Abstract
Objective
T cells contribute to tissue injury in systemic sclerosis (SSc), yet the specific T cell subsets expanded in patients with SSc remain incompletely defined. Here we evaluated specific phenotypes and functions of peripheral helper T (Tph) and follicular helper T (Tfh) cells, which have been implicated in autoantibody production, and assessed their associations with clinical features in a well‐characterized cohort of patients with SSc.
Methods
Mass cytometry of T cells from peripheral blood mononuclear cells of patients with SSc and controls were evaluated using t‐distributed stochastic neighbor embedding visualization, biaxial gating, and marker expression levels. Findings were validated with flow cytometry and in vitro assays.
Results
The frequencies of PD‐1highCXCR5+ Tfh cells and PD‐1highCXCR5− Tph cells were similar in patients with SSc and controls. t‐distributed stochastic neighbor embedding visualization (tSNE) revealed distinct populations within the PD‐1highCXCR5− cells distinguished by expression of HLA–DR and inducible costimulator (ICOS). Among PD‐1highCXCR5− cells, only the HLA–DR+ICOS− cell population was expanded in patients with SSc. Cytometric and RNA sequencing analyses indicated that these cells expressed cytotoxic rather than B cell helper features. HLA–DR+ICOS− PD‐1highCXCR5− cells were less potent in inducing B cell plasmablast differentiation and antibody production than comparator T helper cell populations. HLA–DR+ICOS−PD‐1highCXCR5− cells were significantly associated with the presence and severity of interstitial lung disease among patients with SSc.
Conclusion
Among PD‐1highCXCR5− T cells, a subset of HLA–DR+ICOS− cells with cytotoxic features is specifically expanded in patients with SSc and is significantly associated with interstitial lung disease severity. This potential cytotoxicity appearing in the CD4 T cell population can be evaluated as a prognostic disease biomarker in patients with SSc.
INTRODUCTION
Systemic sclerosis (SSc) is an autoimmune disease characterized by microvascular damage, autoantibody production, and progressive multiorgan fibrosis associated with high morbidity and decreased survival. 1 Although skin thickening is often considered the hallmark clinical feature of this disorder, lung involvement is also common and is one of the main determinants of increased mortality in SSc. 1 Treatment and control of SSc is challenging because the underlying disease process can be subclinical during early phases, and no biomarkers are currently available to reliably identify or quantify the extent of pathologic immune activation.
T cells have been suspected to be primary drivers of the immune response that causes skin and lung injury in SSc, but the specific T cell subsets that are responsible for disease onset or progression remain unclear. 2 , 3 Affected tissue samples obtained from patients with SSc show prominent infiltration of T cells. 4 , 5 Several studies have reported a perturbed repertoire of specific CD4+ T cell subsets in the blood and lesional skin tissues, including Th2 cells, Th17 cells, regulatory T cells, and cytotoxic CD4+ T cells. 2 , 6 , 7 , 8 Cytotoxic CD4 T cells accumulating in SSc skin may directly target endothelial cells to induce microvascular injury and promote tissue fibrosis. 7 B cell–T helper cells, including both follicular helper T (Tfh) cells and peripheral helper T (Tph) cells, have been recently associated with the SSc disease process. 9 , 10 Tfh cells, a subset of CD4 T cells often identified by PD‐1+CXCR5+ expression, are considered the principal T cell population capable of stimulating B cell responses in secondary lymphoid organs. 11 Tph cells, a CD4 T cell subset identified by PD‐1highCXCR5− expression, participate in the stimulation, recruitment, and activation of B cells in peripheral tissue and have been implicated in several autoimmune diseases. 12 Increases in both circulating CXCR5−PD‐1+CD4+ T cells and Tfh cells have been reported in SSc. 9 An independent study observed modest increases in Tfh cells, but not Tph cells, in the circulation of patients with early diffuse cutaneous SSc. 12 Another investigation identified an expanded T cell subset exhibiting a Tph cell phenotype, which included high expression of programmed death 1 (PD‐1) and inducible costimulator (ICOS) without CXCR5 expression, in the circulation of patients with multiple autoimmune diseases, including SSc, using mass cytometry. 12 , 13
Although Tfh cells can be readily identified by expression of CXCR5, a chemokine receptor that promotes their migration into lymphoid follicles, specific markers defining Tph cells are not well established. 14 Most studies have differentiated Tfh and Tph populations using their broader expression of PD‐1 and the presence (Tfh) or absence (Tph) of CXCR5. 15 However, there is substantial heterogeneity in the surface phenotypes of both Tfh and Tph cells, including varied expressions of ICOS, HLA–DR, and T cell immunoreceptor with Ig and ITIM domains (TIGIT). 14 , 15 , 16 These markers may distinguish subpopulations that are functionally distinct.
Here we have used high‐dimensional mass cytometry to quantify and conduct refined phenotyping of PD‐1+ T cells in a large well‐characterized cohort of patients with SSc. We identify a population of PD‐1highCXCR5−CD4+ T cells that express HLA–DR, but not ICOS, that is expanded in patients with SSc and is significantly associated with the presence and severity of interstitial lung disease (ILD). This population shows a cytotoxic profile, in contrast to the phenotype typical of Tph cells, implicating a specific pathogenic function for this expanded T cell subset.
PATIENTS AND METHODS
Study participants
Peripheral blood mononuclear cell (PBMC) samples were obtained from 67 patients with SSc observed at the University of California, San Francisco (UCSF). Additionally, PBMC samples were collected from 15 patients with SSc and 18 healthy controls without a known autoimmune disease recruited from Brigham and Women's Hospital (BWH). All patients with SSc fulfilled the 2013 American College of Rheumatology/EULAR criteria for SSc. 17 Clinical assessment included data on age at onset of Raynaud phenomenon, age at first nonRaynaud phenomenon symptom, sex, race, smoking status, SSc subtype (limited or diffuse), extent of cutaneous involvement measured by the modified Rodnan skin thickness score (MRSS), and autoantibody status. Presence of ILD was confirmed by radiographic evidence of pulmonary fibrosis on high‐resolution computed tomography (HRCT) conducted in patients exhibiting forced vital capacity (FVC) <80% or absolute decline of the FVC (L) >10% over two consecutive assessments. Pulmonary arterial hypertension (PAH) in the UCSF cohort was confirmed by right‐sided heart catheterization, with mean pulmonary artery pressure (mPAP) >25 mm Hg, peripheral vascular resistance >3 Wood units, and pulmonary capillary wedge pressure <15 mm Hg (PAH definition current at the time of testing). Healthy controls were screened for any evidence of autoimmune systemic disease, neoplasm, or lung pathology.
Collection and processing of samples
Blood samples were collected into lithium heparin tubes, and PBMCs were isolated by density centrifugation using Ficoll‐Hypaque in 50‐mL conical tubes. PBMCs were cryopreserved in 10% DMSO‐containing solution. Cryopreserved PBMCs were thawed into complete culture media (RPMI 1640 Medium supplemented with 5% heat‐inactivated fetal bovine serum, 1 mM GlutaMAX, 10 mM HEPES, and penicillin‐streptomycin). Cells were counted, and 0.2 × 106 to 2 × 106 cells from each sample were transferred to a 96‐well conical bottom polypropylene plate for staining.
Generation of mass cytometry data
After the cells were transferred to the plate, the viability staining reagent cisplatin with a dilution of 1:1,000 was added to the cells directly for two minutes and then diluted with culture media. After centrifugation, a human Fc receptor blocking agent was added at a 1:100 dilution with cell staining buffer for 10 minutes, followed by incubation with metal conjugated antibodies for 30 minutes at room temperature. Antibodies were obtained from the Harvard Medical Area cytometry by time‐of‐flight mass spectrometry (CyTOF) Antibody Resource and Core and Fluidigm. Cells were then fixed in 4% paraformaldehyde for 10 minutes before permeabilization with the FoxP3/Transcription Factor Staining Buffer Kit (eBioscience). Cells were incubated in Transcription Factor Fix/Perm Buffer for 30 minutes before barcoding. Cells were then barcoded using the Cell‐ID 20‐Plex Pd Barcoding kit (Fluidigm) and pooled together into one tube. The metal conjugated intracellular antibody mix was then added into the tube, and cells were incubated for 30 minutes. Cells were then fixed with 4% paraformaldehyde for 10 minutes and then washed out and resuspended in a cell staining buffer and left at 4 °C overnight.
The next day, DNA was labeled for 20 minutes with iridium intercalator solution (Fluidigm). Samples were washed and then counted in the presence of EQ Four Element Calibration beads at a final concentration of 1 × 106/mL. Samples were acquired on a Helios CyTOF Mass Cytometer. Mass cytometry data were generated on PBMC samples from patients with SSc and healthy controls using a 39‐marker mass cytometry panel designed to identify T cell subsets. The panel included markers of cell surface and intracellular signaling proteins (Supplementary Table 1). Samples were processed in five batches, with 20 barcoded samples in each batch. Patient samples were analyzed in barcoded batches, with each batch randomized to include samples from both healthy controls and patients with SSc with different degrees of lung involvement (ILD, PAH, neither, or both). The raw flow cytometry standard files were normalized together using bead standard normalization to minimize batch effects and were debarcoded for analysis.
RNA sequencing gene signature analysis
CD4 T cell–specific cytotoxicity and Tfh gene signatures were composed of genes identified in the studies by Hashimoto et al 18 and Locci et al, 19 respectively (Supplementary Table 2). Gene signature scores were calculated using the sum of the transcript per mission (TPM) expression of signature genes within each CD4 T cell subset in individual samples, and the Z score is depicted. Paired t‐tests were used to compare signature scores among T cell subsets.
T and B cell cocultures
Total B cells and CD4+ T cells were isolated from PBMCs using negative selection (Miltenyi Biotec). B cells were stained with CD14‐APC, CD3‐FITC, CD27‐BV421, and CD19‐PE. T cells were stained with ICOS PE‐Cy7, HLA–DR PE, CD4 APC, PD‐1 BV421, CD45RA BV510, and CXCR5 BV605 (BioLegend). Memory B cells were sorted as CD14−CD3−CD19+CD27+ cells on a BD FACSAria Fusion flow cytometry cell sorter. CD4 T cells were sorted as PD‐1−CXCR5−PD‐1highCXCR5+ (Tfh), PD‐1highCXCR5−HLA–DR+ICOS+ (“double positive”), PD‐1high CXCR5− HLA–DR+ ICOS− (“HLA–DR only”), PD‐1highCXCR5−HLA–DR−ICOS− (“double negative”), and PD‐1highCXCR5−HLA–DR−ICOS+ (“ICOS only”). Sorted T cell populations were cocultured with memory B cells at a ratio of 1:10 with 5,000 cells of each T cell subset and 45,000 B cells in a 96‐well plate in the presence of Staphylococcal Enterotoxin B (SEB) (1 μg/mL) for five days. Cells were collected and analyzed by flow cytometry. Total IgG and IgM in culture supernatant were quantified by enzyme‐linked immunosorbent assay (ELISA).
Flow cytometry
Cells from T and B cell cocultures were then stained with CD4‐FITC, CD20‐BV605, CD19‐ACP‐Cy7, CD27‐PECy7, CD38‐PerCPCy5.5, CD138‐APC, CD11c‐PE, IgD‐BV421, and Live/Dead‐BV510. Data were acquired on a BD Fortessa analyzer using FACSDiva software and analyzed using FlowJo. The percentage of plasmablasts (CD19+CD38highCD27+) among B cells was calculated from cocultures using each CD4 T cell population isolated. To validate the findings from mass cytometry, PBMC samples from 21 patients with SSc and 10 noninflammatory controls were stained with a flow cytometry panel to evaluate the expression of granzymes and perforin in CD4 T cells. Total PBMC samples from patients with SSc were incubated with Fixable Aqua Dead‐BV510, CD3‐AF700, CD8‐BV650, CD4‐BUV737, SLAMF7‐BV786, Perforin‐BV711, Granzyme A‐APC, Granzyme B‐FITC, Granzyme K‐PerCPCy5.5, HLA–DR‐PE, CXCR5‐BV605, ICOS APC‐Cy7, and PD‐1‐BV421.
Data analysis
Nonviable cells and CD14+ monocytes were excluded to gate on CD45RO+ memory CD4 T cells. The DNA+ gate was adjusted for each batch separately, and then lineage gating was uniformly applied to all batches. Gating for cell frequencies and marker expression were performed using FlowJo10. Eleven samples with live cell counts of <10,000 were excluded from the final analysis. Surface marker expression was quantified for all proteins in the panel on PD‐1highCXCR5− and PD‐1highCXCR5+ cells comparing patients with SSc and controls using the Mann‐Whitney U‐test. Resulting P values were adjusted for multiple testing using Bonferroni correction. Expanded markers were considered significant if the adjusted P value was less than 0.05. Associations between expanded cell population and lung disease metrics were assessed by Spearman correlation. Paired analysis across multiple conditions was performed using Friedman's test.
RESULTS
Patient population
Demographic data, clinical characteristics, and baseline pulmonary function test (PFT) variables are summarized in Table 1. UCSF patients were mostly White (60%) middle aged (mean ± SD 55 ± 12.3 years, range 43–61 years) women (91%) exhibiting the limited cutaneous phenotype (63%). ILD was present in 43 (65%) study participants, and PAH was present in 15 (22%) study participants. Patients with SSc recruited at BWH (n = 15) exhibited similar features, except they were slightly older (mean ± SD 62.3 ± 10.4 years, range 52–72 years) and had baseline lung volumes indicative of less severe restrictive physiology (mean ± SD FVC predicted 75.7% ± 23.1% vs 84.4% ± 19.3%, respectively).
Table 1.
Demographics and clinical characteristics of patients with SSc from the UCSF and BWH cohorts and healthy controls from the BWH cohort*
| Characteristics | UCSF SSc (n = 67) | BWH SSc (n = 15) | BWH controls (n = 18) |
|---|---|---|---|
| Age, mean ± SD, y | 55 ± 12.3 | 62.3 ± 10.4 | 46 ± 15 |
| Female, n (%) | 61 (91) | 12 (80) | 14 (78) |
| Race, n (%) | |||
| White | 40 (60) | 9 (60) | 8 (44) |
| Hispanic | 5 (7.4) | 1 (6.7) | 4 (22) |
| Black | 8 (12) | 0 (0) | 4 (22) |
| Asian | 9 (13) | 1 (6.7) | 1 (5.6) |
| Other | 5 (7.4) | 4 (26.7) | 1 (5.6) |
| Smoking status, n (%) | |||
| Never | 52 (77.6) | 11 (73) | |
| Current | 0 (0) | 0 (0) | |
| Past | 15 (22.4) | 4 (26.7) | |
| BMI, mean ± SD | 23.8 ± 4.3 | 24.6 ± 4.9 | |
| FEV1, mean ± SD, L | 2.1 ± 0.6 | 2.3 ± 0.5 | |
| FEV1, mean ± SD, % predicted | 77.7 ± 29.3 | 87 ± 17.5 | |
| FVC, mean ± SD, L | 2.6 ± 0.8 | 2.7 ± 0.7 | |
| FVC, mean ± SD, % predicted | 75.7 ± 23.1 | 84.4 ± 19.3 | |
| FEV1/FVC ratio, mean ± SD | 80.2 ± 7.4 | 75.6 ± 25.9 | |
| DLco, mean ± SD, % predicted | 44 ± 22.4 | 54.3 ± 21.3 | |
| eRVSP, mean ± SD, mm Hg | 30.8 ± 15.1 | 29.2 ± 13.4 | |
| MRSS, mean ± SD | 5.4 ± 5.2 | N/A | |
| PAH, n (%) | 15 (22) | 5 (33) | |
| ILD, n (%) | 43 (64) | 10 (66.7) | |
| SSc subtype, n (%) | |||
| Limited | 42 (63) | 8 (53) | |
| DLcopp | 33 (78) | 5 (63) | |
| Diffuse | 25 (37) | 7 (46) | |
| DLcopp | 19 (76) | 5 (71) | |
| ILD severity, n (%) | |||
| Mild (0) | 34 (50) | 6 (40) | |
| Moderate (1) | 20 (30) | 4 (27) | |
| Severe (2) | 13 (19) | 1 (6) | |
| SSc duration, mean ± SD, y | |||
| RP onset | 13.6 ± 9.4 | N/A | |
| First non‐RP symptom | 10.5 ± 7.3 | N/A | |
| Autoantibody status, n | |||
| ANA | 66 | 15 | |
| ACA | 21 | 6 | |
| RNAP3 | 6 | 1 | |
| Scl‐70 | 18 | 5 | |
| U1‐RNP | 8 | 2 | |
| Current immunosuppression, n | |||
| Azathioprine | 3 | 0 | |
| Hydroxychloroquine | 5 | 1 | |
| Mycophenolate mofetil | 29 | 9 | |
| Methotrexate | 3 | 1 | |
| Rituximab | 2 | 1 | |
| Cyclophosphamide | 0 | 0 |
ILD severity: mild (0) = FVCppstand >75%; moderate (1) = FVCppstand ≥55% and ≤75%; severe (2) = FVCppstand ≤ 55. ACA, anticentromere antibody; ANA, antinuclear antibody; BMI, body mass index; BWH, Brigham and Women's Hospital; DLco, diffusing capacity for carbon monoxide; DLcopp. Reduced DLco (<75%); eRVSP, estimated right ventricular systolic pressure; FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity; FVCppstand, forced vital capacity percent predicted standardized; ILD, interstitial lung disease; MRSS, modified Rodnan skin thickness score; N/A, data not available; PAH, pulmonary arterial hypertension; RNAP3, RNA polymerase 3 antibody; RP, Raynaud's Phenomenon; SSc, systemic sclerosis; UCSF, University of California, San Francisco.
Expansion of a specific PD‐1highCXCR5 − CD4 + T cell subpopulation in patients with SSc
We first analyzed mass cytometry data on PBMC samples from 72 patients with SSc and 18 healthy controls with a focus on the CD4 T cell phenotypes. Of note, there were no significant differences in the proportions of CD3+ T cells among live PBMC samples between the two groups (Supplementary Figure 1). For analyses of CD4 T cell populations, DNA+ viable cells were gated as CD3+ and CD4+ T cells, and CD14+ monocytes were excluded. The frequencies of CD4+ and CD8+ cells among T cell subsets were not altered in patients with SSc compared to controls in our cohort (P = 0.15 and P = 0.19, respectively; Figure 1A). 11 The frequencies of PD‐1highCXCR5− cells, representing candidate Tph cells, and PD‐1highCXCR5+ cells, representing Tfh cells, within the memory CD4+ T cell pool were quantified by manual gating, with uniform gates applied across all analyzable samples. Neither PD‐1highCXCR5− nor PD‐1highCXCR5+CD4+ T cells were enriched in patients with SSc compared to controls (Figure 1B). We then explored whether there were phenotypic differences in PD‐1highCXCR5− or PD‐1highCXCR5+CD4+ T cells in patients with SSc compared with controls. A broad analysis of the surface markers in the panel was performed, excluding the lineage markers (CD3, CD4, CD8, CD14, and CD45RO) (Figure 1C). The mean expressions of CD38 and HLA–DR on PD‐1highCXCR5− cells were significantly higher in patients with SSc compared to controls (CD38 1.5 vs 2.7, P = 0.01; HLA–DR 2.5 vs 4.2, P = 0.03). Evaluation of PD‐1highCXCR5+ Tfh cells did not identify differentially expressed surface proteins (Figure 1D).
Figure 1.
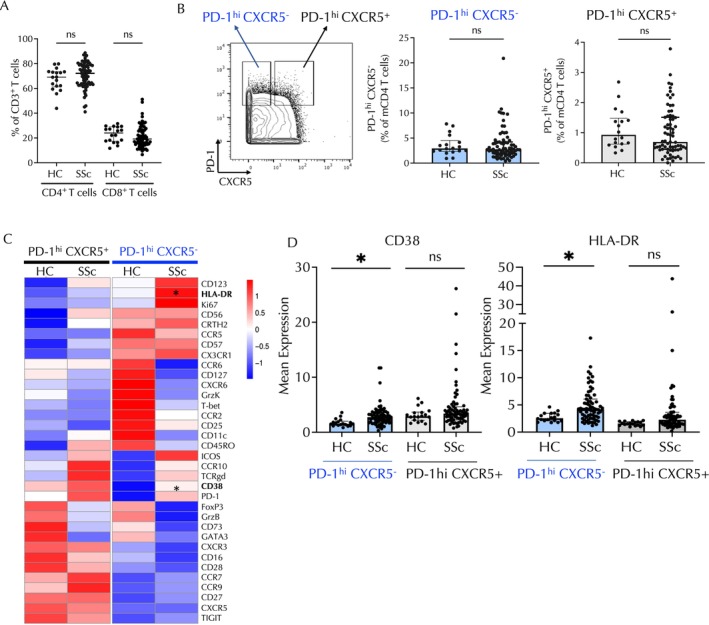
In patients with SSc, the PD‐1highCXCR5−CD4 T cell subset shows enrichment for CD38 and HLA–DR expression. (A) Quantification of CD4+ and CD8+ T cells in patients with SSc compared with HCs using mass cytometry data. (B) Example of gating of PD‐1highCXCR5− and PD‐1highCXCR5+ CD4 T cell subsets. Frequency of PD‐1highCXCR5− and PD‐1highCXCR5+ CD4 T cells in patients with SSc and HCs. (C) Heatmap of row‐normalized expression of the selected markers from the mass cytometry panel. Expression of the markers is shown in PD‐1highCXCR5−CD4 and PD‐1highCXCR5+CD4 T cells in patients with SSc compared with HCs. HLA–DR and CD38 are significantly differentially expressed in PD‐1highCXCR5− cells in patients with SSc compared to HCs. (D) Quantification of expression of CD38 and HLA–DR in PD‐1highCXCR5− and PD‐1highCXCR5+ CD4 T cells. Comparisons were made using the Mann‐Whitney U‐test. Bonferroni correction was applied in panel C. *P < 0.05; **P < 0.01; ***P < 0.001. HC, healthy control; ns, nonsignificant; SSc, systemic sclerosis.
Previous studies conducted on patients with rheumatoid arthritis (RA) and patients with systemic lupus erythematosus (SLE) have highlighted the expression of HLA–DR and ICOS on Tph cells 14 , 20 ; therefore, we evaluated patterns of expression of these molecules on circulating T cells from patients with SSc. Dimensional reduction and visualization by t‐distributed stochastic neighbor embedding revealed partially overlapping expression of HLA–DR and ICOS and highlighted CD38 expression among CD45RO+CD4 T cells, indicating a heterogeneity of marker expression (Figure 2A). HLA–DR expression is preferentially expressed on CXCR5− T cells, as minimal HLA–DR expression was observed in CXCR5+ T cells. ICOS showed variable expression in both PD‐1highCXCR5− cells and PD‐1highCXCR5+ cells (Figure 2A). We next used biaxial gating to quantify the percentage of PD‐1highCXCR5− T cell populations with expression of these markers. Consistent with expression analyses, there was an increase in the proportion of PD‐1highCXCR5− cells that expressed HLA–DR (P = 0.02) or CD38 (P = 0.04) in patients with SSc compared to controls (Figure 2B and C). In contrast, the frequency of PD‐1highCXCR5− that expressed ICOS was not altered in patients with SSc (Figure 2D). Analysis of HLA–DR and ICOS expression on PD‐1highCXCR5− cells revealed that cells with increased expression of HLA–DR but low expression of ICOS (HLA−DR+ICOS−) (P = 0.003) were significantly increased in patients with SSc (Figure 2E), whereas the frequency of cells coexpressing HLA–DR and ICOS was not altered in patients with SSc (Figure 2E). ICOS showed a broad, continuous range of expression in both PD‐1highCXCR5− cells and PD‐1highCXCR5+ cells (Supplementary Figure 2). Hence, a high‐dimensional analysis of potential Tfh and Tph cells revealed a selective expansion of PD‐1highCXCR5− HLA–DR+ICOS− T cells in the circulation of patients with SSc.
Figure 2.
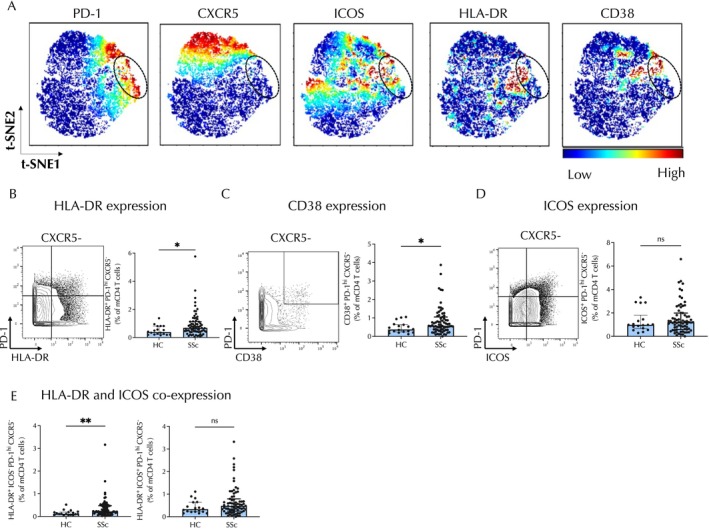
PD‐1highCXCR5−HLA–DR+CD4 T cells are expanded in the circulation of patients with SSc. (A) tSNE plots of memory CD4+ T cells in patients with SSc indicating expression of PD‐1, CXCR5, ICOS, HLA–DR, and CD38. The circle denotes the subpopulation of PD‐1highCXCR5−CD4 T cells. Color indicates level of marker expression. (B–D) Representative biaxial gating and expression of (B) HLA–DR, (C) CD38, and (D) ICOS on CXCR5−PD‐1high cells. (E) Frequencies of CXCR5−PD‐1high cells with positive expression of HLA–DR and negative ICOS expression (left) or with coexpression of the two markers (right). Error bars depict median ± interquartile range. Comparisons were made using Mann‐Whitney U‐tests. *P < 0.05; **P < 0.01. HC, healthy control; ns, nonsignificant; SSc, systemic sclerosis; tSNE, t‐distributed stochastic neighbor embedding.
PD‐1highCXCR5 − HLA–DR + CD4 + cells exhibiting cytotoxic rather than Tfh‐like phenotype
The CD4+PD‐1highCXCR5− population contains Tph cells, which are known as B cell–T helper cells; yet, B cell–T helper lymphocytes, which include both Tph and Tfh cells, typically express ICOS. 14 The lack of ICOS expression suggested the possibility that the PD‐1highCXCR5−HLA–DR+ population expanded in patients with SSc may have a function other than providing help to B cells. To gain insight into alternative functions of this T cell subset, we first explored the expression of other cellular markers in the mass cytometry panel on PD‐1highCXCR5− T cells divided into subpopulations based on the presence of HLA–DR and ICOS. PD‐1highCXCR5−HLA–DR+ICOS− cells displayed selective elevation of a set of surface proteins including CD57 and CX3CR1, suggestive of a cytotoxic T cell phenotype (Figure 3A). 21 , 22 In contrast, other transcription factors, including FoxP3 and GATA‐3, did not show statistically significant differences, and T‐bet expression was significantly lower in patients with SSc (Figure 3A); hence, we chose not to focus on these transcription factors in our analyses.
Figure 3.
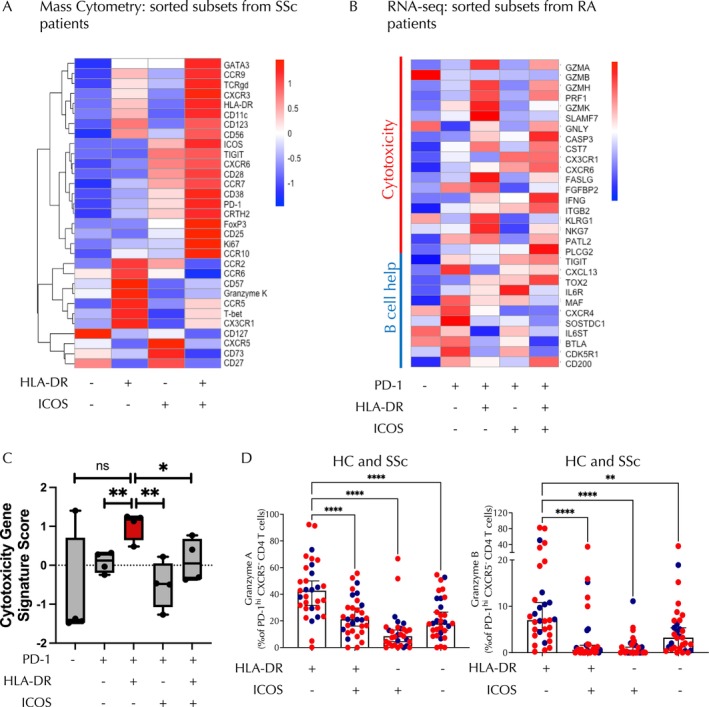
PD‐1highCXCR5−HLA–DR+ICOS−CD4 T cells exhibit a cytotoxic phenotype rather than a follicular helper T–like B cell helper phenotype. (A) Row‐normalized heatmap depicting mean expression of mass cytometry markers on PD‐1highCXCR5−CD4+ T cells divided into four subsets based on expression of HLA–DR and ICOS as indicated. (B) Heatmap showing mean expression of genes associated with cytotoxicity and B cell help derived from bulk RNA sequencing data of CXCR5−CD4 T cell subsets from patients with RA. Subsets are defined by expression of PD‐1, HLA–DR, and ICOS as noted. (C) The cytotoxicity gene signature score is depicted for the CD4 T cell subsets derived from bulk RNA sequencing data of CXCR5−CD4 T cell subsets in patients with RA as depicted in panel B. (D) Quantification of granzyme A expression on PD‐1highCXCR5−CD4 T cells from HCs and patients with SSc from each subset based on expression of HLA–DR and ICOS as indicated. Quantification of granzyme B on PD‐1highCXCR5−CD4 T cells from HC and patients with SSc. Blue dots represent HCs, and red dots represent patients with SSc. Subsets are defined by expression of PD‐1, HLA–DR, and ICOS as noted. Paired t‐test comparisons were made using Friedman's test and Dunn's multiple comparison test. *P < 0.05; **P < 0.01; **P < 0.01; ****P < 0.0001. HC, healthy control; ns, nonsignificant; RA, rheumatoid arthritis; SSc, systemic sclerosis.
To further define the features of this subset, we interrogated a previously reported RNA sequencing data set on PD‐1highCXCR5− cells sorted from blood samples of patients with RA, which were similarly divided into subpopulations based on HLA–DR and ICOS expression. 14 This analysis showed that HLA–DR+ICOS− cells expressed the highest levels of cytotoxicity‐associated genes, including GZMA, GZMH, SLAMF7, FGFBP2, NKG7, and PRF1, whereas their expression of CXCL13, a B cell chemoattractant characteristically expressed by Tph and Tfh cells, was relatively low (Figure 3B). To analyze more comprehensively the cytotoxic potential of HLA–DR+ICOS− cells compared with other subsets, we then applied a CD4 T cell–specific cytotoxic gene signature 18 to this RNA sequencing data set and found that HLA–DR+ICOS− cells exhibit an enhanced cytotoxic signature compared with other PD‐1highCXCR5− populations (Figure 3C and Supplementary Table 2). Intriguingly, this population also showed a decreased Tfh signature compared with the PD‐1highCXCR5−ICOS+ subset (Supplementary Figure 3 and Supplementary Table 2). Next, we conducted flow cytometry analysis in PBMC samples obtained from a subset of the patients with SSc and healthy controls included in the mass cytometry cohort to explore levels of expression of granzyme A and granzyme B, serine proteases that are released by cytotoxic T cells and induce cell death. 23 HLA–DR+ICOS− cells showed increased mean expression of granzyme A and granzyme B compared with other CD4 T cell subsets (Figure 3D). Together, these results indicate that the PD‐1+CXCR5−HLA–DR+ICOS− subset expanded in SSc patients carry a cytotoxic rather than a B cell helper phenotype.
Functional assessment of PD‐1highCXCR5 − HLA–DR + ICOS − CD4 + T cell population
To further explore the functional properties of this T cell population, we performed in vitro T cell and B cell coculture assays to test B cell helper function of different PD‐1highCD4 T cell subsets sorted from blood samples obtained from either controls (n = 11) or patients with SSc (n = 4). PD‐1+CXCR5+ (Tfh) cells effectively induced differentiation of memory B cells into CD38highCD27+ plasmablasts (Figure 4A). Among PD‐1highCXCR5− cells, the HLA–DR−ICOS− population showed similar activity to Tfh cells. In contrast, the HLA–DR+ICOS− subset exhibited, on average, the weakest ability to prompt plasmablast differentiation (Figure 4A). Moreover, quantification of Igs in coculture supernatants revealed that Tfh and PD‐1highCXCR5−HLA–DR−ICOS− cells stimulated the highest IgM and IgG production, whereas PD‐1highCXCR5−HLA–DR+ cells, with or without ICOS expression, stimulated the lowest production (Figure 4B). Of interest, the samples from patients with SSc induced greater B cell production of IgG and IgM in coculture experiments across all T cell subsets compared to controls, although the HLA–DR+ICOS−CD4 T cell subset continued to show a reduced ability to induce IgG and IgM production across the patient and control samples (Figure 4B).
Figure 4.
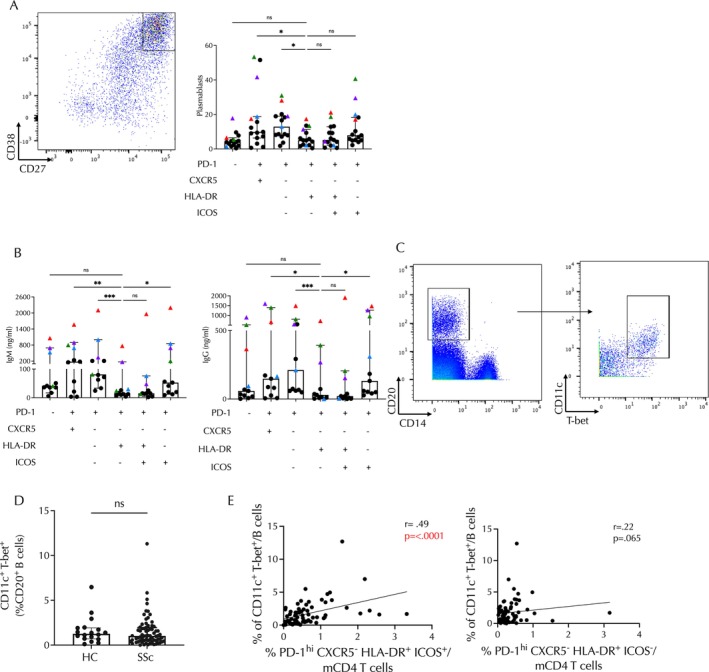
Functional studies and B cell correlative analyses suggest that PD‐1highCXCR5−HLA–DR+ICOS− cells do not behave as B cell helpers. (A) Frequencies of CD38highCD27+ plasmablasts derived from B cell differentiation assays in which memory B cells were cocultured with the indicated subset of CD4 T helper cells. The representative flow plot of plasmablast gating is shown on the left, and frequencies are depicted on the right. (B) Quantification of IgM and IgG in supernatant from T cell–B cell cocultures using the Friedman test. (C) Representative flow cytometry gating for isolation of ABCs defined by CD11c+T‐bet+. (D) Frequencies of ABCs in patients with SSc compared to HCs. (E) Correlation of frequencies of ABCs with PD‐1highCXCR5−CD4 T cells with HLA–DR+ICOS+ expression (n = 71; P < 0.0001, rs = 0.49) (left) or HLA–DR+ICOS− expression (n = 71; P = 0.065, rs = 0.22) (right). In panels A and B, black circles represent T cell subsets derived from collars, whereas colored circles represent individual patient samples. Spearman correlation statistics are shown. *P < 0.05; ***P < 0.001. Comparisons in A and B were made using Friedman's test. **P < 0.01; ****P < 0.0001. ABC, age‐associated B cell; HC, healthy control; ns, nonsignificant; SSc, systemic sclerosis.
It is possible that the HLA–DR+ICOS− population could still be associated with B cell helper function in vivo despite limited helper function in vitro. In patients with RA and patients with SLE, the number of circulating Tph cells is positively correlated with circulating CD11c+ age‐associated B cells (ABCs), in line with their function as B cell helpers. 23 We therefore reasoned that if the HLA–DR+ICOS− subset of PD‐1highCXCR5− cells was also exerting a B cell helper function, the abundance of these cells might also be positively correlated with the abundance of CD11c+ ABCs. Using the mass cytometry data obtained from our cohort of patients with SSc, we quantified CD11c+T‐bet+ ABCs among circulating CD20+ B cells (Figure 4C). Although CD11c+ B cells were not expanded in patients with SSc compared to healthy controls (Figure 4D), the frequency of CD11c+T‐bet+ ABCs and PD‐1highCXCR5−HLA–DR+ICOS+ T cells was positively associated (r = 0.49, P ≤ 0.0001). In contrast, no significant correlation was found between ABCs and the HLA–DR+ICOS− population (r = 0.22, P = 0.065) (Figure 4E). Taken together, the weak B cell helper function in vitro and lack of correlation with ABCs in vivo suggest that the PD‐1highCXCR5−HLA–DR+ICOS−CD4+ T cell population expanded in patients with SSc performs functions other than providing B cell help.
PD‐1highCXCR5 − HLA–DR ICOS − T cell subset and SSc‐ILD
Finally, we assessed whether this expanded CD4 subset was associated with any SSc clinical feature. No significant association was seen between the HLA–DR+ICOS − subset frequency and age (P = 0.24), sex (P = 0.37), body mass index (P = 0.89), race (P = 0.78), MRSS (P = 0.42), or prescriptions of immunosuppressants (P = 0.44). PD‐1highCXCR5 − HLA–DR+ICOS − cells were selectively expanded in patients with SSc‐ILD compared with those without ILD (P = 0.02) or healthy controls (P = 0.001) (Figure 5A). This association of DR+ICOS− cells with ILD remains significant when adjusted for SSc subtype (limited vs diffuse) (P = 0.014). In addition, the frequency of the HLA–DR+ICOS− population was significantly higher in study participants with SSc with more severe ILD (P = 0.017) (Figure 5B). Consistent with this observation, levels of circulating HLA–DR+ICOS−PD‐1highCXCR5−CD4+ T cells showed a weak inverse correlation with both FVC (predicted FVC percentage r = −0.34, P = 0.0043) and diffusing capacity for carbon monoxide (DLco) (predicted DLco percentage r = −0.25, P = 0.038) (Figure 5C). In contrast, PD‐1highCXCR5+Tfh cells showed a weak positive association with FVC (r = 0.31, P = 0.0098) (Figure 5D), and our analyses indicate that there is not a significant correlation of PD‐1highCXCR5 − HLA–DR+ICOS+ cell numbers with the measured lung parameters (Supplementary Figure 4). No association with estimated right ventricular systolic pressure was detected.
Figure 5.
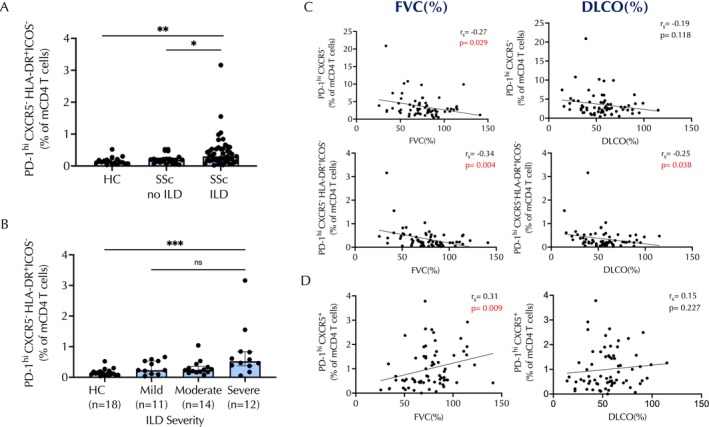
Increased numbers of PD‐1highCXCR5−HLA–DR+ICOS−CD4 T cells correspond with ILD and pulmonary function parameters in patients with SSc. (A) Frequencies of PD‐1highCXCR5−HLA–DR+ICOS−CD4 T cells in HCs, patients with SSc without ILD, and patients with SSc and ILD. (B) Frequencies of PD‐1highCXCR5−HLA–DR+ICOS− CD4 T cells in HCs and patients with SSc subdivided into mild, moderate, or severe ILD severity categories. (C) Frequencies of total PD‐1highCXCR5−CD4 T cells or the PD‐1highCXCR5−HLA–DR+ICOS− subset correlated with FVC or DLco as indicated (PD‐1highCXCR5−/FVC: n = 71, P = 0.029, rs = −0.27; PD‐1highCXCR5−/DLco: n = 71, P = 0.118, rs = −0.19; PD‐1highCXCR5−HLA–DR+ICOS−/FVC: n = 71, P = 0.004, rs = −0.34; PD‐1highCXCR5−HLA–DR+ICOS−/DLco: n = 71, P = 0.038, rs = −0.25). (D) Frequencies of PD‐1highCXCR5+ T cells with respect to FVC (n = 71; P = 0.009, rs = 0.31) or DLco (n = 71; P = 0.227, rs = 0.15) as indicated. *P < 0.05; **P < 0.01; ***P < 0.001. DLco, diffusing capacity for carbon monoxide; FVC, forced vital capacity; HC, healthy control; ILD, interstitial lung disease; ns, nonsignificant; SSc, systemic sclerosis.
DISCUSSION
In this study, we identify a CD4 T cell subset with unique PD‐1highCXCR5 − HLA–DR+ICOS − phenotype that is expanded in peripheral blood samples of patients with SSc and is associated with the presence and severity of ILD. Our analyses revealed that neither Tfh cells nor the broader group of CD4+ cells characterized by CXCR5 − PD‐1high expression, which encompasses Tph cells, were significantly expanded in this cohort of patients with established SSc compared to controls. Rather, a specific PD‐1high subpopulation that expresses HLA–DR but lacks ICOS was selectively associated with SSc‐ILD. This population lacked a transcriptomic signature indicative of B cell helper function as would be expected for Tph and Tfh cells and instead expressed gene and/or protein programs associated with cytotoxicity. 24 These results provide a key insight in helping to dissect the heterogeneity among PD‐1+ T cells in the circulation and suggest that HLA–DR+ICOS − cells within the PD‐1high gate carry a cytotoxic rather than a B cell–T helper cell phenotype.
Recent work has suggested an increase in CXCR5 − PD‐1+ cells in patients with SSc compared to healthy controls. 9 However, in this study, the detection of an expanded CD4 population with a cytotoxic phenotype and its association with presence and severity of ILD support the possibility highlighted by other studies that cytotoxic CD4 T cells may play a significant role in SSc immunopathogenesis. 7 , 25 In fact, evidence has been provided that cytotoxic CD4 T cells, marked by expression of SLAMF7, are increased in the skin of patients with SSc with early diffuse disease. 7 , 12 In particular, granzyme A–positive cytotoxic CD4 T cells infiltrating the skin of patients with SSc exhibit the ability to target endothelial cells and induce microvascular injury. 12 The extent to which the PD‐1highCXCR5 − HLA–DR+ICOS − cells overlap with this cytotoxic population defined by SLAMF7 is not yet clear; however, we expect that these cell populations likely exhibit similar functional properties. Future studies tracking T cell populations using T cell receptor repertoire sequencing may help clarify the clonal and developmental relationships among these cell populations with varied surface phenotypes.
Previous work has shown an association of skin‐derived PD‐1+CXCR5 − /CXCL13+CD4 T cells with SSc‐ILD. 4 In our SSc cohort, the frequency of circulating PD‐1highCXCR5 − HLA–DR+ICOS − CD4 T cells is higher in patients with ILD and more severe lung disease and is inversely associated with the lung function measured by both FVC or DLco. We acknowledge that the cross‐sectional design of our study limits our ability to draw further inferences about the clinical usefulness of this cellular subset as a biomarker. Future prospective studies will define with greater precision whether PD‐1highCXCR5 − HLA–DR+ICOS − CD4 T cells can reliably predict activity and progression of SSc‐ILD as well as response to therapy. Other limitations of our study include demographic variability (ie, age, sex, and race) between the healthy controls and the SSc cohorts as well as broad differences in treatment regimens among patients. In addition, despite multiple attempts, technical limitations have not allowed us to successfully perform cytotoxicity functional assays. Therefore, to confirm the cytotoxic phenotype of PD‐1highCXCR5 − HLA–DR+ICOS − CD4 T cells, we used specific markers such as granzyme A and B as shown in other studies investigating polyclonal CD4 T cell populations. Whether these cytotoxic‐appearing CD4 T cells are implicated in the initiation or progression of SSc‐ILD remains to be investigated. Studies are ongoing to determine to what extent these PD‐1highCXCR5 − HLA–DR+ICOS − CD4 T cells are present in affected SSc target tissues (ie, skin or explanted lungs) and their spatial relationship with key cellular components.
In conclusion, we found that a PD‐1highCXCR5 − ICOS − HLA–DR+ cell population with a cytotoxic phenotype is expanded in the circulation of patients with SSc with ILD and more severe lung disease. This study provides a valuable example of the potential of high‐throughput mass cytometry immunoprofiling to discover cell phenotypes specifically associated with unique disease features.
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Drs Boin and Rao had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design
Elahee, Mueller, Fava, Boin, Rao.
Acquisition of data
Elahee, Mueller, Wang, Marks, Sasaki, Dellaripa, Boin, Rao.
Analysis and interpretation of data
Elahee, Mueller, Wang, Marks, Sasaki, Cao, Boin, Rao.
Supporting information
Disclosure form:
Appendix S1: Supporting Information.
Supported by the Department of Defense Scleroderma Research Program Translational Research Partnership Award SL200016P1 (to Drs Boin and Rao), the Doris Duke Charitable Foundation Clinical Scientist Development Award, the Burroughs Wellcome Fund Career Award in Medical Sciences, and National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), NIH, grant K08‐AR‐072791. Dr Elahee's work was supported by NIAMS, NIH grant T32‐AR‐007530. Dr Mueller's work was supported by the Rheumatology Research Foundation and by NIAMS, NIH, grant T32‐AR‐007530. Dr Rao's work was supported by NIAMS, NIH, grant P30‐AR‐070253.
Drs Elahee and Mueller are co‐first authors and contributed equally to this work. Drs Boin and Rao contributed equally to this work.
Additional supplementary information cited in this article can be found online in the Supporting Information section (http://onlinelibrary.wiley.com/doi/10.1002/acr2.11671).
Author disclosures are available at https://onlinelibrary.wiley.com/doi/10.1002/acr2.11671.
Address correspondence via email to Francesco Boin, MD, at francesco.boin@cshs.org; or to Deepak A. Rao, MD, PhD, at darao@bwh.harvard.edu.
Contributor Information
Francesco Boin, Email: francesco.boin@cshs.org.
Deepak A. Rao, Email: darao@bwh.harvard.edu.
REFERENCES
- 1. Perelas A, Silver RM, Arrossi AV, et al. Systemic sclerosis‐associated interstitial lung disease. Lancet Respir Med 2020;8(3):304–320. [DOI] [PubMed] [Google Scholar]
- 2. O'Reilly S, Hügle T, van Laar JM. T cells in systemic sclerosis: a reappraisal. Rheumatology (Oxford) 2012;51(9):1540–1549. [DOI] [PubMed] [Google Scholar]
- 3. Frieri M, Angadi C, Paolano A, et al. Altered T cell subpopulations and lymphocytes expressing natural killer cell phenotypes in patients with progressive systemic sclerosis. J Allergy Clin Immunol 1991;87(4):773–779. [DOI] [PubMed] [Google Scholar]
- 4. Gaydosik AM, Tabib T, Domsic R, et al. Single‐cell transcriptome analysis identifies skin‐specific T‐cell responses in systemic sclerosis. Ann Rheum Dis 2021;80(11):1453–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kalogerou A, Gelou E, Mountantonakis S, et al. Early T cell activation in the skin from patients with systemic sclerosis. Ann Rheum Dis 2005;64(8):1233–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Frantz C, Auffray C, Avouac J, et al. Regulatory T cells in systemic sclerosis. Front Immunol 2018;9:2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maehara T, Kaneko N, Perugino CA, et al. Cytotoxic CD4+ T lymphocytes may induce endothelial cell apoptosis in systemic sclerosis. J Clin Invest 2020;130(5):2451–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Radstake TRDJ, van Bon L, Broen J, et al. The pronounced Th17 profile in systemic sclerosis (SSc) together with intracellular expression of TGFβ and IFNγ distinguishes SSc phenotypes. PLoS One 2009;4(6):e5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ricard L, Jachiet V, Malard F, et al. Circulating follicular helper T cells are increased in systemic sclerosis and promote plasmablast differentiation through the IL‐21 pathway which can be inhibited by ruxolitinib. Ann Rheum Dis 2019;78(4):539–550. [DOI] [PubMed] [Google Scholar]
- 10. Taylor DK, Mittereder N, Kuta E, et al. T follicular helper‐like cells contribute to skin fibrosis. Sci Transl Med 2018;10(431):1–16. [DOI] [PubMed] [Google Scholar]
- 11. Crotty S. Follicular helper CD4 T cells (TFH). Annu Rev of Immunol 2011;29(1):621–663. [DOI] [PubMed] [Google Scholar]
- 12. Fox DA, Lundy SK, Whitfield ML, et al. Lymphocyte subset abnormalities in early diffuse cutaneous systemic sclerosis. Arthritis Res Ther 2021;23(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Christophersen A, Lund EG, Snir O, et al. Distinct phenotype of CD4+ T cells driving celiac disease identified in multiple autoimmune conditions. Nat Med 2019;25(5):734–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rao DA, Gurish MF, Marshall JL, et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature 2017;542(7639):110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Akiyama M, Suzuki K, Yoshimoto K, et al. Peripheral TIGIT+ T follicular helper cells that produce high levels of interleukin‐21 via OX40 represent disease activity in IgG4‐related disease. Front Immunol 2021;12:651357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ekman I, Ihantola EL, Viisanen T, et al. Circulating CXCR5−PD‐1hi peripheral T helper cells are associated with progression to type 1 diabetes. Diabetologia 2019;62(9):1681–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van den Hoogen F, Khanna D, Fransen J, et al. 2013 classification criteria for systemic sclerosis: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum 2013;65(11):2737–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hashimoto K, Kouno T, Ikawa T, et al. Single‐cell transcriptomics reveals expansion of cytotoxic CD4 T cells in supercentenarians. Proc Natl Acad Sci U S A 2019;116(48):24242–24251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Locci M, Havenar‐Daughton C, Landais E, et al; International AIDS Vaccine Initiative Protocol C Principal Investigators . Human circulating PD‐1+CXCR3−CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity 2013;39(4):758–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Makiyama A, Chiba A, Noto D, et al. Expanded circulating peripheral helper T cells in systemic lupus erythematosus: association with disease activity and B cell differentiation. Rheumatology (Oxford) 2019;58(10):1861–1869. [DOI] [PubMed] [Google Scholar]
- 21. Kared H, Martelli S, Ng TP, et al. CD57 in human natural killer cells and T‐lymphocytes. Cancer Immunol Immunother 2016;65(4):441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gerlach C, Moseman EA, Loughhead SM, et al. The chemokine receptor CX3CR1 defines three antigen‐experienced CD8 T cell subsets with distinct roles in immune surveillance and homeostasis. Immunity 2016;45(6):1270–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bocharnikov AV, Keegan J, Wacleche VS, et al. PD‐1hiCXCR5− T peripheral helper cells promote B cell responses in lupus via MAF and IL‐21. JCI Insight 2019;4(20):e130062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marks KE, Rao DA. T peripheral helper cells in autoimmune diseases. Immunol Rev 2022;307(1):191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fuschiotti P. Current perspectives on the role of CD8+ T cells in systemic sclerosis. Immunol Lett 2018;195:55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Disclosure form:
Appendix S1: Supporting Information.


