Abstract
Viral interference is characterized by the resistance of infected cells to infection by a challenge virus. Mechanisms of viral interference have not been characterized for human parainfluenza virus type 3 (HPF3), and the possible role of the neuraminidase (receptor-destroying) enzyme of the hemagglutinin-neuraminidase (HN) glycoprotein has not been assessed. To determine whether continual HN expression results in depletion of the viral receptors and thus prevents entry and cell fusion, we tested whether cells expressing wild-type HPF3 HN are resistant to viral infection. Stable expression of wild-type HN-green fluorescent protein (GFP) on cell membranes in different amounts allowed us to establish a correlation between the level of HN expression, the level of neuraminidase activity, and the level of protection from HPF3 infection. Cells with the highest levels of HN expression and neuraminidase activity on the cell surface were most resistant to infection by HPF3. To determine whether this resistance is attributable to the viral neuraminidase, we used a cloned variant HPF3 HN that has two amino acid alterations in HN leading to the loss of detectable neuraminidase activity. Cells expressing the neuraminidase-deficient variant HN-GFP were not protected from infection, despite expressing HN on their surface at levels even higher than the wild-type cell clones. Our results demonstrate that the HPF3 HN-mediated interference effect can be attributed to the presence of an active neuraminidase enzyme activity and provide the first definitive evidence that the mechanism for attachment interference by a paramyxovirus is attributable to the viral neuraminidase.
Viral interference is defined as a state induced by an infecting virus that is characterized by the resistance of cells to subsequent infection by a challenge virus (7). Interference can be due to several different mechanisms, one of which is attachment interference. In this situation the interfering virus destroys or blocks the receptors for the superinfecting virus (7). Understanding mechanisms of viral interference can lead to strategies for controlling viral infection.
The envelope of human parainfluenza virus type 3 (HPF3) contains two viral glycoproteins, the hemagglutinin-neuraminidase protein (HN) and the fusion protein (F). Infection of cells by HPF3 is initiated by attachment of the virus to the host cell through interaction of the HN glycoprotein with a sialic acid-containing cell surface receptor. Penetration and uncoating of the virus result from F protein-mediated fusion of the viral envelope with the plasma membrane of the cell, leading to the release of the viral nucleocapsid into the cytoplasm. In the case of HPF3 and other paramyxoviruses, HN as well as F are involved in membrane fusion, and cofunction of the HN and F glycoproteins was found to be necessary for syncytium formation (9, 10, 14, 17). Infection also results in fusion between cells, which involves the interaction of F and HN proteins expressed on the surface of an infected cell with the membrane of an adjacent uninfected cell. By virtue of its neuraminidase moiety, HN also has a receptor destroying potential that plays a role in the spread of infection (11).
Attachment interference has been documented for several paramyxoviruses (Newcastle disease virus [NDV] [3, 4, 5] and Sendai virus [13]) and is proposed to be due to the destruction of viral receptors by the viral neuraminidase. For NDV, it has been suggested that this attachment interference mechanism is due to the destruction of receptors by the neuraminidase of the interfering virus (2). It has been shown by Morrison et al. that the expression of NDV HN results in resistance to viral infection by NDV (16). The question remains as to whether the resistance mediated by the expressed NDV HN was due to HN's neuraminidase activity.
The hypothesis tested in the experiments presented here is that interference by HPF3 is caused by HN depleting or rendering unavailable the cell surface sialic acid receptors for its own binding (17, 18). We have shown in both persistently and acutely infected cells that cell-cell fusion is blocked if viral HN depletes cell surface sialic acid receptors (17, 18). We have also shown that HPF3 fusion and entry can be prevented by exogenous HPF3 viral neuraminidase treatment of cells (18). These data suggested that both continual HN expression in infected cells and exogenous neuraminidase treatment lead to attachment interference via receptor destruction.
To determine whether, for HPF3, continual HN expression results in depletion of the viral receptors and thus prevents entry and cell fusion, we determined whether cells expressing wild-type HPF3 HN are resistant to viral infection. In order to assess whether this resistance is indeed attributable to the viral neuraminidase, we performed experiments using a cloned variant HPF3 HN that has two amino acid alterations in HN leading to the loss of detectable neuraminidase activity (M. Porotto, O. Greengard, N. Poltoratskaia, M. A. Horga, and A. Moscona, unpublished data). We hypothesized that the variant HN, which binds receptor and mediates viral entry but lacks enzymatic activity, would fail to mediate resistance to infection. The use of this neuraminidase deficient HN allows us to address the mechanism whereby HN mediates interference.
MATERIALS AND METHODS
Generation of monoclonal cell lines stably expressing HN-green fluorescent protein (GFP).
The full-length cDNAs encoding either the wild type or the C28a variant of HN were obtained by PCR amplification using Vent Polymerase (New England Biolabs) of the full-length HN cDNA in PUC19 (19) with the primers 5′-CCGGAATTCTCGAATACTGGAAGCACAC-3′ and 5′-CGCGGATCCGCGCTTAACTGCAGCTTTTTGGAATC-3′, which contain the EcoRI and BamHI restriction sites (underlined), respectively. Amplified wild-type HN and C28aHN were digested with EcoRI and BamHI and subcloned using standard cloning techniques (22) into the corresponding sites of pGFP-C3 (Clontech, Palo Alto, Calif.) to obtain the plasmids pwtHN-EGFP and pC28aHN-EGFP. In these plasmids the amplified HNs are fused to the 5′ end of the EGFP gene. The fused genes were sequenced by the dideoxy chain termination method to ascertain that no mutations were introduced during the amplification process and to confirm that the correct open reading frame was maintained. The lentiviral vector pHR′ CMV, derived from insertion of the BglII-XhoI fragment of pcDNA3.0 containing the human cytomegalovirus (CMV) promoter (Clontech) into the BamHI-XhoI site of pHR′ (kindly provided by D. Trono [20]) was used for the expression of the EGFP-tagged HNs. The SnaBI-MfeI fragment containing the 3′- end portion of the human CMV promoter and the complete fused genes was excised from pwtHN-EGFP or pC28aHN-EGFP and inserted into the SnaBI-EcoRI site of a pHR′CMV lentiviral vector. The resulting lentiviral expression vectors, pHRC/EGFP-wtHN and pHRC/EGFP-C28aHN, carry the fused genes under the control of the early CMV promoter. The vector pHRC/EGFP derived from the insertion of the BamHI-NotI fragment containing the EGFP gene from pEGFP-1 (Clontech) into the corresponding sites of pHR′ CMV was used as a control. Lentiviral particles pseudotyped with vesicular stomatitis virus G glycoprotein were derived by transient transfection of 293T cells with either pHRC/EGFP-wtHN or pHRC/EGFP-C28aHN and the transcomplementing plasmids CMVΔR8.2 and pMD.G [20] in a 3:2:1 relative ratio, using Lipofectamine 2000 (Life Technologies, Gaithersburg, Md.) according to the manufacturer's instructions. Viral supernatants were collected 48 and 72 h after transfection and used for infecting naive 293T cells for 24 h in the presence of 10 μg of Polybrene per ml. Clonal populations stably expressing HN-GFP (wild-type and C28a variant HNs) or expressing GFP only were selected by limiting dilution and the levels of expression were analyzed by flow cytometry. All cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 4 mM glutamine, and 100 U of penicillin-streptomycin per ml.
Flow cytometry analysis.
Fifty-percent-confluent monolayers of 293T clonal cell lines stably expressing HN-GFP were washed with phosphate-buffered saline (PBS) without Ca2+-Mg2+ and then detached in PBS–5 mM EDTA at room temperature. An equal volume of DMEM–10% fetal calf serum was then added, and the cells were collected by centrifugation. The cell pellet was washed once with PBS and resuspended in PBS at a concentration of 106 cells/ml. Propidium iodide was added to a final concentration of 1 μg/ml just prior to analysis in order to exclude dead cells. Flow cytometry analysis was performed on a Becton Dickinson FACScalibur flow cytometer.
Confocal laser scanning microscopy.
293T clonal cell lines stably expressing HN-GFP were plated on collagen-coated slides and allowed to attach for 12 h. Monolayers were fixed using 4% formaldehyde in PBS for 5 min. Slides were examined using a Leica TCS-SP (UV) confocal laser scanning microscope (Heidelberg, Germany) equipped with a four-channel spectrophotometer scan head and four lasers (Ar-UV, argon, krypton, and HeNe). For these studies, the cells were illuminated with the l = 488 nm laser line. Images were collected using a ×100 1.4 NA lens. The pinhole size was adjusted such that resultant “optical sections” were approximately 0.25 to 0.5 μm thick. The same gain, offset, pinhole size, and laser intensity settings were maintained for all images collected.
Neuraminidase assay.
The fluorimetric assay of neuraminidase on the surface of suspensions of clonal cell lines expressing HN-GFP was based on the methods of Warner and O'Brien (23) and of Potier et al. (21). Reaction mixtures, containing 100 mM malate buffer (pH 4.75), the indicated concentrations of MUNANA (4-methylumbliferyl-α-d-N-acetylneuraminate), and 3.5 × 106 cells in a total volume of 70 μl were incubated at 37°C for 15 to 20 min. To determine the rate of product formation, which was constant during these periods, samples were taken at four or five time points, mixed with 100 mM methylene diamine to stop the reaction, and read in a Sequoia-Turner fluorimeter at a 365-nm excitation wavelength and a 450-nm emission wavelength. The amount of reaction product denoted by these readings was determined from fluorescence versus concentration curves determined with commercially obtained 4-methylumbelliferone. Enzyme activity is expressed in picomoles of product formed per minute per million cells.
Hemadsorption assay.
Erythrocytes were freshly obtained from whole human blood. Monolayers of cells were washed with medium lacking serum and then incubated with the human erythrocytes for 120 min at 22°C. Nonadherent erythrocytes were removed by washing with cold medium; the extent of erythrocyte adsorption was observed, and the plates were photographed.
Viruses.
Stocks of HPF3 and HPF2 were made in CV1 cells from virus plaque purified four times. Supernatant containing virus was collected 36 to 48 h postinfection and stored at −80°C. The virus titer was determined by a plaque assay using CV1 cells.
Infections.
A total of 5 × 105 cells/well from each cell line were plated on six-well plates and allowed to attach for 12 h. Monolayers of cells were infected with 2 ml of serum-free medium containing HPF3 or HPF2 at multiplicity of infection (MOI) of 0.1. Cells were incubated at 37°C for 90 min. Supernatants containing virus were aspirated, and cells were washed once with 2 ml of PBS to remove unbound virus. Then, 2 ml of fresh medium was added to the monolayers after rinsing, and the cells were reincubated at 37°C, allowing the infection to proceed. Supernatant fluid was harvested after 24, 48, or 72 h, and then the titers were determined using plaque assays.
Plaque assays.
Supernatant fluid from infected or mock-infected cells was serially diluted in serum-free medium, and 100 μl of each serial dilution was added per well to confluent CV1 cell monolayers in 48-well plates. Cells were incubated at 37°C with intermittent rocking. After 90 min, minimum essential medium containing 0.5% agarose was added to the dishes, and incubation continued for 24 h at 37°C. After removal of the agarose overlay, the cells were fixed with methanol for 15 min and immunostained for plaque detection as described previously (15). The number of plaques in the control and experimental wells were counted under a microscope.
Immunoprecipitation and immunoblotting.
A total of 6 × 106 cells from each clonal cell line were collected and centrifuged at 2,000 rpm (Beckman GS-6R centrifuge) at 4°C for 5 min. The cell pellet was washed once with 5 ml of PBS and lysed with 400 μl of gentle lysis buffer (NaCl, 150 mM; Triton X-100, 1% [vol/vol]; HEPES, 20 mM, pH 7.5; EDTA, 1 mM; MgSO4, 1.5 mM; glycerol, 10% [vol/vol]; plus protease inhibitors). Lysates were incubated on ice for 10 min and centrifuged in a Brinkmann Eppendorf centrifuge at 14,000 rpm for 10 min to separate cellular debris. The extracts were subjected to immunoprecipitation as follows: 250 μg of total cellular lysates was incubated with 3 μl of anti-HPF3 serum (Whittaker) in a total volume of 300 μl at 4°C overnight. Then, 50 μl of 50% protein A-agarose was added, and the mixture was incubated for 5 h at 4°C. Samples were washed twice with 1 ml of lysis buffer, and the washed beads were resuspended in 50 μl of loading buffer and boiled for 2 min. The precipitated proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 10% polyacrylamide gels (Laemmli) and transferred to a Zetabind membrane (Cuno) by electroblotting in a Transblot apparatus (Hoeffer Scientific Instruments) for 2 h at 4°C and 100 V (Bio-Rad Tris-glycine transfer buffer; 25 mM Tris, 192 mM glycine; pH 8.3). The membranes were blocked with a solution of 10% dry milk in PBS with 0.05% Tween 20 (PBST) for 1 h, rinsed once with PBST, immunoblotted with polyclonal anti-GFP antibodies (Clontech) in 1% bovine serum albumin (BSA) in PBST (1:1,000 dilution) for 1 h at room temperature, rinsed three times with PBST, and incubated with peroxidase-conjugated protein A (5 μg/μl) (ImmunoPure Recomb Protein A; Pierce) for 1 h at room temperature using a 1:100,000 dilution. Films were developed using Supersignal West Dura Stable Peroxide Buffer (Pierce) according to the manufacturer's instructions.
For Western blot analysis of the infected cells for detection of viral protein synthesis, cells infected with HPF3 were collected 24 h after infection and lysed as described above. The protein concentration per sample was determined using the Bio-Rad protein assay. Equal amounts (20 μg) of protein were loaded per lane and analyzed by SDS-PAGE. Immunoblotting was performed with anti-HPF3 serum (Whittaker) at a 1:1,000 dilution in 1% BSA-PBST for 1 h at room temperature.
RESULTS
Stable expression of HN-GFP fusion proteins: selection of clonal cell lines, quantification of expression levels, and localization of expressed proteins.
In order to address the question of whether expression of HPF3 HN on the cell surface interferes with infection by HPF3, we generated stable cell lines using a lentiviral vector that expresses HN as a fusion protein linked to GFP positioned at the amino terminus of the protein (intracytoplasmic). The fluorescent tag allows for detection of expressed proteins under microscopy with UV light and for quantitation of expression levels by fluorescence-activated cell sorter (FACS) analysis. The generation of stable cell lines expressing HN-GFP fusion proteins is described in detail in Materials and Methods. We selected clonal cell populations stably expressing wild type HN-GFP and C28a variant HN-GFP or else expressing GFP only.
C28a is a neuraminidase-deficient variant of HPF3 virus whose HN molecule differs from the wild type only in two amino acid substitutions in HN. The neuraminidase activity of this variant, C28a, which will be fully described elsewhere (M. Porotto, O. Greengard, N. Poltoratskaia, M.A. Horga and A. Moscona, unpublished data), is insignificant, i.e., <3% of wild-type activity as measured by either the thiobarbituric acid assay or the fluorimetric assay used in our study (21, 23). Analysis of the growth properties of C28a showed a severely decreased release of viral particles from infected cells to the medium (≥6-log-lower titer in the supernatant fluid) which was reversed, resulting in wild-type levels of release, by the addition of Clostridium perfringens neuraminidase to the infected cells after the adsorption period. In cell monolayers infected with C28a, plaques are formed and enlarge, demonstrating that viral entry and virus-mediated cell fusion occur; however, C28a does not spread through the monolayer beyond these individual plaques. The cell lines stably expressing C28a variant HN-GFP bound erythrocytes in a hemadsorption assay (see Fig. 4 below), indicating that C28a variant HN retains sialic acid binding activity. Thus, the variant virus retains its ability to bind receptor, fuse, and enter target cells.
FIG. 4.
Erythrocyte adherence to cell lines GFP#1, WT#1, and C28a#1.1. A 1% solution of human erythrocytes was added to confluent monolayers of cell lines expressing wild-type HN, C28a HN, or GFP. Hemadsorption activity was assessed and photographed after 2 h of incubation at 22°C.
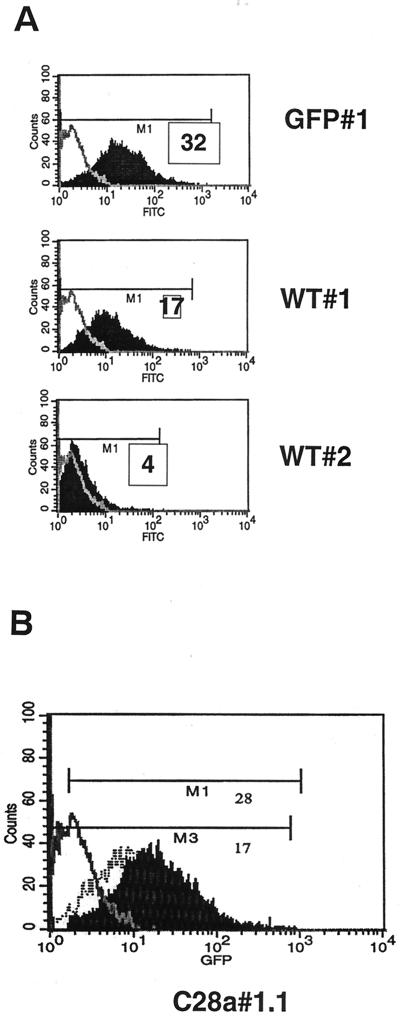
The level of surface expression of HN-GFP was assessed by FACS analysis for each stable cell line. Based on their relative fluorescent intensity as measured by FACS analysis, single cell clone populations were selected: wild-type HN-GFP clone 1 (WT#1), wild-type HN-GFP clone 2 (WT#2), and C28a HN-GFP clone 1.1 (C28a#1.1). A clonal cell line expressing GFP only (GPF#1), was selected as well. As shown in Fig. 1A, the mean fluorescence intensity (MFI) of WT#1 cells was 5- to 10-fold higher than the MFI of WT#2. The level of surface expression of C28a#1.1 was approximately two-fold higher than that of WT#1 (Fig. 1B). The FACS analysis was performed on four separate occasions, and the example shown in Fig. 1 is representative.
FIG. 1.
FACS analysis of clonal cell lines expressing HN-GFP. (A) FACS analysis of WT#1 and WT#2 cell lines. The x and y axes of the histograms represent green fluorescence (fluorescein isothiocyanate) and cell counts, respectively. Fluorescence in the negative control 293T cells is overlaid as a gray line. The relative MFI (M1) for each cell line are indicated. WT#1 cell line displays approximately fivefold more fluorescence than WT#2. (B) FACS analysis of the mutant HN C28a#1.1-expressing cell line. The fluorescence of the negative control 293T and WT#1 cell lines are overlaid as a gray line and a hatched line, respectively. M1 and M3 indicate the mean fluorescent units of C28a#1.1 and WT#1 cells, respectively.
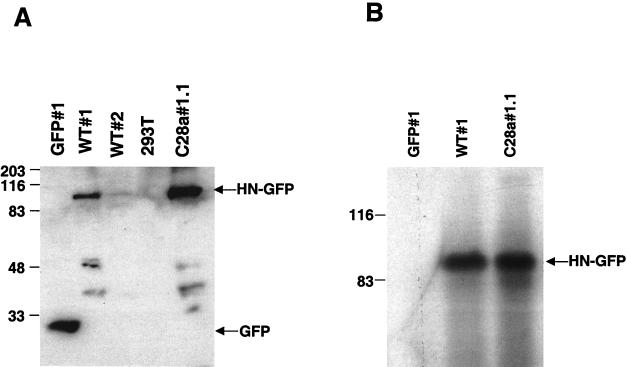
In order to confirm the differences in HN expression levels between clones WT#1, WT#2, and C28a#1.1, we performed Western analysis of the selected clones using anti-GFP antibodies for detection. Figure 2A shows Western blot analysis of cell lysates containing identical quantities of protein per lane (20 μg) from WT#1, WT#2, C28a#1.1 and GFP#1 cells, probed with anti-GFP polyclonal antibodies. Comparison between the 90-kDa bands corresponding to HN-GFP in WT#1, WT#2, and C28a#1.1 confirmed that expression of HN-GFP is at least 10-fold higher in WT#1 than in WT#2 and that expression of variant C28a HN-GFP in C28a#1.1 is higher than that of wild-type HN-GFP in WT#1. These data confirm the expression levels shown by FACS analysis.
FIG. 2.
(A) Western blot analysis of the selected clones using anti-GFP antibodies for detection. Cell lysates containing identical quantities (50 μg) of protein per lane and control were probed with anti-GFP polyclonal antibodies. The lysates are from the cells indicated above the lanes. The 90-kDa band corresponds to the wild-type HN-GFP protein, expressed in different amounts in WT#1 and WT#2. The lower molecular mass bands likely represent degradation products of HN-GFP. (B) Immunoprecipitation of proteins from HN-GFP-expressing cells and control GFP-expressing cells. Cell extracts containing equal amount of protein from WT#1, C28a#1.1, and GFP#1 were subjected to immunoprecipitation using anti-HPF3 serum. The precipitated proteins were resolved by SDS-PAGE in 11% gels, transferred to a Zetabind membrane by electroblotting, and immunoblotted with anti-HPF3 serum. The lysates are from the cells indicated above the lanes. The 90-kDa band corresponds to the expected molecular weight of the HN-GFP fusion protein.
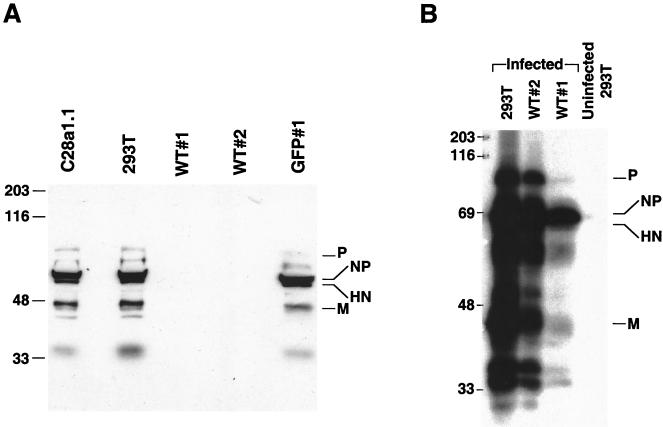
In order to verify that the expressed protein was indeed HN, immunoprecipitation of cell lysates obtained from HN-GFP-expressing cells using anti-parainfluenza 3 antibodies was performed and revealed a protein of the correct size and antigenicity. Cell clones WT#1, C28a#1.1 expressing HN-GFP, GFP#1 cells, and a control of untransfected 293T cells were lysed and immunoprecipitated with polyclonal antiparainfluenza serum. As shown in Fig. 2B, the 90-kDa band corresponds to the expected molecular mass of the HN-GFP recombinant protein.
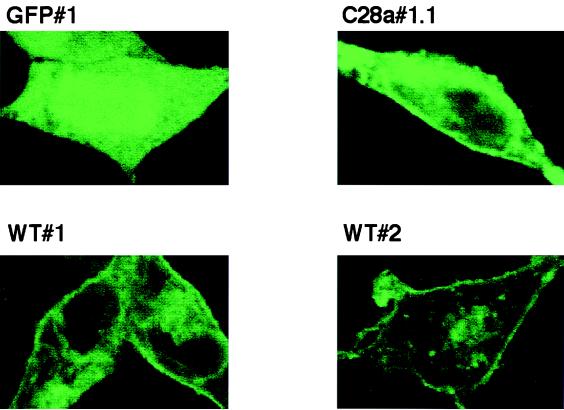
To determine whether the fusion proteins were expressed on the surface, monolayers of GFP#1, WT#1, WT#2, and C28a#1.1 intact cells were visualized by confocal microscopy. As shown in Fig. 3, the HN-GFP fusion proteins expressed by these cell lines clearly localized to the plasma membrane, as evidenced by a ring of fluorescence around the edge of cells WT#1, WT#2, and C28a#1.1, compared to the uniform fluorescence over the entire cell seen in GFP#1 cells.
FIG. 3.
Confocal microscopy of selected cell lines GFP#1, WT#1, WT#2, and C28a#1.1. Clonal cell lines stably expressing HN-GFP were plated on collagen coated slides and allowed to attach. Monolayers were fixed using formaldehyde. Cells were visualized under confocal microscopy. All pictures were taken using the same aperture, time of exposure, laser intensity, and magnification. The HN-GFP fusion proteins localize to the plasma membrane. As a control, cells expressing the GFP protein alone show only cytoplasmic fluorescence.
In order to further confirm the surface expression of HN, we assessed the receptor binding activity of the HN-GFP fusion proteins expressed by the clones using a hemadsorption assay. As shown in Fig. 4, the cell lines expressing wild-type HN-GFP and C28a HN-GFP bound erythrocytes at the cell surface, while the negative control cells expressing GFP alone did not.
Neuraminidase activity of expressed HN.
To assess whether the recombinant HN-GFP molecules retained neuraminidase activity, we applied a fluorescent assay method to cell suspensions. The neuraminidase activities of WT#1, WT#2, and C28a#1.1 were compared to the negative controls of 293T cells and GFP#1. The neuraminidase activities of intact cells (in pmol/min/million cells) were as follows: WT#1, 210 ± 28; WT#2, 20 ± 4.6; 293T, <3; GFP, <3; and C28a#1.1, <3. WT#1 cells exhibited a neuraminidase activity at least six times higher than that of WT#2 cells. The surface neuraminidase activity of C28a#1.1 was undetectable despite having significantly higher levels of HN expression than the WT#1 and WT#2 cell lines. The neuraminidase activity of 293T cells and GFP#1 cells is in the background range. These results indicate that the stably expressed wild-type HNs retained neuraminidase activity and that clone WT#1, which has a higher amount of expression than WT#2, also has significantly higher neuraminidase activity. The assay confirmed that the expressed C28a HN has undetectable neuraminidase activity as described for the variant virus.
The neuraminidase activity of HN is required in order to confer protection from infection with HPF3.
We asked whether stable expression of HN would interfere with subsequent infection with HPF3. In order to address this question, we tested whether the WT#1 and WT#2 cell clones were more resistant to infection with HPF3 than cells not expressing HN. We also sought to determine whether neuraminidase activity is specifically required in order for HN to confer protection from infection by infecting the neuraminidase-deficient cell line C28a#1.1.
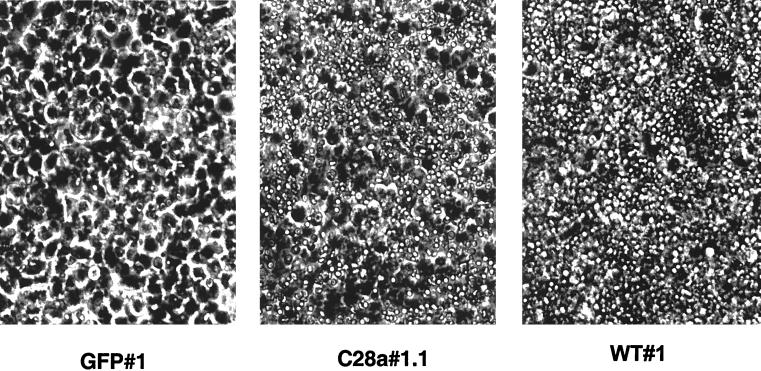
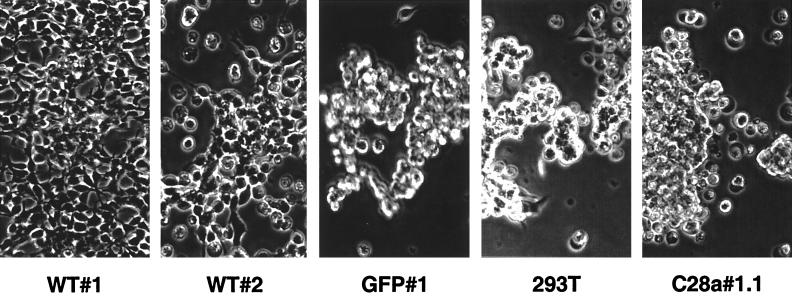
We infected WT#1, WT#2, C28a#1.1, and control GFP#1 and 293T cells with HPF3 virus at an MOI of 0.1 PFU/cell. Cells were visualized 24 and 72 h after infection. At 24 h, mild viral cytopathic effect could be seen in cells expressing C28a#1.1, as well as in control cells. As shown in Fig. 5, 72 h after infection, striking differences were seen between the cytopathic effects in the different cell lines: while WT#1 cells remained mostly intact despite HPF3 infection and WT#2 cells appeared partially protected, the monolayers of C28a#1.1 cells were destroyed by the cytopathic effect of the virus, as were the control GFP#1 and 293T cells. These results indicated that wild-type HN expression confers protection from the cytopathic effect caused by HPF3 during infection and that the degree of protection corresponds to the level of HN expression on the cell surface. This experiment also showed that stable expression of C28a#1.1 did not protect cells from infection with HPF3, indicating that neuraminidase is required for the protection effect.
FIG. 5.
Cytopathic effect in cell lines infected with HPF3. Monolayers of cell lines WT#1, WT#2, GFP#1, 293T, and C28a#1.1 were infected with HPF3 at an MOI of 0.1. Cells were visualized and photographed 72 h after infection. WT#1 cells remained intact, while the monolayers of GFP#1, 293T cells, and C28a#1.1 were destroyed by the cytopathic effect of the virus. WT#2 shows partial protection.
As another approach to assessing the level of protection mediated by wild-type and variant HN, we examined the levels of viral protein synthesis in the cells after HPF3 infection. Western blot analysis of WT#1, WT#2, C28a#1.1, and control GFP#1 and 293T cells was done using HPF3 antiserum for the detection of viral protein synthesis after infection. For this experiment, monolayers of cell lines were infected with HPF3, cells were lysed 24 h after infection, and the extracts containing identical quantities of protein per lane were separated by SDS-PAGE. The samples were probed with anti-HPF3 serum. As shown in Fig. 6A, WT#1 and WT#2 cells support undetectable levels of viral replication after 24 h compared to C28a#1.1 and control cells. This result confirmed that cell lines WT#1 and WT#2, which have detectable neuraminidase on the cell surface, were protected from HPF3 infection, while the expression of the neuraminidase-deficient variant HN did not have an interference effect. The finding of undetectable levels of viral protein synthesis in the WT#2 cells was surprising, since we had shown (Fig. 5) that 72 h after infection, WT#1 cells were mostly intact but WT#2 cells were only partially protected from infection. We therefore analyzed WT#1 and WT#2 lysates 72 h after infection for the presence of viral proteins. As shown in Fig. 6B, at 72 h after infection viral proteins are detectable in WT#2 cells, although far less than in control cells, and some, far diminished, viral protein synthesis is detectable in WT#1 cells as well.
FIG. 6.
Western blot analysis with anti-HPF3 serum of cell lines infected with HPF3. (A) Monolayers of C28a#1.1, 293T, WT#1, WT#2, and GFP#1 cells were infected with HPF3 at an MOI of 0.1 PFU/cell. Cell extracts were collected 24 h after infection. Equal amounts of protein (20 μg) were loaded per lane and analyzed by SDS-PAGE. Immunoblotting was performed with anti-HPF3 serum. The lysates are from the infected cells indicated above the lanes. (B) Monolayers of 293T, WT#2, and WT#1 cells were infected with HPF3 at an MOI of 0.1. Cell extracts were collected 72 h after infection. Equal amounts of protein (20 μg) were loaded per lane and analyzed by SDS-PAGE. Immunoblotting was performed with anti-HPF3 serum. The lysates are from the infected cells indicated above the lanes.
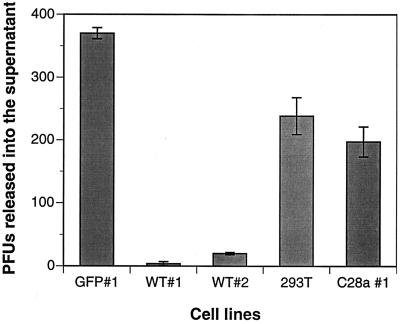
To further estimate the differences in protection between the stably HN-expressing cell lines, plaque assays were used to assess the amount of infectious particles released from the infected cells. The cell clones and control 293T cells were infected with HPF3, and 24 h after infection supernatants from each cell line were collected, plaqued on monolayers of CV1 cells and immunostained. Figure 7 shows the 100-fold decrease in plaque number (PFU) released from the WT#1 cells compared to cells expressing C28a#1.1, control GFP#1, and untransfected 293T cells. WT#2 cells released 10-fold fewer viral particles compared to controls, again indicating partial protection.
FIG. 7.
Effect of HN expression on viral replication. The cell lines were infected with HPF3, and plaque assays were used to assess the amount of infectious particles released by the different cell lines after infection. Cell clones and control 293T cells were infected with HPF3 at an MOI of 0.1. At 24h after infection, supernatants from each cell line were collected, plaqued on monolayers of CV1 cells, and immunostained. Each column is the mean of results of three different experiments done in triplicate. The bars denote the standard deviation.
These results, taken together, indicate that the protection against infection that is conferred by surface expression of recombinant HN requires neuraminidase activity and suggest that the interference observed during HPF3 infection is mediated by HN's neuraminidase.
Specificity of the interference effect.
We determined whether the interference effect seen in the cells expressing the highest levels of wild-type HN-GFP was specific for HPF3. WT#1, WT#2, and C28a#1.1 cells and control cells GFP#1 and 293T were infected with HPF2. We found that the protection from infection conferred by WT#1 extended to HPF2 as well. WT#1 infected with HPF2 showed a reduction of 10- to 100-fold in viral particles released, as measured by plaque assay (data not shown). This result is not surprising in view of our previous data indicating that HPF3 neuraminidase treatment of HPF2-infected cells can block fusion by HPF2 (1).
To confirm that the heterologous interference effect seen with HPF3 did not function to protect against viruses that do not use sialic acid for entry and to ensure that what we observed is not a general antiviral effect, we infected the clonal cell lines with herpes simplex virus type 1 (HSV-1). We found that the cell lines expressing HN were not protected from HSV-1 infection. WT#1 and C28a#1.1 cells and control cells GFP#1 and 293T were infected with HSV-1 in a plaque reduction assay. The same number of HSV-1 plaques were detected on all four cell lines, indicating that no protection was conferred by the expressed proteins (data not shown).
DISCUSSION
Sialoglycoconjugates have been identified as receptors for HPF3 on the basis of their destruction by neuraminidase. This finding has led to the hypothesis that expression of viral neuraminidase on cell surfaces during infection could deplete HPF3 receptors and render cells resistant to infection. This mechanism of viral interference has been proposed to explain the interference phenomenon demonstrated for several paramyxoviruses (2, 3, 5). For NDV, Morrison et al. (16) showed that the expression of cloned NDV HN resulted in resistance to subsequent infection by NDV; however, the mechanism of this interference mediated by HN remained unknown. In this report, we provide definitive evidence that the mechanism for attachment interference by a paramyxovirus is attributable to the viral neuraminidase enzyme, and we render proof of the mechanism of viral interference by HPF3.
Our results demonstrate that the HN-mediated interference effect can be attributed to the presence of an active neuraminidase enzyme activity rather than to the binding functions of HN. The stable expression of wild-type HN-GFP on the membrane in different amounts allowed us to establish a correlation between the level of HN expression, the level of neuraminidase activity, and the level of protection from HPF3 infection: the cell clone WT#1, with the highest levels of HN expression and neuraminidase activity on the cell surface, was found to be resistant to infection by HPF3. WT#2, with lower levels of HN expression and neuraminidase activity, was partially resistant to infection, as evidenced by the slow progression of the cytopathic effect, with significantly less virus released into the supernatant fluid after infection compared to control cells, and undetectable viral protein in the cell lysates at early time points after infection. The expression of a neuraminidase-deficient variant HN, C28a HN-GFP, in the cell clone C28a#1.1, which had levels of surface HN expression higher than WT#1 but no detectable neuraminidase activity, allowed us to correlate neuraminidase activity with protection. C28a#1.1 cells were not protected from infection, despite expressing HN on their surface at levels even higher than the wild-type cell clones. These cells responded to HPF3 infection identically to the control cells, GFP#1 and 293T, showing the same cytopathic effect and amount of viral replication after infection.
The phenomenon of attachment interference has been characterized for several other viruses. For alphaherpes viruses, glycoprotein D (gD) is one of the proteins essential for penetration into cells and mediates interference with infection (8, 12). It has been proposed that cellular expression of the alphaherpesvirus gD interferes with the entry of homologous and heterologous virus by blocking access to ligand-binding sites on gD receptors used for entry (8). A similar mechanism of interference via blockade of HN-binding sites on receptors could be considered for cellular expression of HPF3 HN; however, our results do not support this model since HPF3 neuraminidase enzyme activity is essential for interference. In the case of influenza virus, which also makes use of sialic acid-containing receptors for entry, viral infection mediates interference; however, expression of the hemagglutinin (attachment) protein does not protect against viral infection (16), suggesting that receptor blockade is not important in the interference mechanism for influenza virus.
The biological significance of glycoprotein-mediated interference for HPF3 and other paramyxoviruses remains to be determined. For retroviruses, superinfection exclusion could prevent multiple provirus insertions and promote cell survival, a beneficial state since pathogenesis requires host cell survival. For alphaherpesviruses it has been postulated that gD-mediated interference is important for efficient egress and release of infectious virus by preventing newly enveloped virus from fusing with membranes of the virus-producing cell (6, 12). For HPF3, it has been shown (11) that the neuraminidase activity of HN expressed on the surface allows the virions to be released to begin a new cycle of replication. It has also been shown that the depletion of receptors by the neuraminidase is responsible for the establishment of a persistently infected state (18). Our results confirm that the neuraminidase activity of HN depletes the cell of available receptors, thus protecting it from reinfection by HPF3 and also by HPF2.
Characterization of heterologous interference for HPF3 using the strategies described here will be used in future experiments to determine whether different parainfluenza viruses can use the same receptor for entry. Our results indicate that HPF2 entry was inhibited by heterologous expression of HPF3 HN, suggesting the possibility of a shared receptor. There was no protection of WT#1 cells after HSV infection, showing that the resistance conferred by expressing wt HPF3 HN is not a general antiviral effect.
Our data are consistent with the hypothesis that, for HPF3, neuraminidase activity is required for the establishment of homologous interference. HN expression in itself, without neuraminidase activity, does not suffice to confer resistance to infection. In the model that we favor, the neuraminidase expressed in the cells during expression of wild-type HN would result in depletion of the HPF3 viral receptors, thus preventing entry. The cellular location in which desialidation of cellular receptor molecules takes place is unknown; this process could take place either during transit to the cell surface, at the cell surface, or at both sites. The action of the viral neuraminidase on the sialic acid receptor during infection, or during expression of wild-type HN, could thus be an ongoing process in which most of the available receptors would eventually become desialidated and thereby inactivated for viral entry, rendering the cell resistant to infection.
The expression of C28a (neuraminidase-deficient) HN does not interfere with viral entry since sialidated receptors remain available for attachment. It is possible that the expressed neuraminidase deficient HN remains bound to receptor on the infected cell surface and that neuraminidase may be required to release HN from this receptor. Expressed neuraminidase-deficient HN could thereby sequester receptors, and this sequestered state would remain stable (requiring neuraminidase to be broken). In this way, it is theoretically possible that high enough expression levels of neuraminidase-deficient HN could confer protection by sequestering all the receptors but that the relative amount of these HN molecules expressed in our cells is insufficient to deplete the cells from receptor by solely this mechanism, leaving the unbound receptor molecules, which remain properly sialidated, to mediate viral entry. Ongoing studies are addressing the question of whether C28a HN is bound to sialic acid-containing receptor on the cell surface by virtue of its lack of cleavage activity. These studies will determine whether, as the viral glycoproteins are inserted into the host cell membrane during viral infection, neuraminidase is necessary for the release of the HN molecule from its cellular receptor.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI-31971 to A.M. from the National Institutes of Health, by an Infectious Diseases Society of America Ortho-MacNeill Young Investigator Award to M.A.H., and by Public Health Service grant KO8-AI-01727 to M.A.H. from the National Institutes of Health. Confocal laser scanning microscopy was performed at the Mount Sinai School of Medicine Microscopy Center under the direction of Scott Henderson, supported with funding from an NSF Major Research Instrumentation grant (DBI-9724504).
FACS analysis was performed at the Mount Sinai School of Medicine Flow Cytometry Core Facility, and we thank Hans Snoeck for advice and helpful discussions relating to the FACS analysis.
REFERENCES
- 1.Ah-Tye C, Schwartz S, Huberman K, Carlin E, Moscona A. Virus-receptor interactions of human parainfluenza viruses types 1, 2 and 3. Microb Pathog. 1999;27:329–336. doi: 10.1006/mpat.1999.0313. [DOI] [PubMed] [Google Scholar]
- 2.Baluda M A. Homologous interference by ultraviolet-inactivated Newcastle disease virus. Virology. 1957;7:315–327. doi: 10.1016/0042-6822(57)90044-2. [DOI] [PubMed] [Google Scholar]
- 3.Bratt M A, Rubin H. Specific interference among strains of NDV. I. Demonstration and measurement of interference. Virology. 1967;33:598–608. doi: 10.1016/0042-6822(67)90059-1. [DOI] [PubMed] [Google Scholar]
- 4.Bratt M A, Rubin H. Specific interference among strains of NDV. II. Comparison of interference of active and inactive virus. Virology. 1967;35:381–394. doi: 10.1016/0042-6822(68)90217-1. [DOI] [PubMed] [Google Scholar]
- 5.Bratt M A, Rubin H. Specific interference among strains of NDV. III. Mechanism of interference. Virology. 1968;35:395–407. doi: 10.1016/0042-6822(68)90218-3. [DOI] [PubMed] [Google Scholar]
- 6.Campadelli-Fiume G, Qi S, Avitabile E, Foa-Tomasi L, Brandimarti R, Roizman B. Glycoprotein D of herpes simplex virus encodes a domain which precludes penetration of cells expressing the glycoprotein by superinfecting herpes simplex virus. J Virol. 1990;64:6070–6079. doi: 10.1128/jvi.64.12.6070-6079.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenner F, McAuslan B R, Mims C A, Sambrook J, White D O. The biology of animal viruses. 2nd ed. New York, N.Y: Academic Press, Inc.; 1974. [Google Scholar]
- 8.Geraghty R J, Jogger C R, Spear P G. Cellular expression of alphaherpesvirus gD interferes with entry of homologous and heterologous alphaherpesviruses by blocking access to a shared gD receptor. Virology. 2000;268:147–158. doi: 10.1006/viro.1999.0157. [DOI] [PubMed] [Google Scholar]
- 9.Horvath C M, Paterson R G, Shaughnessy M A, Wood R, Lamb R A. Biological activity of paramyxovirus fusion proteins: factors influencing formation of syncytia. J Virol. 1992;66:4564–4569. doi: 10.1128/jvi.66.7.4564-4569.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu X, Ray R, Compans R W. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J Virol. 1992;66:1528–1534. doi: 10.1128/jvi.66.3.1528-1534.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huberman K, Peluso R, Moscona A. The hemagglutinin-neuraminidase of human parainfluenza virus type 3: role of the neuraminidase in the viral life cycle. Virology. 1995;214:294–300. doi: 10.1006/viro.1995.9925. [DOI] [PubMed] [Google Scholar]
- 12.Johnson R M, Spear P G. Herpes simplex virus glycoprotein D mediates interference with herpes simplex virus infection. J Virol. 1989;63:819–827. doi: 10.1128/jvi.63.2.819-827.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura Y, Norrby E, Nagata I, Ito Y, Shimokata K, Nishiyama Y. Homologous interference induced by a temperature sensitive mutant derived from an HJV carrier culture. J Gen Virol. 1976;33:333–343. doi: 10.1099/0022-1317-33-2-333. [DOI] [PubMed] [Google Scholar]
- 14.Lamb RA. Paramyxovirus fusion: a hypothesis for changes. Virology. 1993;197:1–11. doi: 10.1006/viro.1993.1561. [DOI] [PubMed] [Google Scholar]
- 15.Levin-Perlman S, Jordan M, Brossmer R, Greengard O, Moscona A. The use of a quantitative fusion assay to evaluate HN-receptor interaction for human parainfluenza virus type 3. Virology. 1999;265:57–65. doi: 10.1006/viro.1999.0024. [DOI] [PubMed] [Google Scholar]
- 16.Morrison T G, McGinnes L W. Avian cells expressing the Newcastle Disease virus hemagglutinin-neuraminidase protein are resistant to Newcastle disease virus infection. Virology. 1989;171:10–17. doi: 10.1016/0042-6822(89)90505-9. [DOI] [PubMed] [Google Scholar]
- 17.Moscona A, Peluso R W. Fusion properties of cells persistently infected with human parainfluenza virus type 3: participation of hemagglutinin-neuraminidase in membrane fusion. J Virol. 1991;65:2773–2777. doi: 10.1128/jvi.65.6.2773-2777.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moscona A, Peluso R W. Fusion properties of cells infected with human parainfluenza virus type 3: receptor requirements for viral spread and virus-mediated membrane fusion. J Virol. 1992;66:6280–6287. doi: 10.1128/jvi.66.11.6280-6287.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moscona A, Peluso R W. Relative affinity of the human parainfluenza virus 3 hemagglutinin-neuraminidase for sialic acid correlates with virus-induced fusion activity. J Virol. 1993;67:6463–6468. doi: 10.1128/jvi.67.11.6463-6468.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentivial vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 21.Potier M, Mameli L, Belislem M, Dallaire L, Melanxon S B. Fluorimetric assay of neuraminidase with a sodium 4-methylumbelliferyl-α-d-N-acetylneuraminidase substrate. Anal Biochem. 1979;94:287–296. doi: 10.1016/0003-2697(79)90362-2. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. New York, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Warner T G, O'Brien J S. Synthesis of 2′-(4-methylumbelliferyl)-alpha-d-N-acetylneuraminic acid and detection of skin fibroblast neuraminidase in normal humans and in sialidosis. Biochemistry. 1979;18:2783–2787. doi: 10.1021/bi00580a014. [DOI] [PubMed] [Google Scholar]