Abstract
Background:
As the largest conduit vessel, aorta is responsible for the conversion of phasic systolic inflow from ventricular ejection into more continuous peripheral blood delivery. Systolic distension and diastolic recoil conserve energy and are enabled by the specialized composition of the aortic extracellular matrix. Aortic distensibility decreases with age and vascular disease.
Objectives:
To discover epidemiological correlates and genetic determinants of aortic distensibility and strain.
Methods:
We trained a deep learning model to quantify thoracic aortic area throughout the cardiac cycle from cardiac magnetic resonance images, and calculated aortic distensibility and strain in 42,342 UK Biobank participants.
Results:
Descending aortic distensibility was inversely associated with future incidence of cardiovascular diseases such as stroke (HR 0.59 per SD, P = 3.1E-04). The heritability of aortic distensibility and strain was 22–25% and 30–33%, respectively. Common variant analyses identified 12 and 26 loci for ascending and 11 and 21 loci for descending aortic distensibility and strain, respectively. Of the newly identified loci, 22 were not significantly associated with thoracic aortic diameter. Nearby genes were involved in elastogenesis and atherosclerosis. Aortic strain and distensibility polygenic scores had modest effect sizes for predicting cardiovascular outcomes (delaying or accelerating disease onset by 2–18% per standard deviation change in scores) and remained statistically significant predictors after accounting for aortic diameter polygenic scores.
Conclusions:
Genetic determinants of aortic function influence risk for stroke and coronary artery disease, and may lead to novel targets for medical intervention.
Keywords: Aorta, distensibility, strain, deep learning, cardiovascular disease, genetics
Condensed Abstract
We trained a deep learning model to quantify aortic distensibility and strain from cardiac magnetic resonance imaging in 42,342 UK Biobank participants. Distensibility was inversely associated with stroke risk. Common genetic variant analyses identified 22 distinct loci associated with thoracic aortic distensibility or strain and not diameter. Loci associated with both aortic diameter and aortic strain or distensibility demonstrated an inverse directionality. Transcriptome-wide analyses, rare variant burden tests, and aortic single nucleus RNA sequencing analyses highlighted genes involved in elastogenesis and atherosclerosis. Genetic characterization of aortic function may provide novel targets for medical intervention in aortic disease.
Introduction
Pathogenic insult to the thoracic aorta comes from different influences including hypertension, hypercholesterolemia, aging, valvular dysfunction, and genetic perturbations that interfere with aortic homeostasis. Aortic disease may manifest variably as early or advanced atherosclerotic lesions, intramural hematomas, aneurysms, or even aortic failure in the form of rupture or dissection1. One common early manifestation resulting from all of these predisposing factors to aortic disease is a reduction in aortic motion, or “stiffening”, that can be detected via imaging or hemodynamic consequences2.
Every cardiac cycle the aorta receives the output of ventricular systole to transport blood to target tissues, hence its designation as a “conduit vessel”. However the aorta is not a simple conduit but part of a hemodynamic system that conserves ventricular energy and ensures diastolic flow in the distal vasculature. This “windkessel” function of the aorta occurs when aortic strain stores potential energy during systole which is then utilized during diastole to maintain forward flow3. This property can be estimated noninvasively by measuring aortic distensibility, which indexes aortic strain (the measured relative change in cross-sectional area from diastole to systole) to a noninvasive estimate of central pulse pressure2. In addition to impairing distal blood delivery, aortic stiffening also has deleterious cardiac effects by increasing ventricular afterload and decreasing the coronary flow reserve ratio thereby impairing endocardial blood flow4. Decreased aortic distensibility is seen with aging5, but also prematurely in multiple cardiovascular diseases including hypertension5, atherosclerosis6, aneurysmal disease7, aortic dissection8, and dementia9, and is predictive of non-aortic cardiovascular events and all-cause mortality10. Despite the importance of aortic distensibility in health and disease there is a limited understanding of its basic biologic mechanisms and their relationship to comorbidities.
In this study, we quantified aortic distention from 42,342 participants in the UK Biobank to discover epidemiologic associations with anthropomorphic phenotypes, relationships to other medical disorders, and the underlying human genetic architecture of this fundamental aortic function.
Methods
Study design
All population-level analyses were conducted in the UK Biobank, which is a richly phenotyped, prospective, population-based cohort that recruited 500,000 participants aged 40–69 years in the UK via mailer from 2006–201011. As part of an ongoing imaging substudy, cardiac magnetic resonance imaging was performed for 42,342 participants with 1.5 Tesla scanners (MAGNETOM Aera, Siemens Healthcare)12. Access was provided under UK Biobank application #7089 and approved by the Mass General Brigham institutional review board (protocol 2003P001563).
Statistical analysis
Unless otherwise specified, statistical analyses were conducted with R 3.6.3 (R Foundation for Statistical Computing, Vienna, Austria). PyTorch-based deep learning models were trained with the FastAI library (Supplementary Methods) 13. The fitdistrplus package in R was used to determine skewness and kurtosis during central pulse pressure quality control. Ordinary least squares regression with R was used for continuous-trait association testing, disease PheWAS, and medication adjustment. Logistic regression with R was used for prevalent disease analysis. Cox proportional hazards and accelerated failure time analyses were conducted with the survival package in R 14. Genetic statistical association testing and heritability estimation was performed using linear mixed models with BOLT-LMM 15. LD score regression was performed with ldsc 16. Gene- and pathway-based evidence aggregation was performed with MAGMA v1.09b 17. Multi-trait analysis was conducted with MTAG 1.0.8 18. Bayesian polygenic score derivation was performed with PRScs 19. Rare variant burden testing was performed with REGENIE 20.
Semantic segmentation of thoracic aorta
Segmentation maps were traced for the ascending and descending thoracic aorta manually by a cardiologist (JP). 501 samples were used to train a ResNet34-based UNet deep learning model with fastai v1.0.5913 to infer segmentation of the ascending and descending aorta, compared with 116 samples in our previous work21. Training parameters are detailed in Supplementary Methods.
Diameter, strain, and distensibility
Strain for the ascending and descending thoracic aorta was defined as the change in area (greatest area minus least area throughout the cardiac cycle) divided by the least area. Distensibility was estimated by dividing strain values by Vicorder-based central pulse pressure estimates from the MRI visit, after QC (Supplementary Methods).
Epidemiological analyses
Aortic traits were tested for association with continuous phenotypes with linear models (Supplementary Methods).
Curated cardiovascular diseases for epidemiological analyses and GWAS exclusion criteria were defined based on self report, ICD codes, and procedural codes (Supplementary Table 1). Prevalent cardiovascular diseases were tested for association with aortic phenotypes with linear models. Incident disease analyses were performed with Cox models that evaluated survival to censoring or first disease diagnosis with the R survival package.
Medication adjustment
We grouped medications by the third level of their Anatomical Therapeutic Chemical (ATC) code, which approximately groups medications by therapeutic type (e.g., “beta blocking agents”). We analyzed 2,712 UK Biobank participants who had undergone repeat imaging visits, using a linear model to predict the aortic phenotype at the second visit based on the phenotype at the first visit and starting a medication between the two visits.
PheCode analyses
Using linear models, we tested for association between aortic phenotypes (mean and standard deviation scaled) and all available PheCode-based disease labels, which were derived from ICD-10 codes and OPCS-4 codes. Analyses were also performed by adjusting for all medication classes that achieved P < 0.05 in the univariate analyses described above.
Genotyping, imputation, and genetic quality control
A total of 11,622,901 imputed variants were used for analysis after standard quality control steps (Supplementary Methods). Participants were excluded from genetic analysis if they had a measured aortic diameter > 5cm or a history of aortic aneurysm, dissection, rupture, or procedures documented prior to imaging (Supplementary Table 1).
Heritability and genome-wide association studies
A rank-based inverse normal transformation was applied for aortic diameter, strain, and distensibility prior to heritability estimation and GWAS22. All GWAS were conducted using BOLT-LMM version 2.3.4 15 and adjusted for sex, age at enrollment, age and age2 at the time of MRI, the first 10 principal components of ancestry, the genotyping array, and the MRI scanner’s unique identifier. BOLT-REML v2.3.4 was used to assess SNP-heritability and genetic correlations using directly genotyped variants15. Linkage disequilibrium score regression was conducted on the summary statistics using ldsc version 1.0.1 with its default settings16. We identified lead SNPs for each trait using a clumping and distance-based protocol as detailed in the Supplementary Methods.
Polygenic score analyses
We computed polygenic scores using the software program PRScs in ‘auto’ mode (version sha1@43128be) with a publicly available UK Biobank European ancestry linkage disequilibrium panel19. The aortic strain and distensibility scores were tested for association with the curated cardiovascular diseases described above using accelerated failure time models, as detailed in the Supplementary Methods.
Results
Semantic segmentation of aorta with deep learning
The deep learning model had high accuracy: in a held-out test set of 20 images not used for training, the average Dice score (maximum 1.0, Supplementary Methods) was 0.97 for the ascending aorta and 0.96 for the descending aorta (Supplementary Table 2). The model was applied to all available aortic distensibility images in 42,342 UK Biobank participants. This allowed measurement of aortic diameter and circumferential aortic strain (Central Illustration). Strain was divided by a previously described estimate of central pulse pressure to yield aortic distensibility12,23.
Central illustration: Aortic distensibility measurement, epidemiology, and genetic analysis.
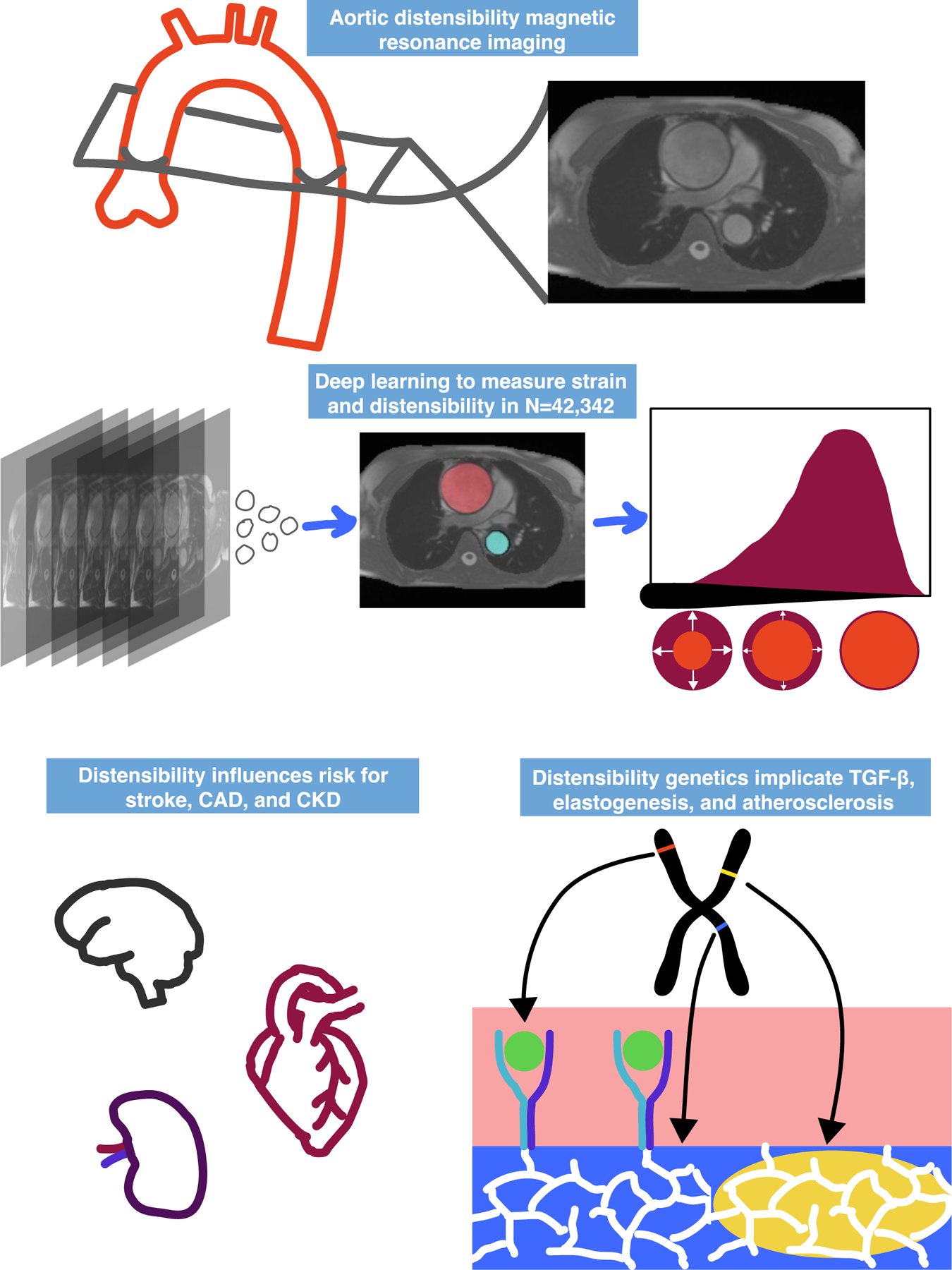
Cardiac magnetic resonance images from 42,342 participants were segmented using a deep learning model to compute aortic strain and distensibility. Disease risk was calculated for measured or genetically predicted strain and distensibility. Genome-wide association studies were performed, revealing links to TGF-β signaling, elastogenesis, and atherosclerosis.
Epidemiological characteristics of aortic strain and distensibility in the UK Biobank
For the 40,577 participants who contributed to at least one GWAS (Supplementary Figure 1), the distribution of ascending and descending aortic strain and distensibility are shown in Supplementary Figure 2. Cross-correlations of traits including diameter, distensibility, strain, and central pulse pressure are shown in Figure 1 Panel A. All central pulse pressure values and related phenotypes (including distensibility) are reported after extensive quality control, described in detail in the Supplementary Note.
Figure 1.
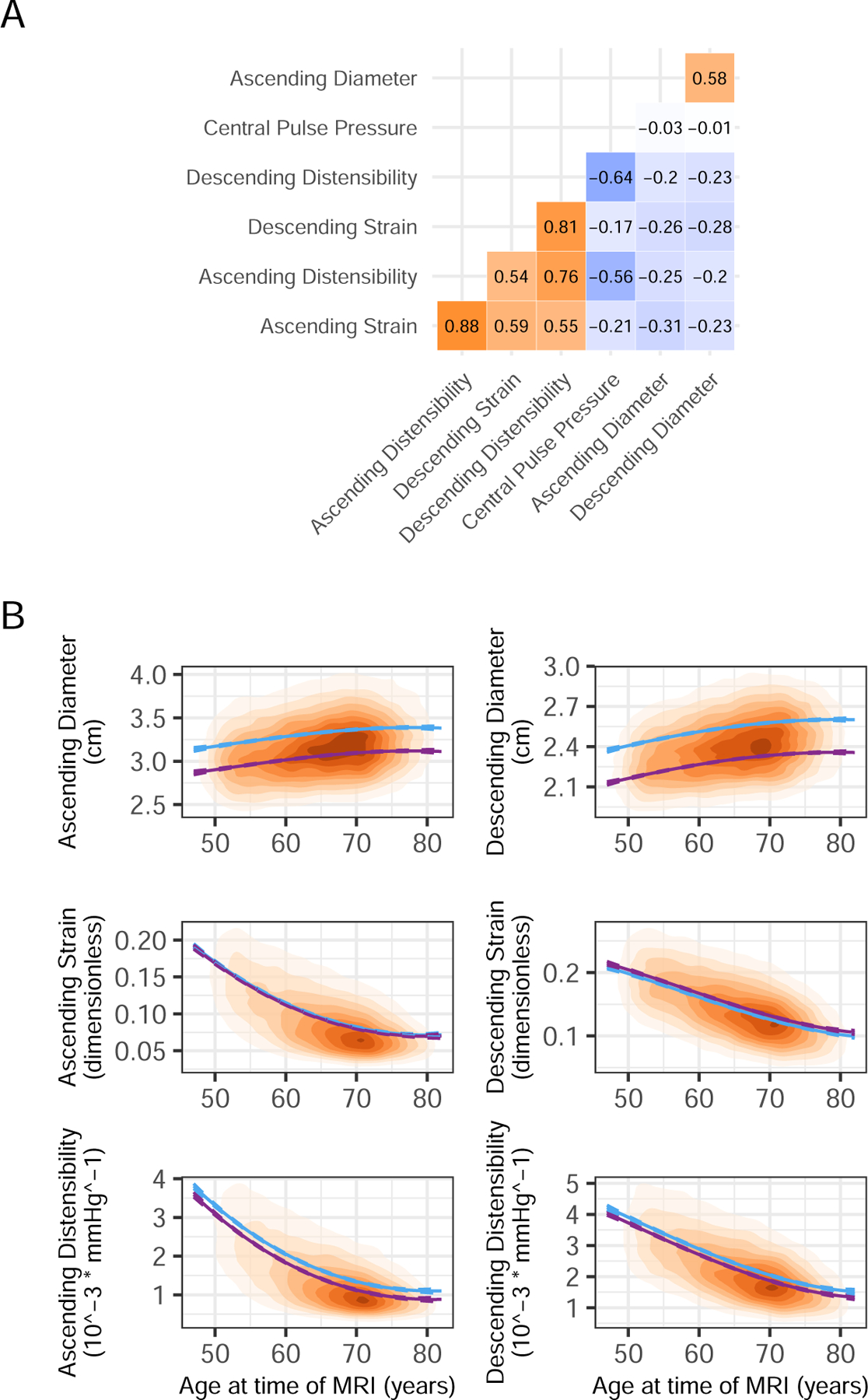
Cross-correlations and age dependencies of aortic parameters
Panel A: Cross-correlation between aortic phenotypes. Panel B: Population density based on age at time of MRI and aortic measurement. Plots are shaded with increasing color intensity by population density. Modeled mean values by age and sex were estimated from a linear model that included sex and a 3 degree-of-freedom basis spline for age (blue for men and in purple for women).
Consistent with previous reports, aortic distensibility was inversely associated with age (Figure 1 Panel B; Supplementary Table 3)24. Distensibility and strain were inversely correlated with anthropometric measurements such as blood pressure, seated height, and heart rate; and blood biomarkers such as triglycerides, glucose, albumin, and hemoglobin (Supplementary Figure 3, Supplementary Table 4). They were positively correlated with mean platelet volume, physical activity, and testosterone, the latter of which was only significant in men but not women (Supplementary Results). Associations with weight were significant but directionally opposed for ascending (0.02 standard deviation [SD] per SD, P = 4.1E-06) and descending (−0.03 SD per SD, P = 3.6E-07) aortic strain.
We assessed the relationships between aortic phenotypes and several cardiovascular diseases (defined in Supplementary Table 1) and their risk factors. Hypertension diagnosed prior to imaging was associated with lower strain and distensibility (Figure 2 left panel). The presence of coronary artery disease (CAD) was associated with increased strain and distensibility, contrary to expectation25. The strongest CAD association was with descending aortic strain, which was 0.24 SD higher in those with a history of CAD (P = 1.6E-26). Previous reports have described that administration of beta receptor antagonists can increase aortic distensibility26. To investigate whether a medication effect might mediate the unexpected CAD-strain observation, we computed the association between aortic phenotypes and medication classes being taken by study participants as defined by ATC codes. The 2,265 participants who reported taking beta blockers had strain and distensibility that were as much as 0.25 standard deviation higher than those of participants not reporting use of beta blockers (P = 2.2E-40; Supplementary Table 5; Supplementary Figure 4). After excluding participants taking beta blockers (leaving 842 of the original 1,550 participants with prevalent CAD), the association effect size of the presence of CAD on descending strain diminished from 0.24 to 0.13, and the association P value diminished by 20 orders of magnitude (from 1.6E-26 to 8.2E-06) (Supplementary Tables 6-7).
Figure 2.
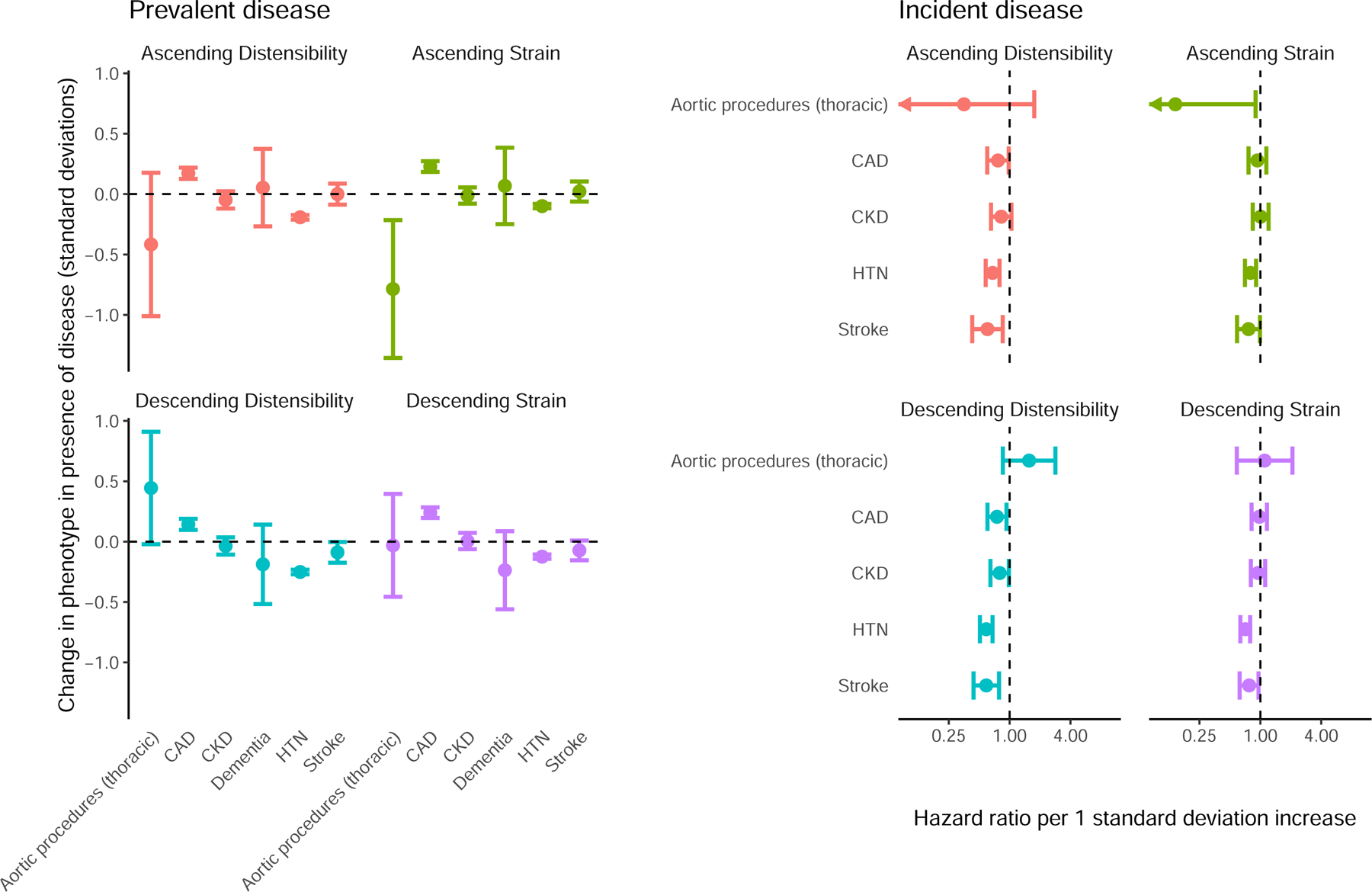
Relationship between strain, distensibility, and disease
Left panel (“Prevalent disease”): the difference in standard deviation normalized aortic phenotypes (y axis) between UK Biobank participants with a history of thoracic aortic procedures, CAD, CKD, Alzheimer’s disease or dementia (“Dementia”), HTN, or stroke occurring prior to MRI, compared to participants without a history of disease (x axis). Right panel (“Incident disease”): hazard ratios for disease incidence (y axis) occurring after MRI per standard deviation increase in the aortic phenotypes (x axis). 95% confidence intervals are represented by error bars.
We produced estimates of the effect of medications on aortic phenotypes: 2,712 participants had undergone two UK Biobank imaging visits; a small number of participants had started taking one or more new medications in between the two visits, allowing an estimate of 43 medication effects on aortic phenotypes at two-tailed P < 0.05 (Supplementary Table 8). For example, beta blockers were associated with a 0.12 SD decrease in ascending aortic diameter (P = 1.2E-04), a 0.31 SD increase in ascending strain (P = 1.3E-04) and a 0.36 SD increase in ascending distensibility (P = 7.3E-04). We conducted PheCode-based association studies before (Supplementary Table 9; Supplementary Figure 5) and after (Supplementary Table 10; Supplementary Figure 6) adjusting the aortic phenotypes to account for the additive effect size of medications (see Supplementary Methods)27. Medications statistically accounted for several of the apparent relationships between disease and aortic phenotypes. For example, accounting for medication use decreased the effect estimate of ischemic heart disease on ascending strain from 0.16 SD (P=3.5E-20) to 0.02 SD (P=0.18); and from 0.10 SD (P=3.5E-08) to 0.00 SD (P=0.80) for ascending distensibility.
Finally, we examined incident cardiovascular diseases that occurred after the aortic phenotypes had been measured. We found greater descending aortic distensibility to be associated with significantly lower chances of being subsequently diagnosed with hypertension (455 incident cases; HR 0.59 per SD increase in descending aortic distensibility; P = 3.8E-13) or stroke (119 incident cases; HR 0.59 per SD increase in descending aortic distensibility; P = 3.1E-04) (Supplementary Tables 11-12; Figure 2 right panel). These associations remained similar in magnitude and statistically significant when adjusting for descending aortic diameter (e.g., stroke HR 0.61; P = 9.2E-04; Supplementary Table 13). They also remained similar after removing participants using beta blockers (Supplementary Table 14).
Heritability of aortic distensibility
Of the 42,342 participants with aortic measurements, 40,577 had genetic data and contributed to at least one genetic analysis (Supplementary Figure 1; Supplementary Table 15). The SNP-heritability using BOLT-REML was 25% for ascending and 22% descending aortic distensibility15. It was 33% for ascending and 30% for descending aortic strain (Supplementary Table 16; cross-trait genetic correlations in Supplementary Table 17 and Supplementary Figure 7).
Common variant genetics of aortic strain and distensibility
We then conducted a GWAS for each aortic trait with BOLT-LMM15. At a commonly used significance threshold of 5E-08, we identified 26 loci associated with ascending aortic strain, 21 with descending aortic strain, 12 with ascending aortic distensibility, and 11 with descending aortic distensibility (Figure 3). There was substantial test statistic inflation (QQ plots in Supplementary Figure 8), but ldsc revealed intercepts near unity and minimal attenuation ratios, consistent with polygenicity rather than confounding (Supplementary Table 18)16. All distensibility-associated loci are shown in Table 1; the full list of strain and distensibility loci is in Supplementary Table 19; and locus plots are in Supplementary Figure 9.
Figure 3.
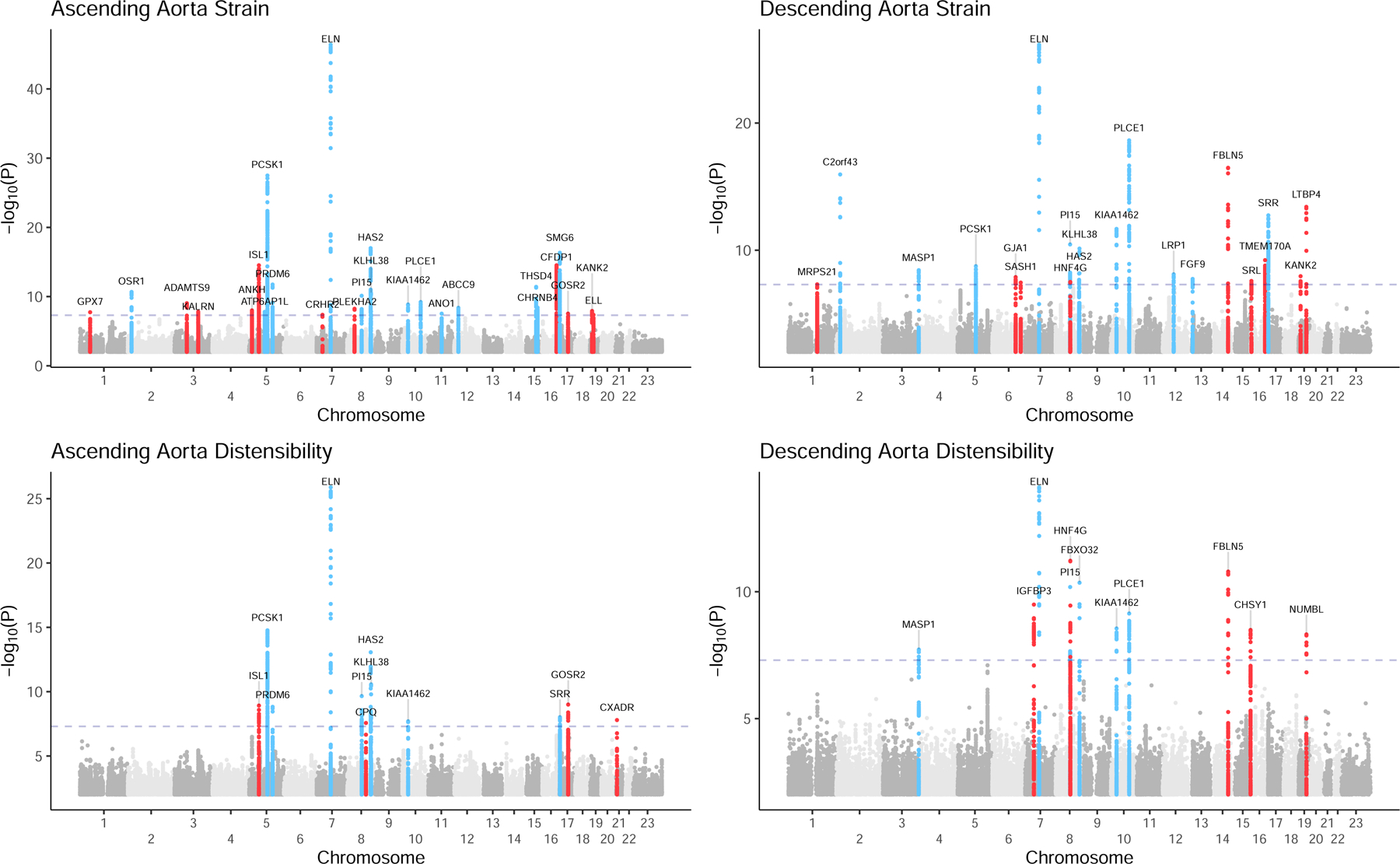
Manhattan plots for genome wide association studies of aortic strain and distensibility
Manhattan plots depicting ascending and descending aortic strain and distensibility. Chromosomal position is on the x axis, and −log10(P value) is on the y axis. Loci with at least one SNP with P < 5E-08 are depicted in color and labeled with the name of the nearest gene. Loci not associated with aortic diameter are red.
Table 1.
GWAS loci for distensibility of the ascending and descending aorta
| Trait | CHR | POS | SNP | Effect Allele | Other Allele | EAF | BETA | SE | P Value | Gene Name | Diameter GWAS |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ascending | 5 | 51200058 | rs10054404 | C | T | 0.48 | −0.039 | 0.006 | 1.20E-09 | ISL1 | Absent |
| Ascending | 5 | 95606712 | rs764443335 | AATG | A | 0.65 | −0.051 | 0.007 | 1.70E-15 | PCSK1 | |
| Ascending | 5 | 122531347 | rs17470137 | G | A | 0.73 | 0.042 | 0.007 | 1.50E-09 | PRDM6 | |
| Ascending | 7 | 73431169 | rs11768878 | A | G | 0.55 | 0.066 | 0.006 | 1.30E-26 | ELN | |
| Ascending | 8 | 75781818 | rs9721183 | C | T | 0.63 | −0.043 | 0.007 | 2.20E-10 | PI15 | |
| Ascending | 8 | 97922051 | rs56252425 | G | T | 0.75 | 0.041 | 0.007 | 2.70E-08 | CPQ | Absent |
| Ascending | 8 | 122649630 | rs563877611 | C | CT | 0.74 | −0.054 | 0.007 | 8.60E-14 | HAS2 | |
| Ascending | 8 | 124607159 | rs34557926 | C | T | 0.63 | 0.043 | 0.006 | 2.70E-11 | KLHL38 | |
| Ascending | 10 | 30169653 | rs10763764 | A | T | 0.42 | −0.036 | 0.006 | 2.00E-08 | JCAD | |
| Ascending | 17 | 2209888 | rs4548913 | G | A | 0.37 | −0.037 | 0.007 | 9.60E-09 | SRR | |
| Ascending | 17 | 45055107 | rs80335285 | A | G | 0.87 | 0.055 | 0.009 | 1.00E-09 | GOSR2 | Absent |
| Ascending | 21 | 18952575 | rs62239946 | G | A | 0.93 | 0.071 | 0.012 | 1.60E-08 | CXADR | Absent |
| Descending | 3 | 186987608 | rs80288719 | T | C | 0.92 | 0.066 | 0.012 | 1.90E-08 | MASP1 | |
| Descending | 7 | 46010100 | rs10260816 | C | G | 0.56 | −0.039 | 0.006 | 3.20E-10 | IGFBP3 | Absent |
| Descending | 7 | 73414589 | rs13231286 | A | C | 0.53 | 0.049 | 0.006 | 7.60E-15 | ELN | |
| Descending | 8 | 75781818 | rs9721183 | C | T | 0.63 | −0.044 | 0.007 | 6.50E-11 | PI15 | |
| Descending | 8 | 76591880 | rs1449544 | A | C | 0.54 | −0.043 | 0.006 | 6.00E-12 | HNF4G | Absent |
| Descending | 8 | 124579985 | rs6470156 | G | A | 0.72 | 0.047 | 0.007 | 4.40E-11 | FBXO32 | |
| Descending | 10 | 30170487 | rs914279 | T | G | 0.42 | −0.037 | 0.006 | 2.80E-09 | JCAD | |
| Descending | 10 | 95910761 | rs79958663 | C | T | 0.83 | 0.051 | 0.008 | 7.20E-10 | PLCE1 | |
| Descending | 14 | 92391955 | rs8023114 | G | A | 0.64 | −0.043 | 0.007 | 1.60E-11 | FBLN5 | Absent |
| Descending | 15 | 101789567 | rs8025126 | C | T | 0.68 | −0.04 | 0.007 | 3.20E-09 | CHSY1 | Absent |
| Descending | 19 | 41188985 | rs113228202 | G | A | 0.99 | −0.175 | 0.03 | 4.80E-09 | NUMBL | Absent |
The lead SNPs from the GWAS for the distensibility of the ascending and descending thoracic aorta. CHR = chromosome; POS = position, GRCh37 coordinates; SNP = the rsID of the variant; EAF = effect allele frequency; SE = standard error. Loci not identified in the thoracic aortic diameter GWAS are highlighted as ‘Absent’ in the ‘Diameter GWAS’ column.
Together, 22 distinct genetic loci were specific to strain and distensibility, after excluding loci found among 110 loci that were associated with ascending or descending thoracic aortic diameter within the same participants (Supplementary Table 20). These 22 loci included 11 associated with ascending strain, nine with descending strain, four with ascending distensibility, and five with descending distensibility (Table 2). A GWAS for central pulse pressure within the same participants yielded five loci; one overlapped with a strain or distensibility locus (rs10260816 near IGFBP3; Supplementary Table 21).
Table 2.
GWAS loci unique to aortic distensibility and strain
| Trait | CHR | POS | SNP | Effect Allele | Other Allele | EAF | BETA | SE | P Value | Gene Name |
|---|---|---|---|---|---|---|---|---|---|---|
| Ascending Aorta Strain | 1 | 53081658 | CT | C | 0.44 | 0.036 | 0.006 | 1.80E-08 | GPX7 | |
| Ascending Aorta Strain | 3 | 64723072 | rs7638565 | A | G | 0.41 | 0.036 | 0.006 | 9.00E-10 | ADAMTS9 |
| Ascending Aorta Strain | 3 | 124026421 | rs146130720 | A | ATAAGT | 0.35 | −0.037 | 0.006 | 1.60E-08 | KALRN |
| Ascending Aorta Strain | 5 | 15023774 | rs9687138 | C | A | 0.83 | −0.045 | 0.008 | 9.90E-09 | ANKH |
| Ascending Aorta Strain | 5 | 51201361 | rs13158444 | T | C | 0.41 | −0.049 | 0.006 | 3.00E-15 | ISL1 |
| Ascending Aorta Strain | 7 | 30727202 | rs75520340 | T | A | 0.97 | −0.092 | 0.017 | 4.30E-08 | CRHR2 |
| Ascending Aorta Strain | 8 | 38761012 | rs75233095 | G | A | 0.83 | −0.046 | 0.008 | 5.70E-09 | PLEKHA2 |
| Ascending Aorta Strain | 16 | 75413242 | rs150284594 | A | AAG | 0.41 | −0.048 | 0.006 | 3.00E-15 | CFDP1 |
| Ascending Aorta Strain | 17 | 45013271 | rs17608766 | T | C | 0.86 | 0.046 | 0.009 | 2.90E-08 | GOSR2 |
| Ascending Aorta Strain | 19 | 11278681 | GT | G | 0.69 | −0.04 | 0.007 | 1.20E-08 | KANK2 | |
| Ascending Aorta Strain | 19 | 18567771 | rs796826122 | GA | G | 0.77 | 0.039 | 0.007 | 4.30E-08 | ELL |
| Descending Aorta Strain | 1 | 150267316 | rs143847321 | C | CAAAAAA | 0.49 | −0.032 | 0.006 | 4.80E-08 | MRPS21 |
| Descending Aorta Strain | 6 | 122089704 | rs58730006 | A | AT | 0.9 | 0.056 | 0.01 | 1.30E-08 | GJA1 |
| Descending Aorta Strain | 6 | 148639909 | rs4574664 | G | T | 0.81 | 0.043 | 0.008 | 3.50E-08 | SASH1 |
| Descending Aorta Strain | 8 | 76591880 | rs1449544 | A | C | 0.54 | −0.034 | 0.006 | 3.10E-08 | HNF4G |
| Descending Aorta Strain | 14 | 92393198 | rs8014161 | T | A | 0.64 | −0.051 | 0.006 | 3.30E-17 | FBLN5 |
| Descending Aorta Strain | 16 | 4281391 | rs11864324 | T | G | 0.78 | −0.037 | 0.007 | 2.60E-08 | SRL |
| Descending Aorta Strain | 16 | 75489254 | CA | C | 0.49 | 0.038 | 0.006 | 6.00E-10 | TMEM170A | |
| Descending Aorta Strain | 19 | 11271714 | rs113783450 | G | A | 0.69 | −0.038 | 0.006 | 1.10E-08 | KANK2 |
| Descending Aorta Strain | 19 | 41117300 | rs34093919 | G | A | 0.99 | −0.203 | 0.027 | 3.90E-14 | LTBP4 |
| Ascending Aorta Distensibility | 5 | 51200058 | rs10054404 | C | T | 0.48 | −0.039 | 0.006 | 1.20E-09 | ISL1 |
| Ascending Aorta Distensibility | 8 | 97922051 | rs56252425 | G | T | 0.75 | 0.041 | 0.007 | 2.70E-08 | CPQ |
| Ascending Aorta Distensibility | 17 | 45055107 | rs80335285 | A | G | 0.87 | 0.055 | 0.009 | 1.00E-09 | GOSR2 |
| Ascending Aorta Distensibility | 21 | 18952575 | rs62239946 | G | A | 0.93 | 0.071 | 0.012 | 1.60E-08 | CXADR |
| Descending Aorta Distensibility | 7 | 46010100 | rs10260816 | C | G | 0.56 | −0.039 | 0.006 | 3.20E-10 | IGFBP3 |
| Descending Aorta Distensibility | 8 | 76591880 | rs1449544 | A | C | 0.54 | −0.043 | 0.006 | 6.00E-12 | HNF4G |
| Descending Aorta Distensibility | 14 | 92391955 | rs8023114 | G | A | 0.64 | −0.043 | 0.007 | 1.60E-11 | FBLN5 |
| Descending Aorta Distensibility | 15 | 101789567 | rs8025126 | C | T | 0.68 | −0.04 | 0.007 | 3.20E-09 | CHSY1 |
| Descending Aorta Distensibility | 19 | 41188985 | rs113228202 | G | A | 0.99 | −0.175 | 0.03 | 4.80E-09 | NUMBL |
Lead SNPs from the GWAS of the strain and distensibility of the ascending and descending thoracic aorta which were not present in GWAS of thoracic aortic diameter. CHR = chromosome; POS = position, GRCh37 coordinates; SNP = the rsID of the variant, where available; EAF = effect allele frequency; SE = standard error.
For all lead SNPs that were associated with either ascending aortic strain or distensibility, the strain- or distensibility-increasing allele was usually directionally associated with decreased aortic diameter, even when the latter was not statistically significant (Supplementary Figure 10).
To prioritize genes for each aortic phenotype, we performed rare variant analysis, multi-marker analysis of genomic annotation (“MAGMA”), a transcriptome wide association study (TWAS), and a single nucleus sequencing analysis (Supplementary Figures 10-15 and Supplementary Tables 22-23, details in Supplementary Results). No genes reached statistical significance in a collapsing analysis of rare germline loss-of-function variants from exome sequencing. Single nucleus sequencing expression analyses showed cell-type specificity in vascular smooth muscle, fibroblast and endothelial cells for several genes prioritized by TWAS. We observed converging evidence from MAGMA and TWAS for ADAMTS7, PRDM6, MASP1, ZNF827, and LDAH in their respective loci.
Polygenic score analyses
We constructed ~1.1 million SNP polygenic scores using PRScs-auto, and surveyed the connection between those scores and cardiovascular diseases using accelerated failure time models in up to 375,184 UK Biobank participants who did not undergo MRI and therefore did not contribute to the polygenic score derivation19. In analyses adjusted for age, sex, and genetic principal components, the polygenic scores were associated with hypertension, CAD, chronic kidney disease (CKD), stroke, and thoracic aortic surgical procedures (Figure 4; Supplementary Table 24). Among the strain and distensibility polygenic scores, the largest effect size was found between the ascending strain polygenic score and risk for thoracic aortic procedures: a 1 standard deviation (SD) increase in genetically predicted ascending aortic strain was associated with a 17.7% delay in disease onset (P = 1.6E-03). Most effect sizes of the strain and distensibility polygenic scores were small; equivalent to delaying or accelerating disease onset by 2–5% per standard deviation change in polygenic score for most of the diseases.
Figure 4:
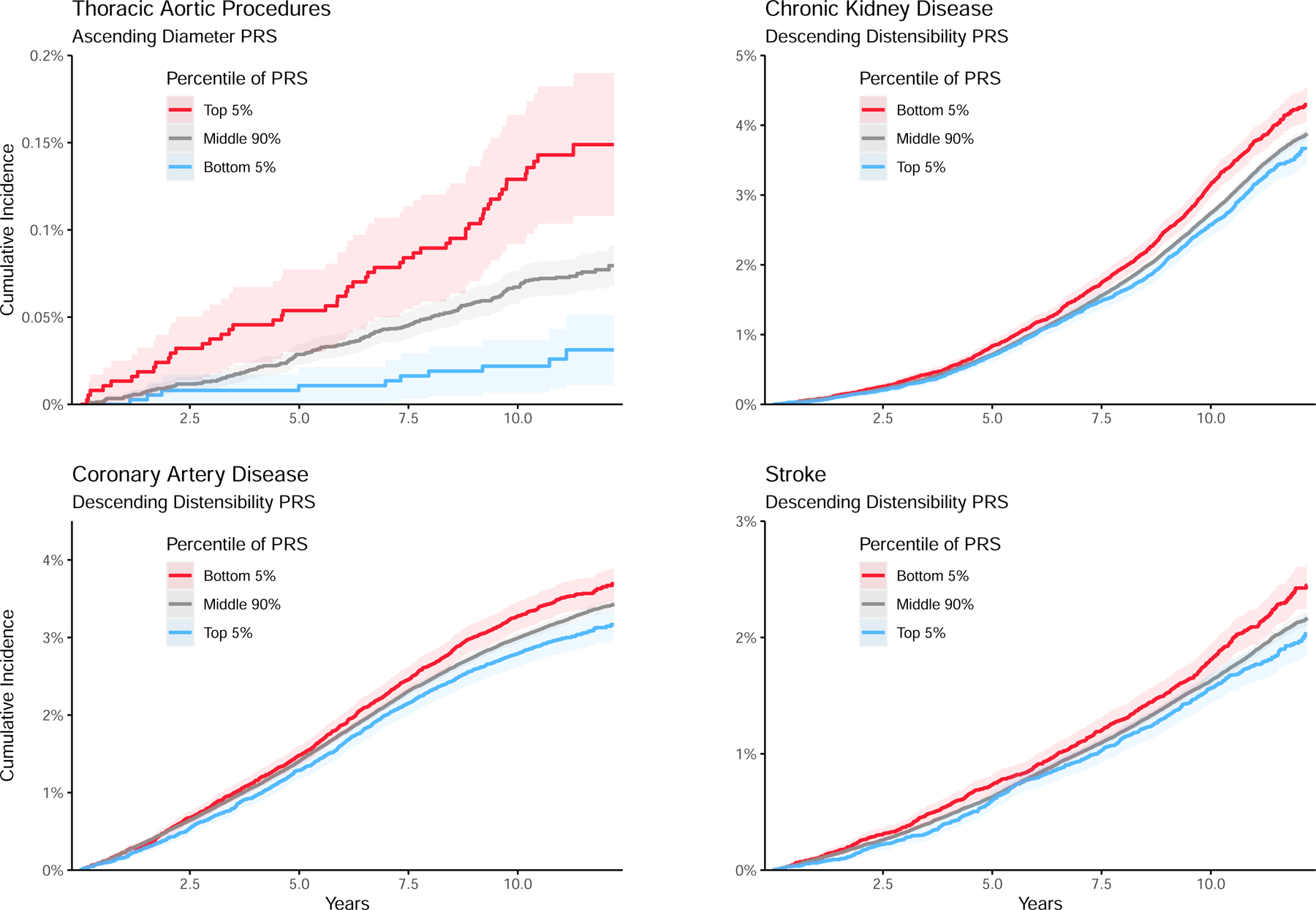
Polygenic scores
Polygenic scores were computed using PRScs. The scores were tested for association with cardiovascular disease endpoints in up to 375,184 UK Biobank participants. Accelerated failure time analysis was pursued with the survreg function in the R survival package, using the log-logistic distribution. Red denotes participants in the top 5% (for diameter) or bottom 5% (for distensibility) polygenic score, while blue denotes the bottom 5% (for diameter) or top 5% (for distensibility).
Among all tested polygenic scores, the largest effect size was found between the ascending aortic diameter polygenic score and risk for thoracic aortic procedures: a 1 SD increase in the score was associated with a 31.5% acceleration in disease onset, P = 8.4E-12. Therefore, we reanalyzed the strain and distensibility scores after adjustment for the aortic diameter polygenic score. The polygenic prediction of descending aortic distensibility remained statistically significantly associated with risk for hypertension, CAD, CKD, and stroke, while the ascending distensibility polygenic score remained significantly associated with hypertension and CKD (Supplementary Table 25).
Discussion
Multiple cardiovascular morbidities have been associated with abnormalities of aortic distensibility, most prominently atherosclerosis, aneurysms, hypertension, and aging. Unlike the static measurement of aortic diameter, distensibility accounts for the dynamic change in aortic cross-sectional area during the cardiac cycle and pulse pressure. Despite the importance of this function in the cardiovascular system, few biologic determinants have been identified. In this study, using a deep learning model that achieved high accuracy, we quantified the thoracic aortic cross-sectional area throughout the cardiac cycle in 42,342 participants to measure strain and distensibility, analyzing their epidemiological properties and genetic architecture.
In epidemiologic and genetic analyses, we found that aortic strain and distensibility provided relevant information in addition to that provided by the static aortic diameter measurement. Greater descending aortic distensibility was associated with lower risk of a future diagnosis of stroke (HR 0.61, P = 9.2E-04), despite accounting for age, sex, and descending aortic diameter. Among UK Biobank participants who did not undergo imaging, we calculated a genetic prediction of descending aortic distensibility, observing that it was associated with more stroke-free time (~3% delay in time to stroke; P = 2E-04), even after accounting for the genetic prediction of descending aortic diameter. Similar observations were made for CAD and chronic kidney disease, underscoring the notion that strain and distensibility add information beyond that which is obtained by assessing only aortic diameter.
A series of analyses highlighted the close relationship between vasoactive medications and aortic distension. We observed a positive association between ischemic heart disease and strain; this unexpected finding was contradicted by survival analyses, which found the expected inverse relationship between distensibility—whether measured or genetically predicted—and CAD risk. We hypothesized that medications could explain the unexpected positive correlation. A small number of individuals had undergone imaging twice and started a beta blocker between those visits (N=53); in that group, we observed a substantial association between medication initiation and aortic strain (effect size equivalent to 0.31 SD; P = 1.3E-04) and distensibility (0.36 SD; P = 7.3E-04) with a relatively smaller effect on diameter (−0.12 SD, equivalent to −0.4 mm; P = 1.2E-04). Statistically, adjusting for medication use accounted for the entirety of the counterintuitive positive association between prevalent ischemic heart disease and ascending strain (from 0.16 SD [P = 3.5E-20] before accounting for medication use to 0.02 SD [P = 0.18] afterwards). Prior observations showed that beta blockade increases aortic distensibility in people with Marfan syndrome26. Here, we observed that beta blockade initiation in a broader population was linked to an increase in strain and distensibility and decrease in diameter, and we extended those observations to several additional medication classes (Supplementary Table 8). Further, the observed effect sizes suggested that vasoactive medications’ effects on aorta may be more readily perceptible by measuring aortic distensibility than diameter alone, with potential implications for monitoring strategies and surrogate endpoints during drug development.
In the genetic analysis, we focused on loci unique to aortic strain and distensibility—and not associated with diameter—since they represent signals not discoverable through diameter alone, and more fundamentally may give insight into diameter-independent aortic wall properties. 22 such loci were identified (Table 2). However, near ADAMTS9, ELL, GOSR2, HNF4G, ISL1, KALRN, LTBP4, and PLEKHA2, the summary statistics revealed at least one nearby SNP with association P < 1E-05 for diameter. Therefore, those loci probably also drive normal variation in diameter, albeit weakly. (The relation between loci associated with strain, distensibility, and diameter are evaluated further in the Supplementary Discussion.)
Among the remaining loci, several associations were interpretable in the context of binding partners or pathways. One example is FBLN5—whose locus was associated with descending aortic strain and distensibility but not diameter. FBLN5 encodes fibulin-5, a mediator of elastogenesis that recruits tropoelastin and elastin cross linking enzymes such as lysyl oxidase like 1 (LOXL1) to microfibrils to direct elastin fiber assembly and maturation28. Homozygous mutations in FBLN5 cause autosomal recessive cutis laxa in humans, a disease of failed elastogenesis29. Fbln5−/− mice have been proposed as a model of vascular aging; our findings lend support to this proposition in humans. Single nucleus sequencing data from aortic tissue identified cell type-specific expression of FBLN5 in vascular smooth muscle cells—the primary cell type responsible for homeostasis of extracellular matrix function in the aorta—and fibroblasts. A notable binding partner of fibulin-5 is latent transforming growth factor beta binding protein 4, encoded by LTBP4, whose locus was also significantly associated with distensibility. LTBP4 also binds to TGF-β, participating in elastogenesis30. While some contributors to extracellular matrix composition (such as ELN) can be detected through their influence on diameter, others such as FBLN5 must be unmasked through a more comprehensive evaluation of the aorta’s dynamic properties.
Several strain- and distensibility-specific loci that were not associated with diameter were found near genes involved in atherosclerosis. A lead SNP specific for descending aortic distensibility was on the transcript of CHSY1, encoding chondroitin sulfate synthase 1—one of the enzymes performing the rate-limiting step in glycosaminoglycan chain elongation. Aortic single nucleus sequencing identified CHSY1 expression in VSMCs and endothelial cells. Expression of this gene is induced in aortic smooth muscle cells by TGF-β via a reactive oxygen species-dependent mechanism31. Rs11992999, associated with diameter and distensibility, is near another gene involved in glycosaminoglycan synthesis, HAS2. Its protein, hyaluronan synthase 2, produces hyaluronic acid—known to modulate the mechanical properties of tissues and affect the proliferation of VSMCs32. Glycosaminoglycans facilitate lipoprotein binding, and thereby promote atherosclerosis31. A lead SNP on the SASH1 transcript, rs4574664, was significantly associated with descending strain but not diameter. SASH1, a member of the TLR4 pathway, has previously been shown to have increased expression in atherosclerotic plaques and to inhibit aortic endothelial cell proliferation33. Finally, two lead SNPs were also expression quantitative trait loci (eQTLs) for expression of their nearest gene. Rs9687138, the lead SNP near the ANKH locus, was associated only with ascending aortic strain but not diameter. This SNP is an eQTL for ANKH: the A allele was associated with increased expression in GTEx v8 and increased strain in our analysis34, consistent with previously established biology wherein ANKH, an inorganic pyrophosphate transport regulator, inhibits vascular calcification35. Rs113783450—an eQTL for KANK2—was also associated with strain and not diameter; its A allele was associated with increased expression and increased strain. KANK2 has previously been identified in atherosclerotic plaques and its expression was negatively correlated with calcification36, consistent with our findings. The association of several atherosclerosis-related loci with strain—but not diameter—suggest that atherosclerosis may be one mechanism by which mechanical properties of the aortic wall become altered in an initially diameter-independent manner. Further study that includes an assessment of vascular calcification will be helpful to refine this biological signal.
In conclusion, we have measured ascending and descending thoracic aortic strain and distensibility in up to 42,342 UK Biobank participants, investigated their epidemiology, and identified 41 significantly associated loci, many of which have established connections to TGF-β signaling, elastogenesis, and atherosclerosis.
Limitations
This study has several limitations. The deep learning models have not been tested outside of the specific devices and imaging protocols used by the UK Biobank and are unlikely to generalize to other data sets without fine tuning. The study population was largely composed of people of European ancestries, limiting generalizability to global populations. Central pulse pressure, used with strain to compute distensibility, was estimated by a validated device and not directly measured; the quality control procedures that we developed for central pulse pressure were heuristic and excluded ~10% of participants from the distensibility analysis. The effect of medication classes on aortic phenotypes was estimated based on a biased sample, because these medications were initiated due to a medical indication—not at random. Due to limited availability of large genotyped cohorts with aortic strain measurements, we have not conducted external replication of the genetic analysis.
Supplementary Material
Clinical Perspectives.
Competency in Medical Knowledge:
Aortic distensibility is predictive of aortopathy, stroke, and hypertension, even when controlling for aortic diameter.
Translational Outlook:
Genetic loci associated with aortic distensibility highlight genes for functional follow-up studies to identify therapeutic targets for aortopathy.
Sources of funding
Dr. Pirruccello was supported by a Sarnoff Scholar Award and National Institutes of Health (NIH) grant K08HL159346. Dr. Lindsay was supported by the Fredman Fellowship for Aortic Disease, the Toomey Fund for Aortic Dissection Research, and NIH R01HL130113. Dr. Ellinor was supported by grants from the National Institutes of Health (R01HL092577, R01HL157635, R01HL139731), from the American Heart Association Strategically Focused Research Networks (18SFRN34110082), and from the European Union (MAESTRIA 965286). Dr. Chou was funded by NIH T32HL007208. Dr. Rämö was funded by a Fellowship from the Sigrid Jusélius Foundation. This work was funded by a collaboration between the Broad Institute and IBM Research. Support for title page creation and format was provided by AuthorArranger, a tool developed at the National Cancer Institute.
Disclosures
Dr. Pirruccello has served as a consultant for Maze Therapeutics and has received research support to the Broad Institute from IBM Research. Dr. Juric is supported by grants from Genentech, Eisai, EMD Serono, Takeda, Amgen, Celgene, Placon Therapeutics, Syros, Petra Pharma, InventisBio, Infinity Pharmaceuticals, and Novartis. Dr. Juric has also received personal fees from Genentech, Eisai, EMD Serono, Ipsen, Syros, Relay Therapeutics, MapKure, Vibliome, Petra Pharma, and Novartis. Dr. Batra receives sponsored research support from Bayer AG and IBM, and has consulted for Novartis and Prometheus Biosciences. Dr. Ellinor has received sponsored research support from Bayer AG, IBM Research, Bristol Myers Squibb and Pfizer; he has also served on advisory boards or consulted for Bayer AG, MyoKardia and Novartis. The Broad Institute has filed for a patent on an invention from Drs. Ellinor, Lindsay, and Pirruccello related to a genetic risk predictor for aortic disease. The remaining authors report no disclosures.
Abbreviations
- ATC
Anatomical Therapeutic Chemical
- CAD
coronary artery disease
- CKD
chronic kidney disease
- eQTL
expression quantitative trait locus
- HR
hazard ratio
- MAGMA
multi-marker analysis of genomic annotation
- SNP
single nucleotide polymorphism
- SD
standard deviation
- TWAS
transcriptome-wide association study
Footnotes
Tweet
Analysis of aortic distensibility from 40,000 UK Biobank MRIs now out in @JACCjournals reveals medication effects; links to coronary disease and stroke; and new genetic loci for strain and distensibility that aren’t found when just looking at static aortic diameter.
Data availability
UK Biobank data are made available to researchers from research institutions with genuine research inquiries, following IRB and UK Biobank approval. GWAS summary statistics and polygenic score weights will be available upon publication at the Broad Institute Cardiovascular Disease Knowledge Portal (http://www.broadcvdi.org ). Single nucleus sequencing data were retrieved from the Broad Institute Single Cell Portal #SCP1265. Aortic diameter, strain, and distensibility measurements will be returned to the UK Biobank for use by any approved researcher. Other data are contained within the article and its supplementary information.
References
- 1.Vilacosta I et al. Acute Aortic Syndrome Revisited: JACC State-of-the-Art Review. J Am Coll Cardiol 78, 2106–2125 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Chirinos JA, Segers P, Hughes T & Townsend R Large-Artery Stiffness in Health and Disease: JACC State-of-the-Art Review. J Am Coll Cardiol 74, 1237–1263 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berne & Levy. Physiology 7th Edition. [Google Scholar]
- 4.Ohtsuka S, Kakihana M, Watanabe H & Sugishita Y Chronically decreased aortic distensibility causes deterioration of coronary perfusion during increased left ventricular contraction. J Am Coll Cardiol 24, 1406–1414 (1994). [DOI] [PubMed] [Google Scholar]
- 5.Resnick LM et al. Direct magnetic resonance determination of aortic distensibility in essential hypertension: relation to age, abdominal visceral fat, and in situ intracellular free magnesium. Hypertension 30, 654–659 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Stefanadis C, Wooley CF, Bush CA, Kolibash AJ & Boudoulas H Aortic distensibility abnormalities in coronary artery disease. Am J Cardiol 59, 1300–1304 (1987). [DOI] [PubMed] [Google Scholar]
- 7.Jeremy RW et al. Relation between age, arterial distensibility, and aortic dilatation in the Marfan syndrome. Am J Cardiol 74, 369–373 (1994). [DOI] [PubMed] [Google Scholar]
- 8.Wilson KA et al. The relationship between aortic wall distensibility and rupture of infrarenal abdominal aortic aneurysm. J Vasc Surg 37, 112–117 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Pase MP et al. Aortic Stiffness and the Risk of Incident Mild Cognitive Impairment and Dementia. Stroke 47, 2256–2261 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Redheuil A et al. Proximal aortic distensibility is an independent predictor of all-cause mortality and incident CV events: the MESA study. J Am Coll Cardiol 64, 2619–2629 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sudlow C et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS medicine 12, e1001779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petersen SE et al. UK Biobank’s cardiovascular magnetic resonance protocol. J Cardiovasc Magn Reson 18, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howard J & Gugger S Fastai: A Layered API for Deep Learning. Information 11, 108 (2020). [Google Scholar]
- 14.Therneau TM & Grambsch PM Modeling Survival Data: Extending the Cox Model. (Springer-Verlag, 2000). doi: 10.1007/978-1-4757-3294-8. [DOI] [Google Scholar]
- 15.Loh P-R, Kichaev G, Gazal S, Schoech AP & Price AL Mixed-model association for biobank-scale datasets. Nature Genetics 50, 906–908 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nature Genetics 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Leeuw CA, Mooij JM, Heskes T & Posthuma D MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Comput Biol 11, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turley P et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nature Genetics 50, 229–237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge T, Chen C-Y, Ni Y, Feng Y-CA & Smoller JW Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nature Communications 10, 1776 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mbatchou J et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat Genet 53, 1097–1103 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Pirruccello JP et al. Deep learning enables genetic analysis of the human thoracic aorta. Nature Genetics 54, 40–51 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J et al. FTO genotype is associated with phenotypic variability of body mass index. Nature 490, 267–272 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hickson SS et al. Validity and repeatability of the Vicorder apparatus: a comparison with the SphygmoCor device. Hypertens Res 32, 1079–1085 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Redheuil A et al. Reduced ascending aortic strain and distensibility: earliest manifestations of vascular aging in humans. Hypertension 55, 319–326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bu Z et al. Ascending Aortic Strain Analysis Using 2‐Dimensional Speckle Tracking Echocardiography Improves the Diagnostics for Coronary Artery Stenosis in Patients With Suspected Stable Angina Pectoris. Journal of the American Heart Association 7, e008802 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Groenink M, de Roos A, Mulder BJ, Spaan JA & van der Wall EE Changes in aortic distensibility and pulse wave velocity assessed with magnetic resonance imaging following beta-blocker therapy in the Marfan syndrome. Am J Cardiol 82, 203–208 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Wu P et al. Mapping ICD-10 and ICD-10-CM Codes to Phecodes: Workflow Development and Initial Evaluation. JMIR Med Inform 7, e14325 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papke CL & Yanagisawa H Fibulin-4 and fibulin-5 in elastogenesis and beyond: Insights from mouse and human studies. Matrix Biol 37, 142–149 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loeys B et al. Homozygosity for a missense mutation in fibulin-5 (FBLN5) results in a severe form of cutis laxa. Hum Mol Genet 11, 2113–2118 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Noda K et al. Latent TGF-β binding protein 4 promotes elastic fiber assembly by interacting with fibulin-5. PNAS 110, 2852–2857 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohamed R et al. Transforming growth factor-β1 mediated CHST11 and CHSY1 mRNA expression is ROS dependent in vascular smooth muscle cells. J Cell Commun Signal 13, 225–233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evanko SP, Angello JC & Wight TN Formation of hyaluronan- and versican-rich pericellular matrix is required for proliferation and migration of vascular smooth muscle cells. Arterioscler Thromb Vasc Biol 19, 1004–1013 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Weidmann H et al. SASH1, a new potential link between smoking and atherosclerosis. Atherosclerosis 242, 571–579 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Lonsdale J et al. The Genotype-Tissue Expression (GTEx) project. Nature Genetics 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen B et al. Wnt1 inhibits vascular smooth muscle cell calcification by promoting ANKH expression. J Mol Cell Cardiol 135, 10–21 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Slenders L et al. Intersecting single-cell transcriptomics and genome-wide association studies identifies crucial cell populations and candidate genes for atherosclerosis. European Heart Journal Open 2, oeab043 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
UK Biobank data are made available to researchers from research institutions with genuine research inquiries, following IRB and UK Biobank approval. GWAS summary statistics and polygenic score weights will be available upon publication at the Broad Institute Cardiovascular Disease Knowledge Portal (http://www.broadcvdi.org ). Single nucleus sequencing data were retrieved from the Broad Institute Single Cell Portal #SCP1265. Aortic diameter, strain, and distensibility measurements will be returned to the UK Biobank for use by any approved researcher. Other data are contained within the article and its supplementary information.


