Abstract
The evolution of human immunodeficiency virus type 1 infection is associated with a shift in the target cell population, driven by variability in coreceptor utilization resulting from diversity in env. To elucidate the potential consequences of these changes for Env-mediated fusion over the course of AIDS, we examined the biological properties of serial viral isolates and determined coreceptor utilization by the products of env cloned from two individuals, followed from the detection of seroconversion throughout the course of their infection. One had a typical course, and the other had an accelerated progression. Early isolates were non-syncytium inducing, and the corresponding Env exclusively utilized CCR5, whereas Env from late phases of infection showed restricted utilization of CXCR4 in both patients. Env from subject SC24, who had a standard progression, demonstrated multitropism, manifested by utilization of CCR3, CXCR4, and CCR5 in the intervening period. In contrast, Env from patient SC51, who experienced early conversion to the syncytium-inducing phenotype, developed dualtropic coreceptor utilization of CCR5 and CXCR4. Genetic analysis of env from each isolate revealed that those with an X4 phenotype formed a distinct subcluster within each subject. Analysis of chimeras constructed from R5 and multispecific env from patient SC24 demonstrated that while the V3 domain played a dominant role in determining coreceptor utilization, sequences in the V4–V5 region also contributed to the latter phenotype. Immunoprecipitation experiments confirmed that the hybrid Env proteins were expressed at similar levels. These experiments demonstrate that progression from the R5 to X4 phenotype may occur through a multi- or dual-tropic intermediate and that multiple domains contribute to this process.
Human immunodeficiency virus type 1 (HIV-1) infection is initiated by entry of the viral genome following merging of the lipid bilayer that circumscribes the virus with the plasma membrane of the target cell. The product of the env gene, which is processed into two noncovalently associated subunits, gp120 and gp41, mediates this membrane fusion process (25). This complex undergoes a series of conformational shifts that result from the interaction of gp120 with cellular receptors and culminate in membrane fusion (38, 53). The association of gp120 with the primary receptor, CD4, results in the exposure of cryptic protein surfaces in Env (40). The finding that human CD4 is necessary but not sufficient to confer sensitivity to infection led to the ultimate recognition that the role of coreceptor is performed by a cadre of serpentine receptors that transmit the signals of chemoattractant cytokines, or chemokines (1, 13, 20, 23, 24, 28). The conformation of Env induced by engagement of CD4 is permissive for interaction with coreceptors, which triggers terminal events in the fusion process, presumably extension of the hydrophobic peptide of gp41 (38). The front-line coreceptor for commonly transmitted strains of HIV-1 is CCR5 (1, 13, 20, 23, 24), and the importance of this function is illustrated by the high degree of resistance to infection of individuals homozygous for a 32-bp deletion in the gene encoding CCR5 (36, 52), who consequently lack a functional receptor.
Whereas the binding of gp120 to CD4 is virtually universal among HIV-1 isolates (18, 37, 39), there is variability in the interaction between coreceptors and this glycoprotein over the course of AIDS (6, 17). Selective pressures exerted by neutralizing antibodies (3, 7, 44, 59, 63) and other immunological responses coupled with imperfect fidelity of reverse transcriptase (4, 47) are likely contributors to the genetic and biological diversification of env (56). This diversity in coreceptor utilization is manifested as the entry block associated with target cell tropism. Env derived from isolates obtained early in the evolution of HIV-1 infection demonstrate exclusive utilization of CCR5, while those arising later often use CXCR4 for membrane fusion, and correspond to macrophage (M)-tropic and T cell-tropic (T-tropic) strains, respectively. The designations of R5 and X4 for M- and T-tropic Env reflect this functional relationship (discussed previously by E. A. Berger, R. W. Doms, E. M. Fenyo, B. T. Korber, D. R. Littman, J. P. Moore, Q. J. Sattentau, H. Schuitemaker, J. Sodroski, and R. A. Weiss, Letter, Nature 391:240, 1998). Although several chemokine receptors have been found to function as coreceptors in vitro, CCR5 and CXCR4 are recognized as the primary coreceptors for HIV-1 infection in vivo at this time, because all available cloned Env have been shown to use either one or both as coreceptors and simultaneous antagonism of these receptors blocks infection of primary cells (70; discussed previously by N. L. Michael and J. P. Moore, Letter, Nat. Med. 5:740–742, 1999).
The molecular determinants specifying HIV-1 tropism have been studied extensively (11, 30, 31, 43, 57, 65, 66). Early reports demonstrated that the V3 region of gp120 is a major, but not the sole, determinant of tropism that determines the target cell repertoire (11, 30, 43, 57, 65, 66). More recently, several laboratories have demonstrated that this region is a key factor differentiating coreceptor utilization as well (13, 15). There is emerging evidence that other regions of gp120 also have a critical role in programming tropism and coreceptor utilization (9, 31, 41, 58). Recent crystallographic studies have provided tremendous insight into the surface of the gp120 core structure that is involved in the association with CD4 and, coupled to mutagenesis experiments, have implicated structures involved in the interaction with coreceptors (35, 46, 67, 68). However, these experiments were limited by the need to delete critical variable loop segments of gp120, including V3 itself. While analysis of serial isolates has provided some insight into changes in env that are associated with shifts in tropism, our understanding of the precise mechanism for the metamorphosis of coreceptor utilization from CCR5 to CXCR4 by Env is limited. In addition, characterization of the utilization of ancillary coreceptors by ENV in vivo or in vitro may further elucidate this process.
Thus far, most of the insights concerning virus tropism and coreceptor utilization have been gained from studies employing laboratory strains and unrelated primary isolates as the starting points for analysis and genetic engineering of variants (5, 12, 13, 49). The present study addresses the question of env evolution during the course of AIDS through characterization of coreceptor utilization by the products of cloned env genes obtained from serial samples from two individuals identified during the earliest stages of primary HIV-1 infection. The subjects were members of an acute-infection cohort in Trinidad and Tobago. The average time from infection to progression to AIDS in this cohort is approximately 5 years, which may be more representative of rates of disease progression in developing nations (2) than of those observed in North America and Europe (42, 48, 50). These patients exhibited a phenotypic switch in their virus isolates from non-syncytium-inducing (NSI) to syncytium-inducing (SI), demonstrating that there is an evolution of coreceptor utilization from an initial M-tropic/R5 strain to one that includes CXCR4. In one case, there was acquisition of multitropic coreceptor utilization (CCR3, CCR2b, and CXCR4 in addition to CCR5), followed by Env that had lost the ability to utilize coreceptors other than CXCR4. However, such multispecific coreceptor utilization was not apparent in infectivity studies with these Env. In the second case there was early conversion to the SI phenotype that was associated with emergence of a dual-tropic Env able to use CCR5 and CXCR4 in both fusion and infection experiments, but promiscuous utilization of other coreceptors was not apparent. Analysis of chimeric Env indicated that elements in the V3 loop made an important contribution to multitropic coreceptor utilization, but sequences within the V4–V5 region also impart this phenotype independent of the V3 sequences. These findings provide critical insights into the utilization of coreceptors during the course of AIDS required to understand the pathogenesis of target cell specificity that will be critical to the design of antagonists of viral entry.
(This work represents partial fulfillment of requirements for a Ph.D. in Biochemistry and Molecular Biology for Zi-xuan Wang.)
MATERIALS AND METHODS
Preparation and phenotypic characterization of low-passage primary virus isolate stocks.
Peripheral blood mononuclear cells (PBMC) were prepared from venous blood obtained from HIV-1-positive study subjects and seronegative donors by standard Ficoll-Hypaque density separation. Low-passage primary virus isolates were prepared by coculturing 5 × 106 freshly prepared (or, in some instances, viably cryopreserved) PBMC from a study subject with phytohemagglutinin-activated seronegative donor PBMC in RPMI 1640 medium supplemented with 20% heat-inactivated fetal calf serum (FCS), 5% interleukin 2, and gentamicin. Twice weekly, half the culture supernatants were harvested and replaced with fresh medium alone (once per week) or medium containing 107 phytohemagglutinin-activated activated donor PBMC (once per week). HIV-1 p24 antigen levels were monitored in culture supernatants twice per week until two successive readings exceeded 30 pg/ml and the levels were increasing. The cells were then pelleted and cocultured in 20 to 30 ml with 1 × 107 to 1.5 × 107 CD8+ cell-depleted PBMC from a pool of 10 seronegative donors that had been prepared and activated as follows. The PBMC from a pool of normal donors were activated for 3 days with a mixture of anti-CD3 (50 ng/ml) and anti-CD28 (100 ng/ml) antibodies in medium supplemented with 10% fetal calf serum, 20 U of recombinant interleukin 2 (Genzyme) per ml, and penicillin-streptomycin (pen/strep) at 37°C in a humidified incubator. The PBMC were depleted of CD8 T cells with anti-CD8-coated magnetic microspheres (Dynal) according to the manufacturer's recommendations. At daily intervals, the cells were pelleted, supernatants were harvested and filtered, and the cells were resuspended in fresh medium. Supernatants were monitored for the presence and amount of viral reverse transcriptase (RT) activity with a micro-RT assay (10) to estimate virus replication. Supernatants containing significant amounts of viral RT activity were subsequently characterized with respect to infectious virus titer in a PBMC-based assay. Virus harvests possessing 103 50% tissue culture infective doses (TCID50)/ml were aliquoted and stored at −70°C until used in biological experiments.
SI phenotypic determinations of the virus isolates were performed in the MT-2 cell line according to established AIDS Clinical Trial Group protocols (33, 34). The MT-2 cell line was obtained from the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program. The MT-2 cells were maintained in log-phase growth by splitting the cultures 1:3 every 3 to 4 days using RPMI 1640 supplemented with 2 mM l-glutamine, pen/strep (100 U/ml and 100 μg/ml, respectively), and 10% fetal bovine serum. Briefly, the assay was performed in duplicate in 96-well flat-bottomed tissue culture plates. A total of 50,000 MT-2 cells were placed in assay wells in 150 μl of medium; 50 μl of virus stock was added to duplicate wells, and 50 μl of medium was added to duplicate control wells. Positive controls were included in each assay. On days 3, 6, 9, and 12, each well was examined microscopically for syncytium formation, and the day that syncytia were first observed was recorded. After inspection of wells, cells were resuspended, and 130 μl of the culture was removed and replaced with flesh medium. Assays continued through 14 days, when the cultures were terminated. Viruses that did not induce syncytium formation within this 14-day period were termed NSI.
Infectivity studies with primary virus isolates.
Virus isolates were titrated on U87 cells and/or HeLa-CD4 cells to assess their ability to use CCR5 and/or CXCR4 to initiate productive infection. U87 cell lines stably expressing CD4 alone (obtained through the AIDS Reagent Program, DAIDS, NIAID, NIH, from HongKui Deng and Dan Littman), CD4 and CCR5, or CD4 and CXCR4 (both gifts from Dan Littman) were seeded in 48-well plates (Costar) 24 h prior to infection. Cells were seeded at 7.5 × 103 cells/well in 150 μl of Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated FCS. The U87-CD4 cells were maintained and initially seeded in medium containing G418 (300 μg/ml), whereas the U87-CD4-CCR5 cells and the U87-CD4-CXCR4 cells were maintained and initially seeded in medium containing G418 (300 μg/ml) and puromycin (1 μg/ml). Medium containing selective agents was removed 24 h later and replaced with 150 μl of DMEM containing 10% FCS. Serial dilutions of virus isolates (150-μl volumes) were added to duplicate wells, bringing the volume of each test well to 300 μl. Cells were fed on days 2, 4, 7, and 10 with fresh DMEM containing 10% FCS. Aliquots of supernatants were taken on days 4, 7, and 11, adjusted to 1% Triton X-100, and assayed for RT activity as previously described (10). Infectious virus titers, expressed as TCID50 per milliliter, were determined by the end-point dilution technique and calculated by the method of Reed and Muench (45).
In addition to the U87-based infectivity studies, we examined the ability of the primary virus isolates to replicate in the HeLa-CD4 indicator cell line (MAGI) originally described by Kimpton and Emerman (32) (obtained from the AIDS Reagent Program, DAIDS, NIAID, NIH, from Michael Emerman). As the HeLa cells express the CXCR4 coreceptor, this analysis scores specifically for the ability of virus isolates to infect via the CXCR4 coreceptor. MAGI cells were seeded 24 h prior to infection in a 48-well plate (Costar) at 2.5 × 104 cells per well in 300 μl of DMEM containing 10% FCS, pen/strep (100 U and 100 μg/ml, respectively), 300 μg of l-glutamine per ml and 12.5 μg of DEAE per ml. Duplicate wells were infected with 100 μl of the appropriate virus. At 24 h after infection, DP178, an HR2 peptide mimetic and inhibitor of gp41-mediated fusion (C. Wild, T. Greenwell, and T. J. Matthews, AIDS Res. Hum. Retroviruses 9:1051–1053, 1993), was added to a final concentration of 5 μg/ml to ensure that infection readouts reflected a single round of replication. The cells were incubated for an additional 48 h and then fixed for 5 min with 1% formaldehyde and 0.2% glutaraldehyde in phosphate-buffered saline (PBS). Cells were stained in situ at 37°C for 50 min with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), 0.4 mg/ml 2 mM MgCl2, 4 mM potassium ferricyanide, and 4 mM potassium ferrocyanide in PBS. Infection resulted in intense nuclear staining, and the number of stained nuclei was interpreted as being equal to the number of infectious virions in the challenge inoculum. Infected cells were enumerated using a charge-coupled device imager and Image Pro software (Georgia Instruments).
Direct sequencing of primary env genes.
The nucleotide sequence of a segment of the HIV-1 env gene that includes the V3 loop of gpl20 was determined from amplification products obtained from proviral DNA templates using primers E100 (5′-ACACATGGAATTAAGCCAGT) and E130c (5′-CCTGCCACATGTTTATAATTTGT). The PCR products were excised from agarose gels, purified, and concentrated. An oligonucleotide designed from sequences at the 5′ end of this segment, E110 (5′-CTGTTAAATGGCAGTCTAGCAGAA), was used to prime the sequencing reaction, which was performed using a Thermo Sequenase radiolabeled terminator cycle sequencing kit (Amersham).
Molecular cloning of primary env genes.
Proviral DNA templates were prepared from activated normal donor PBMC infected with the various primary isolates. Cell pellets were lysed in a buffer containing 10 mM Tris, 1 mM EDTA, 0.001% sodium dodecyl sulfate (SDS), and 0.5% Triton X-100 (pH 8.0). Proteinase K was added to a final concentration of 1 mg/ml, and the lysates were incubated at 60°C for 3 h. The proteinase K was heat inactivated by treating the lysates at 100°C for 10 min. In some cases the DNA was further purified by standard phenol-chloroform extraction techniques. The full-length env gene containing the gp160 open reading frame was amplified by PCR using proviral DNA as templates. A region flanking env was amplified using upstream and downstream primers designed from sequences in tat (5′-TGCTGTTTATCCATTTCAGAATTGG) and nef (5′-TCCAGTCCCCCCTTTTCTTTTAAAAA), respectively. Standard conditions were employed for PCR amplification that included 1.5 mM MgCl2. PCR cycles were preceded by rapid heating to 95°C and consisted of the following intervals: denaturation, 30 s at 95°C; cooling ramp to 55°C, 60 s; annealing, 30 s at 55°C; heating ramp to 72°C; and synthesis, 7 min. Thirty-five amplification cycles were performed using the hot-start method. The 3.3-kb amplification product encompassed tat, rev, vpu, and part of the nef gene in addition to env. The annealing temperature was optimized for each template. The amplification products were molecularly cloned into the pCR2.1-TOPO vector (Invitrogen Corp.).
The open reading frame encoding gp160 was amplified from this cloned template using Pfu DNA polymerase (Stratagene). Restriction sites for EcoRI and XhoI were incorporated into the upstream and downstream primers, respectively. Expression constructs encoding Env were prepared in pcDNA3.1 (Invitrogen Corp.).
In some cases, full-length env was cloned from purified DNA templates as follows. Purified cellular DNA (0.5 μg) was used as the target for amplification of proviral env DNA using a nested PCR approach. Briefly, first-round amplification was performed with the E0 5′-primer (5′-TAGAGCCCTGGAAGCATCCAGGAAGTCAGCCTA) and the Nef 9023 3′-primer (5′-CATTGGTCTTAAAGGTACCTGAGGT), each at 0.1 μM final concentration. Two units of the eLONGase enzyme mix (Gibco-BRL) per reaction was employed for PCR, with 200 μM deoxynucleoside triphosphates (dNTPs) and 1.5 mM MgSO4. An initial denaturation for 3 min at 94°C preceded 40 amplification cycles performed as follows: denaturation for 1 min at 94°C, annealing at 57°C for 1 min, and extension for 5 min at 70°C. These cycles were followed by a final 10-min extension at 70°C. One microliter of the first-round reaction was amplified for 25 cycles in a second-round reaction employing nested primers. Second-round primers were the 5′-ENF primer (5′-AAAGAGCAGAAGACAGTGGCAATGAGAGTGAAGG) used in conjunction with either the 3′-ENR primer (5′-CAATCACACTACTTTTTGACCACTTGCCACCCAT) or the 3′-NL4-38917R primer (5′-AGGTCTCGAGATACTGCTCCCA). The concentrations of all components and reaction conditions for the second-round amplifications were identical to those described above for the first-round PCR except for the number of cycles employed. Amplified env was gel purified and recovered using the QIAEX II gel extraction kit (Qiagen Inc.) and cloned into the pCDNA3.1/V5/His-TOPO expression vector (Invitrogen Corp.).
Throughout the text and figures, individual env clones may be designated either as c.X (for clone no. X) or simply as #X (for clone no. X). For example, QH1503c.1 (a designation appearing in Fig. 1A and B) refers to the same env clone as designation QH1503#1 (a designation appearing in Table 2 and Fig. 2A).
FIG. 1.
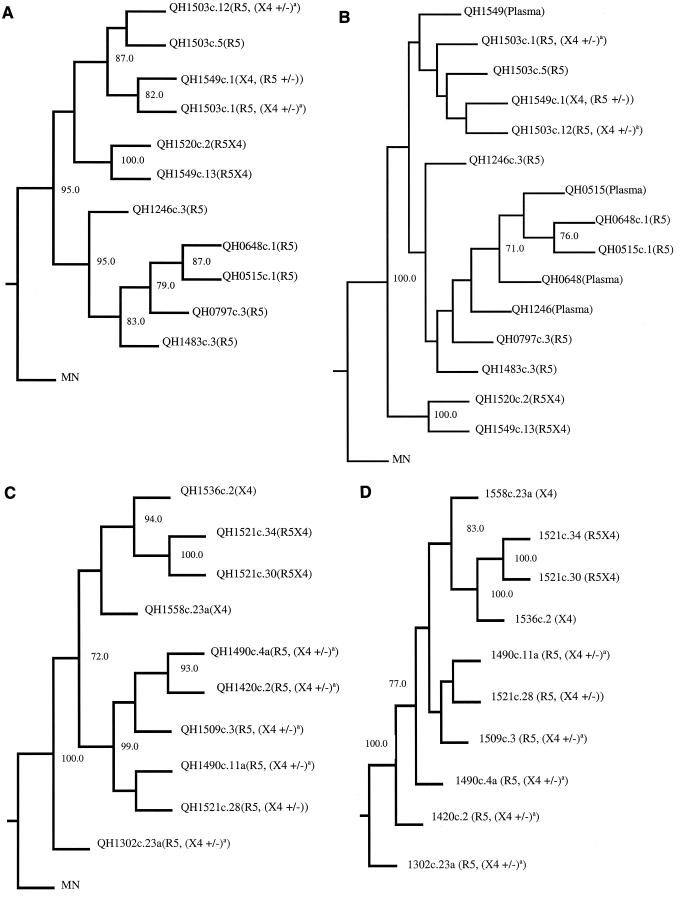
Phylogenetic analyses of env nucleotide sequences cloned from SC24 and SC51 proviral isolate templates and direct sequences from SC24 plasma virus env region. The complete nucleotide sequence of molecular env clones was determined using the Taq Dye terminator cycle sequencing kit analyzed with an ABI 377 automated sequencer. Direct sequences of SC24 plasma viral env regions were similarly determined. All sequences were confirmed by the analysis of sense and antisense DNA strands. The DNA sequences were analyzed and aligned using ABI software. Gaps were inserted into the alignment as needed, and those regions were eliminated from the subsequent analysis. Phylogenetic analysis was performed, and consistency of the branching order of the phylogenetic trees was evaluated as described in Materials and Methods. Bootstrap values in excess of 70% were considered significant. (A) Phylogenetic analysis of full-length proviral env genes from subject SC24. Coreceptor usage of individual env gene products is indicated for each clone. MN represents the sequence of the prototypic clade B HIV-1 MN isolate obtained from the Los Alamos data base and is used as an outgroup to root the phylogenetic tree. (B) Phylogenetic analysis of a 459-bp region that surrounds and includes the V3 loop from direct sequencing of PCR amplified plasma viral env region compared with cloned proviral env genes from subject SC24. (C) Phylogenetic analysis of full-length proviral env genes from subject SC51. Coreceptor usage of individual env gene products is indicated for each clone. The HIV-1 MN isolate is used as an outgroup to root the phylogenetic tree. (D) Phylogenetic tree of the same cloned env genes from SC51 as in C but using the QH1302c.23a cloned env as an outgroup to root the tree. A supercript a indicates that the env clone is predominately R5 in the cell-cell fusion assay (see Fig. 2 and Table 3) but X4 activity was 5- to 10-fold above background levels in cell-cell fusion.
TABLE 2.
Amino acid sequence alignment of HIV-1 envelope V3 loopsa
| Patient and isolate | V3 loop sequence by direct PCR sequencing | env clone | V3 loop sequence |
|---|---|---|---|
| SC24 | ∗ ∗ ∗∗ ∗∗∗ ∗ ∗ | ∗ ∗ ∗∗∗∗∗∗ ∗ ∗ | |
| QH0515 | CTRPNNNTRKSIHIGAGKALYTGEIIGDIRQAHC | QH0515#1 | CTRPNNNTRKSIHIEAGKALYTGEIIGDIRQAHC |
| QH0648 | ––––––N–R–––HI–AGK–L––G––––––––––– | QH0648#1 | ––––––N–R–––HIGAGK–L––G––––––––––– |
| QH0797 | ––––––N–R–––HI–PGK–L––G––––––––––– | QH0797#3 | ––––––N–R–––HIGPGK–L––G––––––––––– |
| QH1020 | ––––––N–R–––HI–PGK–L––G––––––––––– | ||
| QH1246 | ––––––N–R–––HI–PGK–L––G––––––––––– | QH1246#3 | ––––––N–R–––HIGPGK–L––G––––––––––– |
| QH1483 | ––––––N–R–––HI–PGK–L––G––––––––––– | QH1483#3 | ––––––N–R–––HIGPGK–L––G––––––––––– |
| QH1503#1 | ––––––N–R–––HIGPGK–L––G––––––––––– | ||
| QH1503#5 | ––––––N–R–––HIGPGK–L––G––––––––––– | ||
| QH1503#12 | ––––––N–R–––HIGPGK–L––G––––––––––– | ||
| QH1520 | ––––––N–R–––RM–IRR–F––R––––––––––– | QH1520#2 | ––––––N–R–––RMGIRR–F––R––––––––––– |
| HI ?GK L G | |||
| QH1549 | ––––––N–R–––RM–IRR–F––R––––––––––– | QH1549#13 | ––––––N–I–––RMGIRR–F––R––––––––––– |
| ? HI ?GK L G | QH1549#1 | ––––––S–I–––RIGIRR–F––R––––––––––– | |
| SC51 | ∗ ∗ ∗∗∗ ∗ | ∗ ∗ ∗∗∗ ∗ ∗ | |
| QH1302 | CTRPNNNTRKSIHLGAGRALYTGEIIGDIRQAHC | QH1302#23A | CTRPNNNTRKSIHLGAGRALYTGEIIGDIRQAHC |
| QH1420 | ––––––N–––S–––––GR–L–––––I–––––––– | QH1420#2 | ––––––N–––S–––––GR–L––G––I–––––––– |
| QH1490 | ––––––N–––S–––––GR–L–––––I–––––––– | QH1490#4A | ––––––N–––S–––––GK–L––G––I–––––––– |
| ? | QH1490#11A | ––––––N–––S–––––GR–L––R––I–––––––– | |
| QH1509 | ––––––N–––S–––––GR–L–––––I–––––––– | QH1509#3 | ––––––N–––S–––––GR–L––G––I–––––––– |
| ? | |||
| QH1521 | ––––––N–––S–––––GK–L–––––I–––––––– | QH1521#28 | ––––––N–––S–––––GK–L––G––I–––––––– |
| H R? F | QH1521#30 | ––––––N–––H–––––RK–F––G––I–––––––– | |
| QH1521#34 | ––––––N–––H–––––GK–F––G––I–––––––– | ||
| QH1536 | ––––––Y–––H–––––RK–F–––––I–––––––– | QH1536#2 | ––––––Y–––H–––––RK–F––G––I–––––––– |
| N L | |||
| QH1558 | ––––––Y–––H–––––RK–F–––––V–––––––– | QH1558#23A | ––––––Y–––H–––––RK–F––G––V–––––––– |
Nonsynonymous sequence polymorphisms are indicated by assigning two amino acids or ? (if multiple codons are possible) to the same position. Dashes denote that an amino acid is identical to the residue in the first sequence; asterisks indicate the predicated residues are different.
FIG. 2.
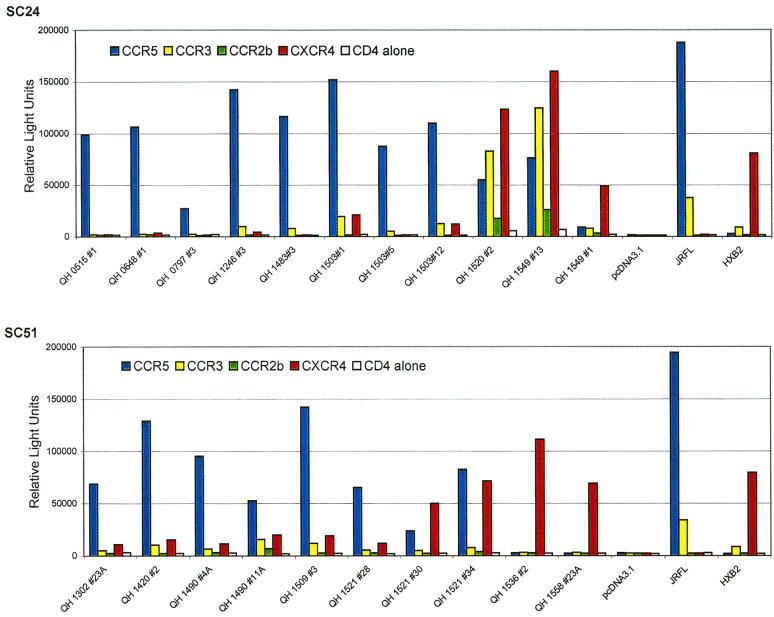
Fusogenic activity of Env encoded by molecular clones from sequential isolates from SC24 and SC51. The abilities of Env glycoproteins to mediate fusion with target cells expressing CD4 and candidate coreceptor was determined using a cell-cell fusion assay with a luciferase reporter gene. QT6 effector cells were prepared by infection with vTF1.1 (encoding T7 polymerase) for 1 h following transfection with Env constructs in pcDNA3.1. JRFL and HXB2 constructs in the same vector were used as positive controls. QT6 target cells were transfected with constructs encoding luciferase under the transcriptional control of a T7 promoter, CD4, and the indicated coreceptors. Effector and target cells were mixed and incubated for 9 h following culture overnight. Fusogenic activity was determined by analysis of luciferase activity in detergent lysates. Data shown are representative of at least three separate experiments yielding similar results, with each determination performed in duplicate.
Sequencing cloned full-length env genes and chimeric env constructs.
Cloned env genes were sequenced completely with an ABI 377 automated sequencer using the Taq Dye terminator cycle sequencing kit (ABI Prism) chemistries. All sequences were determined from both sense and antisense DNA strands to ensure the accuracy of the sequence determinations. Primers used to determine the env sequences were as follows: ENF, 5′-AAAGAGCAGAAGACAGTGGCAATGAGAGTGAAGG; E155R, 5′-CTGTTCTACCATGTCATTTTTCCACATGT; E70, 5′-GGGATCAAAGCCTAAAGCCATGTGTAA; E90, 5′-CACAGTACAATGTACACATGGAAT; E145R, 5′-AGCAGTTGAGTTGATACTACTGG; NL437500, 5′-AAGTAGGAAAAGCAATGTATGCCCCTCCCAT; HXB21299R, 5′-ATGGGAGGGGCATACATT; E180, 5′-GTCTGGTATAGTGCAGCA; E55R, 5′-GCCCCAGACTGTGAGTTGCAACAGATG; E220, 5′-TAACAAATTGGCTGTGGTATATAA; HXB28562R, 5′-CTCGTTACAATCAAGAGTAAGTCTCTCAA; ENR, 5′-CAATCACACTACTTTTTGACCACTTGCCACCCAT; T7, 5′-TAATACGACTCACTATAGGG; and PCR3.1, 5′-TAGAAGGCACAGTCGAGG. An R at the end of a primer name indicates a reverse primer. ENF and ENR were used only in direct sequencing of PCR products. Primers residing in the cloning vector (T7 and PCR3.1) were used to sequence full-length env clones in the immediate 5′ and 3′ regions. Derived DNA sequences were initially analyzed and aligned using software packages from ABI. Gaps were inserted into the alignment as needed, and those regions were eliminated from the subsequent analysis. Phylogenetic analysis was performed, and consistency of the branching order of the phylogenetic trees was evaluated using the SEQBOOT, DNADIST, NEIGHBOR, CONSENSE, and DRAWGRAM modules of the Phylip Package (V3.572) (26, 27). Phylogenetic trees were constructed using neighbor-joining (51), and the stability of the nodes was assessed with the bootstrap value (26). Bootstrap values in excess of 70% were considered significant.
Reverse transcription, PCR amplification, and direct sequencing of plasma virus env regions.
Plasma was prepared from acid citrate dextrose-anticoagulated whole venous blood within 2 h of sample collection and stored at −70°C until analyzed. HIV-1 RNA was extracted with the Roche Amplicor HIV-1 Monitor Assay system (Roche Molecular Systems, Inc., Branchburg, N.J.) as specified by the manufacturer and stored at −70°C. A 25-μl amount of the RNA extract was reverse transcribed and PCR amplified in a total volume of 75 μl with the Perkin-Elmer GeneAmp Gold RNA PCR kit with the following reaction components and conditions: 1X RT PCR Buffer (Perkin-Elmer), 1.75 mM MgCl2, 0.35 mM each dNTP, 10 U of RNase inhibitor (Perkin-Elmer), 10 mM dithiothreitol, and 0.5 μM MG-12 3′-primer [5′-AA(T/C)(T/A)GTCTGGCCTGTACCGTCAGCGT-3′] and 0.5 μM MG-6 5′-primer [5′-GGT(A/G)TCCTTTGA(G/T)CCAATTCCCAT-3′], 2.5 U of AmpliTaq Gold, and 150 U of murine leukemia virus RT enzyme. Reverse transcription was carried out at 42°C for 60 min. Reaction components were heated to 95°C for 10 min, and then PCR was performed with 50 cycles of denaturation at 94°C for 1 min, annealing at 57°C for 1 min, and extension at 72°C for 2 min. A final extension at 72°C for 10 min was employed after the last cycle. The PCR mix (7.5 μl) was transferred to a separate tube for a second round of amplification with nested primers. Reaction conditions and components for the second-round PCR were 1X PCR buffer II (Perkin-Elmer), 1.5 mM MgCl2, 0.2 mM each dNTP, 0.2 μM MG-10 3′-primer [5′-CACTT(C/T)TCCAATTGTCCCTCAT(A/G)TCTCCTCCT-3′] and 0.2 μM MG-7 5′-primer [5′-GTCAGCACAGTACAATGTACACAT-3′], and 2.5 U of AmpliTaq Gold (Perkin-Elmer). Denaturation for 10 min at 95°C preceded 10 cycles of amplification using 95°C for 45 s for denaturation, 59°C for 1 min for annealing, and 72°C for 1.5 min for extension. These were followed by 20 to 40 cycles of amplification using 94°C for 45 s for denaturation, 59°C for 1 min for annealing, and 72°C for 1.5 min for extension. A final extension at 72°C for 10 min was employed. Direct sequencing and phylogenetic analysis of the amplified plasma viral env region were performed as described above for the full-length env genes using the MG-7, MG-10, E110, and HXB21129R (5′-AAATTCCCCTCCACAATT-3′) primers for sequencing.
Genetic engineering of Env chimeras and mutants.
Env chimeras between QH0648#1 and QH1520#2 and between QH0515#1 and QH1520#2 were generated using sites that were present in these env genes, including EcoRI, AccI, SauI, and XhoI. A DraI site was introduced at the 3′ boundary of the V3 region as a silent mutation that did not alter the amino acid programmed by the codons at this position using the Chameleon double-stranded site-directed mutagenesis kit (Stratagene). The identity of all chimeras and mutants was confirmed by DNA sequencing.
Reporter gene assay for analysis of Env-mediated fusion.
The coreceptor utilization of the products encoded by the primary env genes, chimeras, and mutants was determined using a cell-cell fusion assay employing a luciferase reporter gene, as described previously (22, 61, 62), with modifications. Briefly, QT6 effector cells were prepared by infection with vTF1.1, which encodes T7 polymerase, for 1 h following transfection with pcDNA3 constructs containing the primary env clones. Target cells consisted of QT6 cells transfected with pcDNA3 constructs encoding CD4, coreceptors, and a luciferase reporter gene under the transcriptional control of the T7 promoter. The effector and target cell populations were mixed at 16 to 18 h following transfection, and the luciferase activity of cell lysates was determined approximately 9 h later using a LucLite luciferase reporter gene assay kit (Packard). Absolute light units emitted were measured using a Top Count luminometer (Packard).
Production of pseudotyped reporter viruses and infectivity assays.
Stocks of pseudotyped reporter viruses were prepared as follows. 293T cells (a gift from Robert Doms) were maintained in DMEM containing 10% heat-inactivated FCS and 1% pen/strep (Gibco-BRL). 293T cells were seeded at 1.5 × 106 cells per well in a six-well dish (Costar) 1 day prior to transfection. Cells were cotransfected with 10 μg of an env expression vector and 5 μg of plasmid pNL4-3.Luc.R−E− (obtained from the AIDS Research and Reference Reagent Program, DAIDS, NIAID, NIH, from Nathaniel Landau) by the calcium phosphate precipitation method essentially as described (16). Virus-containing supernatants were harvested 48 h posttransfection, mixed with heat-inactivated FCS to bring the concentration to 20% (vol/vol), filtered, and stored at −80°C until used for infection. The ability of cloned env to mediate infection via the CCR5 and/or CXCR4 coreceptor was assessed in U87 astroglioma cells stably expressing the CD4 receptor and either the CCR5 or CXCR4 coreceptor or neither chemokine receptor. U87 cells were seeded at 1.5 × 104 cells per well in a 96-well plate and infected with 100 μl of pseudotyped virus-containing supernatant. Cells were harvested 48 to 72 h postinfection and assayed with either the Promega luciferase assay system (Promega) in a Berthold Lumat 9501 (Wallac) or the LucLite luciferase assay kit (Packard) in a Top Count luminometer (Packard).
Metabolic labeling and immunoprecipitation of Env.
The expression of wild-type and chimeric Env glycoproteins was analyzed by metabolic labeling and immunoprecipitation. Briefly, QT6 or HeLa cells (2 × 105) were seeded in six-well plates and grown to 80 to 90% confluence. Monolayers were infected with the vTF1.1 vaccinia virus, which directs the expression of T7 polymerase, for approximately 1 h and then transfected with the env constructs. These were prepared in the pcDNA3 vector, which contains a T7 promoter downstream of the cytomegalovirus promoter, which is typically employed in mammalian cells. The cells were allowed to incubate for 18 h at 37°C. The transfectants were then depleted of methionine and cysteine by growth in medium lacking these amino acids for 1 h. The cultures were then metabolically labeled in 1 ml of this medium supplemented with 100 μCi of [35S]methionine/[35S]cysteine (NEN Life Science) for 1 h. Complete medium was then added, and the cells were incubated for an additional 1 h at 37°C. Lysates of metabolically labeled cells were clarified by centrifugation in a microcentrifuge and then incubated with HIVIG (obtained from the NIH AIDS Research and Reference Reagent Program) or normal immunoglobulin fractions, followed by protein G-Sepharose (Pharmacia) overnight at 4°C. Immune complexes bound by the protein G-Sepharose were washed, resuspended in SDS sample buffer, and resolved by SDS-polyacrylamide gel electrophoresis (PAGE), and the dried gels were analyzed by autoradiography.
Nucleotide sequence accession numbers.
All sequences described were deposited in the GenBank database under accession no. AF310108 to AF310132.
RESULTS
env genes from sequential primary isolates.
To examine the functional consequences of the diversification in env that occurs during HIV-1 disease, we examined the biological and molecular characteristics of serial virus isolates obtained from two individuals in a cohort of acute seroconverters in Trinidad and Tobago. Cohort subjects were identified by virtue of exhibiting HIV-1 p24 antigenemia in the absence of a positive Western blot, and their status was confirmed by subsequent documentation of seroconversion to HIV-1 antigens. From a cohort of 22 HIV-positive individuals, we examined serial viral isolates and env clones from two subjects whose isolates exhibited a phenotypic switch from NSI to SI during the period of observation. As shown in Table 1, the isolates studied spanned the evolution from NSI to SI, which occurred over an interval of 875 days (∼29 months) for patient SC24, but within a much shorter time frame, 12 months, for the second subject, SC51. Thus far, five subjects in the cohort have evidenced a switch in virus phenotype from an initial NSI isolate to a subsequent SI isolate. The mean time from infection to recovery of an SI isolate from the five subjects in the original cohort has been 31 months (range, 12 to 41 months), which appears to be considerably shorter than that observed in a recent study of nine individuals from the MACS cohort (56), and may reflect a more rapid pace of disease progression in developing versus developed countries. The NSI and SI designations in Table 1 are based on the ability of the virus isolates to induce syncytia in the MT-2 cell line.
TABLE 1.
Sequential HIV-1 primary isolates and phenotypesa
| Patient and isolate | Time after sercoconversion (days) | MT-2 phenotype | Infection assayb
|
|||
|---|---|---|---|---|---|---|
| U87-CD4 | U87-CD4+CCR5 | U87-CD4+CXCR4 | HeLa-CD4 (MAGI) | |||
| SC24 | ||||||
| QH0515 | 0 | NSI | <70 | 1.2 × 103 | <70 | ND |
| QH0648 | 77 | NSI | <70 | 2.7 × 104 | <70 | ND |
| QH0797 | 168 | NSI | <70 | 4.6 × 104 | <70 | 0 |
| QH1020 | 343 | NSI | <70 | 1.4 × 104 | <70 | 0 |
| QH1246 | 511 | NSI | <70 | >2.2 × 105 | <70 | ND |
| QH1483 | 686 | NSI | <70 | 1.0 × 105 | 110 | 0 |
| QH1503 | 777 | NSI | <70 | 1.1 × 105 | <70 | 0 |
| QH1520 | 875 | SI | <70 | 5.5 × 104 | 4.8 × 103 | 4.6 × 103 |
| QH1549 | 1,023 | SI | <70 | 6.3 × 104 | >2.2 × 105 | 1.2 × 104 |
| SC24 090398 | 1,302 | SI | <70 | 4.3 × 104 | 1.4 × 106 | >2.4 × 105 |
| SC51 | ||||||
| QH1302 | 22 | NSI | <70 | 8.5 × 102 | <300 | 0 |
| QH1420 | 99 | NSI | <70 | 1.4 × 104 | <300 | 0 |
| QH1490 | 183 | NSI | <70 | 4.3 × 103 | <300 | 0 |
| QH1509 | 281 | NSI | <70 | 1.7 × 104 | <300 | 0 |
| QH1521 | 357 | SI | <70 | 8.5 × 102 | 6.8 × 103 | 2.8 × 103 |
| QH1536 | 448 | SI | <70 | 2.7 × 103 | 1.3 × 106 | 2.7 × 104 |
| QH1558 | 539 | SI | <70 | 4.7 × 103 | 8.5 × 105 | 1.9 × 104 |
Subjects SC24 and SC51 were identified as experiencing acute HIV-1 infection on the basis of p24 antigenemia in the absence of a positive Western blot (defined as reactivity to two or more HIV-1 gene products).
Infectious titers were determined by the end-point dilution technique, and TCID50 per milliliter was calculated (45). For MAGI assay, titers represent number of infectious centers per milliliter. ND, not determined.
We performed a parallel series of analyses using infection assays in U87-CD4 cells stably expressing either CCR5, CXCR4, or neither coreceptor and HeLa-CD4 cells (6, 32) to confirm that the conversion from NSI to SI corresponded to the emergence of viral species that utilize CXCR4 as a coreceptor. For subject SC24, these studies indeed indicated that recovery of the SI phenotype was associated with the emergence of isolates with the ability to replicate in cells expressing CXCR4 (Table 1). For SC51, the results were somewhat less clear. Although the initial QH1302 isolate was restricted to replication in CCR5-expressing U87 cells, we observed very low levels of infection of U87-CD4 target cells expressing CXCR4 with the three successive NSI isolates (QH1420, QH1490, and QH1509) recovered from 99 to 281 days following seroconversion. However, titers of these isolates in CXCR4-expressing U87-CD4 cells never reached 300 TCID50/ml, and when the isolates were similarly evaluated on the MAGI target cells for infectivity, no evidence for infection of CXCR4-expressing cells was observed (Table 1). In contrast, we easily observed robust levels of replication in both the U87-based CXCR4 targets and the MAGI cells with the three SI isolates from this subject (Table 1), and their infection of MAGI cells was completely blocked by the CXCR4 inhibitor AMD3100 (data not shown). These results are consistent with the view that the SI phenotype is associated with the ability to productively infect via the CXCR4 coreceptor and document that the X4 character of virus isolates recovered from both subjects changed dramatically within a narrow time frame. The change in the X4 character of virus isolates was manifest in samples that were separated by only 98 days for subject SC24 and with samples separated by 76 days for subject SC51 (Table 1). After the initial recovery of virus isolates capable of using the CXCR4 coreceptor for entry, this characteristic became predominant in all subsequent isolates that were recovered from both subjects (Table 1).
Primary env genes were amplified by PCR from proviral DNA templates derived from the successive isolates from these two individuals and molecularly cloned. A total of 128 env clones were isolated and tested for biological activity as described below. Comparison of the nucleotide sequence of molecular clones active in Env-mediated fusion assays (see below) with that obtained from the direct analysis of proviral DNA templates confirmed that the former were representative of the predominant viral species in the isolate (Table 2). For SC24, a similar comparison with plasma virus sequences suggested that the fusion-competent env clones were similar to variants circulating in vivo (Fig. 1B). Some variants showed ambiguities of the predicted primary structure of Env, which were interpreted to be the result of heterogeneity of integrated proviruses. Alignment of the nucleotide sequences of the env clones and translation for the predicted primary structure from patients SC24 and SC51 confirmed that the sequential isolates from each patient were highly related to the others from that individual, consistent with being derived from similar (ancestral) viruses. Divergence at the level of primary structure was noted most commonly in the V1, V2, V3, and V4 regions of gp120. Phylogenetic analysis of the successive env clones confirmed progression along an evolutionary tree (Fig. 1D). We noted a branching of the env sequences from both SC24 and SC51 when there was conversion of phenotype from NSI to SI, with env sequences associated with CXCR4 usage forming a distinct subcluster (Fig. 1A and C). The sequence of the QH1302c.23a env clone from SC51 was an outlier and did not group with either the CCR5- or CXCR4-associated env sequences from this subject (Fig. 1C). In one analysis, this sequence was used as an outgroup to root the phylogenetic analysis for SC51 (Fig. 1D). Inclusion of the QH1302c.23a sequence decreased the bootstrap values at all nodes except that between the 1521c.34 and 1521c.30 sequences, obscuring relationships among the other env clones from this individual.
Coreceptor utilization of primary Env during the evolution of HIV-1 infection.
The utilization of front-line HIV-1 coreceptors by the 128 env-encoded glycoproteins was determined in cell-cell fusion assays. Biological activity (Env-mediated cell-cell fusion) was detected with 40 of these clones (Table 3), and data from 21 representative clones are shown in Fig. 2.
TABLE 3.
Env-mediated fusion activity of molecular clones
| Patient and isolate | Total no. of env clonesa | No. of fusion-competent env clones | Descriptiond |
|---|---|---|---|
| SC24 | 54 | 18 | |
| QH0515 | 3 | 1 | R5 |
| QH0648 | 5 | 1 | R5 |
| QH0797 | 4 | 1 | R5 |
| QH1020 | 4 | 0 | |
| QH1246 | 3 | 1 | R5 |
| QH1483 | 7 | 2 | {1}R5, {1}R5 (X4±)b |
| QH1503 | 10 | 8 | {3}R5, {5}R5 (X4±)b |
| QH1520 | 5 | 2 | {2}R5X4 |
| QH1549 | 13 | 2 | R5X4, X4 (R5±)c |
| SC51 | 74 | 22 | |
| QH1302 | 18 | 2 | R5 (X4±)b |
| QH1420 | 6 | 1 | R5 (X4±)b |
| QH1490 | 8 | 5 | R5 (X4±)b |
| QH1509 | 7 | 2 | R5 (X4±)b |
| QH1521 | 17 | 6 | {3}R5 (X4±)b, {3}R5X4 |
| QH1536 | 3 | 1 | X4 |
| QH1558 | 15 | 5 | X4 |
All env genes were amplified by PCR and cloned into pcDNA3.1 as described in Materials and Methods. Env constructs were tested for coreceptor utilization in cell-cell fusion assays. Primary Env showed a spectrum of M-topic (R5), dual-tropic (R5X4), and T-tropic (X4) activity. Representative clones are depicted in Fig. 2.
Predominantly R5, but X4 activity is 5- to 15-fold over background values with CD4 alone.
Predominantly X4, but R5 activity is 5- to 15-fold over background values with CD4 alone.
Numbers in braces refer to the number of env clones with the corresponding characteristic.
Env encoded by templates isolated from patient SC24 during the early stages of infection (QH0515#1 and QH0648#1 from 0 and 77 days postseroconversion, respectively) utilized CCR5 exclusively. An Env encoded by an NSI strain isolated 511 days following seroconversion, QH1246#3, showed minimal utilization of CCR3 that was above background levels observed in cells expressing CD4 alone. This Env lacked significant activity with CCR2b and CXCR4. Analysis of an Env glycoprotein from an SI isolate taken 875 days following the detection of seropositivity, QH1520#2, showed utilization of CXCR4, CCR3, CCR5, and CCR2b, in order of decreasing fusogenic activity. Two biologically active env were obtained from an SI strain isolated 1,023 days following seroconversion, designated QH1549#1 and QH1549#13. QH1549#13 utilized CXCR4, CCR3, and CCR5 at significant levels and CCR2b at low levels, with relative efficiencies similar to those observed for QH1520#2. In contrast, QH1549#1 showed significant fusogenic activity exclusively with CXCR4. It is noted that in addition to utilization of CXCR4, the HXB2 Env showed a low level of fusogenic activity with CCR3.
Parallel studies were performed with env clones derived by amplification of proviral templates from isolates from patient SC51, who showed conversion from NSI to SI strains early in the course of infection (357 days postseroconversion) and experienced a fulminant clinical course.
The predominant coreceptor utilized by Env encoded by early isolates, which were of the NSI phenotype, was CCR5, and the utilization of CCR2b and CCR3 by these glycoproteins was inconspicuous. They also demonstrated a low level of fusion with target cells expressing CXCR4 that was minimally above the background values established with CD4 alone. Predominant CCR5 utilization with marginal CXCR4 utilization above background was also evident with one clone derived from day 99 (QH1420#2) and two clones from the 183-day isolate (QH1490#4A and QH1490#11A). Env encoded by the molecular clone from the QH1509 provirus used CCR5 in fusion experiments and also showed low-level utilization of CXCR4.
Three env clones were characterized from the isolate taken at 357 days (QH1521), which marked conversion to the SI phenotype (Table 1), although utilization of CCR5 and CXCR4 in infection experiments did not differ dramatically. Two of these clones demonstrated utilization of CCR5 and CXCR4, one in which the latter was the predominant coreceptor (QH1521#30) and one that used them at equivalent efficiencies (QH1521#34). CCR5 was the predominant coreceptor for the third env clone from this isolate (QH1521#28), but fusogenic activity with CXCR4 was minimally above background levels. The SI isolates from the late phases of infection in this patient used CXCR4 in infection experiments, and the Env encoded by molecular clones derived from these templates (QH1536#2 and QH1558#23A) showed high-level, exclusive utilization of CXCR4.
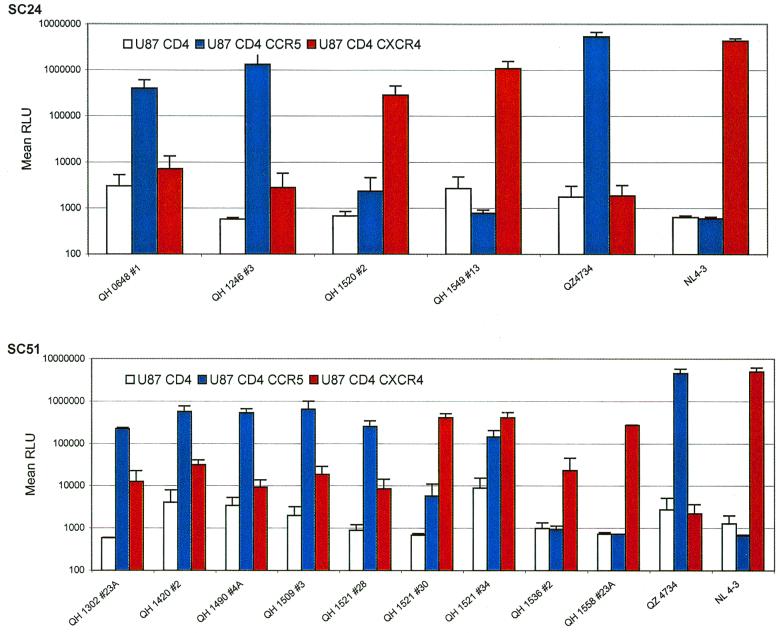
The findings in fusion assays were confirmed in experiments using pseudotyped viruses containing the Env glycoproteins and a luciferase reporter gene to infect target cells programmed to express human CD4 and either CCR5 or CXCR4. Env encoded by proviral templates derived early in the course of infection from patient SC24 utilized CCR5 exclusively, as shown in Fig. 3. The Env encoded by QH1520#2 showed significant utilization of CCR5 and CXCR4 in the cell-cell fusion model, but this was limited to the latter coreceptor in the infection experiments. The Env encoded by QH1549#13 also mediated viral entry only in cells expressing CXCR4. Several Env lacked activity in infection experiments (QH0515#1, QH0797#3, and QH1549#1) and are not depicted in the figure.
FIG. 3.
Analysis of coreceptor utilization by primary Env from patients SC24 and SC51 in infection experiments. The primary env clones were used to derive pseudotyped viruses containing the encoded glycoproteins and a luciferase reporter gene. The 293T cell line was cotransfected with 10 μg of a primary Env expression construct and 5 μg of plasmid pNL4-3.Luc.R−E− using the standard calcium phosphate precipitation method. Virus-containing supernatants were harvested 48 h following transfection and filtered. The ability of cloned env to mediate infection via CCR5 and CXCR4 was assessed in U87 astroglioma cells expressing the CD4 receptor alone or in combination with either coreceptor, as described in Materials and Methods. U87 target cells were infected with pseudotyped virus, harvested 48 to 72 h postinfection, and assayed for luciferase activity as described in Materials and Methods. The results are the means of three independent experiments, each performed in duplicate.
There was good agreement between the coreceptor utilization data from the cell-cell fusion model and infection experiments for the Env encoded by isolates from patient SC51 (Fig. 3). CCR5 was the predominant coreceptor for products of env molecularly cloned from the first four isolates, which were NSI. The utilization of CXCR4 by these Env was consistently above baseline values established for CD4 alone. Subtle differences were evident in the utilization of CCR5 and CXCR4 by the three env clones from 357 days following seroconversion (QH1521), which were similar to those observed in fusion experiments. QH1521#28 showed predominant utilization of CCR5 in both assays, with low levels of fusion and viral entry with CXCR4. QH1521#34 was clearly dual tropic (R5X4) in both types of experiments, and QH1521#30 showed predominant X4 utilization with minimal R5 activity compared to background values established with CD4 alone in infection experiments. These approaches also revealed comparable exclusive utilization of CXCR4 for Env from late in this patient's course, QH1536#2 and QH1558#23A. Viral isolates from the QH1521, QH1536, and QH1558 specimens had limited infectious activity with CCR5-expressing target cells.
Multiple domains of Env contribute to coreceptor specificity.
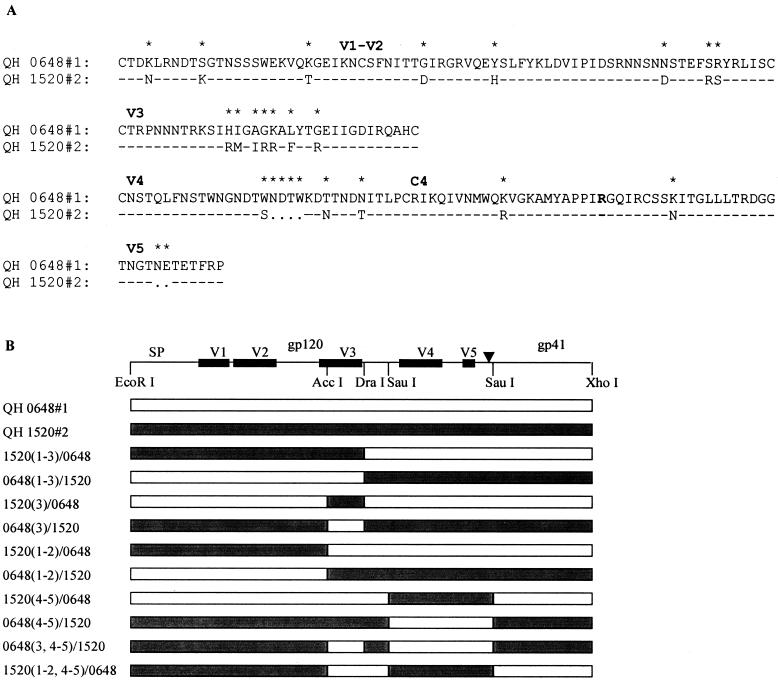
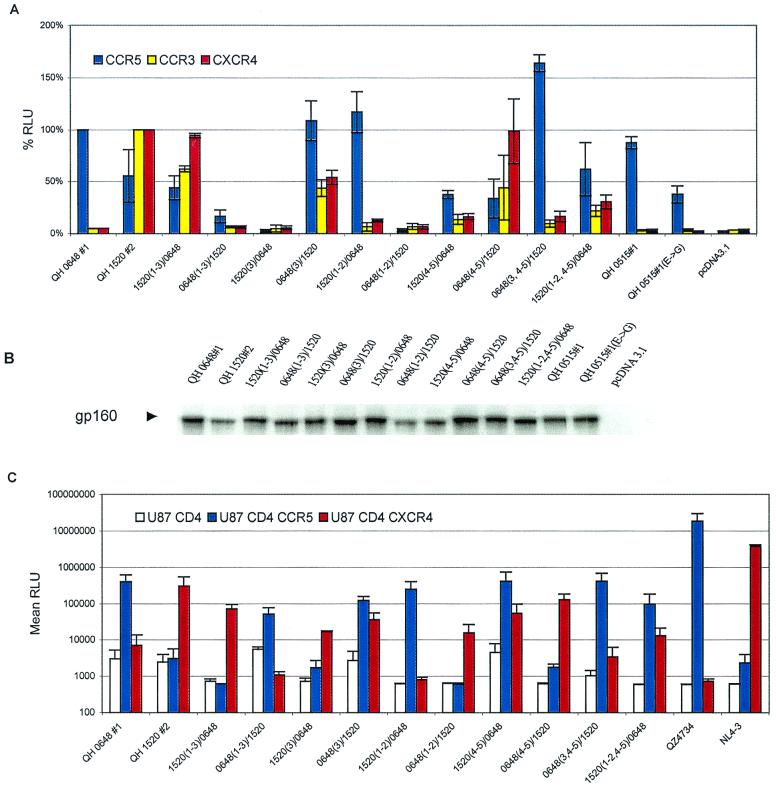
The availability of genetically related env that were derived from successive viral isolates during the evolution of infection in a single patient offered a unique opportunity to study the domains that participate in determining the specificity of coreceptor utilization in vivo. To extend our initial observations, env clones from patient SC24 encoding R5 (QH0648#1) and multitropic (QH1520#1) products (Fig. 4A) were used to generate chimeras, which are depicted in Fig. 4B. As shown in Fig. 5A, whereas the chimeric Env composed of the N-terminal segment of QH1520#2 through the V3 loop [1520(1–3)/0648] demonstrated significant utilization of CXCR4 and CCR3, the fusogenic activity of that extending to the region between the V2 and V3 loops [1520(1–2)/0648] was limited to CCR5. Replacement of the V3 loop of QH1520#2 with that of QH0648#1 [0648(3)/1520] resulted in a multitropic Env with increased CCR5 utilization and mildly decreased use of CXCR4 and CCR3 in comparison to the wild-type QH1520#2 Env. Substitution of QH0648#1 sequences for the V3 loop and the segment containing the V4 and V5 domains of QH1520#2 [0648(3, 4–5)/1520] resulted in coreceptor utilization essentially limited to CCR5. The effect of the V4–V5 segment on coreceptor utilization is further illustrated by the ability of the hybrid Env containing the V1–V2 and V4–V5 domains of QH1520#2 [1520(1–2, 4–5)/0648] to utilize CXCR4 and CCR3 as coreceptors in addition to CCR5. A similar effect was noted with 1520(4–5)/0648, although the total fusogenic activity of this hybrid was less than that observed with the parental Env. Several of the hybrid Env had limited fusogenic activity. These chimeras appeared to be expressed at intermediate [0648(1–3)/1520 and 1520(3)/0648] or low [0648(1–2)/1520 and 1520(4–5)/0648] levels in metabolic labeling experiments (Fig. 5B). All of the hybrids, including those with low levels of fusogenic activity, were expressed at levels similar to or greater than that of the parental QH1520#2 Env (Fig. 5B).
FIG. 4.
Structure of chimeric Env created from R5 and multitropic Env from patient SC24. (A) Alignment of the variable domains and C4 region of Env encoded by QH0648#1 and QH1520#2 molecular clones from isolates obtained in the early (QH0648) and intermediate (QH1520) stages, respectively, of the evolution of AIDS in patient SC24. Dashes denote identical amino acid residues (>95% overall), and dots represent gaps introduced to optimize alignments. Asterisks indicate that the amino acids in that position are different. The R residue that appears in boldface type in the C4 region denotes the position of the R→S conversion highlighted previously (9). (B) Schematic representation of wild-type and chimeric env genes showing restriction endonuclease cleavage sites that were used to genetically engineer the hybrids. Creation of the DraI site by site-directed mutagenesis resulted in the introduction of conservative nucleotide changes that did not alter the primary structure of Env.
FIG. 5.
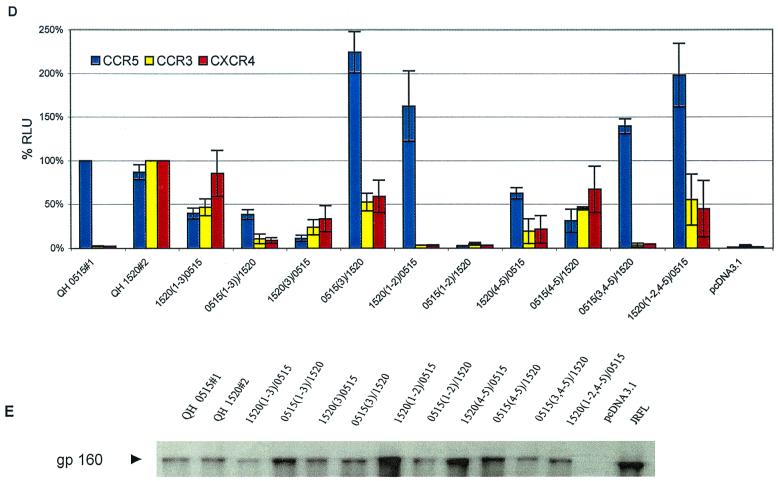
V3 and V4–V5 regions of multitropic Env contribute to coreceptor utilization. (A) The fusogenic activity of the panel of hybrid Env composed of complementary segments of the R5 Env QH0648#1 and the multitropic Env QH1520#2 was determined in the cell-cell fusion assay, as described in Materials and Methods, using assay conditions identical to those described in the legend to Fig. 2. Luciferase intensity is expressed as relative percentages normalizing CCR5 activity to that of QH0648#1 and CCR3 and CXCR4 activity to that of QH1520#2. The data are presented as averages of means of duplicate values from three independent experiments. The structures of the hybrid Env that were tested are depicted in Fig. 4. (B) Expression of wild-type and chimeric Env was analyzed by immunoprecipitation of metabolically labeled cells transiently expressing the cloned env gene. Transfectants were starved for 1 h in medium lacking Met and Cys, labeled with [35S]Met/[35S]Cys for 1 h, and chased with complete medium for 1 h. Detergent lysates were immunoprecipitated with HIVIG or control reagents, as described in Materials and Methods. The components of the immune complexes were resolved by SDS-PAGE and detected by autoradiography. The gp160 precursor glycoprotein is marked with an arrowhead. (C) Ability of the chimeric Env to mediate virus entry was determined in infection experiments using pseudotyped viruses as described in Materials and Methods under conditions identical to those described in the legend to Fig. 3. (D) Contribution of the V3 and V4–V5 domains of QH1520#2, the multitropic Env, was confirmed in fusion assays with a parallel panel of chimeras using the earliest R5 Env from SC24, QH0515#1, which was active in the cell-cell fusion assay but inactive in infection experiments. The cell-cell fusion system was performed as described above and in Materials and Methods. (E) Metabolic labeling experiments confirmed the expression of the hybrid Env glycoproteins, as described above.
To confirm these observations, a parallel array of chimeras were genetically engineered using QH1520#2 and QH0515#1 as the R5 partner in place of QH0648#1. The hybrids 0515(1–3)/1520 and 1520(3)/0515, which correspond to QH0648#1 chimeras that lacked significant fusogenic activity, showed intermediate levels of fusogenic activity in the cell-cell fusion model (Fig. 5D) and expression in metabolic labeling experiments (Fig. 5E). The 0515(1–3)/1520 hybrid showed predominant utilization of CCR5 and activity with CXCR4 and CCR3 that was above background levels established with CD4 alone and greater than that of the parental Env, QH0515#1. The 1520(3)/0515 chimera demonstrated significant utilization of CXCR4 and CCR3 but minimal utilization of CCR5. The contribution of the V4–V5 segment of QH1520#2 to CXCR4 and CCR3 utilization is demonstrated by the 1520(4–5)/0515 and 1520(1–2, 4–5)/0515chimeric Env. In contrast to a previous report highlighting the contribution of an R→S conversion in the C4 region to coreceptor usage (9), both envelopes examined in the current studies possessed an R in this position (Fig. 4). The V1–V2 region of QH1520#2 Env failed to contribute to the multitropic phenotype, as was noted in the panel of chimeras engineered with QH0648#1. All hybrid Env were expressed at levels similar to or greater than those observed for the wild-type parental Env in immunoprecipitation experiments (Fig. 5B and E).
The coreceptor activity of hybrid Env derived from QH0648#1 and QH1520#2 was also tested in infection experiments (Fig. 5C). The 1520(1–3)/0648 Env used CXCR4, and 1520(1–2)/0648 used CCR5, as shown in the cell-cell fusion model. The substitution of V3 sequences in QH0648#1 with those from QH1520#2 was sufficient to confer utilization of CXCR4. Conversely, the substitution of V3 sequences in QH1520#2 with those from QH0648#1 did not significantly alter utilization of CXCR4 but markedly increased the utilization of CCR5. Swaps of the V1 and V2 regions did not modify the coreceptor utilization of QH0648#1 or QH1520#2. Replacement of the V4–V5 region of QH0648#1 with the corresponding sequence from QH1520#2 did not alter CCR5 utilization but was sufficient to confer the ability to use CXCR4. Similarly, the removal of V3 and V4–V5 segments from QH1520#2 and insertion of the analogous domains of QH0648#1 resulted in fusogenic activity with CCR5. The 1520(4–5)/0648 and 1520(1–2, 4–5) hybrids also support the contribution of the V4–V5 domains (of 1520#2) to the multitropic phenotype. These data provide evidence for the participation of the V3 and V4–V5 regions in determining the repertoire of coreceptor utilization for Env-mediated fusion through both gain- and loss-of-function experiments. In contrast, they fail to establish a role for the V1–V2 segment in determining this phenotype.
DISCUSSION
We examined the temporal relationships between the biological, genetic, and functional characteristics of sequential HIV-1 isolates and cloned env genes from two individuals who exhibited evolution in their virus phenotype from an initial NSI isolate to subsequent SI isolates during the course of their disease. The conversion in virus phenotype occurred with dramatically different kinetics in the two subjects and was paralleled by a similar divergence in their clinical courses. Both subjects were enrolled in a study of acute HIV-1 infection and were followed from the earliest ascertainable stages of acute infection throughout their disease course. The initial virus isolates were obtained at a time when each individual exhibited p24 antigenemia but prior to conversion to positive Western blot status. Plasma virus load assessments documented that the initial viral samples from both subjects were obtained at or near the peak of plasma viremia that usually accompanies acute HIV-1 infection. Consistent with prior reports, the initial isolates from each individual were of the NSI phenotype and restricted to replication in target cells expressing CCR5 (6, 17, 54, 60). Subject SC24 exhibited a switch in virus phenotype to SI after approximately 2.4 years, while this switch occurred much more rapidly in subject SC51, as SI variants were recovered after about 1 year of infection. As documented in Table 1, infectivity studies of the isolates from both patients confirmed that recovery of SI isolates coincided with the emergence of strains that infect target cells expressing CXCR4, but also revealed that the SI isolates were composed of differing proportions of subpopulations that used CCR5. However, in both subjects there was a progression with time to populations that were much more efficient in infecting through the CXCR4 receptor rather than CCR5. Characterization of coreceptor usage by the products of cloned env genes demonstrated that the SI phenotype represents the emergence of viral Env capable of using both CXCR4 and CCR5, as well as individual Env restricted to CXCR4, and the persistence of species that use CCR5. These studies amplify the recent findings of Shankarappa et al. (56) but also highlight the possibility that the time frame for the three distinct phases identified during progression by those investigators may be significantly altered in cohorts from developing regions where rates of progression appear to be more rapid.
In addition to the biological studies on the virus isolates, the genetic and functional studies of cloned env genes have provided novel insights on the nature of variants that make up these populations and the evolution of this gene in vivo. For subject SC24, we did not recover individual envelopes that were capable of efficiently infecting via both CCR5 and CXCR4 coreceptors (i.e., dual-tropic Env). This result suggests that the virus quasispecies in these SI isolates are composed of subpopulations that mediate infection through either CXCR4 or CCR5. In contrast to these findings, analysis in the cell-cell fusion model system revealed that the progression from R5 to X4 Env occurred via an intermediate that had multitropic coreceptor usage that included CCR5, CCR3, and CXCR4 as well as CCR2b at levels above background with CD4 alone. As they assess fusogenic activity in different contexts, the fusion assay may be a more sensitive measure of biochemical interactions that can occur between Env and coreceptors, although this surrogate may not predict productive Env-coreceptor interactions in the context of virus infection. In the second subject (SC51), we observed a different picture for the 1-year transition from NSI R5 strains to the development of SI X4 variants. Molecular env clones from the QH1521 isolate of SC51 revealed a mixture in both fusion and infection assays that included R5X4 dual-tropic species in addition to an R5 Env variant. Utilization of CCR3 and CCR2b was not observed in the cell-cell fusion model. Taken together, the results suggest that there may be multiple pathways for HIV-1 to evolve in vivo from an initial CCR5 population to a CXCR4-permissive population during the course of disease. This interpretation must be tempered by the fact that our examination of env clones was not exhaustive, and it is possible that dual-tropic or multitropic Env variants existed in both subjects but were not recovered.
A substantial proportion of HIV-1-infected individuals progress to AIDS without making the phenotypic switch to SI variants (21, 33, 55). In these subjects, the env genes may not evolve to use coreceptors other than CCR5, as suggested by a recent report (21). It is possible that in such individuals, subtle changes in the virus Env may alter the nature or efficiency of its interaction with the CCR5 coreceptor in a way that leads to increased virus replication. Indeed, we have observed an unusual case of an individual who experienced an extremely rapid disease course (time from infection to death from AIDS of less than 7 months), characterized by persistently high virus loads without any evidence for use of CXCR4 (J. F. Demarest, N. Jack, F. R. Cleghorn, M. L. Greenberg, T. L. Hoffman, L. Fantry, J. Edwards, T. R. O'Brien, K. Cao, B. Muhabir, W. A. Blattner, C. Bartholomew, and K. J. Weinhold, submitted for publication). Thus, our findings indicate that R5 NSI HIV variants, R5/X4 SI dual-tropic variants, and X4 SI variants are all capable of eliciting AIDS in the proper host. Moreover, they highlight the fact that CXCR4 usage per se is not a prerequisite for HIV-1 to cause disease and that CCR5 variants can be quite pathogenic.
The evolution of coreceptor utilization during the course of HIV-1 infection represents a key mechanism for extension of the spectrum of target cells. Multiple studies have implicated the V3 loop of gp120 as a major determinant of tropism (8, 11, 30, 31, 43, 57, 65, 66). These findings were extended to demonstrate that this domain plays a dominant role in governing coreceptor utilization (13, 15). Recent crystallographic analysis coupled with mutational evidence has identified amino acid residues in gp120 that are accessible to solvent following association with CD4 and are critical to coreceptor utilization (35, 46, 67, 68). The indirect contribution of the V3 loop is evidenced by the finding that only one of these amino acid residues is located in this domain. Whereas truncation of the V1 and V2 loops of gp120 yielded a variant that retained the ability to bind CD4 and CCR5, further truncation of the V3 loop abolished the ability to bind this coreceptor. Thus, it is likely that the V3 loop controls the availability or conformation of residues that interact with coreceptor structures. Previous studies using genetically engineered hybrid env genes have confirmed the role of the V3 loop and also implicated the V1–V2 region (12, 49). These chimeric Env were typically composed of segments of different molecularly cloned, genetically unrelated laboratory strains of HIV-1. One study employed a parental simian-HIV hybrid that showed exclusive utilization of CXCR4 and a pathogenic isolate that lacked utilization of both front-line coreceptors, CCR5 and CXCR4, and employed CCR2b, CCR3, STRL33, and APJ in infection experiments (29). While other groups have correlated sequence variation in the various domains of sequential isolates with tropism (17, 54), the current study is unique in that it examined multiple molecular clones from serial isolates from the same patients and used these genetically related env to engineer chimeras. In addition to the dominant effect of the V3 loop, our data provide direct evidence that the V4–V5 region can also contribute to coreceptor utilization, which was also demonstrated by Smyth et al. (58) and reported for an R→S conversion in the C4 region by Carrillo and Ratner (9). The R5 and multitropic env examined in these studies did not demonstrate this R→S conversion in the C4 domain (Fig. 4). The changes in amino acid residues between the two R5 Env, QH0515 and QH0648, did not involve any of the residues that have been shown to be essential to coreceptor binding activity but did include deletion of charged amino acids and the loss of a potential N-linked glycosylation site.
The conformation of Env is controlled by many influences, including primary structure and posttranslational modifications. It is also clear that a variety of indirect effects can modify the conformation of this glycoprotein so as to alter its interaction with coreceptors. Therefore, it is not unexpected that multiple types of alterations in primary structure can change the ability of a set of nonlinear residues to assume a conformation that is permissive for utilization of different coreceptors. Our data suggest that during the evolution of HIV-1 infection, genetic variability in sequences encoding the V4–V5 hypervariable regions may contribute to the progression of coreceptor utilization from exclusive employment of CCR5 to the development of a multispecific phase, to the ultimate, exclusive utilization of CXCR4. The current studies, as well as those of Carrillo and Ratner (9), raise the possibility of a direct or indirect interaction between these regions and the V3 loop which is not inconsistent with the crystallographic structures for gp120 that have been determined (35, 67).
To further investigate the basis for the observed switch in coreceptor usage by the virus env genes and to identify the determinants involved, we constructed chimeras from genetically linked Env exhibiting differential coreceptor use that were recovered from subject SC24. Although a significant body of work exists examining Env determinants of coreceptor usage, most of the studies have employed unrelated viruses to construct chimeric Env. We took advantage of the opportunity to use genetically linked viruses that evolved naturally in vivo to ascertain whether this approach might provide additional information about the nature of the determinants involved.
The mechanisms responsible for the diversification of the viral env gene and the interactions driving the in vivo evolution of HIV-1 are imperfectly understood. The high level of virus replication and the error-prone nature of the HIV-1 RT are surely contributing factors but appear insufficient in and of themselves to account for the observed env diversity and the switch in coreceptor usage that often occurs during the course of HIV-1 disease. Such a conclusion is consistent with the relatively stable env genotype observed in progressive disease as the immune system collapses and immune pressure decreases (19). We have also observed a lack of env diversity in a subject (SC50) who exhibited high virus loads but failed to mount significant cellular and humoral responses to HIV-1 infection (Demarest et al., submitted). Thus, it appears that an appropriate selective pressure is required, in addition to the replication characteristics of HIV-1, to bring about diversification in env and alterations in coreceptor usage. What might these pressures be? The β-chemokines are one possibility (14, 54, 69). However, if these were significant sources of selective pressure driving the evolution of the viral env gene, it is difficult to understand why it usually takes years for CXCR4 variants to emerge when so few changes in the V3 loop could effect the switch from CCR5 to CXCR4. In contrast, under the potent selective pressure of the nonnucleoside RT inhibitor nevirapine, escape variants with mutations in the relatively conserved RT gene arise within a matter of weeks (64). In light of these considerations, it may be more likely that pressure exerted by the cellular and/or humoral components of the immune system is more intimately involved with the diversification of the viral env gene. In this context, we note that both study subjects developed neutralizing antibodies to their autologous viruses (M.L.G., unpublished observations). Such reactivities are directed towards the viral env gene, and neutralization escape variants can develop within a period of months. Another subject (SC42) whom we have followed as a member of the cohort of acute seroconverters did not develop an autologous neutralizing antibody response. Despite persistently high levels of viremia in this subject (>105 copies/ml) since the time of infection and continuing for more than 4 years, his virus isolates have remained NSI, indicating an absence of CXCR4-using variants (M.L.G., unpublished observations). The possibility that alterations in coreceptor usage may be linked to escape from immune pressure remains largely unexplored.
ACKNOWLEDGMENTS
We acknowledge Courtney Bartholomew, Noreen Jack, Jeffery Edwards, William Blattner, and Farley Cleghorn and their staff for their tireless efforts in assembling, following, and caring for the Trinidad cohort of acute seroconverters. In addition, we thank Kent J. Weinhold and his laboratory for recovery of primary isolates from patient samples and Derrick Goodman and Rosemary Bennetts for technical assistance with cloning, sequencing, and phylogenetic analysis.
This work was supported in part by grants from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (RO1-AI41346 to S.C.P., RO1-AI40017 to M.L.G., and PO1-AI40237), from the Duke Center for AIDS Research (P30-AI28662), and from the Agnes Brown Dugan Endowment and the Humana Fund for Excellence.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Anzala O A, Nagelkerke N J, Bwayo J J, Holton D, Moses S, Ngugi E N, Ndinya-Achola J O, Plummer F A. Rapid progression to disease in African sex workers with human immunodeficiency virus type 1 infection. J Infect Dis. 1995;171:686–689. doi: 10.1093/infdis/171.3.686. [DOI] [PubMed] [Google Scholar]
- 3.Arendrup M, Nielsen C, Hansen J E, Pedersen C, Mathiesen L, Nielsen J O. Autologous HIV-1 neutralizing antibodies: emergence of neutralization- resistant escape virus and subsequent development of escape virus neutralizing antibodies. J Acquir Immune Defic Syndr. 1992;5:303–307. [PubMed] [Google Scholar]
- 4.Bebenek K, Abbotts J, Roberts J D, Wilson S H, Kunkel T A. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J Biol Chem. 1989;264:16948–16956. [PubMed] [Google Scholar]
- 5.Bieniasz P D, Fridell R A, Aramori I, Ferguson S S, Caron M G, Cullen B R. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjorndal A, Deng H, Jansson M, Fiore J R, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman D R, Fenyo E M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns D P, Desrosiers R C. Envelope sequence variation, neutralizing antibodies, and primate lentivirus persistence. Curr Top Microbiol Immunol. 1994;188:185–219. doi: 10.1007/978-3-642-78536-8_11. [DOI] [PubMed] [Google Scholar]
- 8.Cann A J, Churcher M J, Boyd M, O'Brien W, Zhao J Q, Zack J, Chen I S. The region of the envelope gene of human immunodeficiency virus type 1 responsible for determination of cell tropism. J Virol. 1992;66:305–309. doi: 10.1128/jvi.66.1.305-309.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrillo A, Ratner L. Human immunodeficiency virus type 1 tropism for T-lymphoid cell lines: role of the V3 loop and C4 envelope determinants. J Virol. 1996;70:1301–1309. doi: 10.1128/jvi.70.2.1301-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C H, Weinhold K J, Bartlett J A, Bolognesi D P, Greenberg M L. CD8+ T lymphocyte-mediated inhibition of HIV-1 long terminal repeat transcription: a novel antiviral mechanism. AIDS Res Hum Retroviruses. 1993;9:1079–1086. doi: 10.1089/aid.1993.9.1079. [DOI] [PubMed] [Google Scholar]
- 11.Cheng-Mayer C, Seto D, Levy J A. Altered host range of HIV-1 after passage through various human cell types. Virology. 1991;181:288–294. doi: 10.1016/0042-6822(91)90494-v. [DOI] [PubMed] [Google Scholar]
- 12.Cho M W, Lee M K, Carney M C, Berson J F, Doms R W, Martin M A. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J Virol. 1998;72:2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 14.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV- suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 15.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 16.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 17.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalgleish A G, Beverley P C, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 19.Delwart E L, Pan H, Sheppard H W, Wolpert D, Neumann A U, Korber B, Mullins J I. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J Virol. 1997;71:7498–7508. doi: 10.1128/jvi.71.10.7498-7508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 21.de Roda Husman A M, van Rij R P, Blaak H, Broersen S, Schuitemaker H. Adaptation to promiscuous usage of chemokine receptors is not a prerequisite for human immunodeficiency virus type 1 disease progression. J Infect Dis. 1999;180:1106–1115. doi: 10.1086/314987. [DOI] [PubMed] [Google Scholar]
- 22.Doranz B J, Lu Z H, Rucker J, Zhang T Y, Sharron M, Cen Y H, Wang Z X, Guo H H, Du J G, Accavitti M A, Doms R W, Peiper S C. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta- chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 24.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC- CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 25.Einfeld D, Hunter E. Transport of membrane proteins to the cell surface. Curr Top Microbiol Immunol. 1991;170:107–139. doi: 10.1007/978-3-642-76389-2_4. [DOI] [PubMed] [Google Scholar]
- 26.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 27.Felsenstein J. PHYLIP-phylogenetic inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 28.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 29.Hoffman T L, Stephens E B, Narayan O, Doms R W. HIV type I envelope determinants for use of the CCR2b, CCR3, STRL33, and APJ coreceptors. Proc Natl Acad Sci USA. 1998;95:11360–11365. doi: 10.1073/pnas.95.19.11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 31.Kim F M, Kolson D L, Balliet J W, Srinivasan A, Collman R G. V3-independent determinants of macrophage tropism in a primary human immunodeficiency virus type 1 isolate. J Virol. 1995;69:1755–1761. doi: 10.1128/jvi.69.3.1755-1761.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koot M, Keet I P, Vos A H, de Goede R E, Roos M T, Coutinho R A, Miedema F, Schellekens P T, Tersmette M. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 34.Koot M, Vos A H, Keet R P, de Goede R E, Dercksen M W, Terpstra F G, Coutinho R A, Miedema F, Tersmette M. HIV-1 biological phenotype in long-term infected individuals evaluated with an MT-2 cocultivation assay. AIDS. 1992;6:49–54. doi: 10.1097/00002030-199201000-00006. [DOI] [PubMed] [Google Scholar]
- 35.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 37.Maddon P J, Dalgleish A G, McDougal J S, Clapham P R, Weiss R A, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 38.Matthews T J, Wild C, Chen C H, Bolognesi D P, Greenberg M L. Structural rearrangements in the transmembrane glycoprotein after receptor binding. Immunol Rev. 1994;140:93–104. doi: 10.1111/j.1600-065x.1994.tb00866.x. [DOI] [PubMed] [Google Scholar]
- 39.McDougal J S, Maddon P J, Dalgleish A G, Clapham P R, Littman D R, Godfrey M, Maddon D E, Chess L, Weiss R A, Axel R. The T4 glycoprotein is a cell-surface receptor for the AIDS virus. Cold Spring Harbor Symp Quant Biol. 1986;51:703–711. doi: 10.1101/sqb.1986.051.01.083. [DOI] [PubMed] [Google Scholar]
- 40.Moore J P, Sattentau Q J, Klasse P J, Burkly L C. A monoclonal antibody to CD4 domain 2 blocks soluble CD4-induced conformational changes in the envelope glycoproteins of human immunodeficiency virus type 1 (HIV-1) and HIV-1 infection of CD4+ cells. J Virol. 1992;66:4784–4793. doi: 10.1128/jvi.66.8.4784-4793.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mori K, Ringler D J, Kodama T, Desrosiers R C. Complex determinants of macrophage tropism in env of simian immunodeficiency virus. J Virol. 1992;66:2067–2075. doi: 10.1128/jvi.66.4.2067-2075.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munoz A, Wang M C, Bass S, Taylor J M, Kingsley L A, Chmiel J S, Polk B F. Acquired immunodeficiency syndrome (AIDS)-free time after human immunodeficiency virus type 1 (HIV-1) seroconversion in homosexual men. Multicenter AIDS Cohort Study Group. Am J Epidemiol. 1989;130:530–539. doi: 10.1093/oxfordjournals.aje.a115367. [DOI] [PubMed] [Google Scholar]
- 43.O'Brien W A, Koyanagi Y, Namazie A, Zhao J Q, Diagne A, Idler K, Zack J A, Chen I S. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 44.Poignard P, Sabbe R, Picchio G R, Wang M, Gulizia R J, Katinger H, Parren P W, Mosier D E, Burton D R. Neutralizing antibodies have limited effects on the control of establised HIV-1 infection in vivo. Immunity. 1999;10:431–438. doi: 10.1016/s1074-7613(00)80043-6. [DOI] [PubMed] [Google Scholar]
- 45.Reed L J, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 46.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 47.Roberts J D, Bebenek K, Kunkel T A. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 48.Rosenberg P S, Goedert J J, Biggar R J. Effect of age at seroconversion on the natural AIDS incubation distribution. Multicenter Hemophilia Cohort Study and the International Registry of Seroconverters. AIDS. 1994;8:803–810. doi: 10.1097/00002030-199406000-00013. [DOI] [PubMed] [Google Scholar]
- 49.Ross T M, Cullen B R. The ability of HIV type 1 to use CCR-3 as a coreceptor is controlled by envelope V1/V2 sequences acting in conjunction with a CCR-5 tropic V3 loop. Proc Natl Acad Sci USA. 1998;95:7682–7686. doi: 10.1073/pnas.95.13.7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rutherford G W, Lifson A R, Hessol N A, Darrow W W, O'Malley P M, Buchbinder S P, Barnhart J L, Bodecker T W, Cannon L, Doll L S, Holmberg S D, Harrison J S, Rogers M F, Werdegar D, Jaffe H W. Course of HIV-I infection in a cohort of homosexual and bisexual men: an 11 year follow up study. Br Med J. 1990;301:1183. doi: 10.1136/bmj.301.6762.1183. , 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 52.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 53.Sattentau Q J, Moore J P. The role of CD4 in HIV binding and entry. Phil Trans R Soc Lond B Biol Sci. 1993;342:59–66. doi: 10.1098/rstb.1993.0136. [DOI] [PubMed] [Google Scholar]
- 54.Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng H K, Malnati M S, Plebani A, Siccardi A G, Littman D R, Fenyo E M, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 55.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, de Goede R E, van Steenwijk R P, Lange J M, Schattenkerk J K, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shankarappa R, Margolick J B, Gange S J, Rodrigo A G, Upchurch D, Farzadegan H, Gupta P, Rinaldo C R, Learn G H, He X, Huang X-L, Mullins J I. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shioda T, Levy J A, Cheng-Mayer C. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 58.Smyth R J, Yi Y, Singh A, Collman R G. Determinants of entry cofactor utilization and tropism in a dual-tropic human immunodeficiency virus type 1 primary isolate. J Virol. 1998;72:4478–4484. doi: 10.1128/jvi.72.5.4478-4484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsang M L, Evans L A, McQueen P, Hurren L, Byrne C, Penny R, Tindall B, Cooper D A. Neutralizing antibodies against sequential autologous human immunodeficiency virus type 1 isolates after seroconversion. J Infect Dis. 1994;170:1141–1147. doi: 10.1093/infdis/170.5.1141. [DOI] [PubMed] [Google Scholar]
- 60.van 't Wout A B, Blaak H, Ran L J, Brouwer M, Kuiken C, Schuitemaker H. Evolution of syncytium-inducing and non-syncytium-inducing biological virus clones in relation to replication kinetics during the course of human immunodeficiency virus type 1 infection. J Virol. 1998;72:5099–6107. doi: 10.1128/jvi.72.6.5099-5107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Z, Lee B, Murray J L, Bonneau F, Sun Y, Schweickart V, Zhang T, Peiper S C. CCR5 HIV-1 coreceptor activity. Role of cooperativity between residues in N-terminal extracellular and intracellular domains. J Biol Chem. 1999;274:28413–28419. doi: 10.1074/jbc.274.40.28413. [DOI] [PubMed] [Google Scholar]
- 62.Wang Z X, Berson J F, Zhang T Y, Cen Y H, Sun Y, Sharron M, Lu Z H, Peiper S C. CXCR4 sequences involved in coreceptor determination of human immunodeficiency virus type-1 tropism. Unmasking of activity with M-tropic Env glycoproteins. J Biol Chem. 1998;273:15007–15015. doi: 10.1074/jbc.273.24.15007. [DOI] [PubMed] [Google Scholar]
- 63.Watkins B A, Reitz M S, Jr, Wilson C A, Aldrich K, Davis A E, Robert-Guroff M. Immune escape by human immunodeficiency virus type 1 from neutralizing antibodies: evidence for multiple pathways. J Virol. 1993;67:7493–7500. doi: 10.1128/jvi.67.12.7493-7500.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 65.Westervelt P, Gendelman H E, Ratner L. Identification of a determinant within the human immunodeficiency virus 1 surface envelope glycoprotein critical for productive infection of primary monocytes. Proc Natl Acad Sci USA. 1991;88:3097–3101. doi: 10.1073/pnas.88.8.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Westervelt P, Trowbridge D B, Epstein L G, Blumberg B M, Li Y, Hahn B H, Shaw G M, Price R W, Ratner L. Macrophage tropism determinants of human immunodeficiency virus type 1 in vivo. J Virol. 1992;66:2577–2582. doi: 10.1128/jvi.66.4.2577-2582.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wyatt R, Kwong P D, Desjardins E, Sweet R W, Robinson J, Hendrickson W A, Sodroski J G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 68.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 69.Zagury D, Lachgar A, Chams V, Fall L S, Bernard J, Zagury J F, Bizzini B, Gringeri A, Santagostino E, Rappaport J, Feldman M, O'Brien S J, Burny A, Gallo R C. C-C chemokines, pivotal in protection against HIV type 1 infection. Proc Natl Acad Sci USA. 1998;95:3857–3861. doi: 10.1073/pnas.95.7.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y J, Moore J P. Will multiple coreceptors need to be targeted by inhibitors of human immunodeficiency virus type 1 entry? J Virol. 1999;73:3443–3448. doi: 10.1128/jvi.73.4.3443-3448.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]