Abstract
Nuclear domains called ND10 (nuclear domain 10) are discrete nuclear protein aggregations characterized by a set of interferon-upregulated proteins including Sp100 and PML, where papova-, adeno-, and herpesviruses begin their transcription and DNA replication. Both the alpha- and betaherpesvirus subfamilies disrupt ND10 upon infection by dispersing and/or destroying ND10-associated proteins. We studied the effect of the gammaherpesvirus Epstein-Barr virus (EBV) on ND10 and its spatial distribution in the nucleus of cells during latency and lytic reactivation. In latently infected Burkitt's lymphoma, lymphoblastoid, and D98/HR1 cells, ND10 were intact, as judged by immunofluorescence localization of PML, Sp100, NDP55, and Daxx. Fluorescent in situ hybridization revealed no association between viral episomes and ND10 during latency, implying that the maintenance replication of EBV, which depends on host cell proliferation, occurs independent of ND10. As in mitosis, the EBV genomes were attached to interphase chromosomes, suggesting that they are unable to move freely within the interchromosomal space and thus unable to associate with the interchromosomally located ND10 or other nuclear domains. Upon lytic activation, ND10 became dispersed in cells expressing lytic proteins. Redistribution of ND10 proteins occurred sequentially at different stages of the lytic cycle, with Sp100, Daxx, and NDP55 dispersed before and PML dispersed after the onset of lytic replication. ND10 remnants were retained until the early stages of lytic replication, and replicating EBV genomes were frequently found beside this nuclear domain; the number of replication domains was usually lower than the average latent virus frequency. Thus, latency does not require or induce interaction of EBV with ND10 for transcription and replication, whereas lytic replication triggers dispersion of ND10 proteins and occurs in close association with PML aggregates. The required movement of chromosome-attached latent EBV episomes to ND10 after reactivation from latency might include physical release of the chromosome-bound episomes. Only episomes contacting ND10 after such a release might be able to begin lytic replication.
The eukaryotic nucleus is a highly compartmentalized structure that can be roughly divided into chromosomal domains and interchromosomal space. The latter compartment harbors a variety of discrete structures known as nuclear domains or nuclear bodies, which are composed of defined sets of proteins, sometimes associated with RNAs (reviewed in reference 28). One of these structures, termed PML (promyelocytic leukemia protein) oncogenic domain, PML body, or nuclear domain 10 (ND10), was originally characterized by the presence of the autoantigen of patients with primary biliary cirrhosis, Sp100 (58). Other proteins found in ND10 include NDP55 (4) and PML, which was identified in an oncogenic translocation with the retinoic acid receptor in promyelocytic leukemia and may have functions in major histocompatibility complex gene expression and apoptosis (reviewed in references 36, 49, and 69). Another ND10 protein, Daxx, interacts with a variety of proteins such as the transcription factor Pax3, the centromeric protein CENP-C, and the apoptosis-inducing protein Fas (see reference 23 and references therein).
ND10 have gained increasing interest as nuclear sites where DNA viruses of several families start transcription and replication. Although these viruses apparently enter the nucleus randomly, they transcribe and replicate only in immediate association with ND10. Simian virus 40 (SV40), adenovirus type 5 (Ad5), and two herpesviruses, herpes simplex virus type 1 (HSV-1) and human cytomegalovirus (HCMV), start their replicative cascade at ND10 after entering the cell (reviewed in reference 36). Transfection of protein-free SV40 DNA and infection with Ad5 in the presence of actinomycin D and cycloheximide resulted in the same localization pattern of viral DNA at ND10, thus demonstrating that no viral proteins or transcripts are required for the association with ND10 (22, 60a). The sequence requirements for viral DNA deposition at ND10 are unknown except for SV40, which needs the core origin of replication in the presence of large T antigen to transcribe and replicate at ND10 (60a). It is not yet clear whether viral genomes are directed to ND10 by the cell as part of a defense mechanism, or whether viruses prefer these nuclear domains for optimal replication and transcription. Since it appears that many more viral genomes enter the cell nucleus than are actually transcribed and replicated, not all viral genomes are found at ND10. However, as demonstrated for HCMV, HSV-1, and SV40, viral transcription is seen exclusively at ND10 (22, 24).
ND10 are subject to modifications by a number of factors. Interferon treatment increases the number and size of ND10 by upregulating several ND10 proteins such as PML and Sp100 (16, 17, 29, 41, 54), consistent with the view of this structure as an important part of a cellular defense mechanism against viruses. In contrast, ND10 are disrupted upon heat shock, by exposure to cadmium, or in leukemic cells from patients with acute promyelocytic leukemia, which express a fusion protein of PML with the retinoic acid receptor α (reviewed in references 36 and 49). ND10 proteins can also be dispersed upon viral infection. Ad5, HSV-1, and HCMV have been found either to modify or to completely disrupt ND10 after they have started their transcription at this domain. In Ad5, the viral protein E4ORF3 causes a redistribution of ND10 proteins into elongated tracks (9, 11, 22). HSV-1 (a member of the Alphaherpesvirinae) and HCMV (Betaherpesvirinae) completely disperse ND10 after transcription starts, which in HSV-1 takes place as early as within 2 h of infection (38). For each virus, a single protein can disrupt ND10: the immediate-early gene 1 product ICP0 (IE110) in HSV-1 (38) and IE1 (IE72) in HCMV (24, 27). For the third subfamily, Gammaherpesvirinae, which includes Epstein-Barr virus (EBV), no information about ND10-modifying proteins is available. The underlying biochemical mechanisms for the effects of these proteins on ND10 are unclear, although it has been found that a RING finger domain of ICP0 and a potential Zn-binding motif of IE72 are necessary for the disruption of ND10 (12, 24, 37). Recent studies showed that HSV-1 and HCMV abrogate the modification of PML by SUMO-1 (small ubiquitin-related modifier), which has been demonstrated to be essential for ND10 integrity (reviewed in references 40 and 69). However, when either of the ND10-dispersing proteins was expressed after transfection, a longer time was required for ND10 disruption, suggesting that further viral proteins are necessary for efficient and rapid disruption of ND10 (12, 24, 39). Loss of ICP0 or IE72, i.e., the inability to destroy ND10, reduces the productive infectivity at low multiplicities of infection (15, 55), further supporting the idea that ND10 represent a nuclear organelle involved in an antiviral defense mechanism.
In this study we asked whether EBV might also interact with ND10 and whether this interaction would in any way differ from that of the other herpesvirus subfamily members. Since the great majority of the human population is latently infected with EBV and because this virus is also clearly associated with several types of cancer such as Burkitt's lymphoma and nasopharyngeal carcinoma (reviewed in references 42 and 45), EBV is one of the most extensively studied human viruses. EBV enters the human body by first infecting the oral and pharyngeal epithelia, although its main targets of infection are B lymphocytes, where the virus can persist permanently in a latent state. Infected B cells become immortalized and therefore can proliferate indefinitely. Several types of latency have been described for EBV, ranging from the minimal expression pattern in type I latency, in which EBNA 1 is the only detectable viral protein, to type III latency, in which a set of nine EBV-encoded proteins (EBNA 1 to EBNA 6, LMP1, LMP2A, and LMP2B) is expressed (for reviews, see references 26, 33, and 46). Reactivation from latency, which can occur spontaneously or be induced artificially, is characterized by the expression of lytic genes, which eventually leads to the production of virions and host cell lysis. Two virus-encoded DNA-binding proteins, Zta and Rta, can trigger the switch from latency to lytic cycle replication. The immediate-early protein Zta (also termed ZEBRA, Z, EB1, or BZLF1-protein) is a potent transcriptional activator of multiple viral and cellular genes and has an essential function in lytic DNA replication. Transfection of Zta-expressing plasmids into latently infected cells is sufficient to induce lytic activation of EBV (reviewed in reference 51). The second immediate-early protein, Rta (also termed R or BRLF1-protein), can efficiently disrupt latency in epithelial cells and, to a lesser extent, in B lymphocytes (44, 65). Zta and Rta function cooperatively to stimulate high-level transcription from EBV promoters and induce lytic replication.
Here, we investigate the interaction of EBV with ND10 in two different stages of infection, i.e., during latency and upon lytic activation. We show that EBV leaves ND10 intact and that replication of EBV is not associated with this nuclear domain during latency. In contrast, ND10 are disrupted upon lytic activation, with Sp100, Daxx, and NDP55 being dispersed before and PML being dispersed after the onset of lytic replication. We further show that EBV episomes are attached to interphase chromosomes during latency and thus prevented from diffusing freely within the interchromosomal space. This may account for the absence of EBV from ND10 and the observed exclusion of episomes from SC35 domains. Last, lytic replication takes place in association with PML aggregates.
MATERIALS AND METHODS
Antibodies.
ND10 were visualized using the following antibodies. Monoclonal antibody (MAb) 138 labels NDP55 (4), and MAb 5E10 reacts with PML (56). Polyclonal rabbit serum against PML and rabbit antibodies against Sp100 were obtained from J. Frey (27). Daxx was localized with a MAb raised against human Daxx (23) or a rabbit serum purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, Calif.). SC35 domains (interchromatinic granule clusters) were labeled using a MAb against splicing factor SC35 (13). EBV-encoded proteins were detected using antibodies against Zta (a rabbit serum raised against full-length Zta [31] and MAb AZ69 from Argene Inc., North Massapequa, N.Y.), Rta (MAb 8C12; Argene), and EA-R p85 and EA-D p52/50 (MAbs from ABI, Columbia, Md.). Isotype-matched MAbs of unrelated specificity were used as controls. Secondary antibodies labeled with fluorescein isothiocyanate (FITC), Texas red, or Cy5 were obtained from Vector Laboratories (Burlingame, Calif.) and Jackson Immunoresearch Laboratories (West Grove, Pa.).
Cell culture.
All cells were grown at 37°C in a humidified atmosphere containing 5% CO2. The latently EBV-infected Burkitt's lymphoma cell lines Raji and Akata and the EBV-positive lymphoblastoid cell line CB23 were maintained in RPMI medium containing 10% serum, 2 mM l-glutamine, and antibiotics. D98/HR1 cells, an adherent EBV-positive cell line generated by fusion of an epithelial cell line and P3HR1 Burkitt's lymphoma cells (14), were grown in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum and antibiotics. The Burkitt's lymphoma cell line BJAB and the epidermal carcinoma cell line HEp-2 represent EBV-negative cells which were maintained, respectively, in supplemented RPMI medium and Dulbecco modified Eagle medium.
EBV latency was disrupted by several means. D98/HR1 cells were transfected with a Zta-expressing plasmid (ZtaSRα or Zta-pCDNA3 [66]) using the DOSPER transfection reagent (Boehringer Mannheim, Indianapolis, Ind.); Raji and CB23 cells were grown for 1 to 3 days in medium containing 12-O-tetradecanoylphorbol-13-acetate (TPA; 100 ng/ml) and sodium butyrate (3 mM) (47, 70); and Akata cells were exposed to goat antibodies against human immunoglobulin G (IgG; Boehringer) which had been dialyzed against phosphate-buffered saline (PBS) to remove sodium azide and added to the medium at 10 ng/ml for 1 to 2 days (59). Rta was expressed in HEp-2 cells by transfection with plasmid pRta-RTS2 (48).
Stress-induced chromosome condensation (SICC) was induced as described elsewhere (43). In brief, D98/HR1 cells grown on coverslips were incubated at 41°C for 20 to 30 min while being covered only with a thin film of medium. Cells were then immediately fixed and processed as described below.
Immunofluorescence and microscopy.
Adherent cell lines (D98/HR1 and HEp-2) were grown on coverslips in 24-well plates and fixed in 1% paraformaldehyde (PFA) for 10 min at room temperature. Cells grown in suspension (Burkitt's lymphoma and lymphoblastoid cell lines) were first fixed by suspension in 1% PFA (10 min at room temperature) and then centrifuged onto poly-l-lysine-coated glass slides in a Cytospin 2 (Shandon, Pittsburgh, Pa.) for 5 min at 1,000 rpm. The fixed cells were permeabilized by incubation in 0.2% Triton (20 min on ice), blocked with 10% goat serum (30 min), and incubated with primary (1 h) and FITC-, Texas red-, or Cy5-labeled secondary antibodies (30 min; all solutions in PBS). Finally, cells were stained for DNA with Hoechst 33258 (0.5 μg/ml) or propidium iodide (1 μg/ml) and mounted with Fluoromount G (Fisher Scientific).
Confocal images of cells were obtained using a Leica confocal laser scanning microscope. The two channels were recorded simultaneously if no cross talk could be detected. In the case of strong FITC labeling, sequential images were acquired with more restrictive filters to prevent possible breakthrough of the FITC signal into the red channel. Both acquisition modes resulted in the same images. The Leica enhancement software was used in balancing the signal strength and scanned eightfold to separate signal from noise. Alternatively, cells were analyzed with a Leitz Fluovert inverted microscope equipped with a digital camera. Images were obtained using software from QED Imaging (Pittsburgh, Pa.).
Fluorescence in situ hybridization (FISH).
The following probes were used for the detection of EBV-specific DNA sequences. A mixture of p560oriLyt, EBV-BamC-orip+, and EBV Bam ZR/Sal F fragments (7, 18) was labeled with biotin-16-dUTP (Sigma) by nick translation and used for the detection of single EBV episomes during latency. To detect replicating EBV DNA in D98/HR1 cells transfected with Zta to disrupt latency, we required a probe that met two requirements: first, it was free of any vector and Zta coding sequences, in order to avoid hybridization to transfected plasmid DNA; second, it was small enough to prevent the visualization of single episomes, thus enabling us to observe only EBV DNA that replicates upon induction of the lytic cycle, not latent EBV genomes.
To this end, a 411-bp sequence contained in the repetitive BamHI W fragment of the EBV genome was biotin labeled by PCR as described elsewhere (53), using D98/HR1-DNA as the template. The primers used were 5′-CCATGTAAGCCTGCCTCGAGT-3′ and 5′-TTTAGGCCTAGGTCCGCCATTA-3′, generating a PCR product from positions 22937 to 23347 (numbering according to reference 5). Given a detection limit of our FISH procedure of approximately 9 kb, and assuming that the BamHI W fragment is repeated about 10 times in the EBV genome (5, 21), this PCR probe detected only clusters consisting of more than two episomes (two episomes equal about 8 kb of target sequence, which is not detectable). This probe thus allowed exclusive visualization of replicated EBV after lytic activation (see Results).
Cells were fixed in 4% PFA in PBS (10 min at room temperature) or, when FISH was combined with immunocytochemistry, first immunostained as described above and then refixed in 4% PFA to cross-link bound antibodies. After washing in PBS and permeabilizing in 0.2% Triton (20 min on ice), cells were digested with RNase (Boehringer; 100 μg/ml in PBS for 30 min at 37°C) and equilibrated in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Cells were then dehydrated in an ethanol series (70, 80, and 100% for 3 min each at −20°C), air dried, and immediately covered with the probe mixture containing 50% formamide in 2× SSC, probe DNA (10 ng/μl), 10% dextran sulfate, cot1 DNA (0.5 μg/μl; Gibco BRL), salmon sperm DNA (0.1 μg/μl; Gibco BRL), and yeast tRNA (1 μg/μl; Sigma). Probe and cells were simultaneously heated at 94°C for 4 min to denature DNA and incubated overnight at 37°C.
After hybridization, specimens were washed at 37°C with 50% formamide in 2× SSC (two times for 15 min each), 2× SSC (10 min), and 0.25× SSC (two times for 5 min each). Hybridized probes were labeled with FITC-avidin (Vector Laboratories; 1:500 in 4× SSC plus 0.5% bovine serum albumin for 30 min), and signals were amplified for 30 min using biotinylated antiavidin (1:250; Vector Laboratories), followed by another round of FITC-avidin staining. Finally, cells were equilibrated in PBS and mounted in Fluoromount G.
RESULTS
Latent EBV neither disrupts nor is associated with ND10.
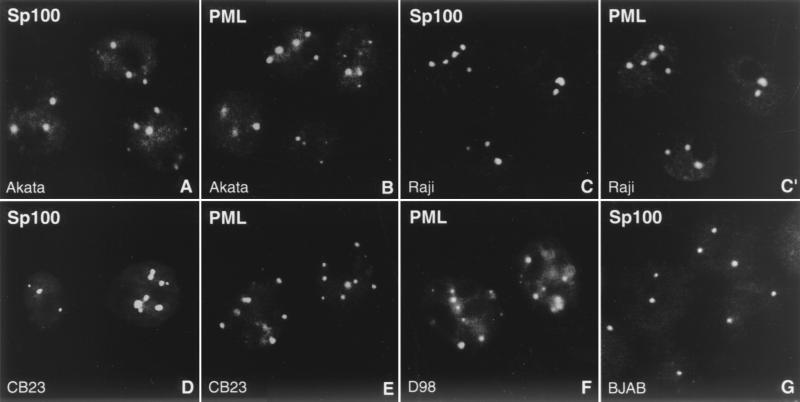
The infection of cells with HSV-1 and HCMV results in the immediate-early dispersion of ND10. We asked whether in latently infected cells EBV and ND10 coexist and whether EBV episomes had any nonrandom intranuclear localization. Though the presence of ND10 in EBV-positive cell lines has previously been described (57), we decided to reinvestigate this issue because Szekely et al. used in most of their experiments the lymphoblastoid cell line IB4, which contains a single integrated copy of the EBV genome instead of free episomes. When we tested the EBV-positive Burkitt's lymphoma cell lines Raji and Akata as well as the latently EBV-infected lymphoblastoid cell line CB23 for the presence of ND10 by performing immunofluorescence against the ND10 proteins Sp100 and PML, almost all cells of each cell line displayed about 2 to 10 clearly labeled aggregates (Fig. 1). As in other cell lines, PML and Sp100, which are both known as marker proteins for ND10, colocalized within the same aggregates (Fig. 1C and C′). To detect any possible changes in number and size of ND10 in EBV-positive cells compared to EBV-free cells, we immunolabeled the EBV-negative Burkitt's lymphoma cell line BJAB with anti-PML and anti-Sp100 (Fig. 1G) antibodies and obtained exactly the same results. Since virtually all cells of the latently infected cell lines harbor EBV episomes, we conclude that the presence of latent EBV neither disrupts ND10 nor changes the number or morphology of this nuclear domain. The Burkitt's lymphoma cell lines have been characterized as type I latency in which the only viral protein expressed is EBNA 1. The CB23 lymphoblastoid cells represent type III latency (6) in which all of the major immortalizing proteins of EBV, including EBNA 2 to 6, LMP1, LMP2A, and LMP2B, are expressed. Thus, we conclude that none of the EBV latency-associated proteins disrupts or modifies ND10 morphology.
FIG. 1.
ND10 are not disrupted during latency. The EBV-positive Burkitt's lymphoma cell lines Akata (A and B) and Raji (C and C′), the latently EBV-infected lymphoblastoid cell line CB23 (D and E), and the EBV-positive fusion cell line D98/HR1 (D98; F) possess intact ND10, as revealed by immunohistochemistry with antibodies against Sp100 (A, C, D, and G) and PML (B, C′, E, and F). C and C′ shows double labeling to demonstrate colocalization of both proteins. As a control, ND10 are visualized in the EBV-negative Burkitt's lymphoma cell line BJAB by staining for Sp100 (G).
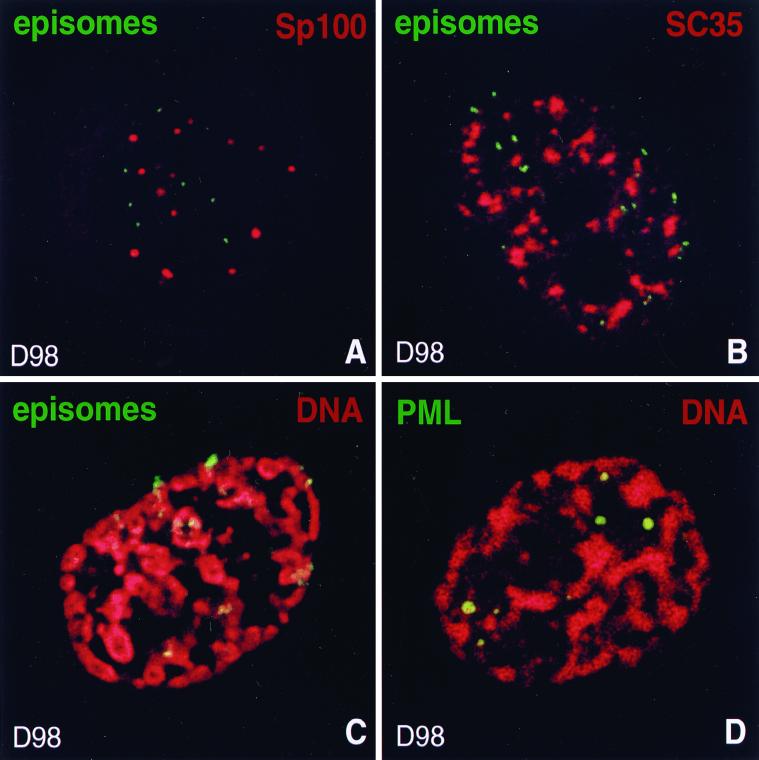
To localize EBV episomes in latently infected cells by FISH, we used D98/HR1 cells, a cell line representing type I latency, that has been generated by fusion of HeLa cells with P3HR1 EBV-infected Burkitt's lymphoma cells (14). Due to its ability to adhere to glass slides as well as its flatness, this cell line proved to be more amenable to in situ hybridization studies. Like Raji, Akata, and CB23 cells, D98/HR1 cells revealed ND10 when stained for PML or Sp100 (Fig. 1F), further confirming our observation that ND10 remain stable in the presence of latent EBV. FISH combined with immunocytochemistry in which ND10 were detected with antibodies to Sp100 and episomes were visualized by hybridization with a probe mixture made from plasmids that contain EBV sequences revealed no specific association of EBV genomes with ND10 in D98/HR1 cells (Fig. 2A). Each nucleus harbored about 5 to 40 episomes which appeared to be randomly distributed within the nucleoplasm; only occasional individual viral genomes were found close to ND10, apparently indicating a nonspecific association. Thus, in latently infected cells EBV is not associated with this nuclear domain, implying that transcription and the cell-mediated replication of EBV during latency occur without any specific association with ND10.
FIG. 2.
Localization of EBV genomes during latency. The upper corner of each image shows the color used for labeling of the respective protein or DNA. (A) EBV episomes are not associated with ND10 during latency. ND10 are shown in a latently infected D98/HR1 cell by labeling Sp100, and episomes were visualized by FISH. (B) D98/HR1 cell during EBV latency stained with anti-SC35 and hybridized with an EBV-specific probe. Episomes are excluded from and only occasionally associated with SC35 domains. (C) Latently infected D98/HR1 cell treated to undergo SICC during interphase. Cellular DNA was stained with propidium iodide to visualize the contracted chromosomes, and in situ hybridization was performed with an EBV-specific probe for the detection of episomes. Episomes are found exclusively closely attached to the condensed interphase chromosomes. (D) As for panel C, but stained for PML to visualize ND10. In contrast to EBV episomes, ND10 are located between the chromosomes and thus occupy a different nuclear space.
EBV episomes are attached to interphase chromosomes during latency and do not occupy the interchromosomal space.
To determine whether other nuclear domains located in the interchromosomal space might recruit or associate with EBV genomes, we tested the spatial interaction of this virus with SC35 domains, which contain the spliceosome assembly factor SC35 and appear to be important for the processing of RNA transcripts (13). Many active genes, both cellular and viral, have been found to be closely associated with these irregularly shaped domains which occupy a relatively large volume within the interchromosomal space (24, 50, 64). If EBV episomes move freely within the interchromosomal space, we would expect to find a certain degree of random colocalization between EBV episomes and the relatively large SC35 domains. Surprisingly, when we stained D98/HR1 cells for SC35 in combination with FISH for EBV, we found EBV episomes always outside of and only occasionally associated with this domain (Fig. 2B). Therefore, EBV must be present in a space not occupied by SC35 domains. Based on findings that EBV is attached to chromosomes in mitosis (19, 61), we tested whether EBV is also closely bound to the interphase chromosomes which may impede its movement. We performed SICC, which is a combination of several stresses including heat shock that leads within 20 to 30 min to a reversible nonmitotic as well as nonapoptotic chromosome contraction (43). This chromosome condensation clearly reveals the interchromosomal space that is not visible when chromatin is unfolded as in normal interphase. When we performed SICC, EBV genomes were exclusively found attached to the chromosomes (Fig. 2C). As has been shown previously, this method does not disrupt the intranuclear organization and leaves organelles located between the chromosomes in place (43). Control D98/HR1 cells after SICC revealed ND10 (in contrast to the episomes) in the interchromosomal space and only occasionally in direct contact with condensed chromosomes (Fig. 2D). Thus, EBV not only associates with mitotic chromosomes, as demonstrated earlier (19, 61), but also is closely attached to chromosomes during interphase. Binding of episomes to interphase and mitotic chromosomes might be the reason why latent EBV does not associate with nuclear domains located in the interchromosomal space.
Lytic activation of EBV induces disruption of ND10.
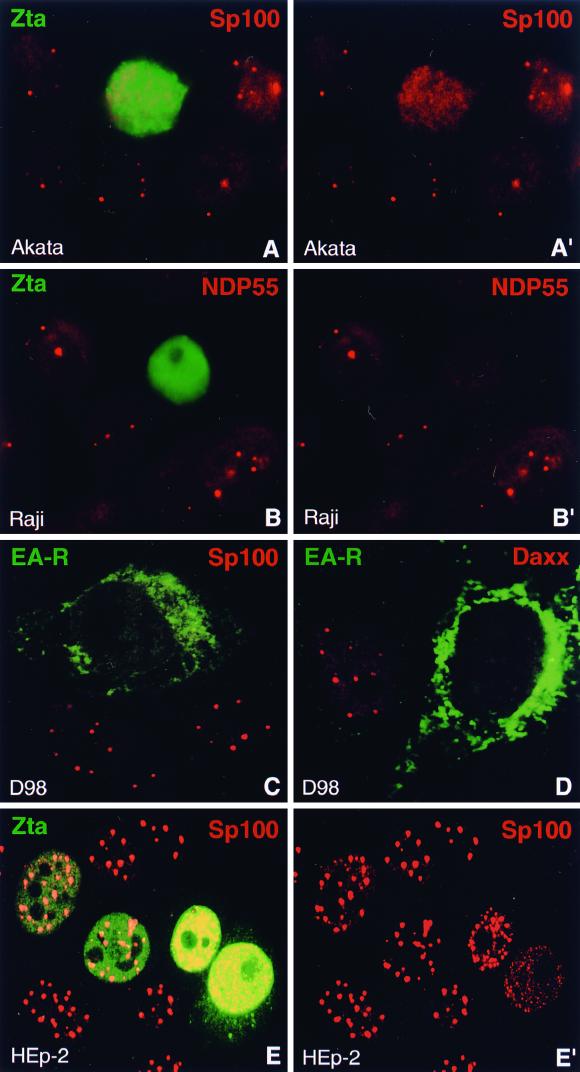
Since EBV was found not to alter ND10 during latency and also not to perform latent replication next to this structure, we wondered whether this behavior might change upon lytic activation. We therefore disrupted latency in Akata cells by incubation with anti-IgG antibodies and in Raji cells by treatment with TPA and butyrate. The cells were then double labeled with antibodies directed to early EBV protein EA-D or Zta to monitor lytic activation and with antibodies against ND10 proteins to control for the integrity of ND10. When we tested for the presence of ND10 using antibodies to Sp100, we never found this structure in Zta- or EA-D-positive cells. In both Akata and Raji cells, Sp100 concentrated in ND10 was completely dispersed upon lytic activation (Fig. 3A and A′). This was not simply a consequence of disintegration of the cell by EBV since cells lacking ND10 displayed an intact nuclear morphology, as judged by Hoechst staining and by labeling other nuclear structures such as nucleoli (not shown). As we show later, dispersion of Sp100 has already occurred shortly before lytic replication starts. When we treated the EBV-negative Burkitt's lymphoma cell line BJAB either with anti-IgG or TPA and butyrate for the same times, no ND10-free cells could be found compared to untreated cells, clearly demonstrating that this effect was actually caused by EBV and not by the treatment with antibodies or chemicals.
FIG. 3.
Lytic activation of EBV disperses the ND10 proteins Sp100, NDP55, and Daxx. (A and A′) Lytic activation of EBV was induced in Akata cells and monitored by staining for the immediate-early protein Zta. Sp100 is no longer concentrated in ND10 in Zta-expressing cells. A′, Sp100 staining only. (B and B′) Lytic activation of EBV in Raji cells, detected by staining of Zta, induces the dispersion of the ND10 protein NDP55. B′, NDP55 staining only. (C) D98/HR1 cells were Zta transfected to induce lytic activation of EBV and stained for the viral protein EA-R and Sp100. EA-R, which is localized in the cytoplasm, allows the identification of cells in which EBV has entered the lytic cycle. Sp100 is no longer concentrated in ND10 after lytic activation. (D) Like Sp100 and NDP55, the ND10 protein Daxx disperses in EA-R-positive D98/HR1 cells. (E and E′) EBV-free HEp-2 cells were transfected with a Zta-expressing plasmid and stained with antibodies against Zta and Sp100. ND10 remain intact at Zta concentrations comparable to those observed after lytic activation, but Sp100 starts to disperse when Zta is expressed at very high levels (cells on right). Note the deposition of Zta in the cytoplasm in the lower right cell. E′, Sp100 staining only.
The same experiment using latently infected CB23 lymphoblastoid cells treated with TPA and butyrate again revealed no ND10 in Zta-positive cells (not shown), indicating that EBV-induced dispersion of ND10 is not restricted to Burkitt's lymphoma cell lines. Finally, we activated EBV in D98/HR1 cells by transfection with Zta-expressing plasmids, which proved to be the only method to induce lytic activation in this cell line. Zta-transfected D98/HR1 cells were grown overnight and then simultaneously stained with antibodies against Sp100 and antibodies directed to the EBV protein Rta or EA-R to identify cells in which EBV became activated. EA-R was found to accumulate in the cytoplasm (34), whereas Rta was concentrated within several nuclear aggregates which we found to be associated with replication domains of EBV DNA (see below). As in Burkitt's lymphoma and lymphoblastoid cell lines, D98/HR1 cells, which expressed lytic proteins, never contained ND10, as observed by staining for Sp100 (Fig. 3C). Thus, in four different latently infected cell lines, Sp100 was no longer concentrated in ND10 when lytic activation had started.
To test the possibility that Zta alone might be able to disrupt ND10, we transfected Zta-expressing plasmids into HEp-2 cells, a cell line not infected with EBV. In most transfected cells, Zta did not affect the integrity of ND10; however, in cells in which Zta was overexpressed at very high levels, we found that Sp100 as well as PML became dispersed, often in the form of numerous small aggregates (Fig. 3E and E′). The high nuclear concentration of Zta necessary to disrupt ND10 did not appear to reflect physiological levels of Zta expressed by EBV. Since in D98/HR1 cells disruption of ND10 occurred at lower Zta levels than in HEp-2 cells, as judged from the staining intensity by immunofluorescence, Zta alone does not seem to play a crucial role in dispersion of ND10 proteins, although it might interact with other EBV proteins to aid in the disruption of ND10. We therefore tested Rta, another immediate-early EBV protein, for its ability to cooperate with Zta in dispersion of ND10 proteins. However, cotransfection of both Zta and Rta into HEp-2 cells did not result in ND10 disruption at lower Zta levels, demonstrating that both proteins do not obviously cooperate to increase or accelerate the dispersion of ND10 associated proteins (data not shown).
We next asked whether other ND10 proteins might behave in the same way as Sp100. After disruption of latency, Akata, Raji, CB23, and D98/HR1 cells were stained for lytic proteins and for the ND10 proteins Daxx and NDP55. As in the case of Sp100, both Daxx and NDP55 were never observed concentrated in ND10 of cells in which latency had been disrupted (Fig. 3B and B′ [NDP55 in Raji cells] and 3D [Daxx in D98/HR1 cells]). In some cells, NDP55 was not detectable in ND10 during latency, which might be a cell cycle-dependent phenomenon. However, we never observed cells displaying both lytic EBV proteins and NDP55-positive ND10 at the same time.
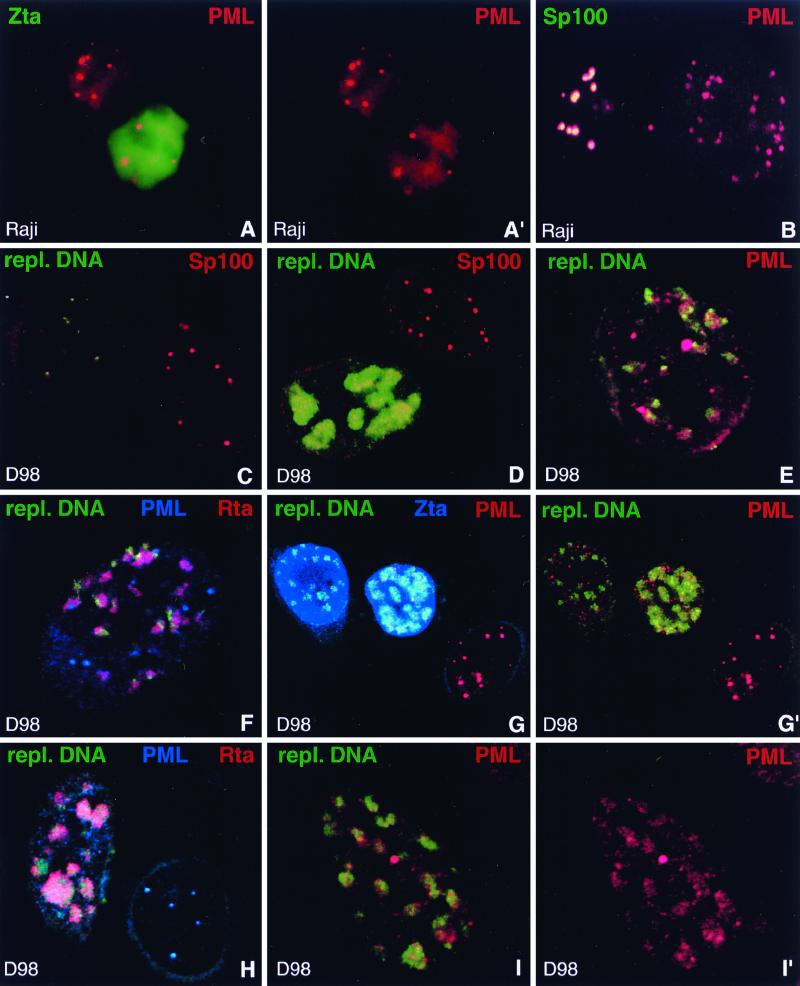
When we finally tested the essential ND10 protein PML for its reaction pattern upon lytic activation, we found that this protein behaved differently than Sp100, Daxx, and NDP55. In all four cell lines tested, PML appeared to become dispersed less easily than the other ND10 proteins. Indeed, about 25% of Raji cells and 40% of Akata cells which stained positive for lytic proteins still contained PML-positive ND10 as in uninduced cells (Fig. 4A and A′). This phenomenon was even more pronounced in the lymphoblastoid cell line CB23, in which 55% of Zta-positive cells still contained PML-positive ND10. Since this was in sharp contrast to the complete absence of ND10-positive cells when stained with antibodies to Sp100, Daxx, or NDP55, we double labeled Raji and Akata cells, after induction of lytic activation, both for Sp100 and PML. As expected, we observed cells with PML-positive ND10 that lacked Sp100, presumably representing cells in which latency had been disrupted (Fig. 4B). D98/HR1 cells behaved in a similar way when lytic activation was induced. Although PML became partially dispersed as judged by a higher nuclear background, a phenomenon that was more obvious in D98/HR1 than in Akata or Raji cells, PML-positive ND10 remained visible in most of the cells that expressed lytic proteins. Thus, proteins recruited to ND10 by PML (23, 30, 68) are removed from this domain before the dispersal of PML. The ND10-dispersing activity of EBV seems therefore to be different from the ND10-disrupting mechanism of other herpesviruses.
FIG. 4.
Behavior of PML upon EBV reactivation compared to Sp100. (A and A′) Raji cells stained for Zta and PML after induction of lytic activation. Unlike other ND10 proteins, PML often remains concentrated in ND10 in Zta-positive cells. A′, PML staining only. (B) Raji cells double labeled with antibodies against PML and Sp100 after disruption of latency to demonstrate release of Sp100 but not of PML from ND10. The left-hand cell shows typical ND10 which contain both proteins, indicating that EBV remained latent. In the right-hand cell, EBV apparently entered the lytic cycle and released Sp100 from ND10 which still contain PML. (C) D98/HR1 cells were Zta transfected to induce lytic activation, stained for Sp100, and probed with a labeled PCR product which recognizes clusters of replicating EBV DNA but not individual episomes. Note the lack of detectable hybridization signal in the right-hand cell, in which no lytic activation occurred. This cell contains Sp100-positive ND10. In contrast, the left-hand cell displays small, i.e., early, foci of replicating EBV DNA. Sp100 is no longer concentrated in ND10 as soon as replicated EBV genomes are detectable. (D) Zta-transfected D98/HR1 cells labeled for Sp100 and hybridized with the probe specific for replicating EBV DNA. The lower cell shows an advanced stage of the lytic cycle, with replicating EBV genomes that form large clusters of DNA. Sp100 remains dispersed and does not associate with the replication domains. In the upper cell, no lytic cycle has been induced and Sp100 is not dispersed from ND10. No replicated EBV DNA is detectable in this cell. (E) A Zta-transfected D98/HR1 cell was stained for PML and probed for replicating EBV DNA. In contrast to Sp100, PML-positive ND10 are still visible after lytic replication has started. Most of the replication domains are found in close association with PML. (F) D98/HR1 cells were transfected with Zta to induce lytic activation and triple labeled for PML and Rta by immunostaining and for replicating EBV genomes by FISH. Rta forms aggregates in close association with the replication domains but without complete colocalization. PML is still visible in the form of several ND10-like structures. (G and G′) Zta-transfected D98/HR1 cells triple labeled for the detection of PML, Zta, and replicating EBV DNA. The replication-specific probe detects EBV genomes only in Zta-positive cells (two leftmost cells). In the right-hand untransfected cell, PML is almost completely aggregated into ND10, whereas PML becomes increasingly dispersed during lytic replication (two leftmost cells). However, even during late replication stages, as represented by the middle cell, PML is still concentrated in several ND10-like foci. In panel G′, Zta staining is omitted. (H) Zta-transfected D98/HR1 cell immunostained for Rta and PML and probed for replicating EBV DNA. In the right-hand cell, which is probably not transfected, no lytic activation was induced. The replication-specific probe fails to recognize EBV-specific genomes in this cell, which displays intact PML-positive ND10. The left-hand cell represents a late stage of the lytic cycle, as judged from the size of the replication compartments. PML foci have almost completely disappeared. In contrast to earlier replication stages as shown in panel F, the Rta aggregations and replication domains show a higher degree of colocalization, suggesting that both components had merged while increasing in size. (I and I′) Zta-transfected D98/HR1 cell hybridized with the replication-specific probe and immunostained for PML. PML is redistributed into the replication domains and colocalizes almost completely with the replicated EBV genomes in the form of diffuse PML accumulations. I′, PML staining only.
Lytic replication starts after dispersion of Sp100 but before disruption of PML.
To visualize EBV DNA during the lytic cycle, FISH was carried out with a probe specific for replicating DNA. This probe, which is complementary to a 411-bp sequence contained in the repetitive EBV W fragment, did not yield a signal strong enough to visualize single episomes during latency but readily detected clusters of more than two EBV genomes after activation (see Materials and Methods). This allowed us to correlate ND10 disruption with the onset of lytic replication. To test the specificity of this probe, we performed FISH on Zta-transfected D98/HR1 cells and compared the results with those obtained with the probe used earlier for the detection of single episomes. In contrast to the probe that allows visualization of individual episomes, the probe specific for replicating DNA yielded signals only in D98/HR1 cells that had been Zta transfected; no signals were found in untransfected cells. When we simultaneously performed FISH and stained for expression of Zta, this probe allowed us to observe signals exclusively in Zta-positive cells, never in cells which did not express Zta (Fig. 4G and G′). Since expression of Zta might not necessarily induce activation of EBV, we labeled D98/HR1 cells for Rta and simultaneously used the replication-specific probe. Only Rta-positive cells displayed in situ signals, thus clearly demonstrating the specificity of this probe for replicating DNA (Fig. 4F and H).
Signals obtained from this probe ranged from several small nuclear dots to large aggregates occupying almost the entire nuclear space, showing early to late stages of replicating EBV DNA (Fig. 4C and D). Typically, we obtained the full range of different stages of lytic replication within the same preparation of Zta-transfected cells. Costaining for Rta showed that this immediate-early transcription factor was associated with the replication domains. Rta usually did not completely colocalize with the EBV genomes at early replication stages but instead was found in aggregates overlapping with the replicated EBV DNA (Fig. 4F). During later stages of the lytic cycle, a closer association between replication compartments and Rta aggregates was observed, eventually resulting in colocalization of both domains (Fig. 4H).
When we tested for Sp100 in Zta-transfected D98/HR1 cells in which replicated EBV DNA had been labeled, we found that all cells which revealed hybridization signals were negative for Sp100. Even when only small clusters of replicated DNA were present, Sp100 had already been completely dispersed (Fig. 4C). Thus, Sp100 disappears from ND10 before lytic replication starts. Based on earlier experiments showing that NDP55, Daxx, and Sp100 are dispersed concomitantly, we conclude that all three proteins are removed from ND10 before the onset of lytic replication. In contrast, analysis of D98/HR1 cells for PML and replicating DNA revealed PML-positive ND10 in cells that displayed clusters of replicated EBV genomes (Fig. 4E and F). PML-positive ND10 were often still visible at advanced stages of the lytic cycle, i.e., when the cells contained large aggregates of EBV genomes, although in most cases there were fewer ND10 than at earlier stages (Fig. 4G and G′). Only some of the cells at late replication stages completely lacked ND10, as indicated by staining for PML (Fig. 4H). To quantify changes in the number of PML-positive ND10 during the progress of lytic replication, we divided lytic EBV replication into three stages according to the size of the replication domains: early (e.g., left-hand cell in Fig. 4C), intermediate (e.g., cell in Fig. 4I), and late (e.g., lower cell in Fig. 4D). We then counted the number of PML-positive ND10 that appeared to have the same size as ND10 during latency. Cells both in the early and intermediate replication stages contained an average of 4 PML-positive ND10, whereas during late replication only an average of 0.5 ND10 was visible. Thus, PML, unlike Sp100, Daxx, or NDP55, begins to disperse shortly after the onset of lytic replication but often remains visible as an ND10 remnant even during later stages of the lytic cycle.
We frequently observed replication domains associated with PML-positive ND10, suggesting that EBV might at least preferentially start to replicate next to this nuclear domain (Fig. 4E). It is unclear whether replication sites not associated with ND10 were in contact with ND10 before dispersion started. In cells in which EBV had been activated, PML did not appear to become degraded but rather was scattered throughout the nucleus, often appearing as small dot-like aggregates reminiscent of stress-dependent dispersion (41). Thus, EBV episomes might start replication at preexisting PML-positive ND10 or might become associated later with fragmented PML aggregates. PML also appeared to become enriched within the replication domains, since diffuse PML staining colocalized with replication sites (Fig. 4I and I′). We conclude that lytic replication of EBV occurs in association with PML, although it remains unclear whether all of the corresponding PML aggregates originate from ND10 or represent dispersed PML targeted to the replication domains.
DISCUSSION
EBV is currently the most amenable herpesvirus for the study of latency and lytic activation because of the availability of cell lines latently infected with this virus. In the present study, we used those cell lines to investigate whether latent and lytic replication occur in association with ND10 and whether both events might compromise the integrity of this nuclear domain. In Burkitt's lymphoma and lymphoblastoid cell lines as well as D98/HR1 cells, we found that ND10 remain intact in latently EBV-infected cells, as judged from the localization pattern of four ND10 proteins, Sp100, PML, Daxx, and NDP55. Undisrupted ND10, as seen by staining for PML, have also been reported for the EBV-positive lymphoblastoid cell line IB4 (57). However, IB4 cells contain a single integrated copy of the EBV genome instead of free episomes and thus might be atypical compared to latently infected cells with numerous unintegrated episomes. In the same study (57), freshly infected B cells were reported to contain undisrupted ND10 as observed by labeling of PML, a behavior which probably reflects the fact that in most infected B cells EBV immediately becomes latent (reviewed in reference 26) and therefore does not affect ND10. Because of the low level of productive infection, it is difficult to analyze the immediate-early events in this virus. However, the effects on ND10 of primary infection of cells with other herpesviruses have been studied in detail in HSV-1 and HCMV. These herpesviruses induce the rapid dispersion of all ND10-associated proteins (10, 12, 25, 27, 38). The effect of latent HSV-1 and HCMV infection on ND10 has not been investigated due to the difficulty in adequately identifying the virus in latently infected cells.
A major difference between ND10 disruption by EBV and ND10 disruption by HSV-1 and HCMV is the different behavior of PML in relation to other ND10 proteins. Whereas PML disperses shortly after infection by HSV-1 and HCMV along with Sp100 (2, 8, 25, 37, 38, 63), induction of the lytic cycle of EBV caused a sequential release of first Sp100, Daxx, and NDP55 and only later of PML. Sp100, along with Daxx and NDP55, dispersed from ND10 before the onset of replication, whereas redistribution and dispersion of PML was observed only after replicating viral DNA was already detectable. In HSV-1-infected cells, PML has been reported to first disperse completely and later to accumulate within the replication compartments identified by labeling of ICP8 (8). This finding appears to be similar to our observations during lytic activation of EBV; however, we did not observe an immediate complete dispersal of PML, since virtually all cells contained PML-positive ND10 when small (i.e., early) replication domains were visible. Thus, it seems unlikely that the PML-positive ND10-like structures observed during lytic replication of EBV have been re-formed after initial dispersion of this protein, although this possibility cannot be completely ruled out. Currently we assume that round and dense PML-positive structures resembling ND10 represent original ND10 stripped of other ND10-associated proteins, whereas diffuse PML aggregations associated with replicating DNA appear to have been formed by recruiting dispersed PML into these compartments.
Raji cells are latently infected with an EBV mutant that is unable to carry out lytic replication due to the deletion of several reading frames (20). Nevertheless, lytic activation causes dispersion of ND10 in Raji cells as rapidly as in other latently infected cell lines, indicating that replication itself is not responsible for ND10 disruption. In HSV-1 and HCMV, a single protein, ICP0 and IE72, respectively, could be identified to be responsible for dispersion of ND10 proteins, although additional viral proteins appear to be necessary for the rapid and sufficient disruption of ND10 (12, 24, 27, 38, 39). Since both proteins represent immediate-early gene products, EBV may also posses an immediate-early protein with similar properties. Zta is an EBV immediate-early protein regulating the switch from latency to the lytic cycle, and we therefore tested whether this transcriptional activator might alone be able to induce ND10 dispersion. When we transfected HEp-2 cells, an EBV-free cell line, with Zta-expressing plasmids, we found Sp100 to remain concentrated in ND10 at what appeared to represent physiological expression levels of Zta. Only when Zta was accumulated at very high nuclear concentrations did ND10 become dispersed. Interestingly, Zta has been reported to interact with the cellular coactivator CREB-binding protein (CBP) (1, 66), which in turn has been found to interact with PML, although it remains controversial whether CBP represents a true ND10 protein (reviewed in reference 40). We did not observe a specific aggregation of Zta at ND10, which should be the case if it bound to CBP or other ND10-associated proteins. Zta alone seems therefore unable to disperse ND10-associated proteins upon lytic activation but might be an accessory factor that cooperates with other proteins in the rapid dispersion of ND10 proteins.
Zta has been described to accumulate, along with EA-D, within the domains of replicating EBV genomes after lytic activation. The redistribution of both proteins into these compartments was found to depend on the lytic replication of viral DNA (60). Our study confirmed the sequestration of EA-D into replication compartments (not shown); however, we observed no enrichment of Zta within the areas of replicating viral DNA in D98/HR1 cells. Since Zta was transfected in these cells, it is possible that the failure to accumulate in punctate domains was a result of overexpression, and that during natural reactivation Zta does accumulate in more restricted domains.
A novel finding in our study is the association of Rta aggregates with replicating EBV DNA. Early replication domains usually did not directly colocalize but rather were closely associated with Rta accumulations. A more complete colocalization between replicating EBV DNA and Rta aggregates was observed at later stages of the lytic cycle, suggesting that both compartments had merged and either that Rta had been sequestered into the growing replication domains or that the replicating EBV genomes developed into the Rta accumulations. Rta might play a crucial role in defining the original site of the replication domains by aggregating next to ND10 and providing an environment conducive for replication of episomes. The formation of prereplication sites by proteins involved in replication is possible in the absence of any replicating viral DNA as demonstrated in HSV-1 (32, 62, 67). Moreover, cells cotransfected with the seven essential replication proteins of HSV-1 were able to form compartments of these replication-associated proteins beside ND10 even in the absence of the viral origin-binding protein or plasmids containing a HSV origin of replication (35).
EBV must replicate at least once per cell cycle to maintain the average number of episomes in cycling cells. The virus probably uses the cell replication machinery, including the controls that limit the number of replicative rounds per cell cycle, since only a limited number of viral proteins are expressed. Replication during primary infection with HSV-1 and HCMV takes place adjacent to ND10, and we had therefore expected to find latent EBV also next to this nuclear domain. However, viral genomes identified by in situ hybridization were not found in association with ND10, irrespective of the cell cycle stage. Since the genomes replicate during the cell cycle of the host cell, we have to conclude that they do so at locations different from ND10.
The integrity of ND10 was no longer maintained when lytic activation of EBV was induced, a finding similar to the events during primary infection by HSV-1 and HCMV. However, PML was not fully dispersed even after EBV replication became noticeable. Use of a probe too short to recognize single latent episomes served to distinguish latent from replicated virus. We often found clusters of replicated EBV DNA in close association with ND10, as seen by staining for PML, and could thus demonstrate that lytic replication of EBV starts at ND10 or its PML remnant, a finding that corresponds to the infectious sequence in HSV-1 (22) and HCMV (3).
EBV reportedly attaches to mitotic chromosomes and thus enters the nucleus after each cell division (19, 61). Retention at that chromosomal position might be the mechanism that prevents EBV transport to or deposition at ND10. Such a block in intranuclear diffusion would require that EBV episomes be retained at such chromosomal sites during interphase when the chromosomes form defined domains of decondensed chromatin. SICC (43) allowed us to test this proposition and showed that EBV was attached to chromosomes even during interphase whereas ND10 was an interchromosomal component of the nucleus like the SC35 domain containing accumulations of splicing factors (52). No EBV genomes were found in those interchromosomal domains either, supporting the idea that EBV episomes are attached to the chromosomes. ND10 also do not move freely in the nucleus (43). To reconcile these observations with the lytic replication of EBV at ND10, we propose that the latent and chromosome-bound EBV episome is physically liberated from its bound state during reactivation from latency and acquires the ability to diffuse or migrate by other means, while gaining full transcriptional competency. Those few viral genomes that come into contact with ND10 start lytic replication. Release from the bound state by stress or other exogenous factors may be the first observable act of reactivation from latency and provide a target to inhibit. The cell culture system of EBV latency may be the system of choice to investigate those mechanisms.
ACKNOWLEDGMENTS
This study was supported by NIH grants AI 41136 and GM 57599 and NSF grant MCB9728398 (P.B.), NIH grant GM 54687-02, the Leukemia/Lymphoma Society, and the American Cancer Society (P.M.L.), and the Human Frontier Science Program and the G. Harold and Leila Y. Mathers Charitable Foundation. NIH Core grant CA-10815 is acknowledged for support of the microscopy and sequencing facility.
We thank J. Frey and N. Stuurman for providing antibodies against PML and Sp100, X. J. Ma and S. Kenney for cell lines (CB23 and D98/HR1), B. Sugden for EBV cosmids, and D. Hayward for plasmid pRta-RTS2.
REFERENCES
- 1.Adamson A L, Kenney S. The Epstein-Barr virus BZLF1 protein interacts physically and functionally with the histone acetylase CREB-binding protein. J Virol. 1999;73:6551–6558. doi: 10.1128/jvi.73.8.6551-6558.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn J H, Hayward G S. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn J H, Jang W J, Hayward G S. The human cytomegalovirus IE2 and UL112–113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10) J Virol. 1999;73:10458–10471. doi: 10.1128/jvi.73.12.10458-10471.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ascoli C A, Maul G G. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, et al. DNA sequence and expression of the B95–8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 6.Benjamin D, Sharma V, Kubin M, Klein J L, Sartori A, Holliday J, Trinchieri G. IL-12 expression in AIDS-related lymphoma B cell lines. J Immunol. 1996;156:1626–1637. [PubMed] [Google Scholar]
- 7.Buell G N, Reisman D, Kintner C, Crouse G, Sugden B. Cloning overlapping DNA fragments from the B95–8 strain of Epstein-Barr virus reveals a site of homology to the internal repetition. J Virol. 1981;40:977–982. doi: 10.1128/jvi.40.3.977-982.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burkham J, Coen D M, Weller S K. ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J Virol. 1998;72:10100–10107. doi: 10.1128/jvi.72.12.10100-10107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carvalho T, Seeler J S, Ohman K, Jordan P, Pettersson U, Akusjarvi G, Carmo-Fonseca M, Dejean A. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chelbi-Alix M K, de The H. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene. 1999;18:935–941. doi: 10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- 11.Doucas V, Ishov A M, Romo A, Juguilon H, Weitzman M D, Evans R M, Maul G G. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 12.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu X D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- 14.Glaser R, Ablashi D V, Nonoyama M, Henle W, Easton J. Enhanced oncogenic behavior of human and mouse cells after cellular hybridization with Burkitt tumor cells. Proc Natl Acad Sci USA. 1977;74:2574–2578. doi: 10.1073/pnas.74.6.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greaves R F, Mocarski E S. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J Virol. 1998;72:366–379. doi: 10.1128/jvi.72.1.366-379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grotzinger T, Jensen K, Will H. The interferon (IFN)-stimulated gene Sp100 promoter contains an IFN-gamma activation site and an imperfect IFN-stimulated response element which mediate type I IFN inducibility. J Biol Chem. 1996;271:25253–25260. doi: 10.1074/jbc.271.41.25253. [DOI] [PubMed] [Google Scholar]
- 17.Guldner H H, Szostecki C, Grotzinger T, Will H. IFN enhance expression of Sp100, an autoantigen in primary biliary cirrhosis. J Immunol. 1992;149:4067–4073. [PubMed] [Google Scholar]
- 18.Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature. 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 19.Harris A, Young B D, Griffin B E. Random association of Epstein-Barr virus genomes with host cell metaphase chromosomes in Burkitt's lymphoma-derived cell lines. J Virol. 1985;56:328–332. doi: 10.1128/jvi.56.1.328-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatfull G, Bankier A T, Barrell B G, Farrell P J. Sequence analysis of Raji Epstein-Barr virus DNA. Virology. 1988;164:334–340. doi: 10.1016/0042-6822(88)90546-6. [DOI] [PubMed] [Google Scholar]
- 21.Henderson A, Ripley S, Heller M, Kieff E. Chromosome site for Epstein-Barr virus DNA in a Burkitt tumor cell line and in lymphocytes growth-transformed in vitro. Proc Natl Acad Sci USA. 1983;80:1987–1991. doi: 10.1073/pnas.80.7.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishov A M, Maul G G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishov A M, Sotnikov A G, Negorev D, Vladimirova O V, Neff N, Kamitani T, Yeh E T, Strauss III J F, Maul G G. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishov A M, Stenberg R M, Maul G G. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J Cell Biol. 1997;138:5–16. doi: 10.1083/jcb.138.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly C, Van Driel R, Wilkinson G W. Disruption of PML-associated nuclear bodies during human cytomegalovirus infection. J Gen Virol. 1995;76:288–293. doi: 10.1099/0022-1317-76-11-2887. [DOI] [PubMed] [Google Scholar]
- 26.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2343–2396. [Google Scholar]
- 27.Korioth F, Maul G G, Plachter B, Stamminger T, Frey J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res. 1996;229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- 28.Lamond A I, Earnshaw W C. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 29.Lavau C, Marchio A, Fagioli M, Jansen J, Falini B, Lebon P, Grosveld F, Pandolfi P P, Pelicci P G, Dejean A. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene. 1995;11:871–876. [PubMed] [Google Scholar]
- 30.Li H, Leo C, Zhu J, Wu X, O'Neil J, Park E J, Chen J D. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol Cell Biol. 2000;20:1784–1796. doi: 10.1128/mcb.20.5.1784-1796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lieberman P M, Berk A J. In vitro transcriptional activation, dimerization, and DNA-binding specificity of the Epstein-Barr virus Zta protein. J Virol. 1990;64:2560–2568. doi: 10.1128/jvi.64.6.2560-2568.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liptak L M, Uprichard S L, Knipe D M. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J Virol. 1996;70:1759–1767. doi: 10.1128/jvi.70.3.1759-1767.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Longnecker R. Molecular biology of Epstein-Barr virus. In: McCance D J, editor. Human tumor viruses. Washington, D.C.: American Society for Microbiology; 1998. pp. 135–174. [Google Scholar]
- 34.Luka J, Miller G, Jornvall H, Pearson G R. Characterization of the restricted component of Epstein-Barr virus early antigens as a cytoplasmic filamentous protein. J Virol. 1986;58:748–756. doi: 10.1128/jvi.58.3.748-756.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukonis C J, Weller S K. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J Virol. 1997;71:2390–2399. doi: 10.1128/jvi.71.3.2390-2399.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maul G G. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays. 1998;20:660–667. doi: 10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 37.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 38.Maul G G, Guldner H H, Spivack J G. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0) J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 39.Maul G G, Ishov A M, Everett R D. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 40.Maul G G, Negorev D, Bell P, Ishov A M. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J Struct Biol. 2000;129:278–287. doi: 10.1006/jsbi.2000.4239. [DOI] [PubMed] [Google Scholar]
- 41.Maul G G, Yu E, Ishov A M, Epstein A L. Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J Cell Biochem. 1995;59:498–513. doi: 10.1002/jcb.240590410. [DOI] [PubMed] [Google Scholar]
- 42.Pagano J S. Epstein-Barr virus: the first human tumor virus and its role in cancer. Proc Assoc Am Physicians. 1999;111:573–580. doi: 10.1046/j.1525-1381.1999.t01-1-99220.x. [DOI] [PubMed] [Google Scholar]
- 43.Plehn-Dujowich D, Bell P, Ishov A M, Baumann C, Maul G G. Non-apoptotic chromosome condensation induced by stress: delineation of interchromosomal spaces. Chromosoma. 2000;109:266–279. doi: 10.1007/s004120000073. [DOI] [PubMed] [Google Scholar]
- 44.Ragoczy T, Heston L, Miller G. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J Virol. 1998;72:7978–7984. doi: 10.1128/jvi.72.10.7978-7984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2397–2446. [Google Scholar]
- 46.Rowe D T. Epstein-Barr virus immortalization and latency. Front Biosci. 1999;4:D346–D371. doi: 10.2741/rowe. [DOI] [PubMed] [Google Scholar]
- 47.Saemundsen A K, Kallin B, Klein G. Effect of n-butyrate on cellular and viral DNA synthesis in cells latently infected with Epstein-Barr virus. Virology. 1980;107:557–561. doi: 10.1016/0042-6822(80)90326-8. [DOI] [PubMed] [Google Scholar]
- 48.Sarisky R T, Gao Z, Lieberman P M, Fixman E D, Hayward G S, Hayward S D. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J Virol. 1996;70:8340–8347. doi: 10.1128/jvi.70.12.8340-8347.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seeler J S, Dejean A. The PML nuclear bodies: actors or extras? Curr Opin Genet Dev. 1999;9:362–367. doi: 10.1016/s0959-437x(99)80054-9. [DOI] [PubMed] [Google Scholar]
- 50.Smith K P, Moen P T, Wydner K L, Coleman J R, Lawrence J B. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J Cell Biol. 1999;144:617–629. doi: 10.1083/jcb.144.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Speck S H, Chatila T, Flemington E. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 1997;5:399–405. doi: 10.1016/S0966-842X(97)01129-3. [DOI] [PubMed] [Google Scholar]
- 52.Spector D L, Fu X D, Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spector D L, Goldman R D, Leinwand L A. Cells—a laboratory manual. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 54.Stadler M, Chelbi-Alix M K, Koken M H, Venturini L, Lee C, Saib A, Quignon F, Pelicano L, Guillemin M C, Schindler C, et al. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene. 1995;11:2565–2573. [PubMed] [Google Scholar]
- 55.Stow N D, Stow E C. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 56.Stuurman N, de Graaf A, Floore A, Josso A, Humbel B, de Jong L, van Driel R. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J Cell Sci. 1992;101:773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- 57.Szekely L, Pokrovskaja K, Jiang W Q, de The H, Ringertz N, Klein G. The Epstein-Barr virus-encoded nuclear antigen EBNA-5 accumulates in PML-containing bodies. J Virol. 1996;70:2562–2568. doi: 10.1128/jvi.70.4.2562-2568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Szostecki C, Guldner H H, Netter H J, Will H. Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J Immunol. 1990;145:4338–4347. [PubMed] [Google Scholar]
- 59.Takada K, Ono Y. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J Virol. 1989;63:445–449. doi: 10.1128/jvi.63.1.445-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takagi S, Takada K, Sairenji T. Formation of intranuclear replication compartments of Epstein-Barr virus with redistribution of BZLF1 and BMRF1 gene products. Virology. 1991;185:309–315. doi: 10.1016/0042-6822(91)90778-a. [DOI] [PubMed] [Google Scholar]
- 60a.Tang Q, Bell P, Tegtmeyer P, Maul G G. Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J Virol. 2000;74:9694–9700. doi: 10.1128/jvi.74.20.9694-9700.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trescol-Biemont M C, Biemont C, Daillie J. Localization polymorphism of EBV DNA genomes in the chromosomes of Burkitt lymphoma cell lines. Chromosoma. 1987;95:144–147. doi: 10.1007/BF00332187. [DOI] [PubMed] [Google Scholar]
- 62.Uprichard S L, Knipe D M. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology. 1997;229:113–125. doi: 10.1006/viro.1996.8430. [DOI] [PubMed] [Google Scholar]
- 63.Wilkinson G W, Kelly C, Sinclair J H, Rickards C. Disruption of PML-associated nuclear bodies mediated by the human cytomegalovirus major immediate early gene product. J Gen Virol. 1998;79:1233–1245. doi: 10.1099/0022-1317-79-5-1233. [DOI] [PubMed] [Google Scholar]
- 64.Xing Y, Johnson C V, Moen P T, Jr, McNeil J A, Lawrence J. Nonrandom gene organization: structural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J Cell Biol. 1995;131:1635–1647. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zalani S, Holley-Guthrie E, Kenney S. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc Natl Acad Sci USA. 1996;93:9194–9199. doi: 10.1073/pnas.93.17.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zerby D, Chen C J, Poon E, Lee D, Shiekhattar R, Lieberman P M. The amino-terminal C/H1 domain of CREB binding protein mediates Zta transcriptional activation of latent Epstein-Barr virus. Mol Cell Biol. 1999;19:1617–1626. doi: 10.1128/mcb.19.3.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhong L, Hayward G S. Assembly of complete, functionally active herpes simplex virus DNA replication compartments and recruitment of associated viral and cellular proteins in transient cotransfection assays. J Virol. 1997;71:3146–3160. doi: 10.1128/jvi.71.4.3146-3160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhong S, Muller S, Ronchetti S, Freemont P S, Dejean A, Pandolfi P P. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–2752. [PubMed] [Google Scholar]
- 69.Zhong S, Salomoni P, Pandolfi P P. The transcriptional role of PML and the nuclear body. Nat Cell Biol. 2000;2:E85–E90. doi: 10.1038/35010583. [DOI] [PubMed] [Google Scholar]
- 70.zur Hausen H, O'Neil F J, Freese U K, Hecker E. Persisting oncogenic herpesvirus induced by the tumor promotor TPA. Nature. 1978;272:373–375. doi: 10.1038/272373a0. [DOI] [PubMed] [Google Scholar]