Abstract
We have made two stocks of a herpes simplex virus 1 mutant lacking intact US5 and US6 open reading frames encoding glycoproteins J (gJ) and D (gD), respectively. The stock designated gD−/+, made in cells carrying US6 and expressing gD, was capable of productively infecting cells, whereas the stock designated gD−/−, made in cells lacking viral DNA sequences, was known to attach but not initiate infection. We report the following. (i) Both stocks of virus induced apoptosis in SK-N-SH cells. Thus, annexin V binding to cell surfaces was detected as early as 8 h after infection. (ii) US5 or US6 cloned into the baculovirus under the human cytomegalovirus immediate-early promoter was expressed in SK-N-SH cells and blocked apoptosis in cells infected with either gD−/+ or gD−/− virus, whereas glycoprotein B, infected cell protein 22, or the wild-type baculovirus did not block apoptosis. (iii) In SK-N-SH cells, internalized, partially degraded virus particles were detected at 30 min after exposure to gD−/− virus but not at later intervals. (iv) Concurrent infection of cells with baculoviruses did not alter the failure of gD−/− virus from expressing its genes or, conversely, the expression of viral genes by gD−/+ virus. These results underscore the capacity of herpes simplex virus to initiate the apoptotic cascade in the absence of de novo protein synthesis and indicate that both gD and gJ independently, and most likely at different stages in the reproductive cycle, play a key role in blocking the apoptotic cascade leading to cell death.
In this report we show that herpes simplex virus 1 (HSV-1) mutants lacking the gene encoding glycoprotein D (gD) and which attach to cell surfaces but cannot initiate productive infection, or initiate an infection with production of gD-deficient progeny, nevertheless induced programmed cell death. The circumstances which led us to initiate these studies were as follows.
This and other laboratories have extensively documented evidence that wild-type HSV-1 blocks programmed cell death induced by exogenous agents and that mutants in early functions induce programmed cell death (2, 3, 17, 18, 22–24, 39). The studies reported from this laboratory began with the observation that a mutant lacking the gene encoding infected cell protein 4 (ICP4)—the major regulatory protein—induced apoptosis in a variety of cell lines, in both caspase 3-dependent and -independent manners (16–18, 23, 24). To test the possibility that apoptosis can be induced by a factor introduced into the cells during viral entry, we analyzed cells infected with a mutant, HSV-(HFEM)tsB7, that blocks release of viral DNA from capsids at the nonpermissive temperature (4). Those studies showed that at the nonpermissive temperature tsB7 induced apoptosis in a cell-type-dependent manner (16). These observations led to two conclusions. First, HSV induced, but also blocked, programmed cell death at multiple steps in the course of the viral replicative cycle. Second, at least one signal for induction of the apoptotic cascade could be expressed in the absence of de novo viral gene expression.
The initiation of infection prior to the release of viral DNA involves attachment of the virion to the cell surface mediated by gC, and possibly gB (20, 35, 38), fusion of the envelope with the plasma membrane mediated minimally by gD but with the involvement of gB, gH, and gL (8, 15, 25, 33); see also references (9) and (35), release of tegument proteins and capsid-tegument proteins from the virion into the cytoplasm, and transport of a capsid-tegument protein to the nuclear pore and of selected tegument proteins (e.g., VP16 or αTIF) to the nucleus (4, 5). In an attempt to sort out which of the components of the nucleus are responsible for the induction of the apoptotic cascade, we decided to start with the initial steps in the attachment and entry of the virus into the infected cell. Studies on HSV entry indicate that in the absence of gD in both the genome and the envelope, the virus attaches but does not penetrate. To test the role of these initial events in the induction of apoptosis, we derived two stocks of viruses. The first, designated gD−/+, was produced by growing a virus lacking the gD gene in cells expressing gD (25). Because this virus contained gD in its envelope, it could enter and produce infectious progeny in both gD-expressing and -nonexpressing cell lines. In the latter, however, the progeny virus remained sequestered in the infected cell. The second stock was derived by passage of gD−/+ virus in cells that did not carry the HSV-1 gene encoding gD. In this instance the virus, designated gD−/−, was recovered from the infected cells. This population of mutant viruses has been shown to attach to cells, but expression of viral genes does not ensue.
We report that both gD−/− and gD−/+ viruses induce programmed cell death in human SK-N-SH cells. In cells carrying the BamHI J fragment of HSV-1 DNA, the gD−/+ viruses recombined with the resident HSV-1 DNA, and several wild-type rescuant were isolated. Analyses of the genes in the immediate environment of US6 encoding gD indicate that gD−/+ viruses lacked in addition to gD a wild-type US5 encoding gJ (25). We report that both gD and gJ delivered in trans block the apoptosis induced by gD−/+ and gD−/− viruses whereas gB, an unrelated gene, or the wild-type baculovirus itself did not. Finally, at multiplicities of infection used in this study, we noted that gD−/− viruses were taken up by vesicles likely to be of endocytic origin and were degraded within a short interval after exposure of cells to the virus.
MATERIALS AND METHODS
Cells.
SK-N-SH, HEp-2 and Vero cells were obtained from American Type Culture Collection (Manassas, Va.) and maintained in Dulbecco's modification of Eagle minimal essential medium (DMEM) containing 10% fetal bovine serum. Insect cell line Sf9 (Spodoptera frugiperda) was obtained from PharMingen (San Diego, Calif.). Unless indicated, cultures were seeded less than 20 h prior to infection and assayed at 60 to 70% confluence. VD60, a cell line carrying the entire BamHI J fragment (25), was a kind gift from D. Johnson. R6 cells were derived from rabbit skin cells in the course of this study. Plasmid pEA102 containing the HSV-1 gD coding sequence under the UL26.5 promoter was constructed as follows. The gD coding sequence was amplified by PCR with primers CTCTTTTGTCTCGAGCGTTCCGGTATGGGG (forward) and GTCAGGTCTGCGGGCTCGAGATGGGACCTT (reverse) to yield a fragment that includes the entire gD coding sequence from 14 bp upstream of the start codon to 32 bp downstream of the stop codon. The purified fragment, excised with XhoI, was filled in and cloned into the HindIII site of pEA101. pEAl01 was derived from pcDNA 3.1(−) by replacement of the cytomegalovirus (CMV) promoter (excised as a 687-bp NruI-NheI fragment) with the UL26.5 promoter, excised as a 887-bp XbaI-BstEII fragment from pRB4090, and cloned in the XbaI-EcoRI sites of the deleted pcDNA 3.1(−). Rabbit skin cells transfected with pEA102 were selected for neomycin G418 resistance, cloned by limiting dilution, and assayed for gD-inducible gD expression, following infection with HSV-2. The cell line was maintained in DMEM supplemented with 5% newborn calf serum and 400 μg of G418 per ml.
Viruses.
HSV-1(F) is the prototype HSV-1 strain used in these laboratories (14). The ICP4 deletion mutant d120 was described elsewhere (13). In the FgDβ mutant, the Escherichia coli lacZ gene replaced portions of US6 and US7 encoding gD and gI, respectively (25). The D10 (gD−/+) stock of FgDβ was kindly provided by D. Johnson. The P6 (gD−/+) virus, kindly provided by P. G. Spear, is a plaque isolate derived from FgDβ in which the deletion in the gI gene has been repaired. Growth of D10 virus in VD60 yielded spontaneous rescuants resulting from recombination between the replicating virus and the resident HSV-1 DNA. Three of these rescuants were plaque purified and tested as described in Results. Virions carrying gD on their envelope provided in trans by growth in complementing cell lines were designated gD−/+. All of stocks of gD−/+ virus used in this study were derived in R6 cells, and titers were determined in these cells. Virions lacking gD in both their genomes and their envelopes were designated gD−/−. The stocks of gD−/− and of HSV-1(F) were prepared by infecting cultures of HEp-2 cells with 10 PFU of gD−/+ and wild-type virus, respectively, per cell.
Baculovirus transfer vectors.
pAc-CMV, kindly provided by M.-T. Sciortino, was derived from the pAcSG2 baculovirus transfer vector (PharMingen) by cloning an XhoI-EcoRI fragment containing the human CMV (HCMV) IE1 promoter/enhancer sequences (provided by L. Hennighausen) in the XhoI-BamHI site of pAcSG2. An EcoRI-BglII fragment encoding gD was amplified by PCR from pEA99 with primers CGGAA-TTCAT-GGGGG-GGGCT-GCCGC-CAG and GAAGA-TCTCT-AGTAA-AACAA-GGGCT-GGTG-CG. The amplified PCR fragment was inserted into the EcoRI-BglII sites of the pAc-CMV polylinker, resulting in the pAc-gD transfer vector. The pAc-gJ transfer vector was constructed by the same strategy as pAc-gD. The primers used for PCR amplification were CGGAATTCATGTCTCTGCGCGCAGTCTGGCATC and GAAGATCTTTTAATATGCTGTTG. Plasmid pRB5252 (29) was digested with EcoRI and BglII. The 1.2-kb fragment containing the α22 gene coding sequences (29) was cloned into the EcoRI-BglII sites in the pAc-CMV polylinker, generating the transfer vector pAc-22. The transfer vector pAc-gB was constructed as follows. The complete gB coding sequence was PCR amplified using primers 5′-GGCAC-GAGGC-CTCCC-CGTAG-TCCCG-CCATG-C (forward) and 5′-CTACG-TGTAC-TCTAG-ATTGT-GGGCA-CC (reverse) to yield a 2,792-bp fragment which included the entire gB open reading frame and 9 bp upstream of the start codon. The fragment was cloned in the StuI-BglII sites of pAc-CMV.
Generation of recombinant baculovirus.
Bac-gD, Bac-gJ, Bac-α22, and Bac-gB were generated using the PharMingen baculovirus expression vector system by cotransfecting transfer vectors pAc-gD, pAc-gJ, pAc-22, and pAc-gB, respectively, along with BaculoGold linearized baculovirus DNA (PharMingen) into Sf9 cells according to the manufacturer's instructions. The viruses were propagated in Sf9 cells grown in 150-cm2 flasks in TNM-FH insect medium (PharMingen). Virus stocks were concentrated by centrifugation at 35,000 × g for 60 min. The pelleted viruses were resuspended in phosphate-buffered saline (PBS) supplemented with 1% fetal bovine serum. Virus titers were determined by plaque assay on Sf9 cells. Bac-XylE is the wild-type baculovirus provided with the PharMingen baculovirus expression vector system.
Exposure of mammalian cells to recombinant baculovirus.
Subconfluent cultures of SK-N-SH cells in 25-cm2 flasks were exposed to 10 to 100 PFU of recombinant baculovirus per cell as stated in Results and incubated at 37°C for 1 h. Then culture medium was then replaced with fresh DMEM containing 10% fetal bovine serum and 2.5 mM sodium butyrate or at concentrations stated in Results and incubated at 37°C for stated time intervals.
Antibodies.
Monoclonal antibodies against gD (clone H170), ICP0 (clone H1083), and gB (clone 1817) were purchased from the Goodwin Cancer Research Institute, Plantation, Fl. The monoclonal antibody to gI (26), the polyclonal antibody R77 to ICP22 (1), and polyclonal antibodies to UL38 and gJ were described elsewhere (11, 37).
Immunblot assays.
Protein concentrations of whole-cell lysates was determined with the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, Calif.). Infected or uninfected cell lysates (50 μg of protein per lane) were electrophoretically separated in 12% denaturing polyacrylamide gels. Proteins were then electrically transferred to a nitrocellulose sheet (Bio-Rad), blocked for 2 h in 5% milk in PBS at room temperature, and then reacted with the primary antibody. The protein bands were visualized by means of an enhanced chemiluminescence detection system (Pierce, Rockford, Ill.) according to the manufacturer's instructions.
Double infection.
Replicate subconfluent cultures of SK-N-SH cells were exposed to 10 PFU of recombinant baculovirus or 100 PFU of Bac-XylE per cell and incubated at 37°C for 2 h. The cells were exposed to either 100 PFU equivalents of gD−/− or 10 PFU of gD−/+ virus per cell and incubated at 37°C for intervals described in Results in medium containing 2.5 mM sodium butyrate.
Induction of apoptosis by TNF-α or Fas ligand.
Cells were exposed to either 10 ng of tumor necrosis factor alpha (TNF-α; Calbiochem, Cambridge, Mass.) per ml of medium for 16 h or 1 μg of anti-Fas monoclonal antibody (immunoglobulin M; Calbiochem) per ml of medium for 12 h and then harvested.
Immunofluorescence.
SK-N-SH cells (5 × 104) were seeded onto four-well glass slides and either exposed to recombinant baculoviruses or doubly infected as described above. Cells were fixed in ice-cold methanol for 20 min at −20°C and then blocked in PBS containing 1% bovine serum albumin at room temperature, rinsed three times with PBS, and reacted for 24 h at 4°C with a 1:2,000 dilution of mouse monoclonal antibody against ICP0 (clone H1083) in PBS. The cells were rinsed five times in PBS, reacted for 1 h with 1:64 dilution of a goat anti-mouse immunoglobulin G conjugated to fluorescein isothiocyanate (FITC) (Sigma, St. Louis, Mo.) in PBS, then rinsed five times with PBS, and mounted in 90% glycerol. Slides were analyzed in a Zeiss confocal fluorescence microscope.
DNA fragmentation assay.
Infected or uninfected cells were collected, washed in PBS, lysed in a solution containing 10 mM Tris-HCl (pH 8.0), 10 mM EDTA, and 0.5% Triton X-100, digested with RNase A (0.1 mg/ml) at 37°C for 1 h, and centrifuged at 12,000 rpm for 25 min in an Eppendorf microcentrifuge to pellet chromosomal DNA. The supernatant fluids were digested with proteinase K (1 mg/ml) at 50°C for 2 h in the presence of 1% sodium dodecyl sulfate, extracted with phenol and chloroform, precipitated in cold ethanol, and subjected to electrophoresis on 1.5% agarose gels containing 0.5 mg of ethidium bromide per ml. Oligonucleosomal DNA fragments were visualized by UV light transillumination. Photographs were taken with the aid of Eagle Eye II (Stratagene).
RESULTS
Experimental design.
To simplify presentation of the data, we have adopted the following specific nomenclature for the viruses used in this study. Wild-type viruses are termed gD+/+ to signify that they contain both the US6 gene encoding gD and gD contained in the envelope; gD−/+ denotes viruses that lack the US6 gene but contain gD made in cells expressing a resident US6 gene; and passage of gD−/+ in cells lacking a resident US6 gene yielded a virus, designated gD−/−, lacking both US6 and gD and shown earlier to be able to attach to the cell surface but not initiate infection (25). Initially we received from D. Johnson a cell line expressing gD (VD60) and the FgDβ (gD−/+) virus. We also received from P. G. Spear a virus [P6 (gD−/+)] derived from FgDβ (gD−/+) but with a restored US7 gene encoding gI. All preparations of gD−/− virus stocks were derived from P6 (gD−/+) virus. The DNA fragment contained in the VD60 cell line exceeded the coding domain of the US6 gene. In consequence, gD−/− stocks contained self rescuants derived by recombination between the resident DNA and the gD−/+ virus. At least three independent rescuants were plaque purified and used as described below. To avoid the generation of self-rescuants, a cell line (R6) was constructed in which the resident HSV-1(F) DNA consisted of the US6 open reading frame fused to the UL26.5 promoter. The R6 cell line expressed gD only after infection with gD−/+ virus and yielded no detectable self-rescuants. All of the gD−/+ stocks used in this study were made in R6 cells. The gD−/− virus was generated from these gD−/+ stocks.
Characteristics of the gD−/− virus.
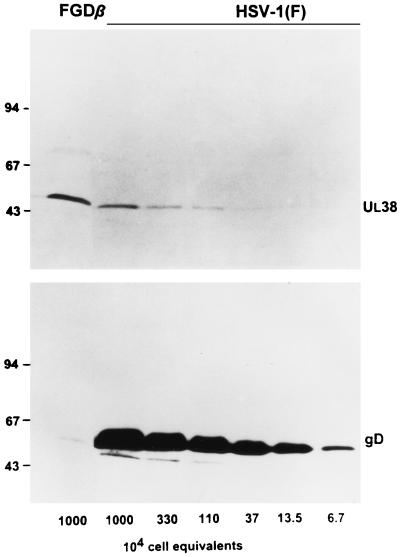
To estimate the relative quantities of gD contained in gD−/− preparations, HEp-2 cells were exposed to 10 PFU of gD−/+ virus made in R6 cells. Total lysates of cells harvested at 18 h after infection was subjected to denaturing electrophoresis in a single lane. Adjacent lanes were loaded with lysates of cells infected with wild-type virus [HSV-1(F)] serially diluted to contain the content of a specific number of cells. The electrophoretically separated polypeptides prepared in duplicate were reacted with a polyclonal antibody to the capsid protein 19C encoded by UL38 and monoclonal antibody to gD. The results (Fig. 1) show that the quantity of gD accumulating in the gD−/− stock was significantly less than 100-fold lower than that contained in HSV-1(F)-infected cells, whereas the amounts of UL38 protein accumulating in cells infected with gD−/− stock was higher than that present in the equivalent amount of HSV-1(F)-infected cells. We estimate on the basis of these results that the amount of gD present in the gD−/− stock containing a lysate from 107 cells was equivalent to that present in 1 × 104 to 2 × 104 cells.
FIG. 1.
Determination of PFU equivalents in a stock of gD−/− mutant virus. Stocks of HSV-1(F) and gD−/− were prepared as described in Materials and Methods. Aliquots from these stocks corresponding to the number of cell equivalents indicated were electrophoretically separated in 10% denaturing polyacrylamide gels and electrically transferred onto a nitrocellulose sheet. The samples were reacted with a UL38-specific polyclonal antiserum and with a gD monoclonal antibody. PFU equivalents for gD−/− stocks were estimated from the ratios of UL38 protein. Sizes are indicated in Mr × 1,000.
gD−/− virus induces annexin V presentation on cell surfaces.
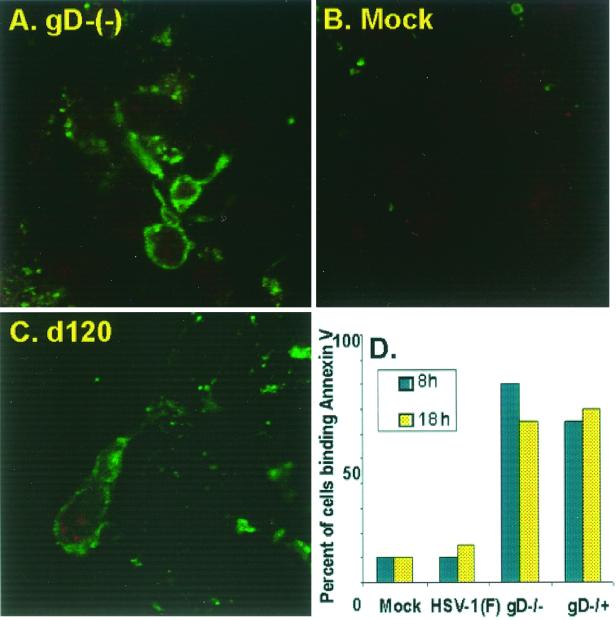
To determine whether both gD−/− and gD−/+ mutant virus stocks induce apoptosis, two experiments were done. In the first, the cells were mock infected or infected with the d120 or gD−/− mutant. The micrographs in Fig. 2 show that the patterns of annexin V binding to cells infected with the d120 mutant were similar to those observed on cells infected with the gD−/− mutant. In the second experiment, (Fig. 2D), the cells were mock infected or infected with HSV-1(F), gD−/−, or gD−/+, and annexin V-positive cells were counted as a percentage of total cells. The results indicate that annexin V bound to the cells as early as 8 h after infection and that the fraction of cells binding annexin V was significantly greater than that binding mock-infected or wild-type virus-infected cells. The slight decrease in the percentage of annexin V-positive cells at 18 h after infection may reflect the loss from the slide surface of cells in more advanced stages of apoptosis. These results are consistent with the hypothesis that both gD−/− and gD−/+ viruses induce apoptosis.
FIG. 2.
(A to C) Digital images of cells infected with gD−/− and dl20. The cells were fixed at 18 h after infection and reacted with annexin V-FITC. The images were collected with a Zeiss confocal microscope as described in Materials and Methods. (D) Quantification of cells with phosphatidylserine on their surface. Uninfected cells or cells infected with HSV-1(F), gD−/−, or gD−/+ were reacted with annexin V-FITC and analyzed in with a Zeiss confocal microscope. The number of annexin V-positive cells as a function of total number of cells was determined at 8 and 18 h after infection. Each data point reflects the average percentage of annexin V-positive cells present in 10 random fields. The data correspond to the average of two independent experiments.
gD−/− and gD+/+ viruses induce DNA fragmentation in SK-N-SH cells.
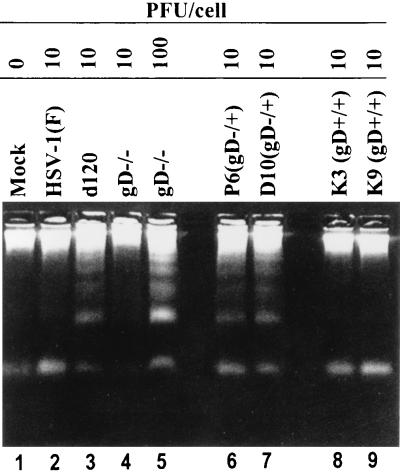
In this series of experiments, SK-N-SH cells were exposed to 10 PFU of HSV-1(F), gD−/+, or two independently derived self-rescuants per cell and to either 10 or 100 PFU equivalents of gD−/− virus per cell. The cells were harvested at 18 h after mock infection or exposure to virus and processed as described in Materials and Methods. As shown in Fig. 3, low-molecular-weight DNA ladders were seen in cells infected with d120 (positive control) and in cells infected with gD−/+ or with 100 PFU equivalents of gD−/−, but not in mock-infected cells, cells infected with the self-rescuants, or cells exposed to 10 PFU of gD−/− virus. We conclude from these results the following. (i) Consistent with the data presented in Fig. 2, both gD−/+ and gD−/− viruses induce fragmentation of DNA consistent with an apoptotic response. (ii) The amount of gD−/− virus required to induce apoptosis was significantly higher than that required to induce apoptosis by gD−/+ virus.
FIG. 3.
Agarose gels containing electrophoretically separated, ethidium bromide-stained low-molecular-weight DNA from SK-N-SH cultures that were mock infected or infected with gD mutant virus. Subconfluent cultures of SK-N-SH cells were either mock infected or exposed to 10 PFU of HSV-1(F), d120, or gD−/+ or 10 or 100 PFU of gD−/− virus per cell. The cells were harvested at 18 h after infection and processed as described in Materials and Methods.
Fate of gD−/− virus.
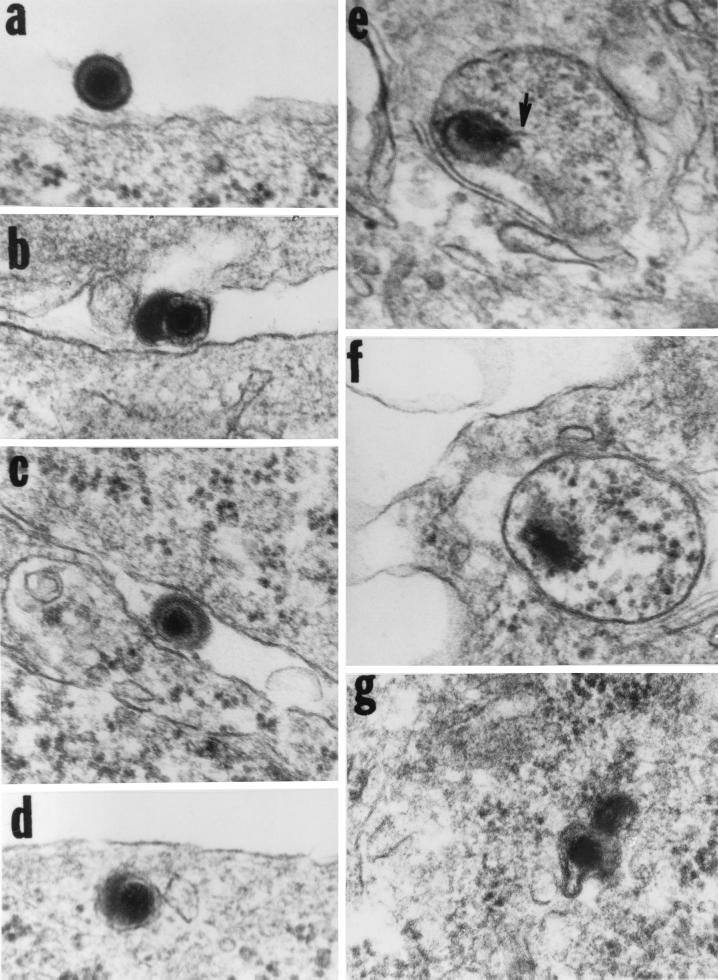
gD−/− viruses have been shown to be able to attach to cell surfaces as a consequence of the interaction of gB and gC with cell surface receptors but to be unable to productively infect cells (24). The rationale of this experiment was based on studies done some time ago showing that whereas cells expressing gD block productive entry of wild-type virus, virus particles are internalized in vesicles likely of endosomal origin and degraded (9). To ascertain the fate of gD−/− virus, SK-N-SH cells were exposed to 100 PFU equivalents per cell and incubated for time intervals ranging from 30 min to 2 h. They were then harvested, fixed, sectioned, and stained for electron microscopy as described in Materials and Methods. The results shown in Fig. 4 were as follows: (i) Virus particles attached to the cell surface (Fig. 4a to d) were readily seen in section of cells harvested at 30 min to 2 h after exposure to virus. (ii) Internalized virus was seen only in cells harvested 30 min after exposure to virus. In all instances (Fig. 4d to g), the virus was present in cytoplasmic vesicles and appeared to be undergoing degradation. As shown below, cells exposed to gD−/− virus do not express α genes, as evidenced by the absence of ICP0.
FIG. 4.
Electron micrographs of thin sections of cells exposed to 100 PFU equivalents of gD−/−. SK-N-SH cells were exposed to 100 PFU equivalents per cell and incubated for 30 min.. They were then harvested, fixed, sectioned, and stained for electron microscopy as described elsewhere (4). The arrow in panel e points to a structure typical of DNA streaming out of damaged capsids.
We conclude from these results that at least a fraction of gD−/− virus is internalized in cells exposed to it.
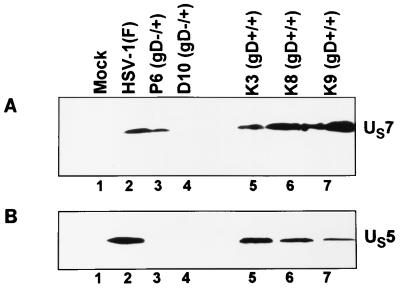
gJ does not accumulate in cells infected with gD−/+ virus.
In principle, to attribute a function to a gene that has been deleted, it is necessary to restore the gene and recover the wild-type phenotype. The failure of self-rescuants (Fig. 3) to induce fragmentation of DNA is only partially convincing evidence since the DNA fragment contained in the VD60 cells extended 3′ and 5′ beyond the domain of the gD gene. The immediate environment of the gD gene contains the genes encoding gJ (US5) and gI (US7). To determine if these genes are expressed, SK-N-SH cells were mock infected or exposed to 10 PFU of wild-type virus or gD−/+ or gD+/+ rescuants per cell. The cells were harvested at 18 h after infection, solubilized, subjected to electrophoresis in denaturing gels, and reacted with a monoclonal antibody to US7 (gI) (Fig. 5A) or polyclonal antibody to US5 (Fig. 5B). The results were as follows: (i) the electrophoretic mobility of gI expressed by P6 (gD−/+) stock could not be differentiated from that of wild-type or rescuant viruses, and (ii) gJ was not detected in cells infected with P6 (gD−/+) or P10 (gD−/+) stocks but was readily detectable in cells infected with wild-type virus or self-rescuants (Fig. 5B). The failure to detect gJ was reproducible in several experiments. We conclude from this experiment that the gD−/+ stocks contain an impaired gJ gene.
FIG. 5.
Electrophoretically separated lysates of cells infected with gD−/+ virus or rescuants and reacted with a monoclonal antibody to gI (A) or polyclonal antibody to gJ (B). SK-N-SH cells were mock infected or exposed (10 PFU/cell) to wild-type virus, P6 (gD−/+), or p10 (gD−/+) or individually to three independently derived gD+/+ rescuants. The cells were harvested at 18 h after infection, solubilized, subjected to electrophoresis in denaturing gels, and reacted with a monoclonal antibody to US7 (gI) or polyclonal antibody to US5 (gJ).
DNA fragmentation associated with apoptosis is blocked by trans complementation of gD−/−.
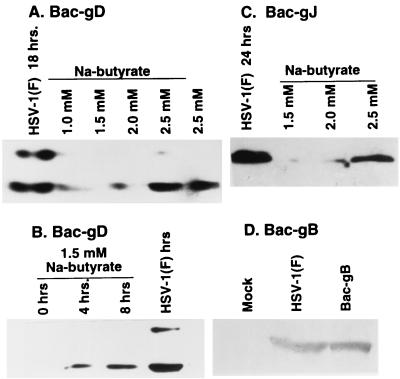
The experiments described above suggested that phenotypes of gD−/− and gD−/+ viruses analyzed in these studies could be due to either gD or gJ. The objective of this series of experiments was to determine whether gJ or gD delivered in trans could block the DNA fragmentation induced by gD−/+ or gD−/− viruses. Two series of experiments were done. In the first, we cloned gD, gJ, and gB driven by the immediate-early HCMV promoter. Figure 6 shows that gD, gJ, and gB were readily expressed in cells exposed to baculoviruses carrying the corresponding gene and treated with sodium butyrate. The positive control consisted of lysates of SK-N-SH cells exposed to HSV-1(F). Although in this experiment the protein accumulation was measured 24 h after exposure to baculoviruses (Fig. 6A, C, and D), the product of the gene carried by baculoviruses could be detected much earlier. As shown in Fig. 6B, gD was readily detected as early as 4 h after exposure of cells to the recombinant baculovirus carrying the gD gene.
FIG. 6.
Electrophoretically separated lysates of cells exposed to baculoviruses carrying the gene encoding gD (A and B), gJ (C), or gB (D) and reacted with the corresponding antibody. SK-N-SH cells were exposed to 10 PFU of HSV-1(F), 10 PFU of recombinant Bac-gD or Bac-gJ, or 18 PFU of Bac-gB per cell. The cells were harvested at 24 h after exposure to HSV-1(F) or recombinant baculovirus (A, C, or D) or at times shown (B), solubilized, subjected to electrophoresis in 10% denaturing polyacrylamide gels, electrically transferred onto a nitrocellulose sheet, and reacted with a polyclonal antibody to gJ or monoclonal antibody to gD or gB.
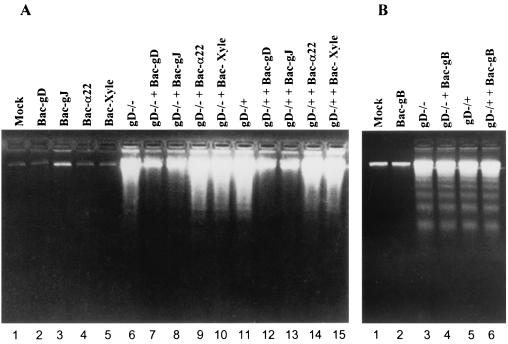
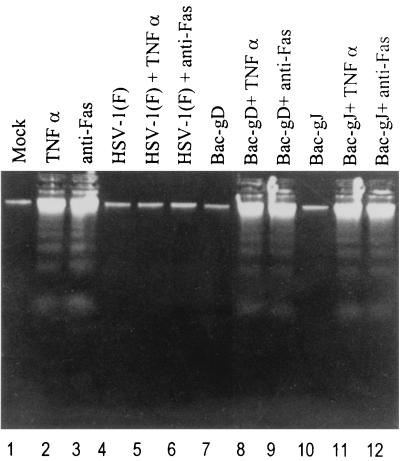
The second series of experiments was designed to determine whether gD or gJ could block fragmentation of DNA induced by gD−/− or gD−/+ viruses. In the experiment illustrated in Fig. 7A, replicate subconfluent cultures of SK-N-SH cells were mock infected, exposed to 10 PFU of recombinant baculovirus per cell as indicated or to 100 PFU of Bac-XylE per cell, or doubly infected with 10 PFU of recombinant baculovirus and either 100 PFU equivalents of gD−/− or 10 PFU of gD−/+ mutant virus per cell. In the experiment illustrated in Fig. 7B, replicate cultures of SK-N-SH cells were exposed to 18 PFU of recombinant baculovirus expressing gB per cell or exposed to both 18 PFU or baculovirus and either 100 PFU of gD−/− or 10 PFU of gD−/+ virus per cell. The cells were harvested 24 h after exposure to virus and processed as described in Materials and Methods. The results were as follows: None of the recombinant baculoviruses induced DNA fragmentation by themselves (Fig. 7A, lanes 2 to 5; Fig. 7B, lane 2). gJ or gD blocked DNA fragmentation induced by gD−/− (Fig. 7A, lanes 7 and 8) or gD−/+ (Fig. 7A, lanes 12 and 13). None of the other recombinant baculovirus were able to block DNA fragmentation induced by the gD−/− or gD−/+ viruses.
FIG. 7.
Agarose gels containing electrophoretically separated, ethidium bromide-stained low-molecular-weight DNA from SK-N-SH cell cultures that were mock infected, exposed to wild-type or mutant virus alone, or exposed to both HSV and recombinant baculovirus. (A) Replicate 25-cm2 flasks of subconfluent cultures of SK-N-SH cells were mock infected, exposed to 10 PFU of recombinant baculovirus per cell as indicated or to 100 PFU of Bac-XylE per cell, or doubly infected with 10 PFU of recombinant baculovirus and either 100 PFU of gD−/− or 10 PFU of gD−/+ mutant virus per cell. (B) Replicate cultures of SK-N-SH cells were exposed to 18 PFU of recombinant baculovirus expressing gB per cell or exposed to both 18 PFU of baculovirus and either 100 PFU of gD−/− or 10 PFU of gD−/+ virus per cell. The cells were harvested 24 h after exposure to virus and processed as described in Materials and Methods.
Recombinant baculoviruses expressing gD or gJ do not block the expression of recombinant viruses under the conditions tested.
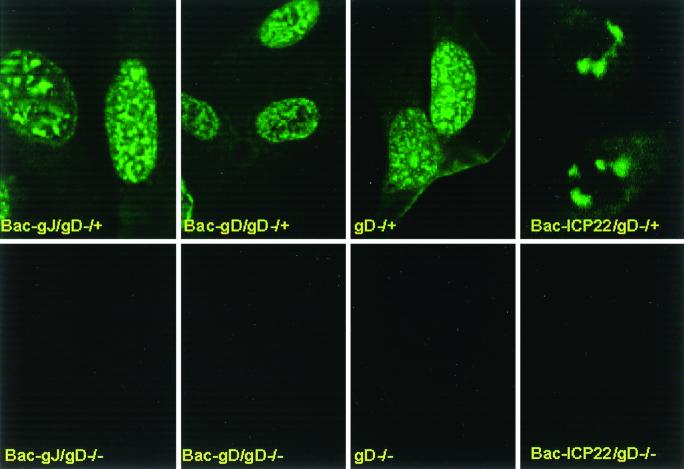
The objectives of the experiment described below were twofold: (i) to determine whether simultaneous exposure of cells to gD−/− virus and baculovirus expressing gD or gJ altered the ability of gD−/− virus to infect cells and express its genes; and (ii) to determine whether expression of gD or gJ altered the capacity of gD−/+ virus to express its genes since this virus was expected to be able to attach, enter cells, and initiate productive infection. SK-N-SH cells grown in microwells were infected with either 100 PFU equivalents of gD−/− or 10 PFU of gD−/+ per cell singly or in combination with 10 PFU of baculovirus expressing gD, gJ, or ICP22. The cells were fixed and reacted with monoclonal antibody to ICP0. The digitized images collected with the aid of a Zeiss confocal microscope are shown in Fig. 8. As expected, cells infected with gD−/− virus singly or in combination with a recombinant baculovirus failed to express ICP0. In contrast, the expression of ICP0 in cells infected with the gD−/+ mutant and either gD or gJ could not be differentiated from that seen in cells infected solely with the gD−/+ mutant.
FIG. 8.
Digitized images of SK-N-SH cells exposed to 100 PFU of gD−/− or 10 PFU of gD−/+ virus per cell alone or doubly infected with gD−/− or gD−/+ as described for Fig. 7 and 10 PFU of baculovirus expressing either gD, gJ, or ICP22 per cell. The cells were fixed at 6 h after exposure to the virus and reacted with an antibody specific for ICP0. The images were collected with a Zeiss confocal microscope as described in Materials and Methods.
gD and gJ do not block fragmentation of DNA induced by anti-Fas antibody or TNF-α.
Earlier studies have shown that HSV-1 blocks apoptosis induced by a number of stimuli, including anti-Fas antibody and TNF-α (16, 21, 22, 34). Jerome at al. (21) published evidence that mutants lacking the US5 encoding gJ failed to suppress apoptosis induced by anti-Fas antibody. In light of the results presented in this report, it was of interest to determine whether either gJ or gD by itself and in the absence of other HSV proteins could block DNA fragmentation induced by TNF-α or anti-Fas antibody. To this end, SK-N-SH cells were mock infected or exposed to 10 PFU of HSV-1(F) or 10 PFU of baculovirus expressing gD or gJ per cell. Replicate cultures were exposed to TNF-α (10 μg/ml) at 2 h after infection or to anti-Fas antibody at 6 h after infection. The cells were harvested 18 h after infection and processed as described in Materials and Methods. The results were as follows: (i) Degradation of DNA was not apparent in mock-infected cells (Fig. 9, lane 1), cells infected with HSV-1(F) (lane 4), or cells infected with baculoviruses expressing gD and gJ (lanes 7 and 10, respectively). (ii) TNF-α or anti-Fas antibody induced degradation of DNA (lanes 2 and 3), and this effect was not suppressed by gD (lanes 8 and 9) or gJ (lanes 11 and 12). We conclude that expression of these genes by themselves is not sufficient to block apoptosis induced by TNF-α or anti-Fas antibody.
FIG. 9.
Agarose gels containing electrophoretically separated, ethidium bromide-stained low-molecular-weight DNA from SK-N-SH cultures that were mock infected or infected and either untreated or exposed to TNF-α or anti-Fas antibody. SK-N-SH cells were mock infected or exposed to 10 PFU of HSV-1(F) or 10 PFU of baculovirus expressing gD or gJ per cell. Replicate cultures were exposed to TNF-α (10 μg/ml) at 2 h after infection or to anti-Fas antibody at 6 h after infection. The cells were harvested 18 h after infection and processed as described in Materials and Methods.
DISCUSSION
The life cycle of resting or dividing cells is tightly regulated. Cells whose genomes are damaged or which fail to meet parameters measured at specific checkpoints are induced to undergo a sequence of events resulting in programmed cell death. Viruses both damage and perturb cellular regulatory cascades, and these events in themselves are sufficient to induce programmed cell death. In some instances, the induction of programmed cell death serves as the preferred environment for viral replication (31). Many viruses, however, block programmed cell death as inimical to viral replication. HSV-1 is an example of a virus that both perturbs the cell and effectively blocks the cell from responding not only to the perturbations induced by the virus but also to the induction of an apoptotic cascade by exogenous agents (3, 16, 17, 23).
This report focused on very early events in the interaction of HSV-1 with its host cell. Our results indicate the following.
(i) gD−/+ stocks used in these studies lack intact genes encoding gD and gJ. Since these stocks were used to produce gD−/− virus in cells that do not carry HSV-1 DNA sequences, the gD−/− mutants also lack both genes.
(ii) Both gD−/− and gD−/+ mutants induce apoptosis in SK-N-SH cells, as shown by binding of annexin V to cell surfaces and degradation of the DNA. Annexin V binding was detected as early as 8 h after exposure of cells to the virus. The two virus stocks differ, however, in two respects. First, gD−/+ stocks infect and replicate in cells that do not carry HSV-1 DNA sequences, whereas gD−/− stocks do not replicate or express de novo viral genes in either cells that carry HSV-1 DNA sequences or those that do not. We have, however, shown that gD−/− virus particles were internalized and contained in vesicles and that they appeared to undergo degradation. These were detected at 30 min after exposure of cells to the virus but not at 1 h after exposure. The second major difference is that we estimate that higher multiplicities of infection are necessary to induce apoptosis with gD−/− virus than with gD−/+ mutants.
(iii) Degradation of cellular DNA characteristic of programmed cell death was blocked in cells exposed to baculovirus expressing gD or gJ and then infected with gD−/− or gD−/+ virus stocks. The genes expressed by baculovirus vectors were driven by the HCMV immediate-early promoter, and gene products were detected as early as 4 h after exposure of cells to recombinant baculoviruses. This is the first evidence (i) that gD or gJ can block apoptosis and (ii) that either gene delivered in trans can block programmed cell death induced by the same null mutant.
(iv) A virus lacking gJ was previously shown to be unable to block apoptosis induced by an exogenous agent, antibody to Fas (21). We show that under the conditions tested in the present study, gJ or gD was not sufficient to block fragmentation of DNA induced by anti-Fas antibody.
Three key issues have arisen in the course of this study: (i) how gD−/− virus induces apoptosis, (ii) how gD−/+ induces apoptosis in light of the evidence that it causes a productive infection resulting in gD−/− progeny, and (iii) the negative regulation of HSV-induced apoptosis exerted independently by gD and gJ. We consider each of these issues separately.
(i) This study stemmed from the observation that tsB7 at the nonpermissive temperature induces the apoptotic cascade in the absence of de novo protein synthesis (16). The two mutually nonexclusive hypotheses that could explain that observation are that in the course of viral replicative cycle one or more viral functions block programmed cell death induced by the attachment of the virus to the cell surface or due to the release of a virion protein into the newly infected cell. The studies with gD−/− virus recapitulate this conclusion. Although gD−/− virus is known to attach to the cell surface and could potentially activate a stress response, we have also shown that the virus is taken up and degraded in vesicles likely to be of endosomal origin.
(ii) The available data do not exclude the possibility that the interaction of HSV with the cell surface induces a stress response that leads to apoptosis in the absence of counteracting viral functions expressed later in infection. Of the various factors that come into play, at least two can be excluded a priori. First, apoptosis was induced irrespective of the presence of gD in the envelope of the virus (gD−/− versus gD−/+); second, the absence of gJ may also not be a factor since in the study of Jerome et al. (21), gJ− virus by itself did not induce the apoptotic cascade.
(iii) The available data also do not exclude the hypothesis that apoptosis is triggered by a factor introduced into cells after infection. The property shared by gD−/− and gD−/+ viruses relevant to apoptosis does include introduction of virion proteins into the cell. For gD−/+ virus, this would constitute the normal sequence of events leading to productive infection. In the case of gD−/− virus, the putative factor responsible for the induction of apoptosis could escape degradation and be released from the endosomic compartment into the infected cell. Since it is likely that only a small amount of this factor would be introduced into the cell by this pathway, it could account for the finding that gD−/− virus requires high PFU equivalents to induce apotosis relative to the gD−/+ virus stock. We further note that entry of gD−/− virus likely in the endocytic-lysosomal compartment may by itself represent a stimulus to apoptosis, as activation of the lysosomal compartment has been shown to induce apoptosis (30).
(iv) gD−/+ virus is the only HSV mutant found to date to induced classical apoptosis and at the same time produce progeny (gD−/− virus), presumably by expressing all genes other than those that that have been specifically deleted. The self-rescuants reported in this study provide evidence that no genes other than gD and gJ relevant to apoptosis are absent from the gD−/+ mutants.
(v) Inasmuch as apoptosis is not caused by the absence of gJ (21), the property shared by the gD−/− and gD−/+ viruses that could trigger apoptosis is the absence of the gene encoding gD and not the absence of gD in the envelope of the virus to which the cells were exposed. It follows therefore that function of gD and gJ required to block apoptosis is expressed by the proteins made de novo after infection.
(vi) There are at least three mechanisms by which the gD could block apoptosis. The first and least likely involves the reported interaction of gD with HveA, a member of the family of TNF receptors (28). HveA in turn interacts with LIGHT, a member of TNF superfamily shown to mediate apoptosis via its interaction with lymphotoxyn-β receptor (32). If gD were to block apoptosis by this pathway, HveA presumably would induce apotosis in the absence of gD. However, HveA is narrowly distributed in human tissues and has been reported not to play a role in LIGHT-mediated apoptosis (28, 32).
The second mechanism would involve interaction of gD with nectins, members of the immunoglobulin superfamily that in uninfected cells function as intercelluar adhesion molecules (12, 19, 27, 36). In contrast to HveA, the nectins are broadly distributed in human cell lines and in human tissues. It is conceivable that in the absence of the gD ligand, disruption of intercellular adhesion by viral gene products during the course of the replicative cycle induces a stress that is remedied either by gD or gJ.
The third potential mechanism may involve interaction of gD, modified by addition of mannose-phosphate with one of the two mannose-phosphate receptors. It has been reported that during productive infection, a small amount of gD becomes phosphorylated by addition of mannose-6-phosphate residues, (6). At the time when this interaction was described, the thought was that the mannose-6-phosphate receptors could act as receptors for virus entry or could sort some of the gD made during infection, modified by addition of phosphate residues (6, 7). The possibility exists, however, that the mannose-6-phosphate receptor interacts with mannose-6-phosphate–gD to block apoptosis rather than to enable virus entry into cells. This latter mechanism may be relevant to the gD-mediated block to apoptosis induced by the gD−/− virus stock, which appears to be taken up in a compartment likely of endocytic origin.
(iv) gJ is dispensable for viral replication at least in cultured cells, and its function is not known. No cellular ligands have been reported. The antiapoptotic stimulus mediated by gJ may differ substantially from that induced by gD. HSV-1 lacking the gJ gene was reported to be unable to block apoptosis induced by anti-Fas antibody, suggesting that gJ plays an essential role in this process (20). Our results indicate that gJ by itself is not able to block apoptosis induced by an exogenous pathway such as Fas ligand but can block apoptosis induced by either gD−/− or gD−/+ viruses. The conclusion to be derived from these data is that gJ acts at a signal transduction or post-signal transduction stage, that it is sufficient to block the apoptotic cascade in the case of gD−/− or gD−/+ viruses, but that it is not sufficient to block apoptosis by Fas ligands under the conditions tested. At least in the case of gD−/− virus, its function is independent from that of gD.
In essence, the results described in this report have shown (i) that viruses impaired in gD and gJ genes can induce programmed cell death and (ii) that in the case of viruses lacking both the gene and the glycoprotein it encodes, apoptosis results in the absence of de novo viral protein synthesis. The association of the missing function with apoptosis is clearly and unambiguously supported by the observation that administration of genes encoding the missing functions in trans blocked apoptosis and in essence reconstructed the phenotype of the wild-type virus. Studies now in progress may shed light on both the mechanism by which the cells are induced to activate the apoptotic cascade and the mechanism by which this cascade is blocked. The findings presented in this report are novel; they reinforce the growing realization that cells can respond by activating an apoptotic cascade to the presence of an invading pathogen and that HSV-1 too has evolved multiple functions to block the cell from doing just that.
ACKNOWLEDGMENTS
We thank David Johnson and Patricia G. Spear for the gD− viruses and cells expressing gD, and we thank Shu-Fen Chou for electron microscopy.
Work done at the University of Chicago was aided by Public Health Service grants CA47451, CA71933, and CA78766 from the National Cancer Institute. Work done at the University of Bologna was supported by grants from Target Project in Biotechnology/CNR, Telethon grant A141, MURST 40%.
REFERENCES
- 1.Ackermann M, Sarmiento M, Roizman B. Application of antibody to synthetic peptides for the characterization of the intact and truncated α22 protein specified by herpes simplex virus 1 and the R325 α22− deletion mutant. J Virol. 1985;56:207–215. doi: 10.1128/jvi.56.1.207-215.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubert M, Blaho J A. The herpes simplex virus type 1 regulatory protein ICP27 is required for the prevention of apoptosis in infected human cells. J Virol. 1999;73:2803–2813. doi: 10.1128/jvi.73.4.2803-2813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aubert M, O'Toole J, Blaho J A. Induction and prevention of apoptosis in human HEp-2 cells by herpes simplex virus type 1. J Virol. 1999;73:10359–10370. doi: 10.1128/jvi.73.12.10359-10370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batterson W, Furlong D, Roizman B. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of viral reproductive cycle. J Virol. 1983;45:397–407. doi: 10.1128/jvi.45.1.397-407.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batterson W, Roizman B. Characterization of the herpes simplex virion-associated factor responsible for the induction of α genes. J Virol. 1983;46:371–377. doi: 10.1128/jvi.46.2.371-377.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunetti C R, Burke R L, Hoflack B, Ludwig T, Dingwell KS, Johnson DC. Role of mannose-6-phosphate receptors in herpes simplex virus entry into cells and cell-to-cell transmission. J Virol. 1995;69:3517–3528. doi: 10.1128/jvi.69.6.3517-3528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunetti C R, Dingwell K S, Wale C, Graham F L, Johnson D C. Herpes simplex virus gD and virions accumulate in endosomes by mannose 6-phosphate-dependent and -independent mechanisms. J Virol. 1998;72:3330–3339. doi: 10.1128/jvi.72.4.3330-3339.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai W, Gu B, Person S. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J Virol. 1988;62:2596–2604. doi: 10.1128/jvi.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campadelli-Fiume G, Arsenakis M, Farabegoli F, Roizman B. Entry of herpes simplex virus 1 in BJ cells that constitutively express viral glycoprotein D is by endocytosis and results in the degradation of the virus. J Virol. 1988;62:159–167. doi: 10.1128/jvi.62.1.159-167.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campadelli-Fiume G, Cocchi F, Menotti L, Lopez M. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev Med Virol. 2000;10:305–319. doi: 10.1002/1099-1654(200009/10)10:5<305::aid-rmv286>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 11.Chang Y E, Menotti L, Filatov F, Campadelli-Fiume G, Roizman B. UL27.5 is a novel γ2 gene antisense to the herpes simplex virus 1 gene encoding glycoprotein B. J Virol. 1998;72:6056–6064. doi: 10.1128/jvi.72.7.6056-6064.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J Virol. 1998;72:9992–10002. doi: 10.1128/jvi.72.12.9992-10002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLuca N A, McCarth A, Shaffer P A. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J Virol. 1985;58:558–570. doi: 10.1128/jvi.56.2.558-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ejercito P M, Kieff E D, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 15.Forrester A, Farrell H, Wilkinson G, Kaye J, Davis Poynter N, Minson T. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J Virol. 1992;66:341–348. doi: 10.1128/jvi.66.1.341-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galvan V, Roizman B. Herpes simplex virus 1 induces and blocks apoptosis at multiple steps during infection and protects cells from exogenous inducers in a cell-type-dependent manner. Proc Natl Acad Sci USA. 1998;95:3931–3936. doi: 10.1073/pnas.95.7.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galvan V, Brandimarti R, Roizman B. Herpes simplex virus 1 blocks caspase-3-independent and caspase-dependent pathways to cell death. J Virol. 1999;73:3219–3226. doi: 10.1128/jvi.73.4.3219-3226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galvan V, Brandimarti R, Munger J, Roizman B. Bcl-2 blocks a caspase-dependent pathway of apoptosis activated by herpes simplex virus 1 infection in HEp-2 cells. J Virol. 2000;74:1913–1938. doi: 10.1128/jvi.74.4.1931-1938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geraghty R J, Krummenacher C, Cohen G M, Eisenberg R J, Spear P G. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 20.Herold B C, WuDunn D, Soltys N, Spear P G. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J Virol. 1991;65:1090–1098. doi: 10.1128/jvi.65.3.1090-1098.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jerome K R, Fox R, Chen Z, Sears A E, Lee H-E, Corey L. Herpes simplex virus inhibits apoptosis through the action of two genes, US5 and US3. J Virol. 1999;73:8950–8957. doi: 10.1128/jvi.73.11.8950-8957.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koyama A H, Miwa Y. Suppression of apoptotic DNA fragmentation in herpes simplex virus type 1-infected cells. J Virol. 1997;71:2567–2571. doi: 10.1128/jvi.71.3.2567-2571.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leopardi R, Roizman B. The herpes simplex virus 1 major regulatory protein ICP4 blocks apoptosis induced by the virus or by hypothermia. Proc Natl Acad Sci USA. 1996;93:9583–9587. doi: 10.1073/pnas.93.18.9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leopardi R, Van Sant C, Roizman B. The herpes simplex virus 1 protein kinase US3 is required for protection from apoptosis induced by the virus. Proc Natl Acad Sci USA. 1997;94:7891–7896. doi: 10.1073/pnas.94.15.7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ligas M W, Johnson DC. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β-galactosidase sequences binds to but is unable to penetrate into cells. J Virol. 1988;62:1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longnecker R, Chatterjee S, Whitley R J, Roizman B. Identification of a novel herpes simplex virus 1 glycoprotein gene within a gene cluster dispensable for growth in cell culture. Proc Natl Acad Sci USA. 1987;84:4303–4307. doi: 10.1073/pnas.84.12.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez M, Aoubala M, Jordier F, Isnardon D, Gomez S, Dubreuil P. The human poliovirus receptor related 2 protein is a new hematopoietic/endothelial homophilic adhesion molecule. Blood. 1998;92:4602–4611. [PubMed] [Google Scholar]
- 28.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 29.Ogle W O, Roizman B. Functional anatomy of herpes simplex virus 1 overlapping genes encoding infected-cell protein 22 and US1.5 protein. J Virol. 1999;73:4305–4315. doi: 10.1128/jvi.73.5.4305-4315.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okaka C, Mizuochi T. Role of macrophage lysosomal enzymes in the degradation of nucleosomes of apoptotic cells. J Immunol. 1999;163:5346–5352. [PubMed] [Google Scholar]
- 31.Ran Z, Rayet B, Rommelaere J, Faisst S. Parvovirus H-1-induced cell death: influence of intracellular NAD consumption on the regulation of necrosis and apoptosis. Virus Res. 1999;65:161–174. doi: 10.1016/s0168-1702(99)00115-x. [DOI] [PubMed] [Google Scholar]
- 32.Rooney I A, Butrovich K D, Glass A A, Borboroglu S, Benedict C A, Whitbeck J C, Cohen G H, Eisenberg R J, Ware C F. The lymphotoxin β receptor is necessary and sufficient for LIGHT-mediated apoptosis of tumor cells. J Biol Chem. 2000;275:14307–14315. doi: 10.1074/jbc.275.19.14307. [DOI] [PubMed] [Google Scholar]
- 33.Roop C, Hutchinson L, Johnson D C. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J Virol. 1993;67:2285–2297. doi: 10.1128/jvi.67.4.2285-2297.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sieg S, Yildirim Z, Smith D, Kayagaki N, Yagita H, Huang Y, Kaplan D. Herpes simplex virus type 2 inhibition of Fas ligand expression. J Virol. 1996;70:8747–8751. doi: 10.1128/jvi.70.12.8747-8751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spear P G. Entry of alphaherpesviruses into cells. Semin Virol. 1993;4:167–180. [Google Scholar]
- 36.Takahashi K, Nakanishi H, Miyahara M, Mandai K, Satoh K, Satoh A, Nishioka H, Aoki J, Nomoto A, Mizoguchi A, Takai Y. Nectin/PRR: an immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with afadin, a PDZ domain-containing protein. J Cell Biol. 1999;145:539–549. doi: 10.1083/jcb.145.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward P L, Ogle W O, Roizman B. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J Virol. 1996;70:4623–4631. doi: 10.1128/jvi.70.7.4623-4631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.WuDunn D W, Spear P G. Initial interaction of herpes simplex virus with cells is binding to heparin sulfate. J Virol. 1989;63:52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou G, Roizman B. Wild-type herpes simplex virus 1 blocks programmed cell death and release of cytochrome c but not the translocation of mitochondrial apoptosis-inducing factor to the nuclei of human embryonic fibroblasts. J Virol. 2000;74:9048–9053. doi: 10.1128/jvi.74.19.9048-9053.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]