Abstract
The distribution of sialic acid (SA) species varies among animal species, but the biological role of this variation is largely unknown. Influenza viruses differ in their ability to recognize SA-galactose (Gal) linkages, depending on the animal hosts from which they are isolated. For example, human viruses preferentially recognize SA linked to Gal by the α2,6(SAα2,6Gal) linkage, while equine viruses favor SAα2,3Gal. However, whether a difference in relative abundance of specific SA species (N-acetylneuraminic acid [NeuAc] and N-glycolylneuraminic acid [NeuGc]) among different animals affects the replicative potential of influenza viruses is uncertain. We therefore examined the requirement for the hemagglutinin (HA) for support of viral replication in horses, using viruses whose HAs differ in receptor specificity. A virus with an HA recognizing NeuAcα2,6Gal but not NeuAcα2,3Gal or NeuGcα2,3Gal failed to replicate in horses, while one with an HA recognizing the NeuGcα2,3Gal moiety replicated in horses. Furthermore, biochemical and immunohistochemical analyses and a lectin-binding assay demonstrated the abundance of the NeuGcα2,3Gal moiety in epithelial cells of horse trachea, indicating that recognition of this moiety is critical for viral replication in horses. Thus, these results provide evidence of a biological effect of different SA species in different animals.
Sialic acid (SA) is a generic term for nine-carbon acidic amino sugars (5-amino-3,5-dideoxy-D-glycero-D-galacto-nonulosonic acid). The amino group is always substituted with either an N-acetyl or N-glycolyl group, yielding N-acetylneuraminic (NeuAc) or N-glycolylneuraminic (NeuGc) acid, respectively (Fig. 1), while the hydroxyl groups can be substituted by acetyl, lactoyl, methyl, sulfate, or phosphate residues. The distribution of specific SAs varies among animal species. For example, bovine, equine, and swine tissues possess both NeuAc and NeuGc, whereas human tissues possess only slight concentrations of NeuGc (less than 0.1% of total SA) (21, 22). Although a difference in the distribution of SAs among different animal species has been recognized for many years (11, 22, 25, 28, 30, 34, 35, 47), its biological significance is largely unknown.
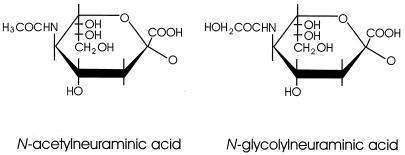
FIG. 1.
Structure of N-acetyl and N-glycolylneuraminic acids. These SA differ at position 5 of the pyranose ring. N-Acetylneuraminic acid is the precursor of N-glycolyneuraminic acid; enzymatic hydroxylation of the former results in the latter.
Influenza A viruses have been isolated from a variety of animals, including humans, pigs, horses, sea mammals, poultry, wild ducks, and other migrating waterfowl (49). Among these animals, wild waterfowl serve as the reservoir from which all influenza viruses are thought to have emerged (49). The causative viruses of the 1957 Asian and 1968 Hong Kong pandemics are in fact reassortants between human and avian influenza viruses (18, 23, 36), illustrating the introduction of avian viral genes into the human population. Despite the apparent common origin of influenza A viruses, their host range is clearly restricted. In experimental infections, avian influenza viruses replicate poorly in primates (3, 27, 38, 39), while human isolates do not replicate efficiently in ducks (13, 19, 50). There are exceptions, however, including an avian influenza virus that was directly transmitted from birds to humans in the Hong Kong outbreak of 1997 (5, 40). Although several genes are known to be involved in host range restriction of influenza viruses (37, 38, 39, 41, 45), the precise contribution(s) of each gene product has not been defined.
The hemagglutinin (HA) of influenza A viruses, a type I membrane glycoprotein, is responsible for binding of the virus to cell surface receptors, or sialyloligosaccharides. Although all influenza viruses recognize oligosaccharides containing a terminal SA, the specificity of the HA towards these molecules differs. Avian and equine influenza viruses preferentially bind the sialic acid-α2,3-galactose (SAα2,3Gal) linkage, while human influenza viruses preferentially bind the SAα2,6Gal linkage (6, 31, 32). Influenza viruses also differ in their recognition of NeuAc, NeuGc, and 9-O-Ac-NeuAc (12). However, the importance of differential recognition of these SA species for host range restriction of influenza virus remains unknown.
Couceiro et al. (7) reported the presence of sialyloligosaccharides reactive with an SAα2,6Gal- but not SAα2,3Gal-specific lectin on the surface of epithelial cells of human trachea. We recently showed that pig trachea contains sialyloligosaccharides reactive with both SAα2,6Gal- and SAα2,3Gal-specific lectins, while duck intestine, the major replicative site of avian influenza viruses, possesses only sialyloligosaccharides reactive with the SAα2,3Gal-specific lectin (16). These findings showed that the availability of receptors (i.e., SAα2,6Gal and SAα2,3Gal moieties) in host animals correlates with the receptor specificity of influenza viruses isolated from the host animals. Although it is highly likely that this phenomenon is controlled by multiple host and viral genes, these data suggest that receptor specificity is one of the important determinants of host range restriction among influenza viruses.
Because NeuGc is barely detectable in humans (less than 0.1% of total SA) (21, 22) but is abundant in horses (more than 97% of total SA in horse erythrocytes) (43), we reasoned that a difference in the recognition of SAs might play an important role in the host range restriction of influenza viruses. Here we show for the first time that the SA distribution in animal species does in fact influence influenza virus host range.
MATERIALS AND METHODS
Viruses.
The viruses used in this study were obtained from repositories at St. Jude Children's Research Hospital and the University of Kentucky. They were propagated in the allantoic cavities of 11-day-old embryonated chicken eggs at 35°C for 2 days.
Genetic reassortment.
Reassortant influenza viruses between A/equine/Kentucky/1/91 (H3N8) and A/Udorn/307/72 (H3N2), R2, or R3 viruses (13) were prepared in chicken embryos as previously described (48). Reassortants were biologically cloned at limiting dilutions in eggs.
Genotyping of reassortant viruses.
The genotypes of the reassortant viruses were determined by partial sequence analysis of each gene using reverse transcription-PCR (17). The sequences of the oligonucleotides used as primers will be supplied upon request.
Immunologic detection of SAα2,3Gal and SAα2,6Gal in animal tissues.
Horse trachea was obtained at a slaughter house in Hokkaido, Japan, and shipped to our laboratory on ice. It was then rinsed with phosphate-buffered saline (PBS, pH 7.2) before being cut into 3-mm3 cubes. The tissue block was embedded in OCT compound (Miles Inc.) and frozen in liquid nitrogen. Sections of each tissue (6 μm) were cut with a microtome cryostat, air dried, and fixed for 15 min with cold acetone before immunostaining.
To identify sialyoligosaccharides reactive with SAα2,3Gal- or SAα2,6Gal-specific lectins (glycan determination kit; Boehringer Mannheim Biochemicals, Mannheim, Germany), we incubated each section with 50 μl of digoxigenin (DIG)-labeled Sambucus nigra (SNA) lectin [1 μg/ml; specific for SAα2,6galactose (Gal)/N-acetylgalactosaminide (GalNac)] or Maackia amurensis (MAA) lectin (5 μg/ml; specific for the SAα2,3Gal) for 1 h at room temperature. After three washes with cold PBS, the sections were incubated with fluorescein- and rhodamine-conjugated anti-DIG antibody (Boehringer Mannheim Biochemicals), respectively, for 1 h at room temperature and then, after three additional washes in cold PBS and buffered glycerol (pH 9.0), were mounted for observation. Control slides were incubated with PBS instead of lectin. All sections were examined with a fluorescence microscope (BH-RFL; Olympus Optics, Tokyo, Japan) equipped with a dark-field condenser and UV excitation.
NeuGc linked to Gal by α2,3 was also immunologically detected using a chicken antiserum against hematoside [GM3 (NeuGc); II3(NeuGc)LacCer] (14, 15) that reacts with NeuGcα2,3Gal in both glycoproteins and glycolipids. The reactivity of this antiserum with tissue sections was tested as described above for lectins except that the secondary antibodies were conjugated with biotin.
SA determination by liquid chromatography.
The molecular species of SA (NeuAc or NeuGc) in gangliosides of epithelial cells in horse trachea was determined by a high-pressure liquid chromatography method using 1,2-diamino-4,5-methylenedioxybenzene (DMB), as described previously (11, 42). Epithelial cells (10 mg) were hydrolyzed with 200 μl of 25 mM sulfuric acid. The hydrolysate was reacted with DMB reagent and heated at 60°C for 2.5 h in the dark to develop the fluorescence of SA. A 10-μl aliquot of the solution was used for these determinations.
Experimental infection of ponies.
The ponies used in these experiments all lacked serological evidence of prior influenza virus infection or vaccination (titers < 10) in hemagglutination inhibition tests for antibodies to H3N8 equine viruses. The different groups of ponies (three per group) were exposed to aerosolized virus (DeVilbiss Ultra-Neb 99 nebulizer) through a face mask for approximately 10 min (4). Each pony received 107 50% egg infections doses (EID50) of virus (in 5 ml). The four groups, each representing a single virus, were isolated from each other by either physical or time barriers; the ponies were kept in individual stalls. Nasopharyngeal swabs were taken daily, beginning just before infection (day 0) and continuing through day 9. The viruses in the swabs were titrated in eggs as described (19).
Virus-binding assay.
To determine the receptor specificity of the viruses, a thin-layer chromatography/virus-binding assay was performed with gangliosides GM3 II3(NeuGc)LacCer, II3(NeuAc)LacCer, and II6(NeuAc)LacCer, as described previously (44).
Nucleotide sequencing.
Viral RNA was isolated as in earlier studies (2), and cDNA was synthesized with reverse transcriptase and random hexamers as described (17). Direct sequencing of the PCR products was done with an autosequencer (Applied Biosystems Inc.) according to the protocol recommended by the company. The sequences of oligonucleotides used as primers will be supplied upon request.
RESULTS
NeuGcα2,3Gal is abundant in the epithelial cells of horse trachea.
We first examined the prevalence of SAα2,6Gal and SAα2,3Gal moieties in epithelial cells in horse trachea, using SA-Gal linkage-specific lectins. SNA lectin, specific for SAα2,6Gal linkages, did not react with horse trachea but did react with pig trachea (positive control) (Fig. 2). By contrast, MAA lectin, specific for SAα2,3Gal linkages, reacted with horse trachea as well as pig trachea. These results establish the predominance of the SAα2,3Gal moiety in the replication site of influenza virus in horses.
FIG. 2.
Predominance of the SAα2,3Gal linkage as detected by lectin staining in horse trachea. The MAA lectin specific for SAα2,3Gal (α2-3; detected with fluorescein isothiocyanate (FITC)-labeled anti-DIG antibody) bound to horse and pig tracheal epithelium, whereas SNA lectin specific for SAα2,6Gal (α2-6; detected with rhodamine-labeled anti-DIG antibody) bound only to the latter. Blue staining is a nonspecific reaction.
NeuGc is the major SA species in horse erythrocytes (97% of total SA) (43), but whether this dominance extends to horse trachea is uncertain, as the prevalence of different SA species varies among organs (11, 34, 35). We therefore examined the relative abundance of NeuAc and NeuGc in the epithelial cells of horse trachea. More than 90% of the SA was NeuGc, while the remaining proportion was NeuAc (Fig. 3). Because the SAα2,3Gal but not the SAα2,6Gal moiety was detected by the lectin assay and the vast majority of SA in epithelial cells were NeuGc, these findings suggested that the major SA-Gal moiety present in horse tracheal epithelial cells is NeuGcα2,3Gal. To directly detect this moiety, we examined the reactivity of thin sections of horse trachea with a chicken antiserum against hemtoside [GM3(NeuGc); II3(NeuGc)LacCer] (14, 15), which contains the NeuGcα2,3Gal structure. This antiserum reacts with both gangliosides and glycoproteins possessing terminal NeuGcα2,3Gal but not with glycoconjugates possessing either NeuAcα2,3Gal or NeuAcα2,6Gal (Y. Suzuki, unpublished data). As shown in Fig. 4, the antiserum bound to epithelial cells lining horse trachea but not with those of chicken trachea, which lack NeuGc (8). These results demonstrate the abundance of the NeuGcα2,3Gal moiety in the epithelial cells of horse trachea.
FIG. 3.
Chromatograms of 1,2-diamino-4,5-methylenedioxybenzene derivatives of NeuAc and NeuGc obtained from the epithelial cells of horse trachea. The standard mixture of NeuAc and NeuGc and epithelial cells from horse trachea was treated as described in Materials and Methods. The fluorescence of SA derivatives was detected at an excitation wavelength of 373 nm and an emission wavelength of 448 nm.
FIG. 4.
Detection of NeuGcα2,3Gal moieties in epithelial cells lining horse trachea. Antiserum specific for NeuGcα2,3Gal but not for NeuAcα2,3Gal or NeuAcα2,6Gal (14, 15) detected this moiety in tracheal samples from horses (FITC staining) but not chickens, which lack NeuGc (8). Blue staining is a nonspecific reaction.
Equine but not human influenza viruses recognize NeuGcα2,3Gal moiety.
Although human influenza viruses have been transmitted to pigs and vice versa (10, 24, 29, 33, 46, 51), human-to-horse transmission remains to be reported. The finding of differences in sialyloligosaccharide species in tracheal epithelial cells between humans (predominantly NeuAcα2,6Gal) and horses (predominantly NeuGcα2,3Gal) suggested that recognition not only of SA-Gal linkages but also of SA species may be responsible for lack of transmission of human viruses to horses and vice versa. We therefore examined the receptor specificity of these viruses. Human influenza viruses preferentially recognized the NeuAcα2,6Gal moiety, whereas equine viruses preferentially recognized NeuAcα2,3Gal over NeuAcα2,6Gal, as has been shown previously (31) (Fig. 5). Interestingly, equine viruses also recognized the NeuGcα2,3Gal moiety, although one virus (A/equine/Miami/1/63 [H3N8]), which has been passaged extensively in embryonated chicken eggs lacking NeuGc, bound poorly to this moiety. Because the predominant SA-Gal moiety differs between the epithelial cells of human and horse trachea (see above), these results suggest a role for HA-sialyloligosaccharide specificity in host range restriction of human and equine influenza viruses.
FIG. 5.
Binding reactivity of influenza A viruses with gangliosides of human and equine influenza viruses. Gangliosides (1 μmol), represented by II6(NeuAc)LacCer (NeuAcα2,6Gal), II3(NeuAc)LacCer (NeuAcα2,3Gal), and II3(NeuGc)LacCer (NeuGcα2,3Gal), were developed in thin-layer chromatography plates. The plates were incubated with virus (28 hemagglutination units at 4°C for 9 h and then processed as described in Materials and Methods. Relative binding reactivity is shown, with the highest activity set at 100%.
NeuGcα2,3Gal recognition is essential for viral replication in horses.
Our recent study showed that the HA of a human influenza virus, A/Udorn/307/72 (H3N2) (Udorn), preferentially recognizes NeuAcα2,6Gal, but not NeuAcα2,3Gal or NeuGcα2,3Gal (T. Ito et al, unpublished data). However, a mutation at position 226 of this HA from Leu to Gln (L226Q) made it preferentially recognize the NeuAcα2,3Gal but not the NeuGcα2,3Gal moiety. Another mutation at position 228, from Ser to Gly, thus L226Q and S228G mutations, made the HA recognize both the NeuAcα2,3Gal and NeuGcα2,3Gal moieties (Table 1). These human virus HAs provided useful tools with which to study the role of HA receptor specificity in the host range restriction of influenza viruses. We therefore designed experiments in which these HAs were used to examine the contributions of receptor specificity and availability of host receptors to host range restriction. To limit the effects of viral gene products to the HA, we made three reassortant viruses possessing these mutant HAs, with the remainder of the genes coming from an equine virus, A/equine/Kentucky/1/91 (H3N8) (Eq/Ky). Udorn-Eq/Ky virus contains the human virus Udorn HA, L226Q-Eq/Ky virus contains the Udorn L226Q mutant HA, and L226Q/S228G-Eq/Ky virus contains the Udorn HA mutant possessing both the L226Q and S228G alterations. We then tested the replicative capacity of these reassortants, as well as the parent Eq/Ky virus, in ponies. As expected, Eq/Ky virus, whose HA recognizes not only the NeuAcα2,3Gal but also the NeuGcα2,3Gal moiety, replicated for up to 1 week (5 days in two ponies and 8 days in one pony), producing titers ranging from 102.7 to 105.3 EID50 on day 2 (mean virus titer, 104.0 EID50) (Table 1). By contrast, Udorn-Eq/Ky virus, whose HA preferentially recognizes the NeuAcα2,6Gal over the NeuGcα2,3Gal moiety, did not replicate in ponies at all, demonstrating an essential contribution of the HA to host range restriction. Interestingly, the L226Q-Eq/Ky virus, whose HA preferentially recognizes the SAα2,3Gal linkage with NeuAc while binding much less avidly to the NeuGc moiety, replicated in one of three ponies tested, but only for 2 days (101.4 EID50 at 1 day and <101.0 EID50 at 2 days postinfection). However, the L226Q/S228G-Eq/Ky virus, whose HA recognizes both NeuAcα2,3Gal and NeuGcα2,3Gal moieties, replicated in all three ponies tested for as long as 1 week (6 days in one pony and 7 days in two ponies), with titers ranging from 101.8 to 103.8 EID50. These findings demonstrate that recognition of the NeuGcα2,3Gal moiety is critical for the efficient replication of influenza A viruses in horses.
TABLE 1.
NeuGcα2,3Gal recognition required for influenza virus replication in horsesa
| Virus | Relative binding reactivity (%) with receptor:
|
No. of virus-positive horses/3 infected on day postinfectionb:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NeuAcα2,6Gal | NeuAcα2,3Gal | NeuGcα2,3Gal | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| Eq/Ky | 0 | 100 | 33 | 0 | 3 | 3 (4.0) | 3 | 3 | 3 | 1 | 0 | 1 | 0 |
| Udorn-Eq/Ky | 100 | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L226Q-Eq/Ky | 72 | 100 | 15 | 0 | 1 (1.4) | 1 (<1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L226Q/S228G-Eq/Ky | 19 | 82 | 100 | 0 | 3 (2.6) | 3 (2.8) | 3 | 2 | 2 | 3 | 2 | 0 | 0 |
Three of the viruses are reassortants possessing the genes of A/equine/Kentuckey/1/91 virus with the exception of the HA: Udorn-Eq/Ky possesses the HA from human A/Udorn/307/72; L226Q-Eq/Ky is the mutant A/Udorn/307/72 HA containing a change at position 226 (Leu to Gln); and L226Q/S228G-Eq/Ky is the mutant A/Udorn/307/72 HA containing changes at positions 226 (Leu to Gln) and 228 (Ser to Gly). The HA receptor specificities of the viruses other than Eq/Ky were previously determined by thin-layer chromatography virus-binding assays with use of gangliosides (16a), while that of Eq/Ky was determined in this study (Fig. 5). The results are presented as relative binding reactivity of virus with linkages shown, with the highest reactivity detected as 100.
Numbers in parentheses are geometric mean virus titers (log10 EID50 per milliliter). Ponies were infected with aerosolized virus (107 EID50). Viruses were recovered daily by taking nasal swabs and inoculating the swab suspensions into embryonated eggs.
DISCUSSION
Although a variety of SAs are found in animals and their distributions differ among animal species, the biological effects of those differences remain largely unknown. Influenza A viruses differ in their ability to recognize SAα2,6Gal and SAα2,3Gal, suggesting a role for HA-SA specificity in host range restriction. Here we demonstrate that NeuGcα2,3Gal is the dominant moiety on the epithelial cells of horse trachea and that a difference in sialyloligosaccharides, NeuAcα2,6Gal (human) versus NeuGcα2,3Gal (horse), on tracheal epithelial cells is a critical factor in differential viral replication. Viruses that failed to recognize the SAα2,3Gal linkage as well as NeuGc did not replicate efficiently in horses. Thus, the absence or low abundance of a particular SA species can serve as a host range barrier for influenza A viruses, providing an example of biological effects related to differences in distribution of different SAs.
Our recent studies showed that many avian influenza viruses recognize the NeuGcα2,3Gal moiety (16a). We also found that the antiserum specific for the NeuGcα2,3Gal moiety reacted with epithelial cells of duck colon crypt, although by chemical analysis the ratio of NeuGc to NeuAc in colon epithelial cells was low (2 versus 98%). These findings would explain the transmission of an avian influenza virus to horses in China, resulting in the death of 20% of the horse population (9).
The results of the current study demonstrate restriction of influenza virus transmission from humans to horses. This finding explains why no human viruses have been isolated from horses, while they have transmitted into pigs multiple times (10, 24, 29, 46, 51). Pig trachea contains both the SAα2,6Gal and SAα2,3Gal moieties (16). In fact, pigs efficiently support the replication of avian influenza viruses (20), which preferentially recognize the SAα2,3Gal moiety (31). These studies emphasize the importance of the availability of receptor sialyloligosaccharides and receptor specificity in the host range restriction of influenza viruses, which likely plays an important role in the evolution of these viruses. Why, then, was an avian virus that binds preferentially to SAα2,3Gal transmitted to humans in Hong Kong in 1997 (5, 26, 40)? It seems that the magnitude of the contribution of receptor specificity to host restriction depends on the combination of host animal and virus strain. In horses, for example, the ability of the virus to recognize a specific receptor is essential for efficient replication, while in humans, the recognition of NeuAcα2,6Gal does not appear to be essential for replication of the virus. In support of this notion, we found that human tracheal epithelial cells possess sialyloligosaccharides reactive with SAα2,3Gal linkage-specific lectin, although oligosaccharides reactive with SAα2,6Gal-specific lectin were more prevalent (P. Gao and Y. Kawaoka, unpublished data), suggesting that the amount of SAα2,3Gal in the epithelial cells of human trachea is sufficient for at least the initiation of viral replication. Whether the same relationship between SAα2,3Gal and SAα2,6Gal linkages extends to other human tissues, such as lung, is unclear. However, the first available human isolates from the 1957 and 1968 pandemics preferentially recognized the SAα2,6Gal moiety, even though their HAs were derived from an avian virus (23, 36); thus, for efficient human-to-human transmission, the viruses may need to recognize SAα2,6Gal more avidly than SAα2,3Gal. In addition to the receptor molecules on epithelial cells, sialyloligosaccharides in body fluids may also play a role in the host range restriction of influenza viruses. Horse serum contains α2-macroglobulin containing 4-O-acetyl-SA. Because influenza virus neuraminidase (NA) cannot cleave this SA from oligosaccharides, glycoproteins containing this SA serve as a potent receptor analog inhibitor. Thus, sialyloligosaccharides in body fluid, as well as those on the cell surface (i.e., receptor), contribute to the evolution of influenza viruses.
Influenza viruses possess both receptor-binding (HA) and receptor-destroying (NA) proteins. Thus, the receptor specificity of the HA and the substrate specificity of the NA may work in concert to determine the efficiency of influenza virus replication. In fact, Baum and Paulson (1) reported that the human influenza virus NA, which was introduced from an avian virus, gradually acquired the ability to cleave the SAα2,6Gal as well as SAα2,3Gal linkage. This finding suggests that viral selection favors enzymes whose substrate specificity matches the receptor specificity of the human virus HA (i.e., SAα2,6Gal specificity).
In this study, we focused on the receptor specificity of the HA as a critical determinant of host range restriction. Future research will need to test the possibility that other genes encoding internal proteins could also contribute to this natural barrier to unimpeded replication. For example, the NP and M genes are responsible for the attenuation of avian viruses in squirrel monkeys (45), and depending on the human influenza viruses used to prepare reassortants with avian viruses, a combination of polymerase genes was found to affect the ability of these reassortant viruses to replicate in squirrel monkeys (38, 39, 41). Thus, the mechanism responsible for host range restriction likely entails a complex interplay of viral and host gene products.
ACKNOWLEDGMENTS
We thank Krisna Wells, Martha McGregor, Lynn Tudor, and Eugene Ferguson for excellent technical assistance and John Gilbert for editing the manuscript.
Support for this work came from the Ministry of Education and Culture of Japan (Y.S. and T.S.), National Institute of Allergy and Infectious Diseases, Public Health Service, research grants (Y.K.), Hatch-Kentucky Agricultural Experiment Station project no. KY014006 (T.M.C.), and the late Paul Mellon (T.M.C.).
REFERENCES
- 1.Baum L G, Paulson J C. The N2 neuraminidase of human influenza virus has acquired a substrate specificity complementary to the hemagglutinin receptor specificity. Virology. 1991;180:10–15. doi: 10.1016/0042-6822(91)90003-t. [DOI] [PubMed] [Google Scholar]
- 2.Bean W J, Sriram G, Webster R G. Electrophoretic analysis of iodine-labeled influenza RNA segments. Anal Biochem. 1980;102:228–232. doi: 10.1016/0003-2697(80)90343-7. [DOI] [PubMed] [Google Scholar]
- 3.Beare A S, Webster R G. Replication of avian influenza viruses in humans. Arch Virol. 1991;119:37–42. doi: 10.1007/BF01314321. [DOI] [PubMed] [Google Scholar]
- 4.Chambers T M, Shortridge K F, Li P H, Powell D G, Watkins K L. Rapid diagnosis of equine influenza by the Directigen FLU-A enzyme immunoassay. Vet Rec. 1994;135:275–279. doi: 10.1136/vr.135.12.275. [DOI] [PubMed] [Google Scholar]
- 5.Claas E C J, Osterhaus A D M E, Van Beek R, De Jong J C, Rimmelzwaan G F, Senne D A, Krauss S, Shortridge K F, Webster R G. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 6.Connor R J, Kawaoka Y, Webster R G, Paulson J C. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 7.Couceiro J N, Paulson J C, Baum L G. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium: the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 1993;29:155–165. doi: 10.1016/0168-1702(93)90056-s. [DOI] [PubMed] [Google Scholar]
- 8.Fujii Y, Higashi H, Ikuta K, Kato S, Naiki M. Specificities of human heterophilic Hanganutziu and Deicher (H-D) antibodies and avian antisera against H-D antigen-active glycosphingolipids. Mol Immunol. 1982;19:87–94. doi: 10.1016/0161-5890(82)90250-4. [DOI] [PubMed] [Google Scholar]
- 9.Guo Y, Wang M, Kawaoka Y, Gorman O, Ito T, Saito T, Webster R G. Characterization of a new avian-like influenza A virus from horses in China. Virology. 1992;188:245–255. doi: 10.1016/0042-6822(92)90754-d. [DOI] [PubMed] [Google Scholar]
- 10.Haesebrouck F, Pensaert M. Influenza in swine in Belgium (1969–1986): epizootiologic aspects. Comp Immunol Microbiol Infect Dis. 1988;11:215–222. doi: 10.1016/0147-9571(88)90040-9. [DOI] [PubMed] [Google Scholar]
- 11.Hara S, Takemori Y, Yamaguchi M, Nakamura M, Ohkura Y. Fluorometric high-performance liquid chromatography of N-acetyl- and N-glycolylneuraminic acids and its application to their microdetermination in human and animal sera, glycoproteins, and glycolipids. Anal Biochem. 1987;164:138–145. doi: 10.1016/0003-2697(87)90377-0. [DOI] [PubMed] [Google Scholar]
- 12.Higa H H, Rogers G N, Paulson J C. Influenza virus hemagglutinins differentiate between receptor determinants bearing N-acetyl-, N-glycolyl-, and N,O-diacetylneuraminic acids. Virology. 1985;144:279–282. doi: 10.1016/0042-6822(85)90325-3. [DOI] [PubMed] [Google Scholar]
- 13.Hinshaw V S, Webster R G, Naeve C W, Murphy B R. Altered tissue tropism of human-avian reassortant influenza viruses. Virology. 1983;128:260–263. doi: 10.1016/0042-6822(83)90337-9. [DOI] [PubMed] [Google Scholar]
- 14.Hirabayashi Y, Suzuki T, Suzuki Y, Taki T, Matsumoto M, Higashi H, Kato S. A new method for purification of anti-glycosphingolipid antibody. Avian anti-hematoside (NeuGc) antibody. J Biochem (Tokyo) 1983;94:327–330. doi: 10.1093/oxfordjournals.jbchem.a134350. [DOI] [PubMed] [Google Scholar]
- 15.Hirabayashi Y, Higashi H, Kato S, Taniguchi M, Matsumoto M. Occurrence of tumor-associated ganglioside antigens with Hanganutziu-Deicher antigenic activity on human melanomas. Jpn J Cancer Res. 1987;78:614–620. [PubMed] [Google Scholar]
- 16.Ito T, Nelson J, Couceiro S S, Kelm S, Baum L G, Krauss S, Castrucci M R, Donatelli I, Kida H, Paulson J C, Webster R G, Kawaoka Y. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Ito T, Suzuki Y, Suzuki T, Takada A, Horimoto T, Wells K, Kida H, Otsuki K, Kiso M, Ishida H, Kawaoka Y. Recognition of N-glycolylneuraminic acid linked to galactose by the α2,3 linkage is associated with intestinal replication of influenza A virus in ducks. J Virol. 2000;74:9300–9305. doi: 10.1128/jvi.74.19.9300-9305.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katz J M, Wang M, Webster R G. Direct sequencing of the HA gene of influenza (H3N2) virus in original clinical samples reveals sequence identity with mammalian cell-grown virus. J Virol. 1990;64:1808–1811. doi: 10.1128/jvi.64.4.1808-1811.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawaoka Y, Krauss S, Webster R G. Avian-to-human transmission of the PB1gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kida H, Yanagawa R, Matsuoka Y. Duck influenza lacking evidence of disease signs and immune response. Infect Immun. 1980;30:547–553. doi: 10.1128/iai.30.2.547-553.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kida H, Ito T, Yasuda J, Shimizu Y, Itakura C, Shortridge K F, Kawaoka Y, Webster R G. Potential for transmission of avian influenza viruses to pigs. J Gen Virol. 1994;75:2183–2188. doi: 10.1099/0022-1317-75-9-2183. [DOI] [PubMed] [Google Scholar]
- 21.Klenk E, Lempfrid H. Uber die Natur der Zellreceptoren fir das Influenzavirus. Hoppe-Seyler's Z Physiol Chem. 1957;307:278–283. [PubMed] [Google Scholar]
- 22.Klenk E, Uhlenbruck G. Uber ein neuraminsaurehaltiges Mucoproteid aus Rindererythrocytenstroma. Hoppe-Seyler's Z Physiol Chem. 1958;311:227–233. [PubMed] [Google Scholar]
- 23.Laver W G, Webster R G. Studies on the origin of pandemic influenza. III. Evidence implicating duck and equine influenza viruses as possible progenitors of the Hong Kong strain of human influenza. Virology. 1973;51:383–391. doi: 10.1016/0042-6822(73)90437-6. [DOI] [PubMed] [Google Scholar]
- 24.Mancini G, Donatelli I, Rozera C, Arangio Ruiz G, Butto S. Antigenic and biochemical analysis of influenza A H3N2 viruses isolated from pigs. Arch Virol. 1985;83:57–167. doi: 10.1007/BF01309913. [DOI] [PubMed] [Google Scholar]
- 25.Martensson E, Raal A, Svennerholm L. Sialic acid in blood serum. Biochim Biophys Acta. 1958;301:24–129. doi: 10.1016/0006-3002(58)90248-8. [DOI] [PubMed] [Google Scholar]
- 26.Matrosovich M, Zhou N, Kawaoka Y, Webster R G. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J Virol. 1999;7:1146–1155. doi: 10.1128/jvi.73.2.1146-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy B R, Hinshaw V S, Sly D L, London W T, Hosier N T, Wood F T, Webster R G, Chanock R M. Virulence of avian influenza A viruses for squirrel monkeys. Infect Immun. 1982;37:1119–1126. doi: 10.1128/iai.37.3.1119-1126.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naiki M. Chemical and immunochemical properties of two classes of globoside from equine organs. Jpn J Exp Med. 1971;41:67–81. [PubMed] [Google Scholar]
- 29.Ottis K, Sidoli L, Bachmann P A, Webster R G, Kaplan M M. Human influenza A viruses in pigs: isolation of a H3N2 strain antigenically related to A/England/42/72 and evidence for continuous circulation of human viruses in the pig population. Arch Virol. 1982;73:103–108. doi: 10.1007/BF01314719. [DOI] [PubMed] [Google Scholar]
- 30.Pettersson S O, Sivertsson R, Sjogren S, Svennerholm L. The sialic acids of hog pancreas. Biochim Biophys Acta. 1958;28:444–445. doi: 10.1016/0006-3002(58)90498-0. [DOI] [PubMed] [Google Scholar]
- 31.Rogers G N, Paulson J C. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983;127:361–373. doi: 10.1016/0042-6822(83)90150-2. [DOI] [PubMed] [Google Scholar]
- 32.Rogers G N, D'Souza B L. Receptor binding properties of human and animal H1 influenza virus isolates. Virology. 1989;173:317–322. doi: 10.1016/0042-6822(89)90249-3. [DOI] [PubMed] [Google Scholar]
- 33.Rota P A, Rocha E P, Harmon M W, Hinshaw V S, Sheerar M G, Kawaoka Y, Cox N J, Smith T F. Laboratory characterization of a swine influenza virus isolated from a fatal case of human influenza. J Clin Microbiol. 1989;27:1413–1416. doi: 10.1128/jcm.27.6.1413-1416.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schauer R. Chemistry, metabolism, and biological functions of sialic acids. Adv Carbohydr Chem Biochem. 1982;40:131–234. doi: 10.1016/s0065-2318(08)60109-2. [DOI] [PubMed] [Google Scholar]
- 35.Schauer R. Sialic acids: chemistry, metabolism, and function. New York, N.Y: Springer; 1982. [DOI] [PubMed] [Google Scholar]
- 36.Scholtissek C, Rohde W, Von Hoyningen V, Rott R. On the origin of the human influenza virus subtype H2N2 and H3N2. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- 37.Scholtissek C, Burger H, Kistner O, Shortridge K F. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology. 1985;147:287–294. doi: 10.1016/0042-6822(85)90131-x. [DOI] [PubMed] [Google Scholar]
- 38.Snyder M H, Buckler-White A J, London W T, Tierney E L, Murphy B R. The avian influenza virus nucleoprotein gene and a specific constellation of avian and human virus polymerase genes each specify attenuation of avian-human influenza A/Pintail/79 reassortant viruses for monkeys. J Virol. 1987;61:2857–2863. doi: 10.1128/jvi.61.9.2857-2863.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snyder M H, Clements M L, De Borde D, Maassab H F, Murphy B R. Attenuation of wild-type human influenza A virus by acquisition of the PA polymerase and matrix protein genes of influenza A/Ann Arbor/6/60 cold-adapted donor virus. J Clin Microbiol. 1987;22:719–725. doi: 10.1128/jcm.22.5.719-725.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 41.Subbarao E K, London W, Murphy B R. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol. 1993;67:1761–1764. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki T, Horiike G, Yamazaki Y, Kawabe K, Masuda H, Miyamoto D, Matsuda M, Nishimura S I, Yamagata T, Ito T, Kida H, Kawaoka Y, Suzuki Y. Swine influenza virus strains recognize sialylsugar chains containing the molecular species of sialic acid predominantly present in the swine tracheal epithelium. FEBS Lett. 1997;404:192–196. doi: 10.1016/s0014-5793(97)00127-0. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki Y, Matsunaga M, Matsumoto M. N-Acetylneuraminyllactosylceramide, GM3-NeuAc, a new influenza A virus receptor which mediates the adsorption-fusion process of viral infection. J Biol Chem. 1985;260:1362–1365. [PubMed] [Google Scholar]
- 44.Suzuki Y, Nakao T, Ito T, Watanabe N, Toda Y, Xu G, Suzuki T, Kobayashi T, Kimura Y, Yamada A, Sugawara K, Nishimura H, Kitame F, Nakamura K, Deya E, Kiso M, Hasegawa A. Structural determination of gangliosides that bind to influenza A, B, and C viruses by an improved binding assay: strain-specific receptor epitopes in sialo-sugar chains. Virology. 1992;189:121–131. doi: 10.1016/0042-6822(92)90687-k. [DOI] [PubMed] [Google Scholar]
- 45.Tian S F, Buckler-White A J, London W T, Reck L J, Chanock R M, Murphy B R. Nucleoprotein and membrane protein genes are associated with restriction of replication of influenza A/Mallard/NY/78 virus and its reassortants in squirrel monkey respiratory tract. J Virol. 1985;53:771–775. doi: 10.1128/jvi.53.3.771-775.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tumova B, Veznikova D, Mensik J, Stumpa A. Surveillance of influenza in pig herds in Czechoslovakia in 1974–1979. 1. Introduction of influenza epidemic A (H3N2) viruses into pig herds. Zentralbl Veterinarmed B. 1980;27:517–523. [PubMed] [Google Scholar]
- 47.Varki A. Diversity in the sialic acids. Glycobiology. 1992;2:25–40. doi: 10.1093/glycob/2.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webster R G. Antigenic hybrids of influenza A viruses with surface antigens to order. Virology. 1970;42:633–642. doi: 10.1016/0042-6822(70)90309-0. [DOI] [PubMed] [Google Scholar]
- 49.Webster R G, Bean W J, Gorman O T, Chambers T M, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webster R G, Yakhno M, Hinshaw V S, Bean W J, Murti K G. Intestinal influenza: replication and characterization of influenza viruses in ducks. Virology. 1978;842:68–278. doi: 10.1016/0042-6822(78)90247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wibberley G, Swallow C, Roberts D H. Characterization of an influenza A (H3N2) virus isolated from pigs in England in 1987. Br Vet J. 1988;144:196–201. doi: 10.1016/0007-1935(88)90053-X. [DOI] [PMC free article] [PubMed] [Google Scholar]