in the presence of nucleoside analogs (unpublished data).
TABLE 3.
Passage of medium-derived virus
| Inoculuma | TaqMan RT-PCR value (ge/culture)b
|
|||
|---|---|---|---|---|
| Cells
|
Media
|
|||
| Well A | Well B | Well A | Well B | |
| Uninfected | 0 | 0 | ND | ND |
| 0.5 ml | 9.4 × 105 | 6.3 × 105 | ND | ND |
| 0.5 ml (10×) | 11.0 × 106 | 3.5 × 106 | 4.4 × 105 | 6.2 × 105 |
Duplicate cultures were infected 3 days after plating with 0.5 ml of medium or a 10-fold concentration of the medium from day 42 of culture from tamarin 12024 hepatocytes. Cultures were harvested day 7 postinfection.
For cell-associated (cells) and secreted viral RNA (media). ND, not determined.
Immunofluorescence of in vitro-infected hepatocytes.
To determine the percentage of cells being infected in these experiments, in vitro-infected hepatocyte cultures were examined by immunofluorescence staining for NS3 using a rabbit anti-GST-NS3 antiserum (Materials and Methods). Hepatocyte cultures from tamarin 12035 grown on glass coverslips were infected with the 12024 day 16 inoculum on day 3 postplating and were harvested 4 days after infection. Staining of infected cultures with the anti-GST-NS3 antiserum revealed a bright green cytoplasmic staining (Fig. 5A). Uninfected cultures stained with the rabbit anti-GST-NS3 (Fig. 5B) did not exhibit specific staining, nor did infected cultures stained with the prebleed serum from the same rabbit prior to immunization with GST-NS3 (Fig. 5C).
FIG. 5.
Immunofluorescence staining for NS3 protein. Hepatocytes grown on glass coverslips were inoculated with GBV-B 3 days postplating and harvested 3 days postinfection unless noted otherwise. GBV-B NS3 protein was detected by immunofluorescence using a rabbit anti-GST-NS3 antiserum and goat anti-rabbit IgG-fluorescein. (A) Infected hepatocytes stained with anti-NS3 (magnification, ×100); (B) uninfected hepatocytes stained with anti-NS3 (×100); (C) infected hepatocytes stained with normal rabbit (prebleed; ×100); (D) lower magnification of infected hepatocytes stained with anti-NS3 (×50); (E) hepatocytes infected with a low multiplicity and harvested 21 days p.i. (×100); (F) higher magnification of infected hepatocytes stained with anti-NS3 (×200).
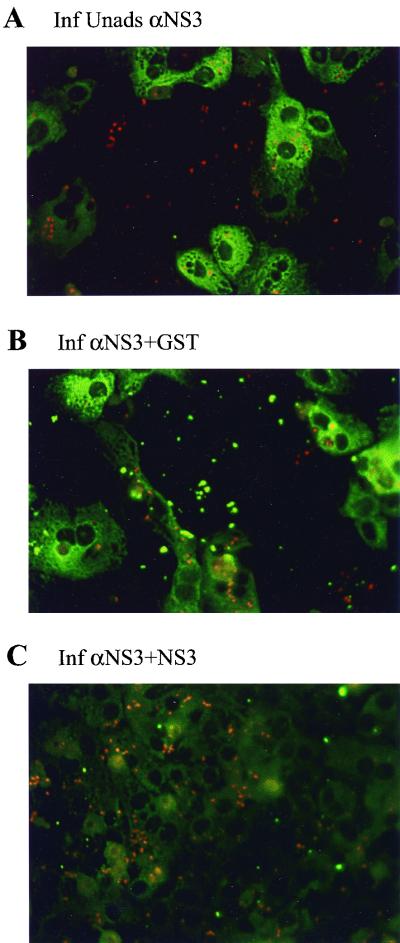
To further demonstrate the specificity of the staining reaction, we performed blocking experiments in which the antiserum was adsorbed with either GST or NS3 lacking GST prior to being used in staining. Unadsorbed antiserum again yielded a bright cytoplasmic staining (Fig. 6A) which was not reduced by prior adsorption of the antiserum with purified GST (Fig. 6B). In contrast, adsorption of the antiserum with purified NS3 completely eliminated the specific fluorescent staining (Fig. 6C).
FIG. 6.
Specificity of rabbit anti-GST-NS3 antiserum in immunofluorescence staining for NS3 protein. Hepatocytes were inoculated with GBV-B and harvested 3 days p.i. Cells were stained for NS3 protein using rabbit anti-GST-NS3 antiserum (A). To demonstrate specificity, the antiserum was adsorbed for 16 h at 4°C with 5 μg of purified GST (B) or NS3 (C) protein prior to being used for immunofluorescence staining. Adsorption with GST had no effect on staining, while adsorption with NS3 eliminated staining.
Lower-magnification photographs of the cultures revealed that a high percentage of cells were infected in some fields (Fig. 5D). Observation of multiple fields from different experiments indicated that approximately 20 to 30% of the cells was positive 3 days p.i. This represents efficient infection, since as described above, the standardized inoculum used in these experiments represented a multiplicity of infection of 0.75 ge per cell. To evaluate the spread of infection over time, cultures were inoculated with a 10-fold-lower multiplicity of infection and were harvested at various times over a 3-week period. In cultures harvested on day 3, approximately 1 to 2% of the cells were positive (data not shown), while in cultures harvested on day 21 almost all cells were positive (Fig. 5E). Despite the extensive spread of infection over a 3-week period, some cells appeared to be resistant to infection. These cells may have lost hepatocyte differentiated functions required for infection, or they may have been protected by the induction of interferon in infected cells. Examination of cultures at higher magnification revealed a cytoplasmic/perinuclear staining suggestive of endoplasmic reticulum localization (Fig. 6F).
DISCUSSION
The past decade has witnessed rapid advances in our understanding of HCV epidemiology, pathogenesis, and molecular biology of replication. The advances in molecular biology of replication have depended on the use of model systems to examine internal ribosome entry site function, polyprotein processing, the functions of individual proteins, and their interactions with the host cell. The primary limiting factor in advances at this time is the lack of an adequate culture system. The development of infectious cDNA clones of HCV have provided a new means to develop tissue culture systems for genetic analysis of replication (3, 11, 13, 29–31); however, this potential has not yet been realized. A single system for the selection of cells that maintain high levels of replication of an HCV replicon expressing the nonstructural proteins has been described (20), but widespread application of this technology has not been achieved. A number of viruses can serve as potential surrogates for HCV, including bovine viral diarrhea virus, a member of the pestivirus family. The use of GBV-B as a surrogate system for HCV is particularly attractive, since it is the virus most closely related to HCV and induces hepatitis in a small-primate model. This study significantly advances the GBV-B model through the development of a tissue culture system with high levels of replication and efficient in vitro infection.
Our previous studies with HCV replication in primary chimpanzee hepatocytes provided some of the most convincing evidence for in vitro replication of HCV; however, this system was limited in many aspects (16, 18). Most importantly, the level of replication of HCV both in vivo and in vitro necessitated the use of highly strand specific RT-PCR assays for negative-strand RNA to document replication (16, 18). The level of replication rarely exceeded the quantity of virus present in inoculum, and the virus remained primarily cell associated. The GBV-B system described in this study was not dependent on strand-specific assays, since cell-associated viral RNA increased as much as 100-fold in comparison to the input inoculum (Fig. 3 and 4 and Table 2), and viral RNA in the medium increased 193-fold between days 1 and 3 p.i. (Fig. 3). In addition, the expression of NS3 could readily be detected by immunofluorescence.
Although viral replication and secretion could be detected for 42 days, the latest time point examined, and cells could be efficiently infected in vitro for 14 days postplating, it is apparent that some cellular requirement for infection and/or replication diminishes with time. The high levels of GBV-B replication coupled with quantitative, real-time TaqMan RT-PCR provide an opportunity to optimize this system to maintain the high levels of replication observed in the first 2 weeks and potentially to define the limiting factors required for replication. Use of the serum-free medium will permit studies on the impact of alterations in the levels of growth factors and hormones on replication as well as the effects of various cytokines.
The recent development of infectious cDNA clones of GBV-B (6; Stanley Lemon, personal communication) also suggests that this system will be useful for genetic analysis of replication. Unlike HCV, it can be anticipated that transfection of primary tamarin hepatocytes with synthetic RNA from an infectious clone will lead to high levels of replication. The availability of infectious clones of both HCV and GBV-B provides an opportunity to make chimeric clones. Chimeric viruses would certainly improve the utility of the system for the screening of HCV antiviral drugs, if such clones are infectious. The failure to develop chimeric clones between different genotypes of HCV (30) suggests that replacement of limited domains containing catalytic motifs would more likely be infectious than those containing global replacements. In contrast, the ability of the dengue virus NS5 protein to provide function in trans to Kunjin virus replicons lacking a portion of the NS5 domain (12) suggests that such replacements are feasible. In addition, the high level of homology of GBV-B with HCV within certain motifs suggests that hybrid clones may not be required for the testing of some antiviral compounds in this system. This approach is supported by data demonstrating that the NS3 protease domain of GBV-B can cleave the NS4A/4B, NS4B/NS5A, and NS5A/NS5B domains of HCV (7, 25). In addition, preliminary data indicate that both interferon and ribavirin are capable of suppressing GBV-B replication in this culture system (unpublished data).
The GBV-B tamarin model will also provide a valuable animal model in which to examine pathogenesis, antiviral therapy, and immune clearance of the virus. The development of a quantitative, real-time TaqMan RT-PCR assay is a significant advancement for the use of this system. Characterization of the infection profile of two animals revealed serum viral RNA titers of 109 ge/ml. Viral clearance occurred in both animals within 12 to 14 weeks. The level of serum viral RNA closely paralleled the rise and fall of the ALT levels. The ALT levels were significantly elevated within 2 weeks p.i. Whether the high levels of GBV-B replication are to some degree cytopathic or whether the early rise in ALT was due to an innate or specific immune response to infected cells was not determined in these studies. No overt cytopathic effect was observed in vitro in infected hepatocytes. This does not exclude a low level of cytopathic effect or a slow but accelerated death of infected cells, since primary cultures of hepatocytes always contain some dying cells. The spread of the infection over 21 days to yield cultures that are uniformly positive for NS3 by immunofluorescence argues against a significant cytopathic effect, but detailed studies on the induction of apoptotic markers should be conducted before it is concluded that no cytopathic consequences of viral infection exist.
Seroconversion for anti-NS3 antibodies did not occur until 8 weeks p.i., and antibody titer rapidly declined to baseline levels in the absence of viral replication. Similar infection profiles for GBV-B have been reported previously (26). This rapid viral clearance, rapid loss of antibody profile is very similar to those that we have recently described for chimpanzees inoculated with HCV (2). Rapid viral clearance was observed in 66% of the infected animals, and rapid loss of antibody occurred in 50% of the animals with viral clearance (2). Whether similar profiles occur in HCV-infected humans is difficult to determine, since the time of infection is rarely known in humans, and all viral markers are lost within months of infection. However, a number of studies support a higher level of viral clearance in humans than previously suspected (4, 8, 14).
At this time, chronic infections with GBV-B have not been reported. The high levels of replication could contribute to a more intense immune response that favors viral clearance. Typically, HCV levels in chimpanzee and human sera are on the order of 106 ge/ml, while serum levels of GBV-B RNA approach 109 ge/ml. Alternatively, there may be other intrinsic differences between the viruses and their hosts that lead to viral clearance. If GBV-B is transmitted in nature in tamarins primarily through a parenteral route with low levels of sexual and maternal transmission, as is observed with HCV in humans, it is difficult to imagine maintenance of the virus in a population without some chronic infections, unless other animals serve as a reservoir for the virus. This of course assumes that tamarins are indeed the natural host for GBV-B. At this time too few animals have been followed long term with sensitive RT-PCR assays to exclude the possibility for a low percentage of chronic infections. The potential exists for the induction of chronic infections through the use of transient immunosuppression with cyclosporin or FK506. Chronically infected animals would certainly be useful in a number of studies including the evaluation of antiviral drugs.
Perhaps one of the most promising potential applications of the tamarin model and the tissue culture system described in this report is development of an in vitro neutralization assay and an animal model for evaluating HCV vaccine approaches. We observed protective immunity in animals that had cleared the infection and were rechallenged, although a very transient low-level viremia was observed. Whether this protection is due to a humoral or cellular immune response remains to be determined. Even if protection was provided by cellular immunity in these animals, the in vitro culture system provides an opportunity to rapidly evaluate the neutralizing potential of serum from small animals immunized with various envelope protein configurations.
ACKNOWLEDGMENTS
This work was supported by grant P51 RR13986 to the Southwest Regional Primate Research Center, by grant RO1 AI49574 from the National Institutes of Health, and by a grant from Schering-Plough Research Institute.
We thank Zhi Hong and Johnson Lau (Schering-Plough) for the purified NS3 protein and many helpful discussions on this project.
REFERENCES
- 1.Alter H, Nakatsuji Y, Melpolder J, Wages J, Jr, Wesley R, Shih J W K, Kim J P. The incidence of transfusion-associated hepatitis G virus infection and its relation to liver disease. N Engl J Med. 1997;336:747–754. doi: 10.1056/NEJM199703133361102. [DOI] [PubMed] [Google Scholar]
- 2.Bassett S E, Brasky K M, Lanford R E. Analysis of hepatitis C virus-inoculated chimpanzees reveals unexpected clinical profiles. J Virol. 1998;72:2589–2599. doi: 10.1128/jvi.72.4.2589-2599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beard M R, Abell G, Honda M, Carroll A, Gartland M, Clarke B, Suzuki K, Lanford R, Sangar D V, Lemon S M. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology. 1999;30:316–324. doi: 10.1002/hep.510300137. [DOI] [PubMed] [Google Scholar]
- 4.Bronowicki J P, Vetter D, Uhl G, Hudziak H, Uhrlacher A, Vetter J M, Doffoel M. Lymphocyte reactivity to hepatitis C virus (HCV) antigens shows evidence for exposure to HCV in HCV-seronegative spouses of HCV-infected patients. J Infect Dis. 1997;176:518–522. doi: 10.1086/517279. [DOI] [PubMed] [Google Scholar]
- 5.Bukh J, Apgar C L. Five new or recently discovered (GBV-A) virus species are indigenous to New World monkeys and may constitute a separate genus of the Flaviviridae. Virology. 1997;229:429–436. doi: 10.1006/viro.1997.8461. [DOI] [PubMed] [Google Scholar]
- 6.Bukh J, Apgar C L, Yanagi M. Toward a surrogate model for hepatitis C virus: an infectious molecular clone of the GB virus-B hepatitis agent. Virology. 1999;262:470–478. doi: 10.1006/viro.1999.9941. [DOI] [PubMed] [Google Scholar]
- 7.Butkiewicz N, Yao N, Zhong W, Wright-Minogue J, Ingravallo P, Zhang R, Durkin J, Strandring D N, Baroudy B M, Sangar D V, Lemon S M, Lau J Y N, Hong Z. Virus-specific cofactor requirement and chimeric hepatitis C virus/GB virus B nonstructural protein 3. J Virol. 2000;74:4291–4301. doi: 10.1128/jvi.74.9.4291-4301.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerny A, McHutchison J G, Pasquinelli C, Brown M E, Brothers M A, Grabscheid B, Fowler P, Houghton M, Chisari F V. Cytotoxic T lymphocyte response to hepatitis C virus-derived peptides containing the HLA A2.1 binding motif. J Clin Investig. 1995;95:521–530. doi: 10.1172/JCI117694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Committee on Care and Use of Laboratory Animals. Guide for the care and use of laboratory animals. Washington, D.C.: Institute of Laboratory Animal Resources, National Research Council; 1996. [Google Scholar]
- 10.Deinhardt F, Holmes A W, Capps R B, Popper H. Studies on the transmission of human viral hepatitis to marmoset monkeys. J Exp Med. 1967;125:673–688. doi: 10.1084/jem.125.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong Z, Beaudet-Miller M, Lanford R E, Guerra B, Wright-Minogue J, Skelton A, Baroudy B M, Reyes G R, Lau J Y. Generation of transmissible hepatitis C virions from a molecular clone in chimpanzees. Virology. 1999;256:36–44. doi: 10.1006/viro.1999.9603. [DOI] [PubMed] [Google Scholar]
- 12.Khromykh A A, Sedlak P L, Westaway E G. trans-complementation analysis of the flavivirus Kunjin ns5 gene reveals an essential role for translation of its N-terminal half in RNA replication. J Virol. 1999;73:9247–9255. doi: 10.1128/jvi.73.11.9247-9255.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolykhalov A A, Agapov E V, Blight K J, Mihalik K, Feinstone S M, Rice C M. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 14.Koziel M J, Wong D K H, Dudley D, Houghton M, Walker B D. Hepatitis C virus-specific cytolytic T lymphocyte and T helper cell responses in seronegative persons. J Infect Dis. 1997;176:859–866. doi: 10.1086/516546. [DOI] [PubMed] [Google Scholar]
- 15.Lanford R E, Butel J S. Antigenic relationship of SV40 early proteins to purified large T polypeptide. Virology. 1979;97:295–306. doi: 10.1016/0042-6822(79)90341-6. [DOI] [PubMed] [Google Scholar]
- 16.Lanford R E, Chavez D. Strand specific rTth RT/PCR for the analysis of HCV replication. In: Lau J Y N, editor. Methods in molecular medicine: hepatitis C. Totowa, N.J: Humana Press, Inc.; 1998. pp. 471–481. [DOI] [PubMed] [Google Scholar]
- 17.Lanford R E, Estlack L E. A cultivation method for highly differentiated primary chimpanzee hepatocytes permissive for hepatitis C virus replication. In: Lau J Y N, editor. Methods in molecular medicine: hepatitis C. Totowa, N.J: Humana Press, Inc.; 1998. pp. 501–515. [DOI] [PubMed] [Google Scholar]
- 18.Lanford R E, Sureau C, Jacob J R, White R, Fuerst T R. Demonstration of in vitro infection of chimpanzee hepatocytes with hepatitis C virus using strand-specific RT/PCR. Virology. 1994;202:606–614. doi: 10.1006/viro.1994.1381. [DOI] [PubMed] [Google Scholar]
- 19.Linnen J, Wages J, Jr, Zhang-Keck Z-Y, Fry K E, Krawczynski K Z, Alter H, Koonin E, Gallagher M, Alter M, Hadziyannis S, Karayiannis P, Fung K, Nakatsuji Y, Shih J W K, Young L, Piatak M, Jr, Hoover C, Fernandez J, Chen S, Zou J-C, Morris T, Hyams K C, Ismay S, Lifson J D, Hess G, Foung S K H, Thomas H, Bradley D, Margolis H, Kim J P. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science. 1996;271:505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- 20.Lohmann V, Körner F, Koch J O, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 21.Muerhoff A S, Leary T P, Simons J N, Pilot-Matias T J, Dawson G J, Erker J C, Chalmers M L, Schlauder G G, Desai S M, Mushahwar I K. Genomic organization of GB viruses A and B: two new members of the Flaviviridae associated with GB agent hepatitis. J Virol. 1995;69:5621–5630. doi: 10.1128/jvi.69.9.5621-5630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohba K, Mizokami M, Lau J Y N, Orito E, Ikeo K, Gojobori T. Evolutionary relationship of hepatitis C, pesti-, flavi-, plantviruses, and newly discovered GB hepatitis agents. FEBS Lett. 1996;378:232–234. doi: 10.1016/0014-5793(95)01441-1. [DOI] [PubMed] [Google Scholar]
- 23.Pilot-Matias T J, Muerhoff A S, Simons J N, Leary T P, Buijk S L, Chalmers M L, Erker J C, Dawson G J, Desai S M, Mushahwar I K. Identification of antigenic regions in the GB hepatitis viruses GBV-A, GBV-B, and GBV-C. J Med Virol. 1996;48:329–338. doi: 10.1002/(SICI)1096-9071(199604)48:4<329::AID-JMV6>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 24.Rijnbrand R, Abell G, Lemon S M. Mutational analysis of the GB virus B internal ribosome entry site. J Virol. 2000;74:773–783. doi: 10.1128/jvi.74.2.773-783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scarselli E, Urbani A, Sbardellati A, Tomei L, De Francesco R, Traboni C. GB virus B and hepatitis C virus NS3 serine proteases share substrate specificity. J Virol. 1997;71:4985–4989. doi: 10.1128/jvi.71.7.4985-4989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlauder G G, Dawson G J, Simons J N, Pilot-Matias T J, Gutierrez R A, Heynan C A, Kniggo M F, Kurpiewski G S, Buijk S L, Leary T P, Muerhoff A S, Desai S M, Mushahwar I K. Molecular and serologic analysis in the transmission of the GB hepatitis agents. J Med Virol. 1995;46:81–90. doi: 10.1002/jmv.1890460117. [DOI] [PubMed] [Google Scholar]
- 27.Simons J N, Leary T P, Dawson G J, Pilot-Matias T J, Muerhoff A S, Schlauder G G, Desai S M, Mushahwar I K. Isolation of novel virus like sequences associated with human hepatitis. Nat Med. 1995;1:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- 28.Simons J N, Pilot-Matias T J, Leary T P, Dawson G J, Desai S M, Schlauder G G, Muerhoff A S, Erker J C, Buijk S L, Chalmers M L, Van Sant C L, Mushahwar I K. Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc Natl Acad Sci USA. 1995;92:3401–3405. doi: 10.1073/pnas.92.8.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanagi M, Purcell R H, Emerson S U, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci USA. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yanagi M, Purcell R H, Emerson S U, Bukh J. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology. 1999;262:250–263. doi: 10.1006/viro.1999.9889. [DOI] [PubMed] [Google Scholar]
- 31.Yanagi M, St. Claire M, Shapiro M, Emerson S U, Purcell R H, Bukh J. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology. 1998;244:161–172. doi: 10.1006/viro.1998.9092. [DOI] [PubMed] [Google Scholar]