Abstract
Vaginal lactobacilli protect against bacterial vaginosis and vaginal candidiasis. They may have probiotic properties and help maintain the balance and health of the vaginal ecosystem while the loss of these bacteria predisposes females to urinary and genital infections. The aim of this study was to investigate the probiotic potential of vaginal Lactobacillus among healthy females in northern Iran. The Lactobacillus strains were isolated from vaginal samples and were identified by sequencing of the 16S rRNA fragment. Functional properties such as tolerance to low pH, H2O2 production, adherence ability to Hela cells and antagonistic activity against Candida albicans was examined. A total of 38 vaginal lactobacilli strains from five species, including Lactobacillus crispatus (n = 13), Lactobacillus gasseri (n = 10), Lactobacillus acidophilus (n = 6), Lactobacillus jensenii (n = 5) and Lactobacillus johnsonii (n = 4), were identified. All of the species showed significant tolerance to low pH over 24 h (p < 0.001). The best adherence ability to Hela cells was seen in Lactobacillus gasseri strains. Nearly 17 of the strains had higher anti-candida activity compared to the other strains. According to the findings, four lactobacilli strains isolated in the vaginal samples of healthy Iranian women had the best probiotic potential.
Keywords: Lactobacillus, Probiotic, Vaginal, Candida albicans, Hela cells
Introduction
Over 50 different microbial species have been identified in the human vagina that promote female health by protecting against pathogens, modulating immunity, preventing infection, and maintaining the pH below 4.7 [1]. A variety of Lactobacillus species have been identified as the predominant biota in the vagina, and over 70% of all bacteria in the vagina of healthy females are identified as lactobacilli [2, 3]. The most common species in the vagina include L. jensenii, L. johnsonii, L. gasseri, L. iners, and L. crispatus, which protect the vaginal mucosa against pathogenic microorganisms, sexually transmitted infections (STIs), bacterial vaginitis, vulvovaginal candidiasis and Neisseria gonorrhoeae [4]. This protection is mainly due to the adherence of lactobacilli to vaginal epithelial cells, thus preventing pathogenic growth. Lactobacillus species use various mechanisms to achieve this property, such as immunomodulation, production of antimicrobial substances such as organic acids, hydrogen peroxide and bacteriocins, and competition with pathogens for food supplies [5, 6]. Also, Lactobacillus species inhibit the adhesion of pathogens to the receptors of epithelial cells through the removal or production of biofilm [7].
Probiotics are known as living microorganisms that, if consumed sufficiently, create positive effects on the health of the host. In the last 15 years, research on probiotic bacteria has increased, and attention has been paid to their use as medicine or in nutrition [8].
Vaginal lactobacilli may have probiotic properties and are known to be responsible for maintaining the balance and health of the vaginal ecosystem, and the loss of these bacteria makes females prone to urinary and genital infections [6]. Therefore, the use of selected lactobacilli may be effective in improving the vaginal microbiota and preventing infection [9]. Researchers have also discussed the effective role of probiotic bacteria in the prevention and treatment of cancer, including uterine cancer, and are further investigating the subject [10]. The ability to inhibit the growth of Hela cells, which originate from the cervical epithelial cells, is one of the anti-cancer properties of Lactobacillus that has been utilized in recent years [11]. In addition, due to the increase in fungal vaginal infections, including Candida albicans in the vagina, new effective therapies are being investigated [12]. A probiotic-like product such as intravaginal lactobacilli as a medicine alone or as a supplement may inhibit Candida albicans infection [13, 14]. Therefore, this study was conducted to examine the probiotic potential of vaginal Lactobacillus, their ability for adherence to Hela cells and their antagonistic activity against Candida albicans among healthy females in northern Iran.
Materials and Methods
Study Groups and Sampling
One hundred healthy females referred to gynecology clinics for routine gynecological consultations in Amol, Iran, were recruited for this study. The inclusion criteria were: Age 22–50 years (premenopausal), no history of sexually transmitted diseases, diabetes, immunodeficiency, vaginitis or vaginal candidiasis infection, and not being menopausal or having vaginal bleeding. The exclusion criteria were: Pregnancy, use of antibiotics or antifungal compounds, vaginal medications or suppositories or contraceptive spermicides over the past 30 days, smoking, high blood pressure and alcohol consumption. A questionnaire was designed to collect the subjects’ demographic information, such as age, history of pregnancy, history of abortion, number of births, type of delivery, and abstinence from antibiotics and antifungals over the 2 months preceding sampling, history of diabetes, and blood pressure. Sterile swabs were used for sampling the exocervix and the side of the vagina; then, the swab was put into MRS broth medium (Liofilchem, Italy) and immediately incubated at 37 °C for 24 h.
Purification of Lactobacillus Isolates
After incubation, the samples were cultured on MRS agar (Liofilchem, Italy) and incubated at 37 °C in an anaerobic jar using Anaerocult C (Merck, Germany) for 24–48 h. Then, gram reaction, morphology, and catalase test were used to identify and select Lactobacillus single colonies grown on MRS agar (Liofilchem, Italy). Total gram-positive bacilli and negative catalase test results were identified as lactobacilli and stored in MRS broth containing 20% (w/v) glycerol at − 20 °C for further investigations. The phenotypic characteristic of some vaginal samples was yeast or cocci, and Lactobacillus did not grow after 48–72 h on MRS agar incubation at 37 °C; therefore, they were excluded from the study.
Molecular Identification of Lactobacillus Isolates
Bacterial DNA was extracted from all the isolated strains using High Pure Template PCR extraction kit (Roche, Germany). The quality and quantity of the extracted DNA were checked with a nanodrop device and electrophoresis on 0.8% agarose gel.
The sequences of the 16S ribosomal RNA intergenic spacer region of phylogenetically related species were retrieved from GenBank (www.ncbi.nlm.nih.gov). DNA fragments encoding 16S rRNA were amplified using specific primer sequences; forward (5′-CTCAAAACTAAACAAAGTTTC-3′) and reverse (5′-CTTGTACACACCGCCCGTCA-3′) primers, which confirmed Lactobacillus genus [15]. The PCR reaction mix with a total volume of 25 µl consisted of distilled water) 17 µl(, buffer )2.5 µl(, Mgcl2) 1.25 µl(, dNTP) 0.4 µl (, 0.25 µl TaqDNA polymerase, 0.8 µl forward, 0.8 µl reverse primer and 2 µl of DNA. A DNA-free vial was used as a negative control. PCR was performed by thermocycler SimpliAmp-ABI (USA) using the following schedule: Initial denaturation at 95 °C for 5 min followed by 35 cycles, each consisting of denaturation at 95 °C for 1 min, annealing at 55 °C for 1 min, extension at 72 °C for 1 min and a final elongation step at 72 °C for 5 min. The PCR product was electrophoresed in a 1% (W/V) agarose gel in TBE buffer at 100 V for 45 min and stained with ethidium bromide, and the gel was visualized with a UV transilluminator (UVitec, UK). A 100-bp molecular mass marker (SinaClon, Iran) was used for assessing the size of the PCR products.
Sequencing
The PCR product was sent to Niagen Noor Company (Tehran, Iran) along with the 16S rRNA primer for sequencing. The chromatograms were edited using Chromas version 3.1 software. After sequencing the amplified fragment of the 16S rRNA gene, the isolated strains were compared with other sequences in GenBank on NCBI website using BLAST software.
Bile Salt Tolerance Assay
For this test, MRS broth medium (Liofilchem, Italy) supplemented with 0.3% Deoxycholate bile salt (Merck, Germany) was used and 100 µl of the full-grown culture isolates were added to 10 ml of this medium. Then, the cells were incubated at 37 °C and optical densities (OD 600 nm) were measured at 1, 2 h, 4 h, and 24 h by a spectrophotometer. The culture medium was used without oxgall as the control. All measurements were performed two times and the average of two replicates was calculated [16, 17].
Tolerance to Low pH
In order to investigate the resistance of lactobacilli, 1 M HCL was added to MRS broth before autoclaving and the sterile MRS broth medium was prepared with pH of 2, 2.5, 3, 4 and 6; then, (1% v/v) full-grown culture isolates were inoculated in these mediums and incubated at 37 °C. Their optical densities (OD 620 nm) were measured at 1, 2, 4, and 24 h by a spectrophotometer. The experiment was repeated twice and each reading represents the average of two replicates [18].
Hemolysis Activity Test
For the hemolysis test, all the Lactobacillus isolates were cultured (in MRS agar medium for 48 h at 37 °C in anaerobic conditions) and then a single colony isolated from Lactobacillus was cultured on blood agar medium (Merck, Germany) containing sheep blood 5% and incubated in anaerobic conditions using Anaerocult C for 24–48 h at 37 °C. The hemolytic activity was visually detected and distinguished as β-hemolysis, α-hemolysis and γ-hemolysis based on the appearance of a clear zone, green halo zone and no zone around the colonies. Streptococcus pyogenes (ATCC 19615) was used as the positive control. After incubation, hemolysis around the isolates was recorded [19].
H2O2 Production
For this test, the strains are cultured on MRS agar containing 0.25 mg/mL of 3,3′,5,5′-tetramethylbenzidine (Merck, Germany) and 0.01 mg/mL of horseradish peroxidase (Sigma-Aldrich, USA) in anaerobic conditions for 48–72 h. The plates were exposed to air for 30 min and hydrogen peroxide was evaluated based on the time required for a blue coloration appearance. The strains were scored as weak (score of 1, time ≥ 20 min), medium (score of 2, time 10–20 min) and strong (score of 3, time ≤ 10 min) H2O2 producer and a score of 0 meant not producing the blue coloration [20, 21].
Evaluation of Antagonistic Activity
Antagonistic Activity of Lactobacilli Against Urinary Pathogens by Spot Agar Assay
To investigate the antimicrobial activity of Lactobacillus isolates, Lactobacillus isolates were first cultured in spot form on MRS agar medium by sterile loop, and then incubated anaerobically at 37 °C for 48 h. Afterwards, E. coli ATCC 25922 and two strains of E. coli isolated from females with urinary tract infections and identified by phenotypic tests were used for this exam. Later, overnight-cultured E. coli strains (approximately 7 × 108 CFU) were mixed in the nutrient agar medium (Merck, Germany) and poured on MRS agar medium which Lactobacillus isolates that had been previously cultured. Afterwards, the plates were incubated in anaerobic conditions at 37 °C for 24 h. Finally, the zone of inhibition of the growth of E. coli around the Lactobacillus isolates was measured [22].
Antagonistic Activity of Lactobacilli Against Candida albicans by Agar Overlay Method
Anticandidal activity was assessed using the modified agar overlay technique; briefly, 0.5 McFarland turbidity of the Candida albicans (ATCC 10231) that had grown on Sabouraud agar medium (Merck, Germany) was prepared, and the isolated lactobacilli were cultured in MRS broth for 24 h at 37 °C in anaerobic conditions. Afterwards, the Lactobacillus strains were cultured in the middle of MRS agar using sterile cotton swabs for culturing the bacteria and were then incubated for 48 h at 37 °C in anaerobic conditions. The Lactobacillus growth was then overlaid with melted MRS agar and the suspension of Candida albicans was streaked over the agar with solidification. After culturing, the plates were placed in the refrigerator for 4 h at a temperature of 4 °C and then in the incubator at 37 °C for 24 h. The plates were later kept at room temperature for 24 h and the antagonistic activity was measured and recorded as follows: ≥ 20 mm was considered sensitive to Lactobacillus, between 10 and 15 mm was semi-sensitive, and less than 5 mm lacked antagonistic activity to Lactobacillus strains [22].
Adherence to Hela Cells
To evaluate the adhesion of lactobacilli to Hela cells, a total of 3 × 105 Hela cells were cultured in DMEM medium with fetal bovine serum 10% at 37 °C with 5% CO2. After 24 h of incubation, the Hela cells were mixed with 1 ml of Lactobacillus (1.5 × 109 cells/ml) at 37 °C for 3 h. The cells were washed twice with phosphate buffered saline and the unattached cells were removed. For the quantitative assessment of bacterial cell adhesion, the remaining cells were washed with phosphate buffered saline and lysed with 1% Triton × 100 and then centrifuged at 2200g for 5 min. The supernatant was discarded, the pellets were re-suspended, and serial dilutions were prepared in phosphate buffered saline and an appropriate dilution was cultured on MRS agar medium for 48 h at 37 °C. The grown colonies were counted and taken as the adherent bacteria. For this test, we used L. brevis ATCC 367 as the control. The following equation was used to calculate the percentage of adherent cells [23].
N1: The amount of adhered bacteria (log10 CFU/ml)
N0: The amount of applied bacteria (log10 CFU/ml)
Adherent cells %: N1/N0 × 100
Statistical Analysis
Data analysis was performed in SPSS, version 21. The antagonistic activity of lactobacilli on Candida albicans and the ability of lactobacilli to bind to Hela cells were examined and the results were then compared using the one-way ANOVA. The one-way ANOVA and repeated-measures analysis were used to compare the pH tolerance and Bile salt tolerance over time and across the Lactobacillus species. In addition, a Chi-square test was used to examine the relationship between the antagonistic activity against Candida albicans and Lactobacillus species.
Results
Isolation of Lactobacillus Strains from the Vagina
The isolated Lactobacillus strains cultured on MRS agar medium were investigated and the phenotype properties, such as colony color (white to milky), were recorded. A total of 38 vaginal Lactic acid bacteria isolated from reproductive-aged women were selected. They were gram-positive, rod shaped, catalase negative. All the isolated vaginal Lactobacillus strains were finally identified by sequencing the 16S rRNA gene and compared with sequences available in GenBank database using BLAST software at NCBI and they were found to belong to five species.
Nucleotide Sequence Accession Number
The NCBI GenBank accession numbers of the spacer sequences used in this study showed in Table 1.
Table 1.
NCBI GenBank accession numbers of Lactobacillus strains sequences
| Accession number | NCBI GenBank name | Name | Isolates lable | NCBI GenBank strain name | Sequence length in bp | BLAST similarity% |
|---|---|---|---|---|---|---|
| OR220801 | Lactobacillus sp | L. crispatus | C2 | C2HZA | 196 | 99 |
| OR220199 | Lactobacillus sp | L. crispatus | C3 | C3HZA | 197 | 98 |
| OR220200 | Lactobacillus sp | L. crispatus | C4 | C4HZA | 215 | 92 |
| OR220201 | Lactobacillus sp | L. crispatus | C5 | C5HZA | 181 | 99 |
| OR220202 | Lactobacillus sp | L. crispatus | C6 | C6HZA | 194 | 99 |
| OR220203 | Lactobacillus sp | L. crispatus | C7 | C7HZA | 195 | 97 |
| OR229087 | Lactobacillus sp | L. crispatus | C8 | C8HZA | 190 | 96 |
| OR229088 | Lactobacillus sp | L. crispatus | C9 | C9HZA | 190 | 96 |
| OR229089 | Lactobacillus sp | L. crispatus | C11 | C11HZA | 185 | 94 |
| OR229090 | Lactobacillus sp | L. crispatus | C12 | C12HZA | 190 | 96 |
| OR220204 | Lactobacillus sp | L. crispatus | C13 | C13HZa | 221 | 92 |
| OR220205 | Lactobacillus sp | L. gasseri | G5 | G5HZA | 212 | 98 |
| OR220195 | Lactobacillus sp | L. gasseri | G6 | G6HZA | 212 | 88 |
| OR220196 | Lactobacillus sp | L. gasseri | G7 | G7HZA | 241 | 97 |
| OR220197 | Lactobacillus sp | L. gasseri | G8 | G8HZA | 238 | 98 |
| OR220198 | Lactobacillus sp | L. gasseri | G10 | G10HZA | 231 | 94 |
| OR220567 | Lactobacillus sp | L. acidophilus | A1 | A1HZA | 197 | 98 |
| OR220568 | Lactobacillus sp | L. jensenii | Je2 | J2HZA | 197 | 98 |
| OR220569 | Lactobacillus sp | L. jensenii | Je3 | J3HZA | 222 | 93 |
| OR220570 | Lactobacillus sp | L. jensenii | Je5 | J5HZA | 195 | 92 |
| OR220564 | Lactobacillus sp | L. johnsonii | Jh1 | Jh1HZA | 185 | 99 |
| OR220565 | Lactobacillus sp | L. johnsonii | Jh3 | Jh3HZA | 237 | 91 |
| OR220566 | Lactobacillus sp | L. johnsonii | Jh4 | Jh4HZA | 235 | 96 |
Demographic Data
The results of the current study showed that the females who were classified in each of these Lactobacillus strains had no significant difference in age (P = 0.618). More details are shown in (Table 2).
Table 2.
Age (mean and SD) of participate based on Lactobacillus species
| Lactobacillus species | Type | Age |
|---|---|---|
| Species | N (%) | Mean ± SD |
| Total | 38 (100) | 37.5 ± 7.1 |
| L. crispatus | 13(32.5) | 35.2 ± 7.4 |
| L. gasseri | 10(25) | 39.2 ± 6.5 |
| L. acidophilus | 6(15) | 36 ± 8.2 |
| L. jensenii | 5(12.5) | 38.8 ± 6.7 |
| L. johnsonii | 4(10) | 40 ± 8.3 |
| F statistics | 0.68 | |
| P value | 0.666 |
A relationship was also found between the type of delivery (natural, caesarean, and both) and the frequency distribution of Lactobacillus strains (=19.12; P = 0.014). Having both delivery methods was observed only in the Lactobacillus johnsonii species group (Appendix 1).
Bile Salt Tolerance Results
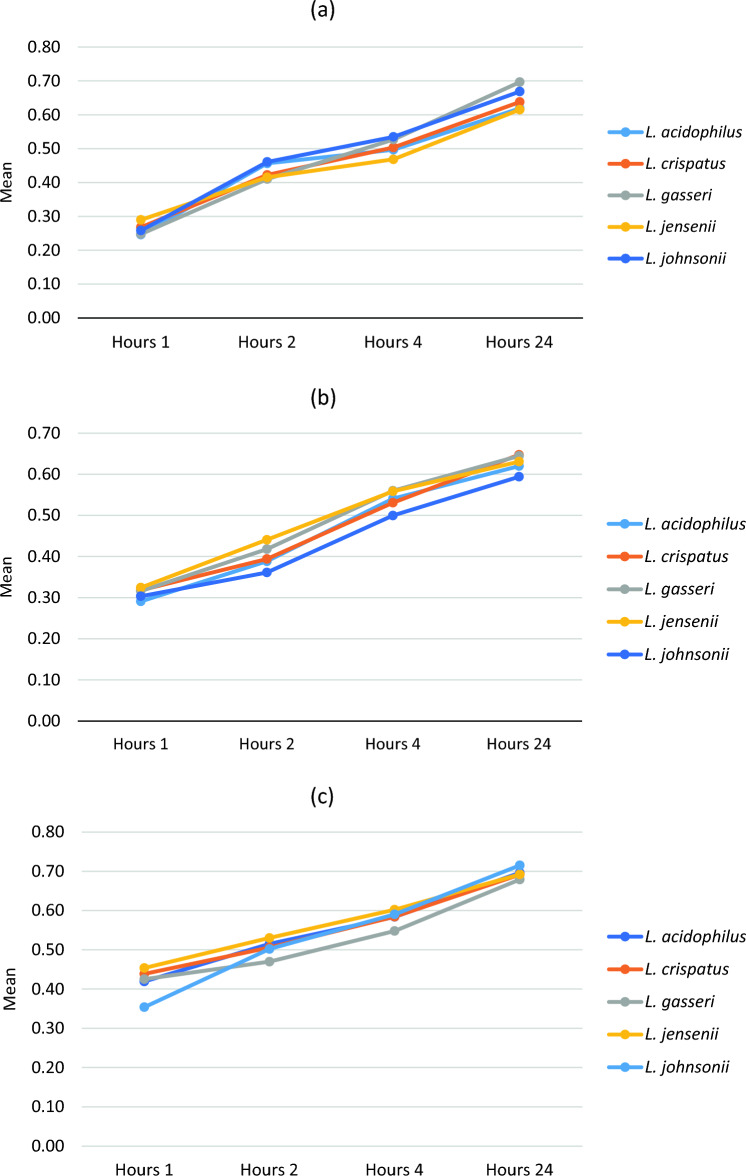
The tolerance of Lactobacillus isolates to bile salt was measured based on the optical properties of the isolates at different time points by a spectrophotometer. The results showed no significant difference among the isolated lactobacilli in terms of bile salt tolerance at each time point (Fig. 1). Nonetheless, all the strains had tolerance to bile salt over time, resulting in their increased growth at the end of 24 h (P < 0.001). The results showed that Lactobacillus crispatus C9 and C10, Lactobacillus johnsonii Jh 4 and Lactobacillus gasseri G5 had the highest growth during the incubation period. By contrast, Lactobacillus jensenii Je 1 and Lactobacillus gasseri G4 showed the lowest viability after 24 h of incubation.
Fig. 1.
Bile salt tolerance according time and concentration (expressed as Mean and SD) of all strain of Lactobacillus specific species in (OD = 600 nm)
Low pH Tolerance Test
The resistance of the isolates at different pH levels was measured based on the turbidity of the culture medium. The survival of the Lactobacillus species showed a significant difference only at pH = 2 and 3 at 0 h (P = 0.038) and 24 h (P = 0.014); however, there was no significant difference between the Lactobacillus species in terms of survival in other pH levels and different hours. Overall, for all the species at all the pH levels, a significant difference was observed in terms of tolerance at different hours, such that tolerance increased significantly as 24 h approached (P < 0.001). According to the results, at the pH of 2.0, Lactobacillus gasseri G5, G7, and G10, Lactobacillus crispatus C12 and Lactobacillus johnsonii Jh2 exhibited the highest viability during 24 h of incubation. The maximum viability at pH of 3.0 was observed in Lactobacillus crispatus C10, C12, and C7 and Lactobacillus johnsonii Jh3, followed by Lactobacillus gasseri G4, which showed the minimum survival rate. More details are shown in (Fig. 2).
Fig. 2.
pH tolerance according to pH levels, time and concentration (expressed as Mean) of all strains of Lactobacillus specific species in (OD = 620nm), a pH = 2, b pH = 2.5, c pH = 3, d pH = 4, e pH = 6, L. acidophilus (n = 6), L. crispatus (n = 13), L. gasseri (n = 10), L. jensenii (n = 5), L. johnsonii (n = 4)
Hemolysis Results
After 24–48 h of incubation of the Lactobacillus strains, the hemolysis zone around the grown bacillus colonies was examined. None of the Lactobacillus strains caused lysis of sheep erythrocytes on blood agar (γ-hemolysis) while complete lysis (β-hemolysis) was observed in the case of the positive controls (Streptococcus pyogenes).
Hydrogen Peroxide Production
The qualitative analysis demonstrated that four strains of Lactobacillus crispatus (C1, C3, C7, and C13) and two strains of Lactobacillus jensenii (Je2 and Je3) showed moderate H2O2 production, and five strains of L. gasseri (G1, G5, G6, G9, and G10) exhibited low ability to produce hydrogen peroxide, and the other strains did not produce any H2O2.
Adherence to Hela Cells Results
The results of the current research revealed that all the strains had the ability to adhere to Hela cells. Moreover, Hela cell adherence showed a significant difference between different Lactobacillus strains compared to the controls (for L. brevis ATCC 367, the average adherence to Hela cells was 95%), but no significant difference was found among the Lactobacillus strains to each other (P value = 0.099), and the highest and lowest adherence percentages were observed in L. gasseri and L. jensenii species, respectively (Table 3). The strains shown below had 75% to 95% adherence ability to Hela cells: Lactobacillus gasseri G1, G2, G3, G4, G5, G6, G7, and G10, Lactobacillus crispatus C1, C3, C4, C5, C7, C12, and C13, Lactobacillus acidophilus A1, A2, A5, and A6, Lactobacillus jensenii Je1 and Lactobacillus johnsonii Jh1.
Table 3.
Adhesion percentage to Hela cells in Lactobacillus species by one way ANOVA
| Lactobacillus sp | Mean ± SD |
|---|---|
| L. acidophilus (n = 6) | 0.73 ± 0.11 |
| L. crispatus (n = 13) | 0.66 ± 0.25 |
| L. gasseri (n = 10) | 0.77 ± 0.20 |
| L. jensenii (n = 5) | 0.46 ± 0.22 |
| L. johnsonii (n = 4) | 0.56 ± 0.21 |
| F statistics | 2.12 |
| P value | 0.099 |
L. brevis ATCC 367 excluded from ANOVA test for comparing the means
Antagonistic Activity of Lactobacillus Isolates Against Candida albicans
The antagonistic activity of Lactobacillus isolates against Candida albicans was examined by measuring the range of non-growth of Candida albicans in the vicinity of Lactobacillus strains, and no significant difference was found between the Lactobacillus species with respect to their antagonistic activity on Candida albicans (P value = 0.930), (Fig. 3). The highest (≥ 20 mm) antagonistic activity against Candida albicans was observed in 17 strains, as below: Lactobacillus crispatus C1, C2, C6, C7, C8, and C12, Lactobacillus gasseri G2, G3, G5, and G6, Lactobacillus jensenii Je2, Je3, and Je4, Lactobacillus acidophilus A1, A4, and A5 and Lactobacillus johnsonii Jh3.
Fig. 3.
Antagonistic activity against Candida albicans in Lactobacillus species (χ2 (df = 8) = 3.067; P value = 0.930)
Antagonistic Activity Against E. coli Strains
The antimicrobial activity of Lactobacillus against different strains of E. coli showed that there was no significant difference between the isolated strains in this regard (P > 0.05). The findings also revealed that among the Lactobacillus species, L. crispatus showed more inhibition of growth compared to the other species. More details are shown in Table 4.
Table 4.
Antagonistic activity against E. coli strains in all strains of Lactobacillus specific species (inhibition zone in mm, expressed as Mean ± SD), L. acidophilus (n = 6), L. crispatus (n = 13), L. gasseri (n = 10), L. jensenii (n = 5), L. johnsonii (n = 4)
| Lactobacillus species |
E. coli strain isolated from urinary infection1 (Mean ± SD) |
E. coli strain isolated from urinary infection 2 (Mean ± SD) |
E. coli ATCC 25922 (Mean ± SD) |
|---|---|---|---|
| L. acidophilus | 7.5 ± 5.9 | 8.7 ± 5.3 | 8.2 ± 6.4 |
| L. crispatus | 9 ± 3.2 | 9.3 ± 3.4 | 10.6 ± 2.6 |
| L. gasseri | 9.9 ± 1.9 | 10.6 ± 1.8 | 10.6 ± 2.2 |
| L. jensenii | 6.6 ± 3.9 | 6.4 ± 3.6 | 7.4 ± 4.3 |
| L. johnsonii | 9.3 ± 2.5 | 10 ± 2.4 | 10 ± 2.8 |
| Total | 8.3 ± 3.9 | 8.8 ± 3.9 | 9.3 ± 4.1 |
| Fa | 0.959 | 1.365 | 1.15 |
| P value | 0.443 | 0.267 | 0.351 |
aF statistics of one-way ANOVA test
Discussion
In this study, L. crispatus and L. gasseri were documented to be the most common vaginal species among women in northern Iran. Martinez et al. determined the most dominant species of Lactobacillus in the vagina to be L. gasseri, L. crispatus and L. jensenii among Mexican women [24]. Moreover, Vitali et al. [25] found that L. acidophilus, L. gasseri, L. vaginalis, and L. iners were the most common species in the vaginal microbiota among healthy Italian women. Overall, various factors, such as individual genetics, nutrition, personal hygiene and sexual activity, can be effective in the colonization of certain species of lactobacilli in the vagina among healthy women [26, 27].
Probiotics are defined as “Live microorganisms that, when administered in sufficient amounts, have beneficial effects on human health [28]. Genitourinary lactobacilli are known as probiotic agents that protect the vagina against invading pathogens that are responsible for urinary tract infections or sexually transmitted diseases [19]. The survival potential of lactobacilli in low pH and bile salts is very important, because probiotics confront a range of gastrointestinal stresses that intensify in the stomach; therefore, resistance to stomach acid and bile salts ensures the successful administration of probiotics [29]. In the study by Kiray et al. [29], it was observed that none of the strains were able to tolerate pH = 2; however, the survival of the strains at pH = 2.5 was equal to 67.6%, and this ratio increased to 89.3% at pH = 3. Our results showed that L. gasseri species had the highest percentage of survival at pH = 2 for 4 h. It has been found that some strains can maintain a high viability in stomach pH and thus maintain sufficient numbers to compete and bind with pathogenic bacteria when they reach the digestive tract [30]. Bile salt tolerance is another essential criterion for selecting probiotic strains. Bile salts play an important and fundamental role in the specific defense of the intestine by destroying microorganisms. Therefore, investigating the viability of the strain in different concentrations of bile salts is necessary to prove the probiotic potential [17, 31]. The resistance of Lactobacilli to different concentrations of salt indicates their higher stability [32]. In our study, Lactobacillus species tolerated 0.3% bile salt for 24 h.
One of the most important features of probiotics is the production of compounds, such as short-chain fatty acids and bacteriocins, which lead to antimicrobial properties against pathogens. In the case of lactobacilli, the presence of compounds such as bacteriocins, hydrogen peroxide and organic acids and competition for nutrients with pathogenic microorganisms lead to antimicrobial properties [21, 29]. Vaginalis Lactobacillus strains control vaginal microorganisms, including Candida albicans, by colonizing the vaginal epithelium and inhibiting the growth of other microorganisms. Therefore, as candidates for vaginal probiotics, Lactobacillus strains are usually tested in vitro for their ability to adhere to the vaginal epithelium and their antimicrobial activity against Candida albicans [29]. The species L. crispatus and L. jensenii are high H2O2 producers compared to L. gasseri [33]. These results are in accordance with the findings of the present study. The production of H2O2 is one of the most important defense mechanisms against the colonization of pathogenic or opportunistic microorganisms by vaginal Lactobacillus [34]. Absence of H2O2 producing Lactobacillus increases the risk of bacterial vaginosis (BV), such as recurrent urinary tract infection by E. coli [22, 32, 34]. Kiray et al. investigated the antimicrobial activity of vaginal probiotics and the antagonistic effect of the strains on pathogenic microorganisms causing urinary tract and genital tract infections. The results of their meta-analysis showed that vaginal lactobacilli are effective in treating bacterial vaginosis (BV) and vulvovaginal candidiasis (VVC) [29]. Hutt et al. showed that L. crispatus species had significantly higher anti-microbial activity compared to L. jensenii, while L. gasseri species showed lower anti-Candida activity compared to L. crispatus. and L. jensenii species [22]. Our results were in line with these findings, showing the antagonist effect of vaginal lactobacilli strains against Candida albicans. Only a small number of Lactobacillus species were found to gather in clusters and prevent the growth of pathogenic agents. The aggregation of lactobacilli species with Candida strains is very important in protecting against vaginal yeast infections, because lactobacilli can create a small environment around pathogens through co-aggregation, and by synthesizing large amounts of inhibitory factors, they can prevent the pathogen microorganisms from distribution [33, 35]. Moreover, one of the most important tests in potential probiotics is the adhesion of the probiotic microorganism to mucin and human epithelial cells. The attachment of lactobacilli to vaginal epithelial cells is defined as the first step in the formation of a barrier to prevent the colonization of opportunistic pathogenic microorganisms [34, 36]. Various models, including Caco-2 and HT29 cells, have been used to evaluate the ability of bacteria to attach to cells in vitro [13, 22]. In the present study, Hela cells were used to evaluate the adhesion capacity of vaginal lactobacilli. As reported in previous studies, different species of lactobacilli have different abilities to attach to the cells [37]. Similarly, a lectin-like protein in the cell wall of L. plantarum may play a role in adhesion [38]. In pervious literature, Lactobacillus surface factors, such as lipoteichoic acid of L. johnsonii and S-layer protein of L. crispatus, were reported to play roles in adhesion to epithelial cells. They may be involved in sticking to epithelial cells. Therefore, different mechanisms could exist for different Lactobacillus strains to adhere to epithelial cells [13, 39]. In the present study, the highest percentage of adhesion to Hela cells was seen in L. gasseri species; in contrast, Mousavi et al. [23] reported that L. rhamnosus species adhered to the epithelial cells to a greater degree compared to other Lactobacillus species and L. gasseri, which showed weak adherence.
Conclusion
Based on the results, each Lactobacillus strain isolated from the vagina of healthy Iranian women had different probiotic properties. In particular, four Lactobacillus strains (C7, C12, G5 and G10) exhibited stronger potential probiotic characteristics such as tolerance to a low pH, H2O2 production, adherence to Hela cells and better antagonistic activity against Candida albicans and E. coli strains; there fore, they are effective in maintaining vaginal health and preventing infections of the genitourinary system. Each vaginal strain is specific and needs to be clinically well investigated through in vitro and in vivo studies.
Acknowledgements
Authors would like to thank staff of Amol Pasture Institute.
Appendix
See Table 5.
Table 5.
The relationship between delivery types and Lactobacillus species frequencies for study sample
| Delivery types | Total | |||
|---|---|---|---|---|
| Only natural | Only caesarean | Both | ||
| n (%) | n (%) | n (%) | ||
| L. acidophilus | 2 (33.3) | 4 (66.7) | 0 (0) | 6 (100) |
| L. crispatus | 5 (38.5) | 8 (61.5) | 0 (0) | 13 (100) |
| L. gasseri | 5 (50.0) | 5 (50.0) | 0 (0) | 10 (100) |
| L. jensenii | 3 (60.0) | 2 (40.0) | 0 (0) | 5 (100) |
| L. johnsonii | 1 (25.0) | 1 (25.0) | 2 (50) | 4 (100) |
| Total | 16 (42.1) | 20 (52.6) | 2 (5.3) | 38 (100) |
= 19.12; P value = 0.014
Author Contributions
All authors contributed to the study conception and design and experiments. Material preparation, performed experiments, data collection and analysis were performed by HZ, RIA, MR, HK. The first draft of the manuscript was written by HZ. FZ Supervised, directed and managed the study. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of Data and Materials
Not applicable.
Code Availability
Not applicable.
Declarations
Conflict of interest
The authors declare that there are no conflict of interest.
Ethical Approval
The study protocol was approved by the Ethics Committee of Amol Azad University (ID: IR.IAU.AMOL.REC.1400.051). At the beginning, all study participants were given adequate information about the study. Then informed consent was obtained.
Consent to Participate
The procedure was explained to all the recruited participate who signed written informed consent forms.
Consent for Publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nami Y, et al. Assessment of probiotic potential and anticancer activity of newly isolated vaginal bacterium Lactobacillus plantarum 5BL. Microbiol Immunol. 2014;58:492–502. doi: 10.1111/1348-0421.12175. [DOI] [PubMed] [Google Scholar]
- 2.Srinivasan S, Fredricks DN. The human vaginal bacterial biota and bacterial vaginosis. Interdiscip Perspect Infect Dis. 2008 doi: 10.1155/2008/750479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmadi M, et al. Analysis of cervical lesions for presence of HSV-2 and HPV-16 and HPV-18 in Iranian patients by PCR. Horm Mol Biol Clin Investig. 2017;31:20170019. doi: 10.1515/hmbci-2017-0019. [DOI] [PubMed] [Google Scholar]
- 4.Chee WJY, Chew SY, Than LTL. Vaginal microbiota and the potential of Lactobacillus derivatives in maintaining vaginal health. Microb Cell Fact. 2020;19:203. doi: 10.1186/s12934-020-01464-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santos CM, et al. Selection of Lactobacillus strains as potential probiotics for vaginitis treatment. Microbiology. 2016;162:1195–1207. doi: 10.1099/mic.0.000302. [DOI] [PubMed] [Google Scholar]
- 6.Borges S, Silva J, Teixeira P. The role of lactobacilli and probiotics in maintaining vaginal health. Arch Gynecol Obstet. 2014;289:479–489. doi: 10.1007/s00404-013-3064-9. [DOI] [PubMed] [Google Scholar]
- 7.Petrova MI, et al. Lactobacillus species as biomarkers and agents that can promote various aspects of vaginal health. Front Physiol. 2015;6:81. doi: 10.3389/fphys.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kligler B, Cohrssen A. Probiotics. Am Fam Phys. 2008;78:1073–1078. [PubMed] [Google Scholar]
- 9.Witkin SS, Linhares IM. Why do lactobacilli dominate the human vaginal microbiota? BJOG Int J Obstet Gynaecol. 2017;124:606–611. doi: 10.1111/1471-0528.14390. [DOI] [PubMed] [Google Scholar]
- 10.Champer M, et al. The role of the vaginal microbiome in gynaecological cancer. BJOG Int J Obstet Gynaecol. 2018;125:309–315. doi: 10.1111/1471-0528.14631. [DOI] [PubMed] [Google Scholar]
- 11.Motevaseli E, Dianatpour A, Ghafouri-Fard S. The role of probiotics in cancer treatment: emphasis on their in vivo and in vitro anti-metastatic effects. Int J Mol Cell Med. 2017;6:66. doi: 10.22088/acadpub.BUMS.6.2.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson MD. Changing patterns and trends in systemic fungal infections. J Antimicrob Chemother. 2005;56:i5–i11. doi: 10.1093/jac/dki218. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, et al. The S-layer proteins of Lactobacillus crispatus strain ZJ001 is responsible for competitive exclusion against Escherichia coli O157: H7 and Salmonella typhimurium. Int J Food Microbiol. 2007;115:307–312. doi: 10.1016/j.ijfoodmicro.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Chen C-C, Walker WA. Probiotics and prebiotics: role in clinical disease states. Adv Pediatr. 2005;52:77–113. doi: 10.1016/j.yapd.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Dubernet S, Desmasures N, Guéguen M. A PCR-based method for identification of lactobacilli at the genus level. FEMS Microbiol Lett. 2002;214:271–275. doi: 10.1111/j.1574-6968.2002.tb11358.x. [DOI] [PubMed] [Google Scholar]
- 16.Gu XC, et al. Cloning and analysis of bile salt hydrolase genes from Lactobacillus plantarum CGMCC No. 8198. Biotechnol Lett. 2014;36:975–983. doi: 10.1007/s10529-013-1434-9. [DOI] [PubMed] [Google Scholar]
- 17.Corzo G, Gilliland S. Bile salt hydrolase activity of three strains of Lactobacillus acidophilus. J Dairy Sci. 1999;82:472–480. doi: 10.3168/jds.S0022-0302(99)75256-2. [DOI] [PubMed] [Google Scholar]
- 18.Khalil R, et al. Evaluation of the probiotic potential of lactic acid bacteria isolated from faeces of breast-fed infants in Egypt. Afr J Biotechnol. 2007;6:939–949. [Google Scholar]
- 19.Al Kassaa I, et al. Identification of vaginal lactobacilli with potential probiotic properties isolated from women in North Lebanon. Microb Ecol. 2014;67:722–734. doi: 10.1007/s00248-014-0384-7. [DOI] [PubMed] [Google Scholar]
- 20.Pino A, et al. Detection of vaginal lactobacilli as probiotic candidates. Sci Rep. 2019;9:3355. doi: 10.1038/s41598-019-40304-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eschenbach DA, et al. Prevalence of hydrogen peroxide-producing Lactobacillus species in normal women and women with bacterial vaginosis. J Clin Microbiol. 1989;27:251–256. doi: 10.1128/jcm.27.2.251-256.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hütt P, et al. Characterisation of probiotic properties in human vaginal lactobacilli strains. Microb Ecol Health Dis. 2016;27:30484. doi: 10.3402/mehd.v27.30484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mousavi E, et al. In vitro adherence of Lactobacillus strains isolated from the vaginas of healthy Iranian women. J Chin Med Assoc. 2016;79:665–671. doi: 10.1016/j.jcma.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 24.Martínez-Peña MD, Castro-Escarpulli G, Aguilera-Arreola MG. Lactobacillus species isolated from vaginal secretions of healthy and bacterial vaginosis-intermediate Mexican women: a prospective study. BMC Infect Dis. 2013;13:1–9. doi: 10.1186/1471-2334-13-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vitali B, et al. Dynamics of vaginal bacterial communities in women developing bacterial vaginosis, candidiasis, or no infection, analyzed by PCR-denaturing gradient gel electrophoresis and real-time PCR. Appl Environ Microbiol. 2007;73:5731–5741. doi: 10.1128/AEM.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nami Y, Haghshenas B, Khosroushahi AY. Molecular identification and probiotic potential characterization of lactic acid bacteria isolated from human vaginal microbiota. Adv Pharm Bull. 2018;8:683. doi: 10.15171/apb.2018.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta S, Kakkar V, Bhushan I. Crosstalk between vaginal microbiome and female health: a review. Microb Pathog. 2019;136:103696. doi: 10.1016/j.micpath.2019.103696. [DOI] [PubMed] [Google Scholar]
- 28.Organization WH. Food and Agriculture Organization of the United Nations. Probiotics in food. Health and nutritional properties and guidelines for evaluation. FAO Food Nutr Pap. 2006;85:2. [Google Scholar]
- 29.Kiray E, Kariptas E, Azarkan SY. Evaluation of vaginal lactobacilli with potential probiotic properties and biotherapeutic effects isolated from healthy Turkish women. PSM Microbiol. 2019;4:56–70. [Google Scholar]
- 30.Nami Y, et al. Probiotic potential and biotherapeutic effects of newly isolated vaginal Lactobacillus acidophilus 36YL strain on cancer cells. Anaerobe. 2014;28:29–36. doi: 10.1016/j.anaerobe.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 31.Gu X-C, et al. Cloning and analysis of bile salt hydrolase genes from Lactobacillus plantarum CGMCC No. 8198. Biotechnol Lett. 2014;36:975–983. doi: 10.1007/s10529-013-1434-9. [DOI] [PubMed] [Google Scholar]
- 32.Starling S (2013) Global probiotics market to grow 6.8% annually until 2018
- 33.Dover S, et al. Natural antimicrobials and their role in vaginal health: a short review. Int J Probiotics Prebiotics. 2008;3:219. [PMC free article] [PubMed] [Google Scholar]
- 34.Reid G, Dols J, Miller W. Targeting the vaginal microbiota with probiotics as a means to counteract infections. Curr Opin Clin Nutr Metab Care. 2009;12:583–587. doi: 10.1097/MCO.0b013e328331b611. [DOI] [PubMed] [Google Scholar]
- 35.Otero MC, Nader-Macías ME. Lactobacillus adhesion to epithelial cells from bovine vagina. Commun Curr Res Educ Top Trends Appl Microbiol. 2007;20007:749–757. [Google Scholar]
- 36.Kumherová M, et al. Novel potential probiotic Lactobacilli for prevention and treatment of vulvovaginal infections. Probiotics Antimicrob Proteins. 2021;13:163–172. doi: 10.1007/s12602-020-09675-2. [DOI] [PubMed] [Google Scholar]
- 37.Sengupta R, et al. The role of cell surface architecture of lactobacilli in host-microbe interactions in the gastrointestinal tract. Mediat Inflamm. 2013 doi: 10.1155/2013/237921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vladareanu R, et al. New evidence on oral L. plantarum P17630 product in women with history of recurrent vulvovaginal candidiasis (RVVC): a randomized double-blind placebo-controlled study. Eur Rev Med Pharmacol Sci. 2018;22:262–267. doi: 10.26355/eurrev_201801_14128. [DOI] [PubMed] [Google Scholar]
- 39.Kinoshita H, et al. Cell surface Lactobacillus plantarum LA 318 glyceraldehyde-3-phosphate dehydrogenase (GAPDH) adheres to human colonic mucin. J Appl Microbiol. 2008;104:1667–1674. doi: 10.1111/j.1365-2672.2007.03679.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.