Abstract
AIM
To address the challenges of data labeling difficulties, data privacy, and necessary large amount of labeled data for deep learning methods in diabetic retinopathy (DR) identification, the aim of this study is to develop a source-free domain adaptation (SFDA) method for efficient and effective DR identification from unlabeled data.
METHODS
A multi-SFDA method was proposed for DR identification. This method integrates multiple source models, which are trained from the same source domain, to generate synthetic pseudo labels for the unlabeled target domain. Besides, a softmax-consistence minimization term is utilized to minimize the intra-class distances between the source and target domains and maximize the inter-class distances. Validation is performed using three color fundus photograph datasets (APTOS2019, DDR, and EyePACS).
RESULTS
The proposed model was evaluated and provided promising results with respectively 0.8917 and 0.9795 F1-scores on referable and normal/abnormal DR identification tasks. It demonstrated effective DR identification through minimizing intra-class distances and maximizing inter-class distances between source and target domains.
CONCLUSION
The multi-SFDA method provides an effective approach to overcome the challenges in DR identification. The method not only addresses difficulties in data labeling and privacy issues, but also reduces the need for large amounts of labeled data required by deep learning methods, making it a practical tool for early detection and preservation of vision in diabetic patients.
Keywords: diabetic retinopathy, multisource-free, domain adaptation, pseudo-label generation, softmax-consistence minimization
INTRODUCTION
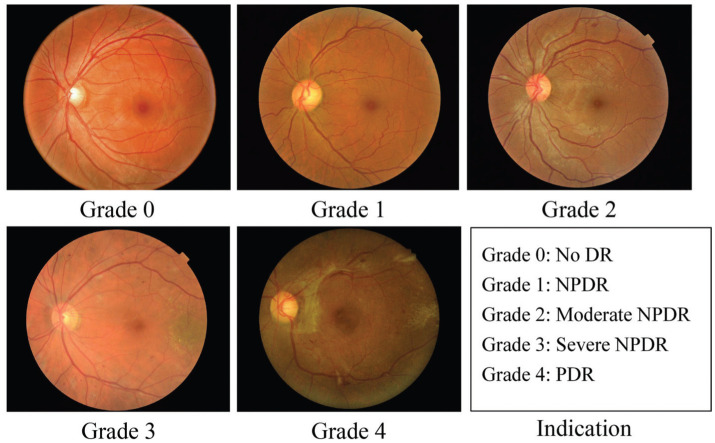
Diabetic retinopathy (DR) is a prevalent disease worldwide[1]–[4]. It is a metabolic disorder related to blood sugar and considered to be a primary cause of blindness in diabetic patients[5]–[6]. Early screening and diagnosis of DR are crucial in mitigating the risk of vision loss. DR is categorized into five stages: no DR, mild non-proliferative DR (NPDR), moderate NPDR, severe NPDR and proliferative DR (PDR), as shown in Figure 1. Detecting and classifying DR from color fundus photograph (CFP) images, especially at an early stage, is challenging.
Figure 1. Example CFP images from different DR stages.
Task 1: Non-referable (grade 0, 1) vs referable (grade 2, 3, 4) DR classification; Task 2: Normal (grade 0)/abnormal (grade 1–4) DR classification. DR: Diabetic retinopathy; PDR: Proliferative diabetic retinopathy; NPDR: Non-proliferative diabetic retinopathy; CFP: Color fundus photograph.
In general, DR is often considered to be the significant symptom of diabetes[7]–[8]. DR can be broadly classified into two stages, namely, early DR and advanced DR. 1) NPDR is also commonly referred to as early DR. At this stage, new retinal vessels do not proliferate, and early DR can be further classified into mild, moderate, and severe non-proliferative stages. During the mild stage, DR often involves small bleeding spots or small microhemangiomas, while the moderate stage may exhibit yellowish-white punctate hard exudates. In the severe stage, cotton-like and soft exudates may be observed in the retina, along with more blocked retinal blood vessels. 2) Advanced DR is characterized by proliferative retinopathy, which damages the proliferation of the new vessels in the retina. New retinal vessels are fragile and may leak into the clear area, resulting in severe vision loss. Moreover, this may damage the optic nerve, leading to glaucoma. Extracting effective features for DR is challenging due to the complex and diverse characteristics of fundus. In clinical practice, DR is often treated as two recognition tasks, referred to as referable DR classification and normal/abnormal DR recognition (Figure 1). Identifying referable DR can effectively detect patients with vision-threatening lesions (more than mild), which can help save their vision. Abnormal DR recognition aims to detect the growth of abnormal blood vessels in the retina, thereby providing early treatment options for abnormal patients. As both tasks are significant, this study focuses on addressing them using deep learning technology.
Deep learning-based methods have recently gained popularity in various fields such as DR, breast cancer, and cardiology, as evidenced by several studies[9]–[10]. Many automated algorithms have been proposed for the diagnosis and segmentation of DR in retinal images, including the detection of red lesions and hemorrhages. Deep learning-based methods have demonstrated significant advantages in DR diagnosis[8],[11]. Among various deep learning-based algorithms, convolutional neural network-based algorithms are widely employed in medical image classification and have shown remarkable performance on CFP images. Typically, a convolutional neural network (CNN) is used to extract features from retinal images, which is effective for classification and segmentation. Deep learning methods have the potential to efficiently process large retinal image datasets.
Moreover, developing an effective and reliable diagnostics system to tackle this problem has its own set of unique challenges. Existing deep learning techniques, despite their commendable success in the medical field, suffer from serious drawbacks. These limitations generally stem from difficulties in large-scale dataset labeling, complicated and expensive data collection processes, and often strong restrictions from patient data protection regulations. Therefore, the need for building a system that's efficient, cost effective, and respects individual data privacy protocols becomes crucial. Transfer learning—a subdomain of machine learning, has illustrated excellent proficiency in hastening the learning process and strengthening the performance of the diagnosis systems for DR when labeled data is scanty. Typically, in a transfer learning problem, valuable information is gleaned from the “source” domain to enhance the learning accuracy of new models designated the “target” domain where labels are close to none or non-existent.
Nonetheless, when it comes to transfer learning implementation in datasets holding confidential personal data as seen in the domains of retinal image analysis, accessing relevant source-domain data is a deal breaker due to several rigors in the likes of privacy protection constraints, ethical restrictions, legal implications, and data access rights collectively inhibiting unbounded data sharing. Such vital challenges have necessarily sprung the call for scientific perspectives outside the confines of conventional transfer learning. Rightfully so, working with transfer learning domains where source data access is restricted, we face, what in scientific spheres is called a source-free domain adaptation (SFDA) problem. Intuitively, SFDA circumvents the need for access to the source domain data, overcoming the limitations of learning by applying inferences that map learned labeled data to shifting target data into distinct classes. This demonstrates the unique motivations underlying the implementation of a “source-free domain adaptation approach” that effectively leverages large volumes of unlabeled data, marking significant progress towards an advanced diagnostic tool for DR.
To address the challenges associated with labeling constraints, privacy concerns, and domain adaptation techniques, our research puts forward a novel SFDA framework tailored for DR identification. It not only ensues operability with large volumes of unlabeled dataset, but also minimizes data sharing hazards therein. The distinctive aspects and novel contributions of our approach laps into introducing the term of softmax-consistence minimization to exploit more discriminative information from reducing domain discrepancies, launching pseudo labels based multi-source models. Compared to existing DR identification models, this work aims to leverage previously labeled datasets and unlabeled target data in a transductive manner. By doing so, we can exploit the existing knowledge while minimizing the data labeling expense and data sharing risks. The motivations and contributions are concluded as following:
Motivation
This study is driven by three major motivations, with each addressing critical aspects within the field of applying deep learning for DR identification.
1) The need for robust domain adaptation: striking variances and underlying complexities across multi-domain platforms pose significant challenges to model effectiveness and optimal performance. In acknowledging this issue, there arises an earnest need for cogent yet flexibility-durable domain adaptation methodologies.
2) Data privacy issues and restrictions: medical data—ones utilized in DR identification—are often encumbered by privacy and ethical limitations. This directly leads to challenges accessing or utilizing essential source-domain data. Herein lies one of the motivations for our study - propagating a solution with SFDA.
3) Utilization of pseudo labels for unlabeled data: owing to the impediment of acquiring large-scale labeled data, we introduce a synthetic stand-in—“pseudo labels” for unlabeled data. This trifles the necessity for extensive real-world data and hence facilitates not just efficient, but more importantly effective DR identification.
Taken together, these considerations reinforce our approach's assortment of existing performance limitations and offer advancements in the process and accuracy of the task at hand DR—identification.
Contribution
Based on the above analysis, we propose a novel framework, softmax-consistence minimization and multisource-model pseudo label generation-based (SMPL) algorithm. The major contributions of this study are as follows:
1) We introduce the term of softmax-consistence minimization to decrease discrepancies between data from various domains. This process helps to minimize the distances between the intra-class samples in both source and target domains while maximizing the distances between samples from different classes.
2) We present a multi-source-model pseudo-label generator, which enhances the model's performance. Besides, this paper represents a preliminary investigation into the application of source-data-free domain adaptation techniques for retinal image classification tasks.
3) We conduct extensive experiments using several typical DR datasets to demonstrate the effectiveness of our proposed framework.
The following sections of this paper detail the proposed methodology (section 2), follow up with an in-depth analysis of the implementation, several typical datasets, and the resulting experiments (section 3), and conclude the study (section 4).
MATERIALS AND METHODS
To address the problem of source-free unsupervised domain adaptation in DR identification, we propose a novel framework that includes the terms of SMPL.
In SFDA of DR identification, assume a source domain , where denotes the ith fundus image sample from the source domain, and represents the DR label of the ith source sample. Ns is the sample number of the source data. In source-free settings, we denote as n different source DR identification models, trained from the same source fundus images. Given a target domain without DR labels, Nt is the number of the target fundus images.
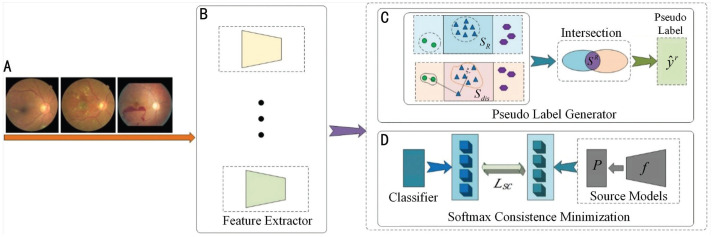
The goal of this framework is to predict the target DR label of by domain adaptation from source models without access to any source fundus images Ds in training stage. An illustration of the proposed SMPL is presented in Figure 2. In detail, this paper first introduces a multisource-model pseudo-label generator to output the synthetic pseudo labels for each target fundus image, which are jointly determined by the pre-trained multiple source models . Then a softmax-consistence minimization loss is deployed to further improve the identification ability of the target model with the help of SFDA.
Figure 2. Overview of the proposed SMPL approach.
A: Target domain data; B: Feature extractors; C: Process of generating pseudo labels; D: Process of minimizing softmax consistence between the source models (feature extractors of source domain and the classifier of source domainP) and the target model.SMPL: Softmax-consistence minimization and multisource-model pseudo label generation-based algorithm.
Multisource-Model Pseudo-Label Generator
In our framework, pseudo-labels are obtained based on an ensemble of multiple source identification models , which can be useful for promoting the target model θT. Because the extracted features from multiple source models have more powerful ability to represent the fundus images than single model's feature, this paper employ several different models to learn the representative features for each sample. The process of generating multisource-model pseudo labels is as follows:
1) Construct one set using category prediction probability. Given a sample from the target domain, the features are extracted by the multisource models, as shown in Figure 2B. The DR category prediction probability can be obtained by the one-hot encoding of feature .
Here, we select with the max as the reliable candidate reference point in Figure 2C. Inspired by the idea of the k-nearest neighbor, we select k points as one set SR as follows:
| (1) |
2) Construct the set based on the distances between features. The category center mR is computed using the category prediction probability and feature as follows:
| (2) |
The target fundus image is classified as Sdis if the distance between and mean mR of the ith class is shorter than the distance between and mean . The process of classifying the target samples was as follows:
| (3) |
To guarantee the accuracy of the DR pseudo-label generation, we selected common elements of both sets SR and Sdis, whose operation can be defined as follows:
| (4) |
3) Optimization.For all samples, we generate their pseudo labels during model training as follows:
| (5) |
where d(·,·) represents the distance between and DR category center (nR is the sample number of SR). Then, we designed the DR classification loss for all target samples to enhance the identification ability:
| (6) |
where R is the total number of classes in Eq. (6).
Benefit from the proposed multisource-model pseudo-label generator, the obtained pseudo labels are more confident than those from the single source DR identification model, and the feature extractor have excellent capability on feature learning for fundus images.
Softmax-Consistence Minimization Loss
Since source fundus data is unavailable, the multisource-model pseudo-labeling process may encounter uncertainty/error classification when target samples lack ground-truth DR labels. To address this obstacle, we propose the softmax-consistence minimization method, presented in Figure 2D. This technique reduces feature distances between fundus samples of the same class and increases the distances between samples from different DR classes in both the source and target domains. The essence of this method is to minimize the discrepancy between the source and target domain by adjusting the softmax values of the target samples to align with that of the source domain, hence improving the model's performance in classifying the target samples correctly.
We input the target fundus image into each source model θS, adjusting the softmax values of to align with those of the source data. However, we seek to decrease the entropy of through the use of a trainable target DR identification model. The loss function for softmax-consistence minimization is defined as follows:
| (7) |
where σ(·) denotes the softmax function. θS(·) and θT(·) denote source and target models, respectively. represents the label of the rth category.
The implementation of the softmax-consistence minimization term ensures that the model maintains a consistent distance measure between data samples across different domains. This is integral to the success of our multi-source domain adaptation model, as it ensures a harmonious alignment of data in a multi-domain setup, hence promoting the correct classification of the unlabeled target domain samples.
Combining the loss terms (6) and (7), the total loss in our model is formulated as follows:
| (8) |
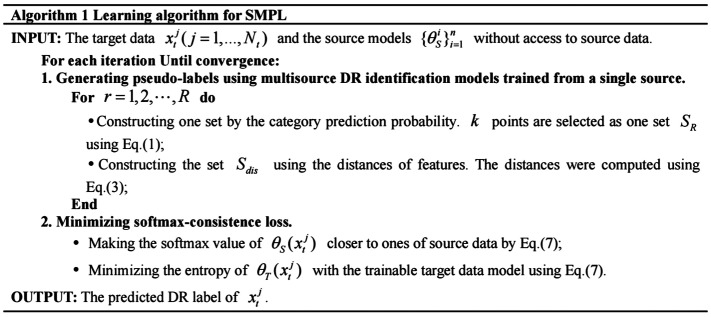
The procedure for training our DR identification model is summarized in Figure 3.
Figure 3. Learning algorithm for SMPL.
SMPL: Softmax-consistence minimization and multisource-model pseudo label generation-based algorithm.
RESULTS
Datasets and Settings
Dataset description
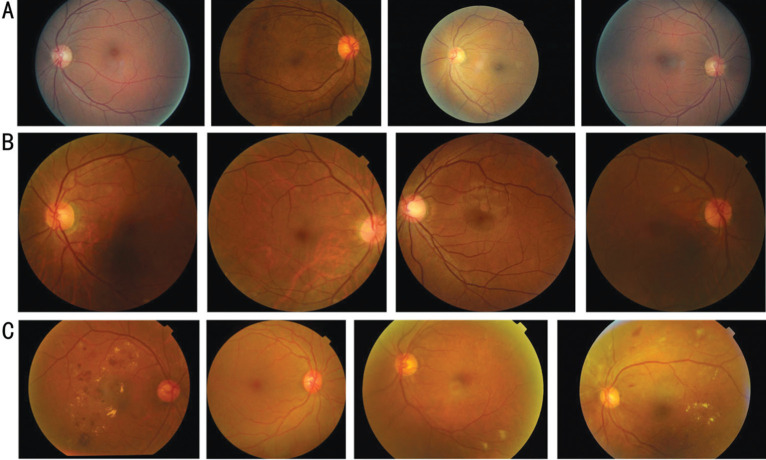
For our experiments, we chose three retinal image datasets, one of which is EyePACS[12]–[13]. This dataset is currently the largest public dataset for detecting DR and consists of 35 126 training images and 53 576 testing images, as demonstrated in Figure 4A. Each image is graded on a scale of 0 to 4. However, EyePACS is a particularly challenging dataset due to variations in image resolution, lighting, and quality. We should note that only the training set was used to train the source domain model in our experiment.
Figure 4. Samples from three DR datasets.
A: Samples from the EyePACS dataset; B: Samples from the DDR dataset; C: Samples from the APTOS2019 dataset. DR: Diabetic retinopathy.
Besides, we employ a high-quality DDR dataset[14] in our experiments, which was derived from 84 grade-A tertiary hospitals. This dataset included 13 673 retinal fundus images collected between 2016 to 2018. These images were captured using single-view imaging as shown in Figure 4B, and were desensitized for non-commercial use. The images were obtained from 9598 patients with an average age of 54, with 48.23% of the images belonging to male patients and 51.77% to female patients. For our experiments, we used samples labeled with five grades and divided them into training and testing sets.
The APTOS dataset[15] was released as part of the Kaggle blindness detection challenge in 2019 by the Asia Pacific TeleOphthalmology Society. The dataset comprises 3662 high-resolution fundus images captured using several types of clinical cameras, taken in varying conditions and environments over an extended period. The images are graded on a scale of 0 to 4 to denote the severity level of DR, where 0 signifies no DR and 4 denotes PDR, with mild, moderate, and severe grades in between. Examples of fundus photographs from this dataset are illustrated in Figure 4C.
Baseline methods
We compared SMPL with several traditional domain adaptation approaches such as DANN[16], DAN[17], JAN[18], ADDA[19], and CDAN[20] to validate the effectiveness of the proposed methods on DR classification. Specifically, DANN[16] implemented that the domain adaptation predictions must be made based on confused features between source and target domains, which is achieved in neural network architectures; DAN[17] generalized deep convolutional neural network to the domain adaptation, by embedding hidden representations of all task-specific layers in a reproducing kernel Hilbert space where the mean embeddings of different domain distributions can be explicitly matched; JAN[18] learned a joint adaptation networks by aligning the joint distributions of multiple domain-specific layers across domains based on a joint maximum mean discrepancy criterion; ADDA[19] designed an adversarial discriminative domain adaptation method that combines discriminative modeling, untied weight sharing, and a GAN loss to align the domain shifts; CDAN[20] presented a conditional adversarial domain adaptation method conditions the adversarial adaptation models on discriminative information conveyed in the classifier predictions.
Evaluation metric
We employed a range of performance metrics[21], including accuracy, F1-score, area under the curve (AUC), recall, sensitivity, and specificity, to evaluate the effectiveness of our approach and other competing methods. Additionally, recall and precision were utilized to measure the model's ability to retrieve referable diabetic retinopathy (RDR) images and accurately classify them[21]. The accuracy and F1-score were also used to assess the model's capability to distinguish positive samples across all categories.
Furthermore, binary classifiers' detection capability is often presented through receiver operator characteristic (ROC) curves[22]. These curves plot the true positive rate (TPR) against the false positive rate (FPR) to show the effect of classifier more comprehensively.
Data preparation
Due to difference in brightness, resolution, and other aspects among fundus images collected from various hospitals using different cameras, they were preprocessed to minimize their impact on the training process. Specifically, the following steps were employed. 1) Image clipping: As original images had varying resolutions, we resized them to a specific range. In this study, images having length or width greater than 1024 pixels were proportionally resized to 1024 pixels. Otherwise, their original configuration was left untouched. Moreover, we observed black areas surrounding the fundus images, leading to possible interference. Thus, we cropped these black areas to fill the images with color fundus except for the corner areas; 2) Image augmentation: To offset the uneven quality among the images and to highlight blood vessels, as well as lesion areas, we applied data augmentation using Graham's algorithm[23]:
| (9) |
where I is the input fundus image. G(ρ) represents a Gaussian filter with standard deviation ρ. ⊗ denotes the convolution operation. Parameters λ, ω, ρ, and δ were set to 4, -4, 128, and 30, respectively.
Experimental setting
To promote model convergence, we initialized the model parameters with ImageNet[5] and fed the images, which were resized to 224×224 pixels into the network. We standardized the model using the mean and standard deviation of ImageNet. During the training stage, we implemented rotation and random horizontal flips for data augmentation. Besides, we optimized the source and target domain models with 40 epochs using the SGD optimizer. The learning rates of the source and target domain models were set to 0.01 and 0.005, respectively. We set k=0.1 and α=1 as the trade-off parameters for all datasets. The batch size was 32, and we measured feature vectors using cosine distance. The PyTorch platform was used to implement the algorithms, which were trained using four NVIDIA RTX2080-Ti GPUs. To make it easier to repeat the experiments, further parameter settings can be found in https://github.com/Jieming1022/SMPL.
Referable Diabetic Retinopathy
We use the term RDR[21] in this paper to refer to fundus images that display moderate NPDR, severe NPDR, or PDR. Images displaying no DR or mild DR are categorized as non-referable. Our model was compared against other models, including DANN[16], DAN[17], JAN[18], ADDA[19], and CDAN[20]. To conduct our experiments, we set the EyePACS dataset as the source domain dataset and the APTOS2019 dataset as the target domain dataset. We utilized all labeled data in the source domain and all unlabeled data in the target domain.
Our experimental evaluation focused on a diverse set of metrics including accuracy, precision, recall (sensitivity), specificity, and F1-score, which served to paint a comprehensive model performance. For the critical task of RDR detection, what stands notable in model's creations outperform existing benchmarks. The deployed algorithm demonstrated a brilliant performance with an achieved accuracy of 0.9044, precision of 0.8258, very high recall of 0.9691, specificity 0.8602, and an outstanding F1-score index reaching 0.8917. These metrics, notably comprehensive, starkly validates the new frontier in convergence proficiencies witnessed, robustly perpetuating stronger cases in favor of our user quality argument assessment process alike.
Comparing our model with existing domain adaptation techniques as demonstrated in Table 1[16]–[20], the superb performance of our model is evident with regards to the prime metrics affecting prediction quality—accuracy, recall (sensitivity), and F1-score, it appears forerunning. Benefitted substantially from the multi-SFDA method developed in our study, accurate and efficient predictions of RDR within target domains with unlabeled data is attainable, significantly mitigating the data labeling complications associated with preserving data privacy. In executing in-depth and comprehensive tests, we found that our model obtained compelling results, surpassing compared models at least 2.02% in accuracy, 16.82% in sensitivity, and 4.28% in F1-scores; effectively denoting advancements in DR identification resulting from the application of our model. More drastically, this model surpasses the performance of compared models by successfully minimizing the computational biases posed by various-domain integration, concurrently advancing the robustness of RDR predictions to seek unknown data by driving-up generalizeability.
Table 1. Detection results for referable diabetic retinopathy with different domain adaptation methods.
| Method | Accuracy | Precision | Recall/sensitivity | Specificity | F1-score |
| DANN[16] | 0.7812 | 0.8580 | 0.5528 | 0.9375 | 0.6724 |
| DAN[17] | 0.8842 | 0.9030 | 0.8009 | 0.9411 | 0.8489 |
| JAN[18] | 0.7891 | 0.9231 | 0.5245 | 0.9701 | 0.6690 |
| ADDA[19] | 0.8525 | 0.8454 | 0.7794 | 0.9025 | 0.8111 |
| CDAN[20] | 0.8162 | 0.8521 | 0.6624 | 0.9214 | 0.7454 |
| Ours | 0.9044a | 0.8258a | 0.9691a | 0.8602a | 0.8917a |
The source domain dataset: EyePACS. The target domain dataset: APTOS2019. aBest results.
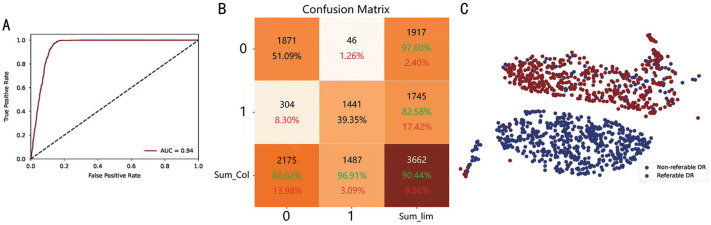
In addition, we plotted the ROC curve (Figure 5A) to show the effectiveness of our proposed model in detecting RDR. As shown in the plot, our model performs remarkably well. The x-axis indicates the FPR, which refers to the rate of misclassifying non-referable data as referable RDR data. The y-axis represents the TPR, which refers to the rate of accurately detecting true referable RDR data. False positives occur when anomalous data are incorrectly classified as referable, while they are, in reality, non-referable.
Figure 5. Visualization of the non-referable/referable DR identification task.
A: ROC curve; B: Confusion matrix; C: tSNE plot. The green color denotes correct matching ratio and the red one represents the wrong matching rate. DR: Diabetic retinopathy; ROC: Receiver operator characteristic; tSNE: t-distributed Stochastic Neighbor Embedding.
Normal/abnormal detection
In the experiments, we regarded images as abnormal DR when the fundus images were mild, moderate, severe, and PDR. No DR images were set to normal fundus images. In this section, we evaluate the methods from the DDR source domain dataset to the target domain dataset APTOS2019. Our model also illustrated similar advancements upon application to the task of normal/abnormal detection (NAD) in DR assessment from related medical imaging. As shown in Table 2, our approach achieves the highest accuracy (0.9792), precision (0.9821), recall/sensitivity (0.9768), specificity (0.9817), and F1-score (0.9795) when evaluated on the DDR source domain dataset and the APTOS2019 target domain dataset.
Table 2. Results for normal/abnormal with different domain adaptation methods.
| Method | Accuracy | Precision | Recall/sensitivity | Specificity | F1-score |
| DANN[16] | 0.9642 | 0.9652 | 0.9273 | 0.9657 | 0.9459 |
| DAN[17] | 0.9696 | 0.9760 | 0.9639 | 0.9756 | 0.9699 |
| JAN[18] | 0.9057 | 0.9554 | 0.8541 | 0.9590 | 0.9017 |
| ADDA[19] | 0.8339 | 0.9595 | 0.7022 | 0.9695 | 0.8109 |
| CDAN[20] | 0.7908 | 0.9245 | 0.6397 | 0.9463 | 0.7562 |
| Ours | 0.9792a | 0.9821a | 0.9768a | 0.9817a | 0.9795a |
The source domain dataset: DDR. The target domain dataset: APTOS2019. aBest results.
Comparing numerous domain adaptation methods applied for NAD classification, our method has outperformed all others. From a data-driven perspective, our model's performance has manifested significant superiority in all evaluated metrics. Specifically, the accuracy of our model reached 0.9792, significantly higher than the second-best performing model, DAN[17], which achieved an accuracy of 0.9696. This suggests that our method is more successful in distinguishing between normal and abnormal classes. Our model also exhibited excellent precision, scoring 0.9821, indicating that our model is more effective in avoiding false positives, thereby rendering its positive predictions more reliable. When considering recall or sensitivity, the advancements suggest that our model was more proficient in identifying true positives from the entire set of actual positives. Furthermore, our model with a specificity of 0.9817 and superior F1-score of 0.9795, reveals that our model is more skillful at correctly identifying negatives, thereby reducing the risk of false alarms and achieves an ideal balance in avoiding both false positives and negatives. In conclusion, our multi-SFDA method has proven to be effective in DR identification, overcoming challenges related to data labeling, data privacy, and the need for large amounts of labeled data. The superior performance of our method makes it a practical tool for the early detection of DR, thereby contributing to the preservation of vision in diabetic patients.
Confusion Matrix and Analysis
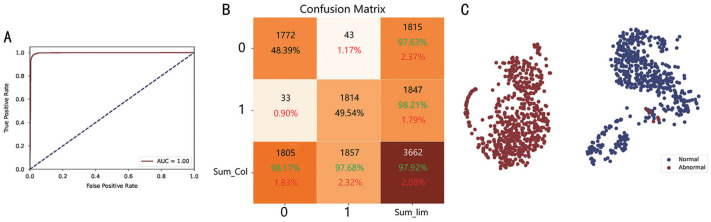
To visualize the performance of our DR identification model and evaluate the potential of the classifier on the test dataset, we utilized a confusion matrix for RDR and NAD tasks, presented in Figures 5B and 6B, respectively.
Figure 6. Visualization of the normal/abnormal DR identification task.
A: ROC curve; B: Confusion matrix; C: tSNE plot. The green color denotes correct matching ratio and the red one represents the wrong matching rate. DR: Diabetic retinopathy; tSNE: t-distributed Stochastic Neighbor Embedding.
The confusion matrix shows the correctly identified outcomes on the diagonal, while the wrong outcomes lie on the off-diagonal elements. The model that performs better will have fewer off-diagonal elements. In the RDR task, 51.09% of the images were non-referable, while 39.35% were referable DR. On the other hand, in the NAD task, 48.39% of the images were normal, and 49.54% were abnormal DR. As shown by the results of the confusion matrices, the majority of the samples were correctly identified.
Ablation Study
Recent research studies[24]–[25] indicate that deep learning algorithms can acquire transferable features for new domain adaptation to some degree. However, the transferability of features in higher layers of the SMPL network decreases significantly with an increase in different domains, as most deep learning features eventually transfered from general to specific types. In this section, we leverage ablation analysis to explore the impacts of the various components in our SMPL. Our method consists of two essential components, including softmax-consistence minimization (SCM) and multisource-model pseudo-label generator (PLG). We present the experimental results for the RDR and NAD tasks to verify the different components of our proposed approach in Tables 3 and 4.
Table 3. Ablation experiments of non-referable/referable DR on APTOS 2019.
| Source | Method | Accuracy | Precision | Recall | Specificity | F1-score |
| DDR | S only | 0.8820 | 0.7787 | 0.9913 | 0.8074 | 0.8722 |
| DDR | S+SCM | 0.8276 | 0.7569 | 0.8480 | 0.8138 | 0.7999 |
| DDR | S+SCM+PLG | 0.9033 | 0.8221 | 0.9724 | 0.8561 | 0.8909 |
| EyePACS | S only | 0.8967 | 0.8160 | 0.9630 | 0.8515 | 0.8834 |
| EyePACS | S+SCM | 0.9011 | 0.8272 | 0.9563 | 0.8634 | 0.8871 |
| EyePACS | S+SCM+PLG | 0.9044 | 0.8258 | 0.9691 | 0.8602 | 0.8917 |
SCM: Softmax-consistence minimization; PLG: Pseudo-label generator. S denotes the source domain, and the S+SCM+PLG employs the beta distribution and cosine distance metric.
Table 4. Ablation experiments of normal/abnormal DR on APTOS 2019.
| Source | Method | Accuracy | Precision | Recall | Specificity | F1-score |
| DDR | S only | 0.9672 | 0.9452 | 0.9930 | 0.9407 | 0.9685 |
| DDR | S+SCM | 0.9765 | 0.9831 | 0.9704 | 0.9828 | 0.9767 |
| DDR | S+SCM+PLG | 0.9792 | 0.9821 | 0.9768 | 0.9817 | 0.9795 |
| EyePACS | S only | 0.9647 | 0.9758 | 0.9542 | 0.9756 | 0.9649 |
| EyePACS | S+SCM | 0.9505 | 0.9895 | 0.9122 | 0.9900 | 0.9493 |
| EyePACS | S+SCM+PLG | 0.9683 | 0.9926 | 0.9445 | 0.9928 | 0.9680 |
DR: Diabetic retinopathy; SCM: Softmax-consistence minimization; PLG: Pseudo-label generator. S denotes the source domain, and the S+SCM+PLG employs the beta distribution and cosine distance metric.
To ensure a fair comparison, we conducted evaluations of our approach using the same hyperparameters and number of training iterations. In Table 3, we observed that for the RDR task (DDR/EyePACS to APTOS2019), using only the source model in our SMPL resulted in an accuracy of 88.2% (89.67%). However, when two components were incorporated, the accuracy increased to 90.33% (90.44%). Similarly, Table 4 demonstrates that for the NAD task (DDR/EyePACS to APTOS2019), using only the source model in our SMPL resulted in an accuracy of 96.72% (96.47%). But when two components were included, the accuracy increased to 97.92% (96.83%), and their training steps are also visualized in Figure 7. This indicates the necessity of utilizing these two modules in our model. The SCM loss aids in minimizing the distances between the intra-class samples from the source and target domains while also maximizing the distances between inter-class samples. Meanwhile, the multisource-model pseudo-label generation approach enhances DR identification performance by employing pseudo-labels more effectively.
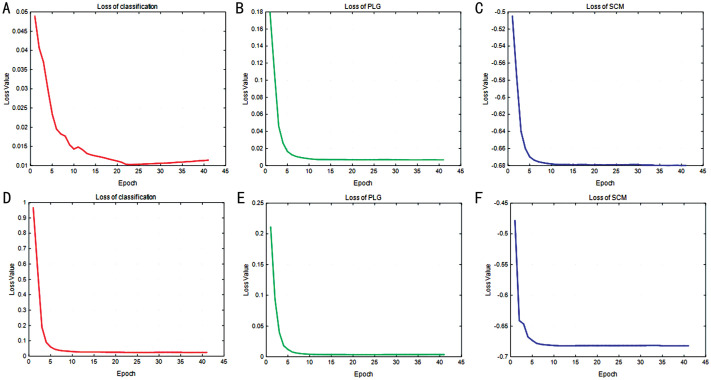
Figure 7. Convergence curve of non-referable/referable DR and normal/abnormal DR tasks.
A–C: The loss of classification, PLG, and SCM for the RDR task; D–F: The loss of classification, PLG, and SCM for the NAD task. DR: Diabetic retinopathy; RDR: Referable diabetic retinopathy; SCM: Softmax-consistence minimization; PLG: Pseudo-label generator; NAD: Normal/abnormal detection.
Performance Analysis
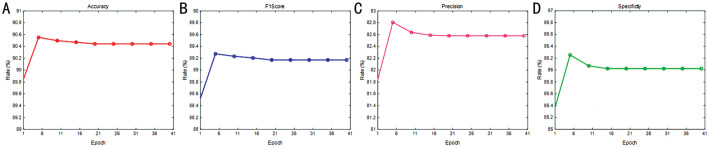
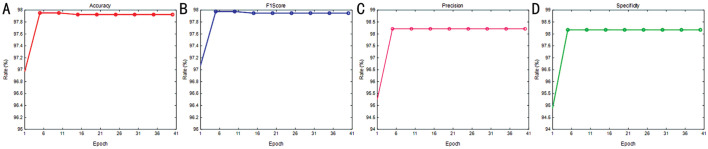
In this section, we examine the impact of training epochs on model performance. Figures 8 and 9 display the results for the RDR and NAD tasks, respectively. The evaluation of each task utilized four metrics, including accuracy, F1-score, precision, and specificity to generate a performance curve. Interestingly, the results indicate that stability was achieved for the first 11 epochs, though several rates yielded the optimal results. As illustrated in Figure 7, the convergence of loss was not stable. Consequently, we set the number of epochs for our model to 40 to optimize the identification performance.
Figure 8. Rate vs epoch curve of non-referable/referable diabetic retinopathy.
A: Accuracy; B: F1-score; C: Precision; D: Specificity.
Figure 9. Rate vs epoch curve of normal/abnormal diabetic retinopathy.
A: Accuracy; B: F1-score; C: Precision; D: Specificity.
Convergence Analysis
This section proves the convergence of loss over epochs. Figure 7 demonstrates that for both the RDR and NAD tasks, the model loss quickly reached convergence. As the number of training epochs increased, the losses of all six terms decreased uniformly. Following 40 epochs in our approach, the model achieved remarkable stability. As a result, we configured the epoch of SMPL to 40 for the experiments.
DISCUSSION
In this paper, we proposed a multi-SFDA method for the identification of DR using medical image analysis. Our approach addresses the challenges faced by deep learning methods, such as the need for a large amount of labeled data, data labeling difficulties, and data privacy concerns.
By integrating multiple source models trained on a single source data, we are able to generate synthetic pseudo labels for the unlabeled target domain. This allowed us to leverage the knowledge from different source models and improve the performance of DR identification in the target domain.
Furthermore, we introduced a softmax-consistence minimization term to minimize the distances between the same classes in the source and target domains, while maximizing the distances between different classes. This helped to enhance the discriminative power of our model and improve its ability to accurately classify different stages of DR.
To evaluate the effectiveness of our SMPL method, we conducted experiments using three CFP datasets: APTOS2019, DDR, and EyePACS. The results demonstrated the superiority of our approach in terms of DR identification accuracy compared to existing methods.
According to the above analysis, our approach addresses deep learning challenges, including the need for abundant labeled data, labeling complexities, and privacy concerns. By merging multiple source models trained on single-source data, we generate synthetic pseudo-labels for unlabeled target domains, enabling us to leverage varied source model insights and enhance DR identification performance in the target domain. It presents an innovative solution to expand the application scenarios of artificial intelligence models, improve their generalization ability, and pave the way for diagnosis of DR. However, this study still faces several constraints, including the need for more computational power and model training costs during the training process of multi-source domain models, the negative impact of noisy data in the target domain, and issues related to poor data collection quality.
In terms of clinical applicability, our method has the potential to be integrated into existing ophthalmological diagnostic systems to provide a more accurate and efficient diagnosis of DR. This could reduce the burden on healthcare professionals by automating the initial screening process, and allow for earlier detection and treatment of DR, potentially reducing the risk of vision loss in diabetic patients. Considering potential real-world application scenarios, our method could be utilized in remote or under-served areas where access to qualified ophthalmologists is limited. By using our method, healthcare workers in these areas could conduct initial screenings for DR using fundus photographs, and refer patients with positive results to specialists for further examination and treatment. Our method could also be integrated into telemedicine platforms, providing an efficient and cost-effective solution for DR screening on a large scale.
However, there are certain limitations to our study. Firstly, the computational requirements for training the multi-source domain models can be demanding, which may limit its application in resource-constrained settings. Additionally, the presence of noisy data in the target domain can impact the performance of our method.
Furthermore, the quality of data collection in medical imaging datasets may vary, which can affect the reliability of our approach. Lastly, while our method shows promise for DR identification, its generalizability to other retinal diseases or imaging modalities needs further exploration.
In our future work, we will focus on addressing these aforementioned challenges to improve the efficiency and accuracy of model applications. We aim to significantly broaden the scope of model applications, achieve widespread screening and diagnosis of DR, and ultimately enhance the quality of life for DR patients.
Footnotes
Foundations: Supported by the Fund for Shanxi “1331 Project” and Supported by Fundamental Research Program of Shanxi Province (No.202203021211006); the Key Research, Development Program of Shanxi Province (No.201903D311009); the Key Research Program of Taiyuan University (No.21TYKZ01); the Open Fund of Shanxi Province Key Laboratory of Ophthalmology (No.2023SXKLOS04); Shenzhen Fund for Guangdong Provincial High-Level Clinical Key Specialties (No.SZGSP014); Sanming Project of Medicine in Shenzhen (No.SZSM202311012); Shenzhen Science and Technology Planning Project (No.KCXFZ20211020163813019).
Conflicts of Interest: Zhang GH, None; Zhuo GP, None; Zhang ZX, None; Sun B, None; Yang WH, None; Zhang SC, None.
REFERENCES
- 1.Huang SQ, Li JN, Xiao YZ, Shen N, Xu TF. RTNet: relation transformer network for diabetic retinopathy multi-lesion segmentation. IEEE Trans Med Imaging. 2022;41(6):1596–1607. doi: 10.1109/TMI.2022.3143833. [DOI] [PubMed] [Google Scholar]
- 2.Tan TN, Wong TY. Diabetic retinopathy: looking forward to 2030. Front Endocrinol. 2023;13:1077669. doi: 10.3389/fendo.2022.1077669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sivaprasad S, Sen S, Cunha-Vaz J. Perspectives of diabetic retinopathy-challenges and opportunities. Eye (Lond) 2023;37(11):2183–2191. doi: 10.1038/s41433-022-02335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silva PS, Zhang DA, Jacoba CMP, et al. Automated machine learning for predicting diabetic retinopathy progression from ultra-widefield retinal images. JAMA Ophthalmol. 2024;142(3):171–177. doi: 10.1001/jamaophthalmol.2023.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galappaththige CJ, Kuruppu G, Khan MH. Generalizing to unseen domains in diabetic retinopathy classification; 2024 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV); Waikoloa, HI, USA. IEEE; 2024. pp. 7670–7680. [Google Scholar]
- 6.Hou XH, Wang LM, Zhu DL, et al. China National Diabetic Chronic Complications Study Group Prevalence of diabetic retinopathy and vision-threatening diabetic retinopathy in adults with diabetes in China. Nat Commun. 2023;14(1):4296. doi: 10.1038/s41467-023-39864-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo YA, Copado IA, Yonamine S, et al. The relationship between health insurance status and diabetic retinopathy progression. Ophthalmol Sci. 2024;4(3):100458. doi: 10.1016/j.xops.2023.100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das D, Biswas SK, Bandyopadhyay S. A critical review on diagnosis of diabetic retinopathy using machine learning and deep learning. Multimed Tools Appl. 2022;81(18):25613–25655. doi: 10.1007/s11042-022-12642-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen B, Fang XW, Wu MN, et al. Artificial intelligence assisted pterygium diagnosis: current status and perspectives. Int J Ophthalmol. 2023;16(9):1386–1394. doi: 10.18240/ijo.2023.09.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu JG, Shen JX, Jiang Q, Wan C, Zhou F, Zhang SC, Yan ZP, Yang WH. A multi-modal fundus image based auxiliary location method of lesion boundary for guiding the layout of laser spot in central serous chorioretinopathy therapy. Comput Biol Med. 2023;155:106648. doi: 10.1016/j.compbiomed.2023.106648. [DOI] [PubMed] [Google Scholar]
- 11.Yang WH, Shao Y, Xu YW, Ophthalmic Imaging and Intelligent Medicine Branch of Chinese Medicine Education Association Expert Workgroup of Guidelines on Clinical Research Evaluation of Artificial Intelligence in Ophthalmology (2023) Guidelines on clinical research evaluation of artificial intelligence in ophthalmology (2023) Int J Ophthalmol. 2023;16(9):1361–1372. doi: 10.18240/ijo.2023.09.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diabetic retinopathy detection. Kaggle. 2015. https://kaggle.com/competitions/diabetic-retinopathy-detection .
- 13.Cuadros J, Bresnick G. EyePACS: an adaptable telemedicine system for diabetic retinopathy screening. J Diabetes Sci Technol. 2009;3(3):509–516. doi: 10.1177/193229680900300315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T, Gao YQ, Wang K, Guo S, Liu HR, Kang H. Diagnostic assessment of deep learning algorithms for diabetic retinopathy screening. Inf Sci Int J. 2019;501(C):511–522. [Google Scholar]
- 15.Karthik M, Sohier D. APTOS 2019 Blindness Detection. Kaggle. 2019 https://kaggle.com/competitions/aptos2019-blindness-detection . [Google Scholar]
- 16.Ganin Y, Ustinova E, Ajakan H, et al. Domain-adversarial training of neural networks. Journal of Machine Learning Research. 2016;17(59):1–35. [Google Scholar]
- 17.Long MS, Cao Y, Wang JM, Jordan MI. Learning transferable features with deep adaptation networks; Proceedings of the 32nd International Conference on International Conference on Machine Learning, Volume 37; July 6–11, 2015; Lille, France. ACM; 2015. pp. 97–105. [Google Scholar]
- 18.Long MS, Zhu H, Wang JM, Jordan MI. Deep transfer learning with joint adaptation networks; Proceedings of the 34th International Conference on Machine Learning–Volume 70; August 6–11, 2017; Sydney, NSW, Australia. ACM; 2017. pp. 2208–2217. [Google Scholar]
- 19.Tzeng E, Hoffman J, Saenko K, Darrell T. Adversarial discriminative domain adaptation; 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); Honolulu, HI, USA. IEEE; 2017. pp. 2962–2971. [Google Scholar]
- 20.Long MS, Cao ZJ, Wang JM, Jordan MI. Conditional adversarial domain adaptation; Proceedings of the 32nd nternational Conference on Neural Information Processing Systems; December 3–8, 2018; Montréal, Canada. ACM; 2018. pp. 1647–1657. [Google Scholar]
- 21.Chetoui M, Akhloufi MA. Explainable diabetic retinopathy using EfficientNET. Annu Int Conf IEEE Eng Med Biol Soc. 2020;2020:1966–1969. doi: 10.1109/EMBC44109.2020.9175664. [DOI] [PubMed] [Google Scholar]
- 22.Mazurowski MA, Tourassi GD. Evaluating classifiers: relation between area under the receiver operator characteristic curve and overall accuracy; 2009 International Joint Conference on Neural Networks; Atlanta, GA, USA. IEEE; 2009. pp. 2045–2049. [Google Scholar]
- 23.Graham B. Kaggle diabetic retinopathy detection competition report. Vol. 22 University of Warwick; 2015. [Google Scholar]
- 24.Franco-Barranco D, Pastor-Tronch J, González-Marfil A, Muñoz-Barrutia A, Arganda-Carreras I. Deep learning based domain adaptation for mitochondria segmentation on EM volumes. Comput Methods Programs Biomed. 2022;222:106949. doi: 10.1016/j.cmpb.2022.106949. [DOI] [PubMed] [Google Scholar]
- 25.Guan H, Liu M. Domain adaptation for medical image analysis: a survey. IEEE Trans Biomed Eng. 2021;69(3):1173–1185. doi: 10.1109/TBME.2021.3117407. [DOI] [PMC free article] [PubMed] [Google Scholar]