Highlights
-
•
The related lncRNA–miRNA–mRNA ceRNA axis of RILI following irradiation in a mouse model were constructed.
-
•
2044 differentially expressed lncRNAs, 191 differentially expressed miRNAs, and 2230 differentially expressed mRNAs were identified.
-
•
BNIP1 contributing to radiation-induced cellular apoptosis in RILI.
Keywords: Radiation, Lung injury, lncRNA, miRNA, mRNA, BNIP1
Abstract
Background
Radiation-induced lung injury (RILI) is a serious complication of radiation therapy, and it is mediated by long non-coding RNAs (lncRNAs).
Study design and methods
Mouse lung tissues were examined using RNA-Seq and RNA-Seq libraries 72 h after the administration of 6 Gy of X-ray irradiation. The target mRNAs were functionally annotated and the target lncRNA-based miRNAs and target miRNA-based mRNAs were predicted after irradiation to establish the lncRNA-miRNA-mRNA ceRNA axis.
Results
The analyses showed that relative to unirradiated controls, 323 mRNAs, 114 miRNAs, and 472 lncRNAs were significantly up-regulated following irradiation, whereas 1907 mRNAs, 77 miRNAs, and 1572 lncRNAs were significantly down-regulated following irradiation. Voltage-gated ion channels, trans-membrane receptor protein tyrosine kinases, and vascular endothelial growth factor have all been associated with dysregulated miRNA-mRNA relationships. KEGG pathway analysis of the dysregulated miRNA-mRNA targets revealed involvement in pathways associated with the hedgehog signaling pathway-fly, ErbB signaling, VEGF signaling, axon guidance, and focal adhesion. KEGG analysis of differentially expressed showed enrichment of mRNAs in primary immunodeficiency, the intestinal immune axis for IgA production, hematopoietic cell lineages, systemic lupus erythematosus, and Th1 and Th2 cell differentiation. Finally, the ceRNA network revealed that BNIP1 was a critical mRNA modulated by the most significant upregulation of lncRNA E230013L22Rik.
Conclusion
In summary, the lncRNA-miRNA-mRNA ceRNA axis of RILI was constructed following irradiation in a mouse model. RNA dysregulation in the early stage of RILI may lead to severe complications at a later stage, with BNIP1 contributing to radiation-induced cellular apoptosis in RILI.
Introduction
Radiation therapy (RT) is commonly utilized in cancer treatment because of its advantages in improving patient survival, minimizing recurrence, and complementing the efficacy of other cancer-related procedures [1,2]. However, radiation-related damage can cause a wide range of complications, including the development of secondary cancers [3,4]. The lung is very vulnerable to RT damage, which can result in radiation-induced lung injury (RILI). Radiation fibrosis and radiation pneumonitis are two dose-limiting toxicities of lung cancer radiotherapy [5]. Consequently, the utilization of elevated radiation doses for lung cancer patients requiring radiation therapy is restricted due to issues associated with RILI [6].
Compelling data indicates that DNA damage caused by radiation leads to widespread changes in gene transcription throughout the genome [7]. The stability and organ specificity of noncoding RNAs facilitates their use as robust diagnostic and therapeutic agents [8]. Long non-coding RNAs (lncRNAs) are between 200 and 100 000 nucleotides (nt) long and do not encode proteins; they are increasingly used for diagnosis and therapy [9,10]. LncRNAs modulate numerous cellular processes and are ubiquitous in multiple tumors [11,12]. MicroRNAs (miRNAs) are single-chain noncoding RNAs of 22 nt in length that modulate gene expression [13]. Interactions between lncRNAs and miRNAs or messenger RNAs (mRNAs) are known to contribute to radiation-induced damage.
In clinical environments, non-invasive screening for tissue injury indicators is essential [5]. Recent research about radiation bioassays indicates that the efficacy of utilizing multiple markers surpasses that of depending solely on a single marker [14]. A thorough inquiry that advances knowledge of RILI has the potential to result in improved strategies for RILI prevention and management. Potentially identifying targetable networks for countermeasures against radiation damage and predicting toxicity in normal tissues, modifications in RNA expression induced by radiation may facilitate the development of more precise radiotherapeutic strategies [7].
This study investigated the genes that are susceptible to RILI in a mouse model and utilized RNA sequencing to compare the genomic expression of irradiated and non-irradiated lung tissues. We also assessed the differences in Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways induced by irradiation to identify differentially expressed (DE) lncRNAs, DE genes, and intercellular RT-modulated networks. Finally, the lncRNA-miRNA-mRNA ceRNA axis of radiation-induced RILI in mice was established.
Methods
Mouse model of RILI
Beijing Unilever Laboratory Animal Co. Ltd. provided the four-week-old C57BL mice, which were kept in an animal center with a 12-hour light/dark cycle. There was no restriction on their access to food or water. The animals were separated into two groups (n = 3), namely the irradiated and non-irradiated groups. The irradiated mice received a single dose of 6 Gy (800 cGy/min, radiation distance 40 cm) X-ray, while the non-irradiated mice received sham irradiation. Our prior research indicated that changes in the radiation-sensitive elements of lung tissue took place 72 h following radiation exposure, while pathological alterations in lung tissue manifested six days after radiation (Wang et al., 2022). After 72 h, the animals were sacrificed and their lung tissues were harvested for analysis. The animal protocols received ethical approval from Zaozhuang University.
RNA isolation and sequencing
The lung samples were frozen in liquid nitrogen and then pulverized into a fine powder. The total RNA was then extracted using TRIzol reagent (Invitrogen, USA) following the supplied protocols. The RNA quality and quantity were determined using an Agilent Bioanalyzer with the RNA6000 Nano Lab Chip (Agilent Technologies, USA) and the DeNovix DS-11 nanodrop spectrophotometer (DeNovix, USA), respectively. Subsequently, we employed the SEQuoia RiboDepletion rRNA Removal Kit (Bio-Rad, USA) to eliminate ribosomal RNA. Following this, we fragmented the RNA samples into smaller fragments and performed reverse transcription to cDNA using real-time quantitative polymerase chain reaction (RT-qPCR). Subsequently, the cDNAs obtained were utilized for U-labeled second-strand DNA synthesis, and an A-base was added to the blunt ends of each strand. Finally, following library enrichment, RNA sequencing was conducted by the Shanghai Jikai Gene Medical Technology Co. Ltd.
Analysis of differential expression
DE lncRNAs, miRNAs, and mRNAs were identified using the “limma” package in R, with 3 replicates per group. The standards were as follows: Radiation group/control group |fold change (FC)|≥2.0 (Zhang et al., 2020) and Q-value < 0.05 (Lai, 2017), using three biological replicates for each group.
Pathway enrichment analyses
GO and KEGG enrichments of DE mRNAs were analyzed with Phyper based upon hypergeometric distributions. For considerably enriched terms and rectified networks, a strong Q-value (Q-value < 0.05) was employed. The relationship between miRNA-mRNA was validated and predicted using miRanda-3.3a and RNAhybrid v2.0. [15]. The relationship between miRNA-lncRNA was verified and predicted using the Blastn (BLAST 2.2.28+) [16].
Generation of the lncRNA-miRNA-mRNA ceRNA axis
The lncRNA-miRNA-mRNA axis was generated using the “GDCRNATools” package in R [17], and was based on the concept that lncRNAs serve as miRNA sponges (ceRNAs) to modulate mRNA activities [18]. The network visulaization was done using Cytoscape v3.6.0 [19].
Exploration and verification experiments
Western blotting
Mlg2908 cells (BNCC364345) were purchased from the Beijing Beina Chuanglian Biotechnology Institute. Mlg2908 cells were treated with irradiation before harvesting at 3, 6, 12, 24, 48, and 72 h. A mixture of 50 mL NP-40 lysis buffer (20 mM Tris–HCl, pH 7.4, 137 mM NaCl, 2 mM Na2EDTA, 1 % NP-40, 10 % Glycerol) containing 1 tablet cocktail protease inhibitor was utilized to lyse the harvested cells. An estimated 100 mL of lysis buffer per 106 cells was utilized. Phenylmethanesulfonyl fluoride (PMSF) was introduced into NP-40 lysis buffer before utilization, with the ultimate concentration being established at 1 mM. The NP-40 lysis buffer was utilized to lyse the cell. The BCA (bicinchoninic acid assay) protein assay reagent was used to measure the protein content. After loading 60 µg of proteins onto SDS-PAGE, the proteins were moved to the PVDF membrane. The primary antibody was diluted and then the secondary antibody coated with horseradish peroxidase was diluted 1:1000 and incubated on the membrane. Then, using ECL (enhanced chemiluminescence, Shanghai, China), the protein bands were visible. Using ImageJ software, the intensity (area X density) of each band on the western blots was determined. The computed area was reduced by the background.
RT-qPCR
The process of isolating total RNA was carried out utilizing a TRIzol-based RNA isolation procedure provided by TAKARA (Japan). The RNA sample was analyzed using the Nanodrop 2000C device produced by Thermo Scientific, USA. Subsequently, a quantity of 2 µg of RNA was subjected to reverse transcription using the SweScript RT I First Strand cDNA Synthesis Kit, following the guidelines provided by the manufacturer. The miRNA was transcribed in reverse using the cervical ring method, utilizing 2 × SYBR Green qPCR Master Mix (High ROX). The PCR cycling conditions for BNIP1, E230013L22Rik, mmu-miR-1966–5p, mmu-miR-1970c-3p, mmu-miR-188–5p, mmu-miR-5110, mmu-miR-3083b-5p were as follows: an initial denaturation step at 95 ℃ for 1 min, followed by 40 cycles of denaturation at 95 ℃ for 15 s, annealing at 60 ℃ for 30 s, and extension at 72 ℃ for 45 s. The primers were designed and synthesized by Wuhan Servicebio Biotechnology Co., Ltd. in Wuhan, China. The sequences of the several primers utilized are listed as follows.
Oligonucleotides
| Gene | Sequence |
|---|---|
| M-BNIP1-S | AACAGTCAGCAAGGGAGCAAGA |
| M-BNIP1-A | AGTTCTGCCTTCTCCAGGTTGT |
| M-GAPDH-S | CCTCGTCCCGTAGACAAAATG |
| M-GAPDH-A | TGAGGTCAATGAAGGGGTCGT |
| M-E230013L22Rik-S | TCTGTACCGGAGTGTGAACG |
| M-E230013L22Rik-A | GGAAGGCGCTCTGGAACTTT |
| mmu-miR-1966-5p-RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGACTCTCT |
| mmu-miR-1966-5p-S | ACACTCCAGCTGGGAAGGGAGCTGGCTCAGGAG |
| mmu-miR-1970c-5p-RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCAAAGCAT |
| mmu-miR-1970c-5p-S | ACACTCCAGCTGGGTGTGTCACTGGGGTTAT |
| mmu-miR-188-5p-RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCCTCCAC |
| mmu-miR-188-5p-S | ACACTCCAGCTGGGCATCCCTTGCATGGT |
| mmu-miR-3083b-5p-RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCTCCGAA |
| mmu-miR-3083b-5p-S | ACACTCCAGCTGGGAAGGCTGGGAATGTTT |
| mmu-miR-5110-RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAATTCCAC |
| mmu-miR-5110-S | ACACTCCAGCTGGGGGAGGAGGTAGAGGGTGGT |
| U6-S | CTCGCTTCGGCAGCACA |
| U6-A | AACGCTTCACGAATTTGCGT |
| Universal primer-A | TGGTGTCGTGGAGTCG |
S: Upstream primer; A: Downstream primer.
Flow cytometry
After radiation, Mlg2908 cells were extracted one or two days later. Following a PBS wash, the cells were treated following the directions provided by the Annexin V-FITC apoptosis kit (Beyotime, China). The cells were resuspended at a concentration of 1–5 × 106 cells/mL in 195 µL of binding buffer. Incubated for 10 to 15 min at ambient temperature with 5 µL PI and 10 µL Annexin V. Flow cytometry was utilized to distinguish apoptosis by observing green fluorescence, cell death by red fluorescence, and the absence of fluorescence from viable cells.
Immunohistochemistry
6 days after radiation, mouse lung tissue was taken on ice, fixed with 4 % paraformaldehyde for 3 h, and paraffin-embedded, sliced, dewaxed, and hydrated, after antigen recovery by high temperature and high pressure, the samples were treated with buffer containing bovine serum protein, and primary antibody BNIP1 was incubated at 4 ℃ (1: 200, Proteintech, Cat No.15964-1-AP) overnight, and incubated the secondary antibody at room temperature (1:200, Beyotime, A2077) for 20 min. The reaction was then visualized with a 3, 3-diaminobenzidine (DAB) peroxidase substrate kit (DA1010, Solarbio, China). The experiment was repeated three times for each group.
Hematoxylin and eosin staining
6 days after radiation, lung tissue from mice was taken on ice and embedded in paraffin. Cut the embedded tissue blocks into 5–8 µm slices and place the slices on the slide. After the xylene dewaxing treatment, the slices were stained with hematoxylin for 5 min and eosin for 3 min. The slices were placed in ethanol of different concentrations successively for dehydration treatment, and then transparent with xylene. Finally, the sections were sealed on a slide using a tablet sealant and observed and recorded with a microscope. Repeat three times for each set.
Statistical analysis
Data were analyzed using GraphPad Prism 8.0 and ImageJ, and are presented as mean ± standard deviation (SD). The t-tests were used for the comparison of two groups, and the One-way ANOVA was used for the double comparison of multiple groups. P < 0.05 was set as the significance threshold.
Results
Identification of De lncRNA, miRNA, and mRNA
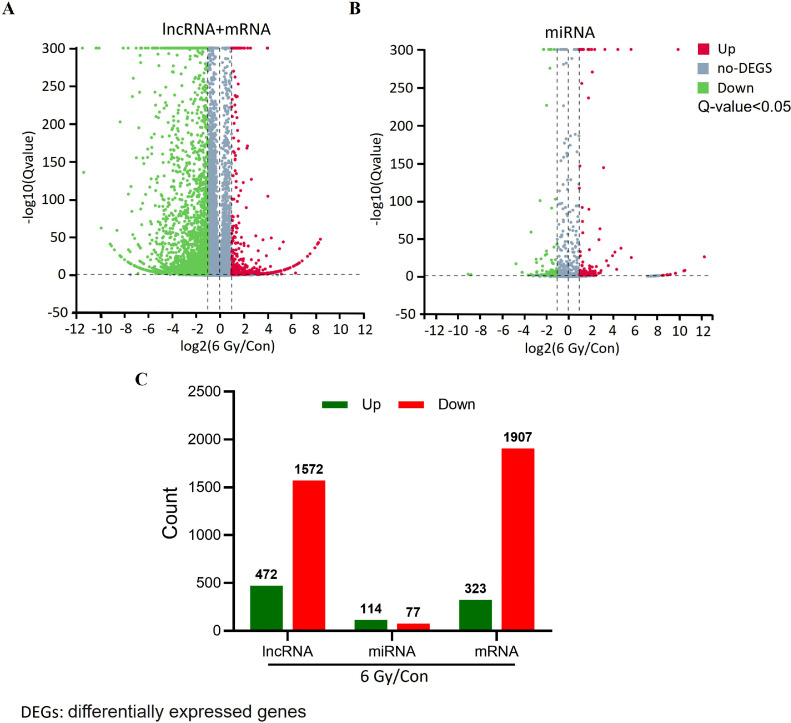
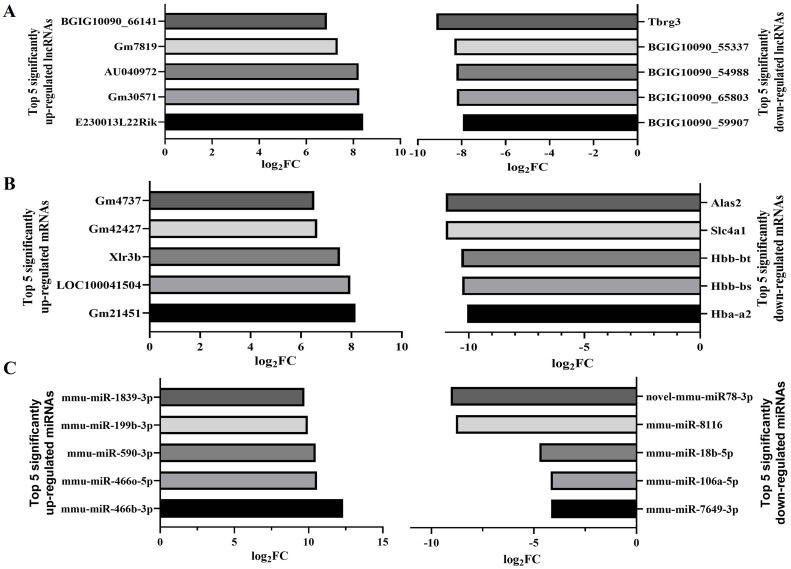
To provide a better grasp of the expression patterns associated with RILI, we performed comprehensive analyses of lncRNA and mRNA expression across the RILI samples. We used log2|FC| ≥ 1 and Q-value < 0.05 as the criteria for determining differential expression. Relative to the controls, we identified 4274 DE genes in the irradiated tissues, among which 3479 were downregulated and 795 highly expressed. 472 lncRNAs and 323 mRNAs were up-regulated, and 1572 lncRANs and 1907 mRNA were down-regulated after radiation damage (Fig. 1A). When we performed the study using miRNAs, we discovered that 114 miRNAs showed a significant increase in expression, while 77 miRNAs showed a significant decrease in expression (Fig. 1B). The top five DE lncRNAs, mRNAs, and miRNAs are shown in Fig. 2.
Fig. 1.
Volcano plot of differentially expressed (DE) lncRNAs, miRNAs, and mRNAs. A. Volcano plot of lncRNA and mRNA profiling between control and 6 Gy-irradiated cells. B. Volcano plot of DE miRNA between control and 6 Gy-irradiated cells. Upregulated genes are colored red, downregulated genes are colored green; and genes showing no change are colored gray. C. Numbers of DE lncRNAs, miRNAs, and mRNAs. Q-value < 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 2.
The five top-upregulated and downregulated lncRNAs, miRNAs, and mRNAs (6 Gy/Con). A. The five top-upregulated and downregulated lncRNAs. B. The five top-upregulated and downregulated mRNAs. C. The five top-upregulated and downregulated miRNAs.
Venn analysis
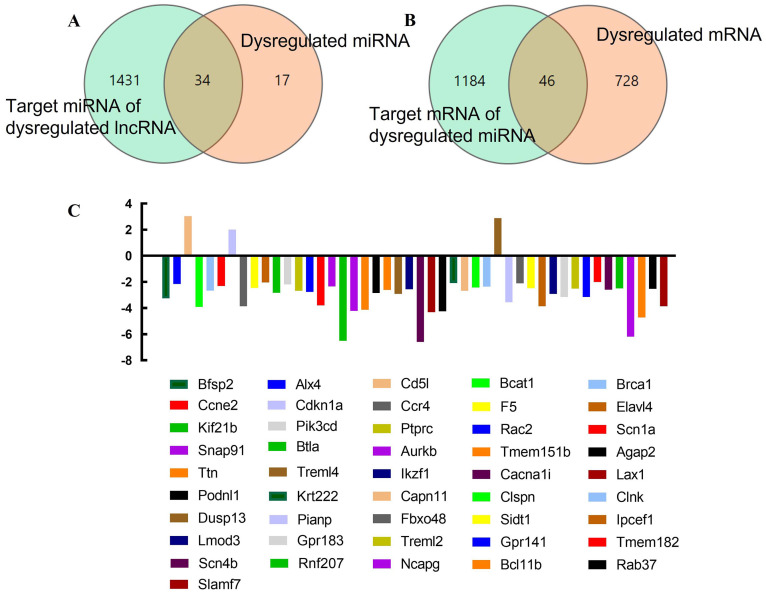
Subsequently, we investigated the physiological functions of the dysregulated lncRNAs in RILI by utilizing the ceRNA axis. The cut-off criteria were defined as Log2|FC| ≥ 2 and Q-value < 0.05. All dysregulated lncRNAs were selected to identify their related sponging miRNAs and target mRNAs. We finally identified 34 target miRNAs of the DE lncRNAs and 46 target mRNAs of the DE miRNAs (Fig. 3AB). The 46 target mRNAs were Bfsp2 Alx4, Cd5l, Bcat1, Brca1, Ccne2, Cdkn1a, Ccr4, F5, Elavl4, Kif21b, Pik3cd, Ptprc, Rac2, Scn1a, Snap91, Btla, Aurkb, Tmem151b, Agap2, Ttn, Treml4, Ikzf1, Cacna1i, Lax1, Podnl1, Krt222, Capn11, Clspn, Clnk, Dusp13, Pianp, Fbxo48, Sidt1, Ipcef1, Lmod3, Gpr183, Treml2, Gpr141, Tmem182, Scn4b, Rnf207, Ncapg, Bcl11b, Rab37, and Slamf7 (Fig. 3C). Additionally, we identified the intersection of targeted mRNAs of 34 miRNA and DE mRNAs, including, 29 DE mRNAs (S-Fig. 1AB), which were Cacna1i, Btla, Ncapg, Lax1, Aurkb, Tmem151b, Bcat1, Pianp, Gpr141, Lmod3, Treml4, Agap2, Kif21b, Ptprc, Capn11, Brca1, Ikzf1, Rab37, Rnf207, Sidt1, F5, Snap91, Pik3cd, Alx4, Fbxo48, Krt222, Elavl4, Cdkn1a, and Dusp13 (S-Fig. 1C).
Fig. 3.
Differentially expressed lncRNAs, miRNAs, and mRNAs and their relationships. A. Differentially expressed (DE) miRNAs, the target miRNAs of DE lncRNAs, and their interactions. The green circle shows the 1465 target miRNAs of DE lncRNAs, and the orange circle shows the 51 DE miRNAs. B. DE mRNAs, the target mRNAs of DE miRNAs, and their interactions. The green circle represents the 1230 target mRNAs of DE miRNAs, and the orange circle refers to the 774 DE mRNAs. C. Names of 46 target mRNAs of DE miRNAs. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
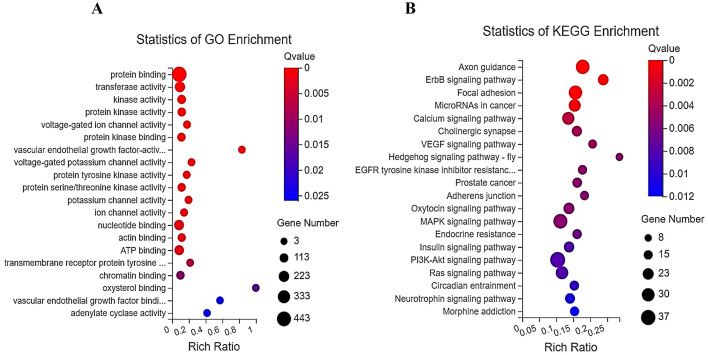
GO and KEGG network analyses
Fig. 4A illustrates the results of the GO study on the target mRNAs of all DE miRNAs. We found the main activities affected by dysregulated miRNA-mediated mRNA, including those related to VEGF, transmembrane receptor protein tyrosine kinases, and voltage-gated ion channels. KEGG network analysis of the target mRNAs of the dysregulated miRNAs showed enrichment in pathways associated with the hedgehog signaling pathway-fly (Btrc, Spop, Sufu, Grk6, Cul3, Gsk3b, Spopl, and Csnk1a1), ErbB signaling (Nrg2, Camk2d, Abl1, Abl2, Akt2, Bad, Camk2a, Cdkn1a, Eif4ebp1, Erbb2, Myc, Nras, Pik3cd, Shc1, Stat5b, Cblb, Pak6, Map2k7, Mapk1, and Gsk3b), VEGF signaling (Akt2, Bad, Cdc42, Nras, Pik3cd, Ppp3r2, Rac2, Vegfa, Pla2g4c, Mapk1, Pla2g4f, and Spnk2), axon guidance (Camk2d, Abl1, Srgap1, Camk2a, Cdc42, Cfl2, Cxcr4, Epha5, Epha6, Ephb2, Plxnb2, Neo1, Nras, Nrp1, Ntn1, Pik3cd, Prkcz, Plxna2, Ppp3r2, Rac2, Sema3a, Sema4c, Sema4f, Sema6b, Pak6, Sema4f, Sema6b, Pak6, Sema3g, Wnt5a, Ablim1, Ssh2, Srgap3, Mapk1, and Gsk3b), and focal adhesion (Rapgef1, Itga1, Akt2, Xiap, Bad, Cdc42, Col4a1, Col4a3, Erbb2, Vegfd, Flt1, Flt4, Lama5, Pdgfra, Pik3cd, Itgb4, Rac2, Shc1, Pak6, Thbs1, Vcl, Vegfa, Vwf, Zyx, Mylk2, Itga8, Mapk1, Flnb, Ppp1r12b, Gsk3b, and Col6a5) (Fig. 4B). In addition, we assessed the KEGG networks of differentially expressed mRNAs. We found that these mRNAs were enriched in pathways related to primary immunodeficiency, the intestinal immune axis for IgA synthesis, hematopoietic cell lineages, systemic lupus erythematosus, and Th1 and Th2 cell differentiation (S-Fig. 2).
Fig. 4.
GO and KEGG analyses of target mRNAs for differentially expressed (DE) miRNAs. A. GO analysis of target mRNAs of DE miRNAs. B. KEGG analysis of target mRNAs of DE miRNAs. The 20 top mRNAs are displayed based on the Q-value of individual analyses.
Establishment and verification of the ceRNA axes
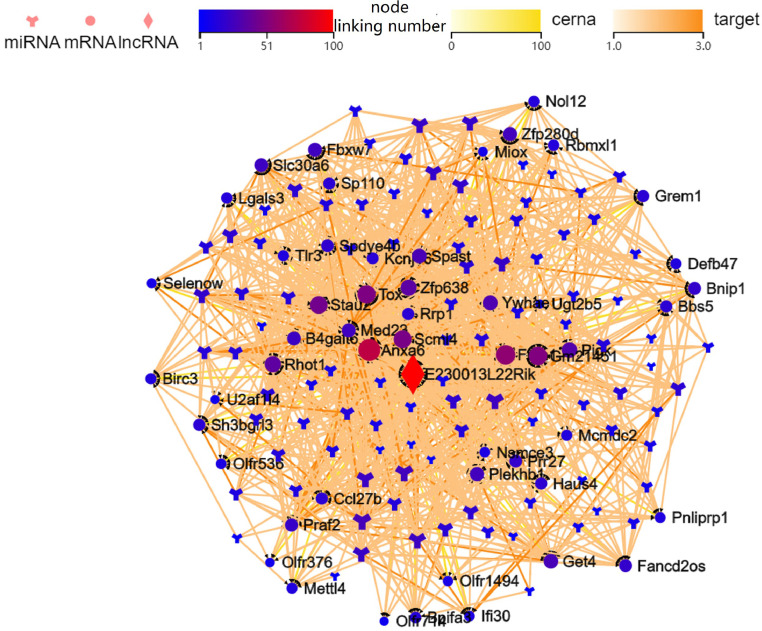
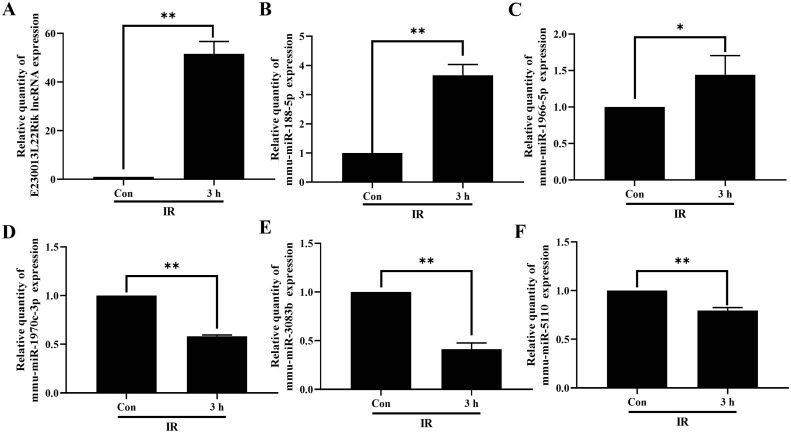
For constructing the lncRNA-miRNA-mRNA ceRNA axis in RILI, we selected DE mRNAs associated with vascular endothelial growth factor identified in the GO analysis. In all, 690 lncRNA-miRNA-mRNA axes were generated with the most up-regulated lncRNA (E230013L22Rik) and down-regulated lncRNA (BGIG10090_59907) were linked to 50 miRNAs and 50 mRNAs. A visual lncRNA-miRNA-mRNA ceRNA axis was then generated based on the filtered RNAs (Fig. 5). We observed multiple pathways associated with BNIP1, namely, E230013L22Rik-mmu-miR-1966–5p-Bnip1, E230013L22Rik-mmu-miR-1970c-3p-Bnip1, E230013L22Rik-mmu-miR-188–5p-Bnip1, E230013L22Rik-mmu-miR-5110-Bnip1, and E230013L22Rik-mmu-miR-3083b-5p-Bnip1. Finally, the ceRNA axes for the most substantially downregulated lncRNA, Tbrg3 (S-Fig.3), and the most significant DE mRNAs, including Alas (downregulated) and GM21451 (upregulated) (S-Fig. 4). 3 h after radiation of Mlg2908 cells, RT-qPCR results showed that E230013L22Rik, mmu-miR-1966–5p, and mmu-miR-188–5p were up-regulated, and mmu-miR-1970c-3p, mmu-miR-5110, and mmu-miR-3083b-5p were down-regulated (Fig. 6).
Fig. 5.
The lncRNA-miRNA-mRNA ceRNA axis was generated using the most upregulated lncRNA E230013L22Rik. The circle indicates mRNAs, the diamond denotes lncRNAs, and the Y-shape represents miRNAs.
Fig. 6.
Validation of RNA expression. RNA expression level of E230013L22Rik(A), mmu-miR-188–5p(B), mmu-miR-1966–5p(C), mmu-miR-1970c-3p(D), mmu-miR-3083b-5p(E), mmu-miR-5110(F) in Mlg2908 cells 3 h after irradiation. *P < 0.05, **P < 0.01.
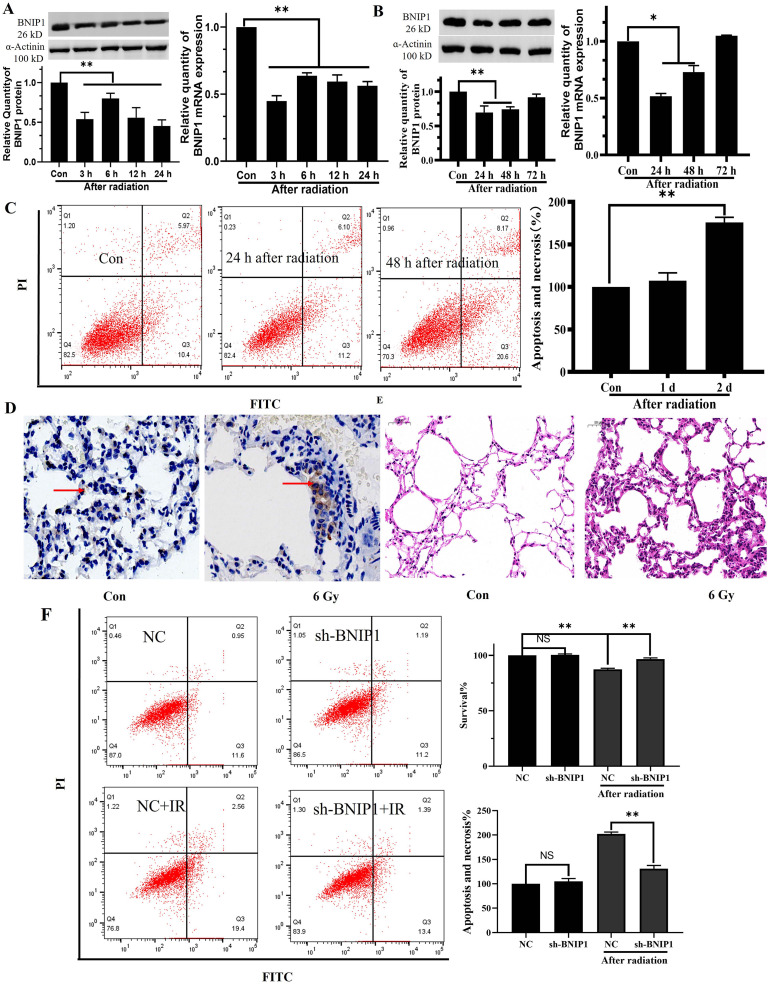
Expression of BNIP1 in lung fibrocytes and lung tissues of mice after radiation
The BNIP1 content was reduced 24 h after radiation, indicating that the irradiated cells were in a self-adaptive protective condition. The irradiation Mlg2908 cells showed considerably lower mRNA and protein levels of BNIP1 relative to the controls (Fig. 7A). Compared with the control group, the expression of BNIP1 decreased at 24 h and gradually increased from 24 h to 72 h after radiation. The changes in protein and mRNA were consistent (Fig. 7B). Flow cytometry demonstrated no evident apoptosis on the first day after radiation, but considerably elevated levels of apoptosis and lowered survival rates on the second day following radiation (Fig. 7C). Lastly, immunohistochemistry showed that the BNIP1 levels in the mouse lungs were markedly increased on the sixth day after irradiation (Fig. 7D). After six days of radiation exposure, the lung tissue in the affected mice showed a considerable drop in integrity, accompanied by a notable increase in the alveolar interstitium. As a consequence, there was a reduction in the volume of the alveoli (Fig. 7E). There was no obvious apoptosis in Mlg2908 cells 2 days after transfection with sh-BNIP1. After transfection with sh-BNIP1, apoptosis decreased 2 days after irradiation compared with the irradiation group alone (Fig. 7F).
Fig. 7.
BNIP1 levels were reduced in the early post-radiation period. A. BNIP1 protein and mRNA expression at different time points within 24 h in Mlg2908 cells following irradiation. B. BNIP1 protein and mRNA levels at different time points from 24 h to 72 h in Mlg2908 cells following irradiation. C. Apoptosis of Mlg2908 cells 1 d and 2 d after radiation, shown by flow cytometry. D. Immunohistochemical analysis of BNIP1 contents in mouse lungs 6 d after irradiation. E. Pathological changes in mouse lungs 6 d after irradiation. F. Flow cytometry showed the survival and apoptosis of Mlg2908 cells 2 days after sh-BNIP1 transfection and 2 days after radiotherapy. *P < 0.05, **P < 0.01.
Discussion
RILI is a multifaceted process that involves diverse cellular and molecular associations that result in tissue fibrosis, necrosis, atrophy, and vascular injury [5,20]. Novel biomarker discovery is essential, as evidenced by recent studies in the area of radiation bioassays [14]. Prognosis improvement for RILI patients may be possible with early prediction and injury identification, which can also significantly lower medical expenses associated with treating secondary diseases. An increasing body of research indicates that lncRNAs function as ceRNAs by binding to miRNAs and controlling target gene and mRNA expression [21]. The ability to obtain more accurate results has improved because of the rapid development of microarray and RNA-Seq technologies. Here, we compared irradiated and non-irradiated cells using RNA-Seq to examine RNA transcriptome changes linked to RILI. We found 4274 DE genes (795 up-regulated and 3479 down-regulated) in comparison to controls. According to the data reported in Fig. 5, E230013L22Rik, the most significantly elevated lncRNA, was chosen to determine its targeted miRNA, which was then targeted to mRNA for network mapping. The changes of E230013L22Rik and mmu-miR-1970c-3p, mmu-miR-5110, and mmu-miR-3083b-5p showed an up-down trend. The changes of mmu-miR-1966–5p mmu-miR-188–5p and BNIP1 showed an up-down relationship. But E230013L22Rik, mmu-miR-1966–5p and mmu-miR-188–5p were upregulated, mmu-miR-1970c-3p, mmu-miR-5110, The changes between mmu-miR-3083b-5p and BNIP1 did not strictly follow the up-down relationship, which may be due to the restriction of other molecules in the middle, resulting in the incomplete targeting relationship between them. The most frequently associated mRNA with miRNA, BNIP1, was discovered.
BNIP1 (Bcl2 Interacting Protein 1) is a protein of 228 amino acids localized to the endoplasmic reticulum [22]. It is known to modulate multiple cellular processes, such as apoptosis, autophagy [23], the formation and maintenance of the endoplasmic reticulum network structure [24], and mitochondrial dynamics [25]. RNF185 functions as an E3 ubiquitin ligase within mitochondria, and BNIP1 is classified as one of its substrates. Through ubiquitinating BNIP1 on the mitochondrial membrane, RNF185 controls mitochondrial homeostasis by facilitating its engulfment in autophagosomes and subsequent degradation [26]. Fibroblasts from humans with disproportionately short stature show retarded end-stage formation and/or clearance of autophagosomes, and BNIP1 is known to be associated with autophagy in bone development [23]. The mitochondrial targeting domain (MTD) in BNIP1 is a necrosis-promoting motif termed B1MLM. The B1MLM peptide in combination with a cell-penetrating peptide can induce necrosis in a manner like MTD conjugated with eight arginine residues (R8: MTD) (Park et al., 2019; Seo et al., 2009). BNIP1 possesses a complete Bcl2-homologous domain 3 (BH3), and its overexpression promotes apoptosis [27]. After its association with the anti-apoptotic protein Bcl2, the BH3 domain of BNIP1 triggers cellular apoptosis in a manner dependent on BAX. Furthermore, BNIP1 controls the connection between apoptosis and endoplasmic reticulum fusion; it can detect defects in vesicular fusion at the membrane, thereby inducing apoptosis [28]. BNIP1 also stimulates the mitochondrial apoptosis network by promoting mitochondrial fracture. A decrease in BNIP1 expression has the potential to decrease both the end-folding endoplasmic reticulum signal and the caspase-9 cleavage induced by RNF186. This indicates that the relationship between endoplasmic reticulum stress and mitochondrial apoptosis is regulated by BNIP1. Thus, BNIP1 facilitates the transduction of pro-apoptotic signals by mitochondria [26,29]. BNIP1 may be a TRIB3-interacting protein and immunoprecipitation data have also confirmed that this protein interacts with TRIB3 in radiation-resistant MDA-MB-231 cells (Lee et al., 2019).
Our findings indicated that the pro-apoptotic factor BNIP1 was strongly downregulated within 24 h after irradiation, and the expression was still downregulated at 48 h after radiation, compared with the control group. However, flow cytometry showed that cell apoptosis was significantly enhanced 48 h after radiation. Although BNIP1 is a pro-apoptotic factor, its downregulation 48 h after radiation did not cause a reduction of apoptosis, suggesting the question of what causes the increase in apoptosis. This may be related to the modulation of other apoptotic proteins. The Bcl2 family of proteins consists of both pro-apoptotic (Bax, Bak, Bad, Bid, Puma, Bim, and Noxa) and anti-apoptotic (Bcl2, Bcl-xL, Bcl-w, and Mcl-1) factors. Cellular apoptosis is predominantly regulated by the intricate homeostasis between pro-apoptotic and anti-apoptotic factors; therefore, it is probable that overexpression of pro-apoptotic factors or downregulation of anti-apoptotic factors could ultimately induce cellular apoptosis. By studying the expression changes of apoptosis-related proteins such as BNIP1 during radiotherapy, we preliminarily confirmed the important role of BNIP1 in radiation lung injury, providing data for the study of radiation injury protection, and providing a new therapeutic target for the prevention and treatment of clinical radiation lung injury.
Similar to other studies, we demonstrated an essential role of VEGF in RILI development [5,30,31]. The results of our GO analysis of target mRNAs indicated a substantial change in VEGF activity. Additionally, DE mRNAs have been associated with hematopoietic cell lineages, cytokine-cytokine receptor interactions, primary immunodeficiency, systemic lupus erythematosus, the intestinal immune axis for IgA synthesis, and cytokine-cytokine receptor associations, according to KEGG analysis. This is in agreement with observations regarding lung injury induced by chest irradiation [6], which indicated enrichment in Th1 and Th2 cell differentiation, and hematopoietic cell lineage following cellular irradiation.
Furthermore, there are key radiation damage-related genes (such as Akt, Myc, and Bad) that were differentially expressed in major enrichment networks. Bad is a protein that induces cell death and is a member of the Bcl2 family. Phosphorylation of Bad enhances cell survival in certain types of cells [32]. Compared to the control group, embryos that express Bad exhibit a much higher susceptibility to apoptosis produced by IR [33]. Similarly, in this study, we demonstrated that following irradiation, the ErbB axis, focal adhesion, VEGF network, and Bad gene were differentially regulated.
Activation of protein kinase B (Akt) is known to protect cells against ionizing radiation-induced apoptosis [34,35]. In response to multiple growth factors and ionizing radiation, Akt is strongly activated [26]. Moreover, the erythroblastic oncogene B (ErbB) axis strictly modulates the phosphatidylinositol 3-kinase (PI3K)/Akt network to regulate cell proliferation, migration, differentiation, apoptosis, and cell motility. Recent findings indicate that the PI3K/Akt pathway influences the development of resistance to radiotherapy by increasing the expression of VEGF and promoting the formation of capillary-like tubules in different types of cancers [36].
In this study, we have shown that the Akt gene, which is expressed at varying levels, is found in the ErbB axis, focal adhesion, and the VEGF networks. Myc is a protooncogene responsible for encoding transcription factors that are involved in cellular growth, proliferation, apoptosis, and angiogenesis. Aberrant Myc expression is a hallmark of radiation-related angiosarcoma [37], and it often serves as a diagnostic marker for the delineation of RT-AS from other radiation-related vascular lesions [38]. Here, we demonstrated the differential expression of Myc in the ErbB axis of irradiated cells. The interaction between Myc and members of the ErbB family could be further explored in the future, and how this interaction affects cell growth and differentiation.
Although remarkable progress has been made in predicting lncRNA-miRNA-mRNA ceRNA axis using tools such as miRanda-3.3a, RNAhybrid v2.0, Blastn, studies still face some limitations that should not be ignored. There are some errors and false positive rates in the prediction, which affect the accuracy and reliability of the data. Therefore, further experimental studies are needed to verify the accuracy of the prediction results. Our research relies on publicly available data. While these public data sets provide us with a great deal of information, there are some inherent potential biases. Therefore, when using these tools to predict lncRNA-miRNA-mRNA ceRNA axis, we need to treat publicly available data with caution and combine multiple data sources for comprehensive analysis as far as possible.
Conclusion
In summary, we employed both RNA-Seq and microarrays to identify DE lncRNAs, miRNAs, and mRNAs. We also generated and assessed a lncRNA-miRNA-mRNA ceRNA axis. The ceRNA network revealed that BNIP1 was a critical mRNA modulated by the most significant upregulation of lncRNA E230013L22Rik. RNA dysregulation in the early stage of RILI may lead to severe complications at a later stage, with BNIP1 contributing to radiation-induced cellular apoptosis in RILI. Based on our FEA, VEGF activity was found to be crucial for radiation-induced RILI, and it utilized the proposed ceRNA axis. Furthermore, several critical genes associated with radiation damage exhibited differential regulation, including Akt, Myc, and Bad. Further investigation, including the present report, into the effects of associated ceRNA networks could potentially be advantageous for RILI interventions.
Data availability
All data are presented within this article.
Source of funding
This work was supported by the Natural Science Foundation of China (NSFC) (Approval Nos. 81773358, 11705158, and U1504824).
Consent to publish
All authors approve of the publication of this article.
CRediT authorship contribution statement
Qing-hua Yu: Validation, Methodology, Formal analysis. Shu-yan Duan: Software, Resources, Data curation. Xue-kun Xing: Methodology. Xin-ming Fan: Validation, Resources. Nan Zhang: Methodology, Investigation, Formal analysis. Gui-yuan Song: Validation, Software, Methodology, Investigation, Formal analysis. Yong-jian Hu: Methodology, Formal analysis. Fei Wang: Software, Data curation. Tian-zhu Chao: Methodology. Li-tao Wang: Data curation. Ping Xu: Writing – review & editing, Writing – original draft, Validation, Project administration, Funding acquisition.
Declaration of competing interest
The authors have no conflicts of interest to disclose.
Acknowledgments
We convey our heartfelt appreciation to the Shanghai Jikai Gene Medical Technology Co., Ltd. China for their assistance with sequencing. Thanks to Professor Cheng-shi Ding for assisting in flow cytometry to detect cell apoptosis.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2024.102007.
Appendix. Supplementary materials
References
- 1.Brizel D.M. Pharmacologic approaches to radiation protection. J. Clin. Oncol. 2007;25:4084–4089. doi: 10.1200/JCO.2007.11.5816. [DOI] [PubMed] [Google Scholar]
- 2.Buergy D., Lohr F., Baack T., Siebenlist K., Haneder S., Michaely H., Wenz F., Boda-Heggemann J. Radiotherapy for tumors of the stomach and gastroesophageal junction–a review of its role in multimodal therapy. Radiat. Oncol. 2012;7:192. doi: 10.1186/1748-717X-7-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darby S., McGale P., Peto R., Granath F., Hall P., Ekbom A. Mortality from cardiovascular disease more than 10 years after radiotherapy for breast cancer: nationwide cohort study of 90 000 Swedish women. BMJ. 2003;326:256–257. doi: 10.1136/bmj.326.7383.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dracham C.B., Shankar A., Madan R. Radiation induced secondary malignancies: a review article. Radiat. Oncol. J. 2018;36:85–94. doi: 10.3857/roj.2018.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giuranno L., Ient J., De Ruysscher D., Vooijs M.A. Radiation-induced lung injury (RILI) Front. Oncol. 2019;9:877. doi: 10.3389/fonc.2019.00877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y., Zou L., Yang X., Chu L., Ni J., Chu X., Guo T., Zhu Z. Identification of lncRNA, MicroRNA, and mRNA-associated CeRNA network of radiation-induced lung injury in a mice model. Dose Response. 2019;17 doi: 10.1177/1559325819891012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aryankalayil M.J., Martello S., Bylicky M.A., Chopra S., May J.M., Shankardass A., MacMillan L., Sun L., Sanjak J., Vanpouille-Box C., Eke I., Coleman C.N. Analysis of lncRNA-miRNA-mRNA expression pattern in heart tissue after total body radiation in a mouse model. J. Transl. Med. 2021;19:336. doi: 10.1186/s12967-021-02998-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayupe A.C., Reis E.M. Evaluating the stability of mRNAs and noncoding RNAs. Methods Mol. Biol. 2017;1468:139–153. doi: 10.1007/978-1-4939-4035-6_11. [DOI] [PubMed] [Google Scholar]
- 9.Arun G., Diermeier S.D., Spector D.L. Therapeutic targeting of long non-coding RNAs in cancer. Trends. Mol. Med. 2018;24:257–277. doi: 10.1016/j.molmed.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 11.Mendell J.T. Targeting a long noncoding RNA in breast cancer. N. Engl. J. Med. 2016;374:2287–2289. doi: 10.1056/NEJMcibr1603785. [DOI] [PubMed] [Google Scholar]
- 12.Schmitt A.M., Chang H.Y. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo J.W., Wang X., Yang Y., Mao Q. Role of micro-RNA (miRNA) in pathogenesis of glioblastoma. Eur. Rev. Med. Pharmacol. Sci. 2015;19:1630–1639. [PubMed] [Google Scholar]
- 14.Coy S.L., Cheema A.K., Tyburski J.B., Laiakis E.C., Collins S.P., Fornace A., Jr. Radiation metabolomics and its potential in biodosimetry. Int. J. Radiat. Biol. 2011;87:802–823. doi: 10.3109/09553002.2011.556177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNa. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y., Ye W., Zhang Y., Xu Y. High speed BLASTN: an accelerated MegaBLAST search tool. Nucleic. Acids. Res. 2015;43:7762–7768. doi: 10.1093/nar/gkv784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li R., Qu H., Wang S., Wei J., Zhang L., Ma R., Lu J., Zhu J., Zhong W.D., Jia Z. GDCRNATools: an R/Bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC. Bioinformatics. 2018;34:2515–2517. doi: 10.1093/bioinformatics/bty124. [DOI] [PubMed] [Google Scholar]
- 18.Guo L.L., Song C.H., Wang P., Dai L.P., Zhang J.Y., Wang K.J. Competing endogenous RNA networks and gastric cancer. World J. Gastroenterol. 2015;21:11680–11687. doi: 10.3748/wjg.v21.i41.11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smoot M.E., Ono K., Ruscheinski J., Wang P.L., Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding N.H., Li J.J., Sun L.Q. Molecular mechanisms and treatment of radiation-induced lung fibrosis. Curr. Drug Targets. 2013;14:1347–1356. doi: 10.2174/13894501113149990198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou R.S., Zhang E.X., Sun Q.F., Ye Z.J., Liu J.W., Zhou D.H., Tang Y. Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in squamous cell carcinoma of tongue. BMC Cancer. 2019;19:779. doi: 10.1186/s12885-019-5983-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishiwaki Y., Masai I. β-SNAP activity in the outer segment growth period is critical for preventing BNip1-dependent apoptosis in zebrafish photoreceptors. Sci. Rep. 2020;10:17379. doi: 10.1038/s41598-020-74360-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holling T., Bhavani G.S., von Elsner L., Shah H., Kausthubham N., Bhattacharyya S.S., Shukla A., Mortier G.R., Schinke T., Danyukova T., Pohl S., Kutsche K., Girisha K.M. A homozygous hypomorphic BNIP1 variant causes an increase in autophagosomes and reduced autophagic flux and results in a spondylo-epiphyseal dysplasia. Hum. Mutat. 2022;43:625–642. doi: 10.1002/humu.24368. [DOI] [PubMed] [Google Scholar]
- 24.Nakajima K., Hirose H., Taniguchi M., Kurashina H., Arasaki K., Nagahama M., Tani K., Yamamoto A., Tagaya M. Involvement of BNIP1 in apoptosis and endoplasmic reticulum membrane fusion. EMBO J. 2004;23:3216–3226. doi: 10.1038/sj.emboj.7600333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryu S.W., Choi K., Yoon J., Kim S., Choi C. Endoplasmic reticulum-specific BH3-only protein BNIP1 induces mitochondrial fragmentation in a Bcl-2- and Drp1-dependent manner. J. Cell Physiol. 2012;227:3027–3035. doi: 10.1002/jcp.23044. [DOI] [PubMed] [Google Scholar]
- 26.Tang F., Wang B., Li N., Wu Y., Jia J., Suo T., Chen Q., Liu Y.J., Tang J. RNF185, a novel mitochondrial ubiquitin E3 ligase, regulates autophagy through interaction with BNIP1. PLoS ONE. 2011;6:e24367. doi: 10.1371/journal.pone.0024367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Handschuh L., Wojciechowski P., Kazmierczak M., Lewandowski K. Transcript-level dysregulation of BCL2 family genes in acute myeloblastic leukemia. Cancers. 2021;13 doi: 10.3390/cancers13133175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishiwaki Y., Yoshizawa A., Kojima Y., Oguri E., Nakamura S., Suzuki S., Yuasa-Kawada J., Kinoshita-Kawada M., Mochizuki T., Masai I. The BH3-only SNARE BNip1 mediates photoreceptor apoptosis in response to vesicular fusion defects. Dev. Cell. 2013;25:374–387. doi: 10.1016/j.devcel.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 29.Wang P., Wu Y., Li Y., Zheng J., Tang J. A novel RING finger E3 ligase RNF186 regulate ER stress-mediated apoptosis through interaction with BNip1. Cell Signal. 2013;25:2320–2333. doi: 10.1016/j.cellsig.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 30.Trott K.R., Herrmann T., Kasper M. Target cells in radiation pneumopathy. Int. J. Radiat. Oncol. Biol. Phys. 2004;58:463–469. doi: 10.1016/j.ijrobp.2003.09.045. [DOI] [PubMed] [Google Scholar]
- 31.Wirsdörfer F., Jendrossek V. The role of lymphocytes in radiotherapy-induced adverse late effects in the lung. Front. Immunol. 2016;7:591. doi: 10.3389/fimmu.2016.00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.She Q.B., Ma W.Y., Zhong S., Dong Z. Activation of JNK1, RSK2, and MSK1 is involved in serine 112 phosphorylation of Bad by ultraviolet B radiation. J. Biol. Chem. 2002;277:24039–24048. doi: 10.1074/jbc.M109907200. [DOI] [PubMed] [Google Scholar]
- 33.Jette C.A., Flanagan A.M., Ryan J., Pyati U.J., Carbonneau S., Stewart R.A., Langenau D.M., Look A.T., Letai A. BIM and other BCL-2 family proteins exhibit cross-species conservation of function between zebrafish and mammals. Cell Death. Differ. 2008;15:1063–1072. doi: 10.1038/cdd.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng Z., Gan Y.H. Activating PTEN by COX-2 inhibitors antagonizes radiation-induced AKT activation contributing to radiosensitization. Biochem. Biophys. Res. Commun. 2015;460:198–204. doi: 10.1016/j.bbrc.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 35.Park H.S., Yun Y., Kim C.S., Yang K.H., Jeong M., Ahn S.K., Jin Y.W., Nam S.Y. A critical role for AKT activation in protecting cells from ionizing radiation-induced apoptosis and the regulation of acinus gene expression. Eur. J. Cell Biol. 2009;88:563–575. doi: 10.1016/j.ejcb.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Shiojima I., Walsh K. Role of Akt signaling in vascular homeostasis and angiogenesis. Circ. Res. 2002;90:1243–1250. doi: 10.1161/01.res.0000022200.71892.9f. [DOI] [PubMed] [Google Scholar]
- 37.Sheu T.G., Hunt K.K., Middleton L.P. MYC and NOTCH1-positive postradiation cutaneous angiosarcoma of the breast. Breast. J. 2021;27:264–267. doi: 10.1111/tbj.14171. [DOI] [PubMed] [Google Scholar]
- 38.Kuba M.G., Xu B., D'Angelo S.P., Rosenbaum E., Plitas G., Ross D.S., Brogi E., Antonescu C.R. The impact of MYC gene amplification on the clinicopathological features and prognosis of radiation-associated angiosarcomas of the breast. Histopathology. 2021;79:836–846. doi: 10.1111/his.14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are presented within this article.