Abstract
The envelope glycoproteins of human cytomegalovirus (HCMV) virions are incompletely characterized. We have analyzed complex formation between glycoprotein M (gM or gpUL100) and a second glycoprotein. gM-homologous proteins are conserved throughout the herpesvirus family and represent type III membrane proteins containing multiple hydrophobic sequences. In extracellular HCMV particles, gM was found to be complexed through disulfide bonds to a second protein with an apparent molecular mass of 50 to 60 kDa. The 50- to 60-kDa protein was found to be derived from reading frame UL73 of HCMV strain AD169. UL73-homologous genes are also conserved within herpesviruses. When transiently expressed by itself, the UL73 gene product consisted of a protein of 18 kDa. However, in the presence of gM, the UL73 gene product was posttranslationally modified to the 50- to 60-kDa species. Thus, gM and the UL73 gene product, which represents the gN homolog of herpesviruses, form a disulfide-linked complex in HCMV virions. Transient expression of gM and gN followed by fluorescence imaging with monoclonal antibodies against either protein demonstrated that complex formation was required for transport of the proteins from the endoplasmic reticulum to the Golgi and trans-Golgi compartments. Finally, we tested the gM-gN complex for reactivity with sera from HCMV-seropositive donors. Whereas most sera failed to react with either gM or gN when expressed alone, 62% of sera were positive for the gM-gN complex. Because a murine monoclonal antibody reactive with gN in the gM-gN complex efficiently neutralizes infectious virus, the gM-gN complex may represent a major antigenic target of antiviral antibody responses.
Human cytomegalovirus (HCMV) infects the majority of the population worldwide. Fortunately, this infection usually remains clinically asymptomatic in immunocompetent individuals. However, in immunocompromised hosts, such as infants infected in utero, transplant recipients, or patients with AIDS, HCMV infections can be life threatening (1). Identification and characterization of HCMV proteins that elicit a protective response will allow a more thorough understanding of the control of infection and, eventually, the design of strategies to limit disease in the infected host.
HCMV is the most genetically complex member of the family of human herpesviruses. The laboratory-adapted strain AD169 encodes more than 200 open reading frames within its genome of 230,000 bp (10). Low-passage clinical isolates contain an additional 15 kb of sequence potentially encoding an additional 20 proteins (9). Among herpesviruses, HCMV has the largest coding capacity for glycoproteins; at least 57 potential glycoproteins are encoded by AD169 and an additional 15 by clinical isolates (9, 10).
The structural glycoproteins of the viral particle remain incompletely defined. To date three major glycoprotein complexes (designated gCI to -III) have been identified. The gCI complex is formed by homodimeric molecules of glycoprotein B (gB or gpUL55) linked by disulfide bonds (4, 6). gH (gpUL75), gL (gpUL115), and gO (gpUL74) together represent the gCIII complex. Again, the individual components are covalently linked through disulfide bonds (16, 31). gCII has been reported to consist of a heterogeneous family of glycoproteins with molecular masses between 39 and more than 200 kDa (25). Components of the gCII complex have been identified as heparin binding proteins of the envelope, yet very little else is known about the components or the characteristics of this complex (23, 24). A 48-kDa polypeptide within this complex was identified as the product of open reading frame UL100 (26). The gene product of UL100 was first described by Lehner et al. (30) as a structural component of HCMV virions and termed the integral membrane protein. Subsequently, proteins from homologous reading frames were identified in other herpesviruses, including the glycoprotein termed gM in the herpes simplex virus. Following the recommendations for nomenclature of HCMV glycoproteins, we will therefore designate the UL100 gene product of HCMV as gM.
The gM-homologous proteins of herpesviruses represent type III membrane proteins containing multiple (six to eight) transmembrane sequences. They constitute components of the mature virus particle (3, 12, 39, 40, 46). Functions of gM have been described in fusion and/or entry and cell-to-cell spread (28, 38). gM has been reported to be nonessential for the in vitro replication of alphaherpesviruses (herpes simplex virus type 1 [HSV-1] and pseudorabies virus [PRV]) and the gammaherpesvirus equine herpesvirus type 1 (2, 12, 38). However, viral mutants lacking gM replicate to lower titers in tissue culture than the wild-type viruses and exhibit reduced virulence in animal models or the natural host (11, 31, 34).
For a number of alpha- and gammaherpesviruses, complex formation of gM with other viral proteins has been described. One protein in the complex has been identified as gN, a protein which is also conserved throughout the herpesvirus family (19, 29). The various gN homologs have been reported to be small type I membrane proteins with molecular masses in the range of 7 to less than 20 kDa (20, 29, 46). Previous studies have suggested that gN is nonessential for the in vitro replication of alphaherpesviruses PRV, varicella-zoster virus, and bovine herpesvirus type 1 (BHV-1) (19, 41, 46). Mutant PRVs lacking gN replicate in noncomplementing cell lines to slightly reduced titers, and their penetration into susceptible cells is considerably delayed (19, 33). Despite the conservation, description of the gN homologs indicates a variety of processing differences, e.g., lack of glycosylation in the proteins from BHV-1 and varicella-zoster virus (32, 41). Also, complex formation with gM can be mediated through both noncovalent interactions and covalent disulfide bonds (19, 29, 46). Viruses containing double mutations in the gM and gN genes have not been described so far; however, the absence of gN from virions derived from a gM-negative mutant PRV indicates that both proteins can be absent from infectious virions (19). In contrast to the results described above, gM has been shown to be an essential protein for in vitro replication of HCMV (15). Whereas considerable effort has been devoted to understanding the molecular functions of gM-gN proteins, very little is known about their immunogenicity during natural infection. As glycoprotein components of the viral envelope, they are potentially capable of inducing neutralizing antibodies.
We investigated the complex formation of gM of HCMV. We report here that gM associates with a second protein of 50 to 60 kDa through disulfide bonds and that both molecules represent major constituents of mature virions. The 50- to 60-kDa protein is encoded by the UL73 reading frame of HCMV AD169, which represents the gN homolog of herpesviruses. Processing of recombinant-derived gN appears to differ only slightly from that of the native protein expressed in virus-infected cells. In the absence of gM, the UL73 gene product is a 18-kDa polypeptide which, in the presence of gM, acquires complex modifications resulting in the formation of a 50- to 60-kDa glycoprotein. Fluorescence imaging experiments indicated that complex formation between gM and gN was required for the transport of both proteins from the endoplasmic reticulum (ER) to more distal parts of the secretory pathway. Finally, we have provided evidence that the gM-gN complex represents a highly immunogenic structure for the humoral immune response during natural infection.
MATERIALS AND METHODS
Cell culture and virus production.
HCMV strain AD169 was propagated in primary human foreskin fibroblasts grown in minimal essential medium (Gibco BRL, Glasgow, Scotland) supplemented with 5% fetal calf serum (FCS), glutamine (100 mg/liter), and gentamicin (350 mg/liter). Virions were isolated via glycerol-tartrate gradient centrifugation as described previously (44). 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco BRL) supplemented with 10% FCS, glutamine, gentamicin, and 50-μg/ml G418 (Geneticin; Gibco BRL). Cos-7 cells were passaged in DMEM supplemented with 10% FCS and antibiotics as described above.
MAbs.
Monoclonal antibody (MAb) IMP 91-3/1 was produced by standard procedures. Briefly, a fusion protein (termed IMP12) containing amino acids (aa) 300 to 372 of gM fused to aa 1 to 375 of β-galactosidase was expressed in Escherichia coli using the vector pROS as described previously (13). The recombinant protein was purified, and the viral polypeptide was released by cleavage with blood coagulation factor Xa. The gM-specific polypeptide was then purified by fast protein liquid chromatography. It was injected intramuscularly into the hind limbs of adult BALB/c mice by using complete Freund's adjuvant. Following two boosts with 50 μg of protein emulsified in incomplete Freund's adjuvant, cells from the draining lymph nodes were fused with the myeloma cell line Sp20 according to standard procedures. Wells containing hybridoma cells were screened by indirect immunofluorescence using HCMV-infected cells. The remaining MAbs that were used in this study have been described previously: gB-specific MAb 27-287 (43) and gp65-specific MAb 14-16A (5). Anti-Flag M2 was purchased from Sigma (Deisenhofen, Germany).
Plasmids.
Plasmid pcIMP was constructed by inserting a 1.5-kb NciI fragment (nucleotides 144928 to 146466 of the AD169 sequence) containing the entire UL100 reading frame into the vector pcDNA3 (Invitrogen, Carlsbad, Calif.). The fragment was derived from the plasmid pHM9 (30). pcUL73flag was constructed by inserting the entire UL73 open reading frame (417 bp) into the vector pcDNA3. To insert the FLAG epitope at the 3′ end of the UL73 coding sequence, the following primers were used: 73-forward (5′ACCGAATTCATGGAGTGGAACACACTAGTATTAGG 3′) and 73-reverse (5′TCACTTGTCATCGTCGTCCTTGTAGTCCCATAGCCTTTGGTGGTGGTTGC3′), in which the stop codon was moved to the end of the sequence encoding the FLAG epitope. The FLAG epitope consists of the sequence DYKDDDK, which is recognized by antibody M2 (Sigma).
DNA transfection.
293T cells were transfected with DNA using Lipofectamine Plus reagent (Gibco BRL) according to the manufacturer's suggestion, except that the transfection mixture consisted of 6.5 μg of DNA, 1.5 ml of DMEM, and 46 μl of Lipofectamine Plus reagent. The mixture was added to a petri dish (10-cm diameter) which had been seeded with 3 × 106 cells 2 days earlier. After 48 to 55 h, cells were harvested, washed three times with phosphate-buffered saline (PBS), and stored at −20°C until used. For imaging experiments which used transfected cells, Cos-7 cells were grown on 13-mm glass coverslips and transfected using calcium chloride as described (39).
SDS-PAGE and immunoblotting.
To avoid the formation of high-molecular-weight aggregates after boiling, extracellular HCMV particles were incubated in sample buffer containing 15 mM Tris-Cl (pH 6.8), 8 M urea, 4% (wt/vol) sodium dodecyl sulfate (SDS), 2% (vol/vol) β-mercaptoethanol, 10% (vol/vol) glycerol, and 0.01% bromophenol blue for 2 to 3 h at room temperature. The separation of proteins was carried out by using conventional 8 to 10% polyacrylamide gel electrophoresis (PAGE), except that solutions for stacking and separating gels contained 3 M urea and 0.5% (vol/vol) Triton X-100 (final concentrations). All solutions containing urea were prepared fresh. Gel electrophoresis and transfer of samples to nitrocellulose membranes were carried out by standard procedures. For the detection of antigen, antibodies were diluted in PBS containing 0.1% Tween 20. Antibody binding was detected using either horseradish peroxidase-coupled anti-murine immunoglobulin and the enhanced chemiluminescence detection system (Pharmacia Biotech, Freiburg, Germany) or alkaline phosphate-coupled anti-murine immunoglobulin and 5-bromo-4-chloro-3-indolylphosphate–Nitro Blue Tetrazolium. Removal of N-linked oligosaccharides was carried out using recombinant peptide:N-glycosidase F and endoglycosidase H (New England Biolabs, Beverly, Mass.) according to the manufacturer's specifications, except that denaturation of samples was carried out at room temperature for 2 to 3 h.
Immunoprecipitations.
All immunoprecipitations were carried out using biotinylated antigen preparations. Extracellular virions were biotinylated using Sulfo-NHS-Biotin (Pierce, Rockford, Ill.) according to the manufacturer's suggestions. Briefly, 650 μg of HCMV particles was resuspended in 500 μl of PBS, 270 μg of biotin was added, and the mixture was incubated for 30 min at room temperature with gentle agitation. Particles were pelleted and resuspended in 30 ml of DMEM and centrifuged for 70 min at 92,000 × g at 10°C. The pellet was resuspended in 150 μl of PBS and used for immunoprecipitation. To biotinylate transfected cells, 1.2 × 107 cells were labeled using 250 μg of biotin in a reaction volume of 500 μl. For immunoprecipitations, virions or transfected cells were treated with buffer A (50 mM Tris-Cl [pH 8.0], 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.1 mM phenylmethylsulfonyl fluoride) for 20 min at 4°C with gentle agitation. Lysates were cleared by centrifugation (30,000 × g, 10 min, 4°C) and incubated with protein A-Sepharose CL-4B (Sigma) precoated with MAb for 2 h at 4°C with gentle agitation. Samples were washed three times with buffer A, and precipitated proteins were dissociated from the protein A-Sepharose by incubating samples for 2 to 3 h at room temperature in sample buffer with or without 2-mercaptoethanol. The precipitated proteins were then analyzed by SDS-PAGE as described above. Precipitated proteins were detected in immunoblots by using streptavidin peroxidase and the enhanced chemiluminescence system (Amersham). In experiments in which MAb 14-16A was used as the precipitating antibody, protein A-Sepharose was precoated with rabbit anti-mouse immunoglobulin M (IgM) (Dako, Hamburg, Germany) for 3 h at 4°C, washed once with buffer A, and resuspended in the same buffer.
Immunofluorescence.
Transfected cells were harvested, washed with PBS, spotted on glass coverslips, air dried, and fixed for 10 min with −20°C acetone. Human sera were diluted 1:50 in PBS–0.1% Tween 20 and incubated for 45 min at 37°C with the cells. After three additional washes with PBS, 0.1% Tween 20 fluorescein isothiocyanate-conjugated rabbit anti-human IgG (Dako) was added for 60 min at 37°C. Cells were washed twice with PBS and counterstained with Evans blue (0.001%).
Imaging of transiently expressed gM and gN.
Cos-7 cells were grown on 13-mm glass coverslips in 24-well plates and were transfected with expression plasmids for gM(UL100), gNFLAG(UL73), or gN(UL73) or were cotransfected with both gNFLAG and gM by using calcium chloride as described previously (42). Forty-eight hours after transfection, the coverslips were fixed for 30 min at room temperature in 2% paraformaldehyde freshly prepared in PBS (pH 7.4). Following several rinses with PBS, the cells were permeabilized in cold PBS containing 0.05% NP-40 and 0.002% SDS for 5 min at 4°C. The coverslips were then rinsed several times with PBS and blocked by incubation in PBS supplemented with 20% normal goat serum for 60 min at room temperature. The coverslips were rinsed, and then primary antibody was added and the coverslips were incubated for 60 min at 37°C. After rinsing, fluorochrome-conjugated secondary antibody diluted in 20% normal goat serum was added and the coverslips were incubated for 60 min at 37°C. The wells were washed and postfixed in 2% paraformaldehyde. Following mounting in Slow Fade (Molecular Probes, Eugene, Oreg.), the coverslips were viewed under a Leitz Dialux epifluorescence microscope at a magnification of ×50 and the images were captured with a digital camera (Photometrics, Tucson, Ariz.). The images were processed with Image Pro software (Media Cybernetics, Silver Spring, Md.). Deconvolution was accomplished with Hazebuster (Vaytek, Fairfield, Iowa).
The antibodies which were used to identify cell markers in this study included (i) anticalreticulin for the ER (Affinity BioReagents, Golden, Colo.), (ii) anti-ERGIC53 for the ER-Golgi intermediate compartment (ERGIC) (Peter Hauri, University of Basel, Basel, Switzerland), (iii) anti-p115(TAP) for ERGIC (Elizabeth Sztul, University of Alabama, Birmingham), (iv) anti-GM130 for the Golgi (Elizabeth Sztul), and (v) Texas red-conjugated wheat germ agglutinin (WGA) for the trans-Golgi network (TGN) (Molecular Probes). These reagents were described in a recent publication (42).
RESULTS
Isolation and characterization of a MAb specific for gM.
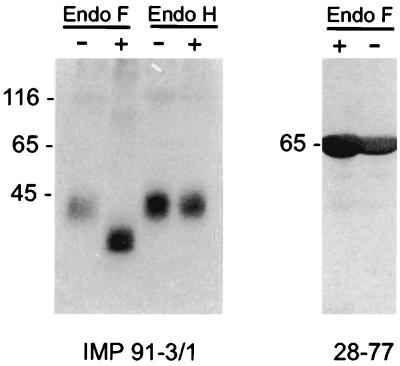
To further define the gCII complex, we initially generated gM murine MAbs by immunizing mice with an E. coli-produced polypeptide which consisted of aa 300 to 372 of gM fused to aa 1 to 375 of β-galactosidase (termed IMP12). Immunization induced a readily detectable antibody response to gM, as measured by indirect immunofluorescence of HCMV-infected fibroblasts (data not shown). Several antibody-producing hybridoma cell lines were isolated, and antibodies from one line (designated IMP 91-3/1) were further characterized. The specificity of MAb IMP 91-3/1 was established in immunoblots using fusion proteins derived from the UL100 open reading frame. Antibody IMP 91-3/1 specifically reacted with fusion protein IMP12 and not with sequences derived from other regions of gM, other HCMV proteins, or the vector-encoded β-galactosidase moiety (data not shown). The MAb IMP 91-3/1 was nonreactive in the enzyme-linked immunosorbent assays and indirect immunofluorescence assays using Epstein-Barr virus (EBV)- or HSV-infected cells or cell lysates (data not shown). When extracellular virions were used in immunoblot assays, IMP 91-3/1 reacted with a protein of 42 to 45 kDa, which was in agreement with our previous results using a polyclonal antiserum against gM (Fig. 1) (30). When virions were treated with endoglycosidase F and analyzed by immunoblotting, MAb IMP 91-3/1 reacted with a protein with an estimated molecular mass of about 35 to 40 kDa. A control digestion using a nonglycosylated viral protein, pp65(UL83), indicated that the decrease in size of gM was not secondary to nonspecific proteolytic activity of the endoglycosidase (Fig. 1). Treatment with endoglycosidase H failed to alter the migration of the protein, indicating that virion gM contained complex, N-linked sugars (Fig. 1).
FIG. 1.
gM of HCMV is a structural protein containing complex carbohydrates. Extracellular virus particles were purified by centrifugation through a glycerol-tartrate gradient. Lysates were digested with endoglycosidases (+) or left untreated (−), separated by SDS-PAGE, and analyzed in immunoblots using MAb IMP 91-3/1 (gM specific). As a control for nonspecific proteolytic activity, pp65 was precipitated from virions using MAb 28-77 and treated with endoglycosidase F under identical conditions. Molecular weight (in thousands) is on left of each gel.
gM forms a disulfide-linked complex with a second protein of 45 to 60 kDa.
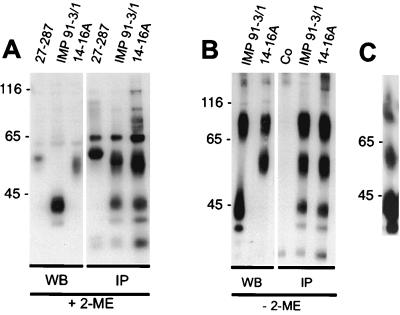
To analyze potential complex formation between gM and other viral proteins through intermolecular disulfide bonds, virions were solubilized in the presence and absence of the reducing agent 2-mercaptoethanol and analyzed by immunoblotting using MAb IMP 91-3/1. In the presence of 2-mercaptoethanol, a single protein migrating at 42 to 45 kDa was detected by MAb IMP 91-3/1 (Fig. 2A). MAb 14-16A, which has previously been shown to react with a highly glycosylated envelope protein with an estimated molecular mass of 65 kDa (5), reacted with protein(s) which migrated between 50 and 60 kDa (Fig. 2A). As a control, MAb 27-287 detected the transmembrane component of gB, which migrated with an estimated molecular mass of 58 kDa (Fig. 2A). When the same virion preparations were subjected to SDS-PAGE in the absence of the reducing agent, MAb IMP 91-3/1 reacted with two proteins which migrated between 70 and 100 kDa and at 45 kDa (Fig. 2B). MAb 14-16A reacted with a 50- to 60-kDa protein and 70- to 100-kDa proteins which comigrated with the 70- to 100-kDa proteins detected by MAb IMP 91-3/1 (Fig. 2B). These results suggest that gM was complexed to a second protein through both disulfide bonds and nonconvalent interactions and that MAb 14-16A was reactive with this second protein.
FIG. 2.
Viral gM and gp65 form a disulfide-linked complex. Lysates from extracellular virions were subjected to Western blotting (WB) or immunoprecipitation (IP) using MAbs specific for gM (IMP 91-3/1), gB (27-287), and gp65 (14-16A), respectively. SDS-PAGE was performed following disruption of immune precipitates of the virion lysates in the presence (A) or absence (B) of the reducing agent 2-mercaptoethanol (2-ME). The 70- to 100-kDa protein from nonreduced samples was cut from the gel and chromatographed a second time under reducing conditions (C). Co represents a control immunoprecipitation without primary antibody. Molecular weight (in thousands) is on left of each panel.
When lysates from extracellular virions were subjected to immunoprecipitation with IMP 91-3/1 followed by SDS-PAGE under reducing conditions, gM and an additional protein of 50 to 60 kDa were precipitated (Fig. 2A). A protein which comigrated with pp65 was also present in the immunoprecipitates; however, the presence of this protein was thought to be secondary to nonspecific binding of the pp65, as a similar band was also seen in the immunoprecipitates generated with the anti-gB MAb 27-287 (Fig. 2A). Similarly, MAb 14-16A specifically precipitated proteins with estimated molecular masses of 50 to 60 kDa and 42 to 45 kDa from virions (Fig. 2A). This MAb also precipitated a band comigrating with pp65 as well as lower-molecular-mass proteins, which migrated more rapidly than did the 45-kDa gM (Fig. 2A). If the immunoprecipitated samples were analyzed in the absence of β-mercaptoethanol, three major bands of 42 to 45, 50 to 60, and 70 to 100 kDa were precipitated by both MAbs IMP 91-3/1 and 14-16A (Fig. 2B). Again, both MAbs precipitated lower-molecular-mass proteins, which appeared not to be disulfide linked to these complexes (Fig. 2B). These data suggest that gM was complexed through disulfide bonds to a second protein with a 50- to 60-kDa molecular mass and that this complex likely accounted for the 70- to 100-kDa species. Together, these data also suggest that gM was complexed with the virion envelope protein identified by MAb 14-16A and that these two virion proteins were present in the virion envelope, both as monomers and as a disulfide-linked complex. To directly examine this possibility, the gel slice containing the 70- to 100-kDa species from samples separated under nonreducing conditions was treated with 2-mercaptoethanol and electrophoresed in a second gel. Although the 70- to 100-kDa species was incompletely dissociated with this treatment, reduction of disulfide bonds resulted in the appearance of both the 50- to 60-kDa and 42- to 45-kDa proteins (Fig. 2C). Taken together, these results strongly suggest that the gM envelope protein was complexed with a second virion protein of 50 to 60 kDa within the envelope of HCMV.
Cotransfection of plasmids expressing the UL73 and gM open reading frames results in the formation of a protein complex recognized by MAb 14-16A.
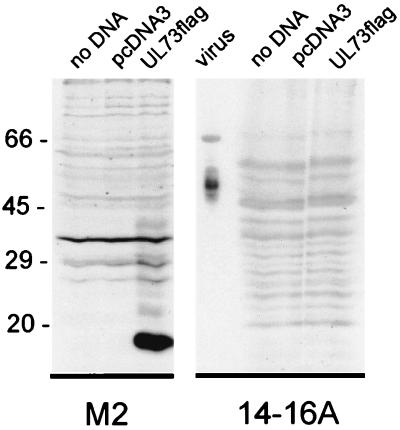
gM molecules from other herpesviruses, including BHV and PRV, have been shown to form a complex with a second glycoprotein, termed gN. The UL73 open reading frame of HCMV encodes the gN homolog. The predicted gene product of UL73 is a protein of 138 aa, which has an expected molecular mass, without posttranslational modifications, of approximately 15 kDa. To monitor expression of the protein product of UL73, the expression plasmid pc73FLAG was constructed by fusing the FLAG epitope to the carboxyl terminus of the UL73 open reading frame. Expression of this fusion protein could then be detected by reactivity with the M2 MAb. Following transfection of the pc73FLAG into 293T cells, cell lysates were analyzed by immunoblotting. By use of the M2-specific antibody, a protein of 18 kDa and several faint bands between 20 and 35 kDa were detected (Fig. 3). Although we initially believed that gN could represent the second protein in the gM complex of HCMV, we failed to detect any reactivity of MAb 14-16A for the forms of UL73FLAG transiently expressed in this transfection (Fig. 3).
FIG. 3.
The UL73 reading frame is translated into an 18-kDa protein in transfected cells. 293T cells were transfected with DNA encoding UL73 (UL73FLAG) or the vector (pcDNA3) or left untreated (no DNA). After 48 h cell lysates were analyzed by immunoblotting. The blots were developed with either the anti-FLAG MAb (M2) or MAb 14-16A. Lysates from extracellular HCMV virions were included as control for MAb 14-16A reactivity (virus).
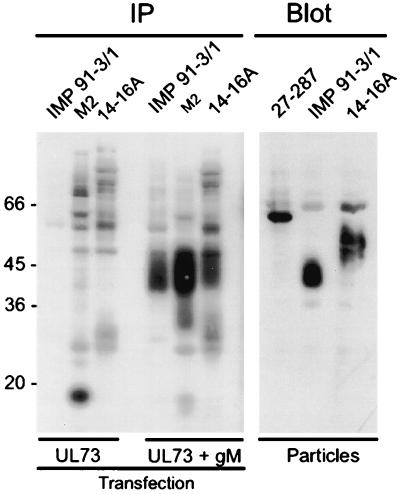
The correct folding, transport, and/or modification of at least one protein partner of the HCMV envelope glycoprotein complex consisting of gH-gL-gO has been suggested to require the simultaneous expression of all three proteins (16, 31). Based on this model, we next used immunoprecipitation experiments to investigate the expression and posttranslational modifications of the respective proteins and/or complexes which formed following cotransfection of 293T cells with gM- and gN-expressing plasmids. In cells transfected only with the plasmid for UL73FLAG, an 18-kDa protein could be precipitated using the M2-specific antibody (Fig. 4). In agreement with our previous findings, no specific signal was obtained with either antibody IMP 91-3/1 or 14-16A (Fig. 4). In contrast to these results, a significantly reduced amount of the 18-kDa species was precipitated from cells cotransfected with UL73FLAG and gM (Fig. 4). As expected, a protein of 42 to 45 kDa was precipitated with antibody IMP 91-3/1, indicating that gM was expressed in these transfected cells (Fig. 4). The most abundant protein precipitated from these cell lysates by antibody M2 had an apparent molecular mass of approximately 40 to 52 kDa, based on its diffuse migration (Fig. 4). A protein of similar size was also precipitated by using antibody 14-16A (Fig. 4). These data indicate that cotransfection of the genes encoding gM and UL73 resulted in the production of a diffusely migrating 42- to 55-kDa protein recognized by antibody 14-16A as well as antibody M2. The estimated molecular mass of the protein recognized by MAb 14-16A in transfected cells was less than the estimated mass of the corresponding protein in virions, suggesting that the protein expressed in transfected 293T cells was modified incompletely compared to the mature form of the protein (Fig. 4). Findings consistent with this interpretation have been provided by studies in virus-infected human fibroblasts and transformed astrocytoma cells (27). In this study, Kari and coworkers demonstrated a marked difference in the carbohydrate modifications, particularly O-linked modifications, of HCMV gB and proteins of the gCII complex (27). MAb 14-16A also recognized the complex formed between gM and the product of the UL73 open reading frame without the carboxyl-terminal FLAG epitope, indicating that the presence of the FLAG epitope was not required for MAb 14-16A reactivity (data not shown). These data were most consistent with the interpretation that gM and the product of the UL73 open reading frame formed a complex when coexpressed in cells and that the complex formation allowed intracellular transport and posttranslational modifications of the protein encoded by the UL73 open reading frame. The recognition of the complex between gM and the UL73-encoded protein by MAb 14-16A suggested that this antibody was specific for a determinant which required a posttranslation modification of the UL73-encoded protein present in the mature glycoprotein complex. Moreover, the finding that the UL73 gene product formed a complex with gM provided evidence that the UL73 gene encoded the gN homolog of HCMV. Carbohydrate modifications of gN were determined by digestion of the protein in virions as well as of the protein expressed in cotransfected cells with a number of different glycosidases. However, with the exception of a reduction in size by 4 to 5 kDa following digestion with endoglycosidase F, the molecule appeared resistant to conventional endoglycosidase treatment (data not shown).
FIG. 4.
Cotransfection of gM and UL73 DNA results in a 14-16A-reactive protein. 293T cells were transfected with a plasmid for UL73FLAG DNA (UL73) or a mixture of plasmids for UL73 plus gM (UL73 + gM). Cell lysates were analyzed in immunoprecipitations (IP) using MAbs specific for gM (IMP 91-3/1), the FLAG epitope of UL73 (M2), or gp65 (14-16A). Controls included an immunoblot (Blot) using extracellular virus particles as antigen and MAbs specific for gB (27-287), gM (IMP 91-3/1), and gp65 (14-16A).
The glycoprotein complex of gM and gN is recognized by MAb 14-16A.
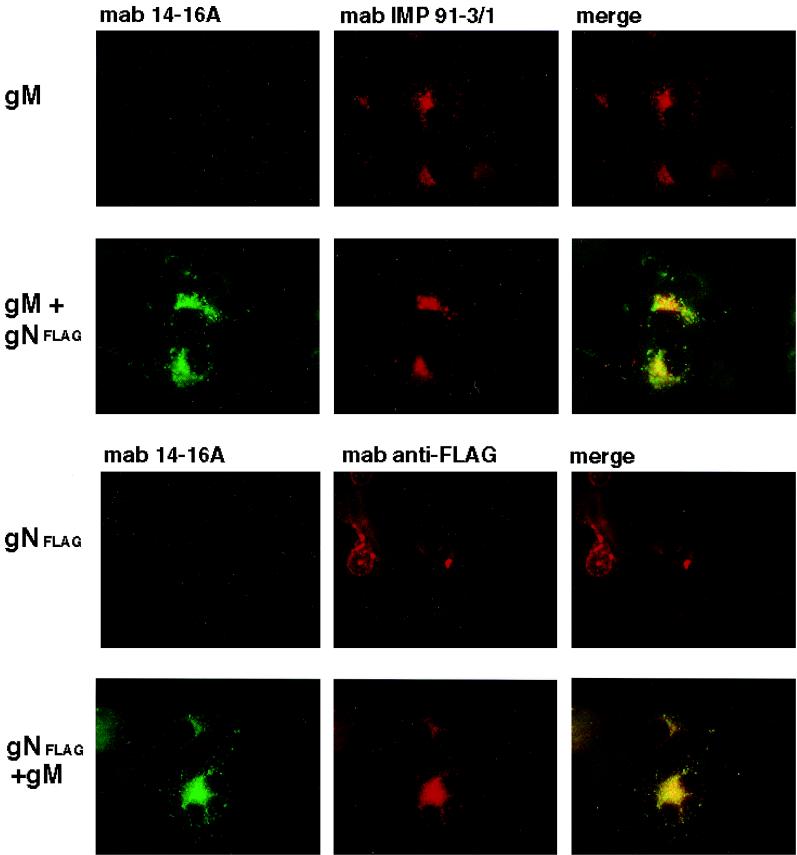
To further confirm that gN formed a complex with gM and to define the intracellular compartment in which this association occurred, we utilized transient expression of gM and gN in Cos-7 cells, followed by fluorescence imaging with antibodies directed against the viral protein or against cellular markers of the secretory pathway. When Cos-7 cells were transfected with an expression plasmid for UL100 (gM) or UL73FLAG (gN), we could not detect reactivity with antibody 14-16A (Fig. 5). In contrast to these results, when plasmids for gM and gNFLAG were cotransfected into Cos-7 cells, we detected prominent staining with MAb 14-16A, which also demonstrated nearly complete colocalization with the signal from the M2 MAb reactive with the protein encoded by UL73FLAG (Fig. 5). Although there was considerable overlap between the signals from MAbs 14-16A and IMP 91-3/1, this overlap was consistently less than the nearly complete colocalization between the signals from MAbs 14-16A and M2 present in cells cotransfected with plasmids for both gM and gNFLAG (Fig. 5). These results provide further support for the interpretation that antibody 14-16A was directed at a determinant on gN, the protein product of UL73. Although the nature of this antibody binding site remains undefined, this result further confirms that MAb 14-16A recognized a conformation-dependent determinant on gN which was formed following complex formation with gM, perhaps secondary to posttranslational processing of the molecule (such as the addition of carbohydrate modifications).
FIG. 5.
The gM-gN complex in transfected cells is recognized by MAb 14-16A. Cos-7 cells were cotransfected with plasmids for gM and gNFLAG, and protein expression was assayed by reactivity with MAb 14-16A, IMP 91-3/1, or the anti-FLAG MAb M2. Reactivity with MAb 14-16A was detected with a fluorescein isothiocyanate-conjugated anti-mouse IgM secondary antibody, while reactivity with MAb IMP 91-3/1 or M2 was detected with a Texas red-conjugated anti-mouse IgG antibody. The appearance of yellow indicates colocalization of the signal from the antibody. Note the nearly complete overlap of the signals from MAbs 14-16A and M2.
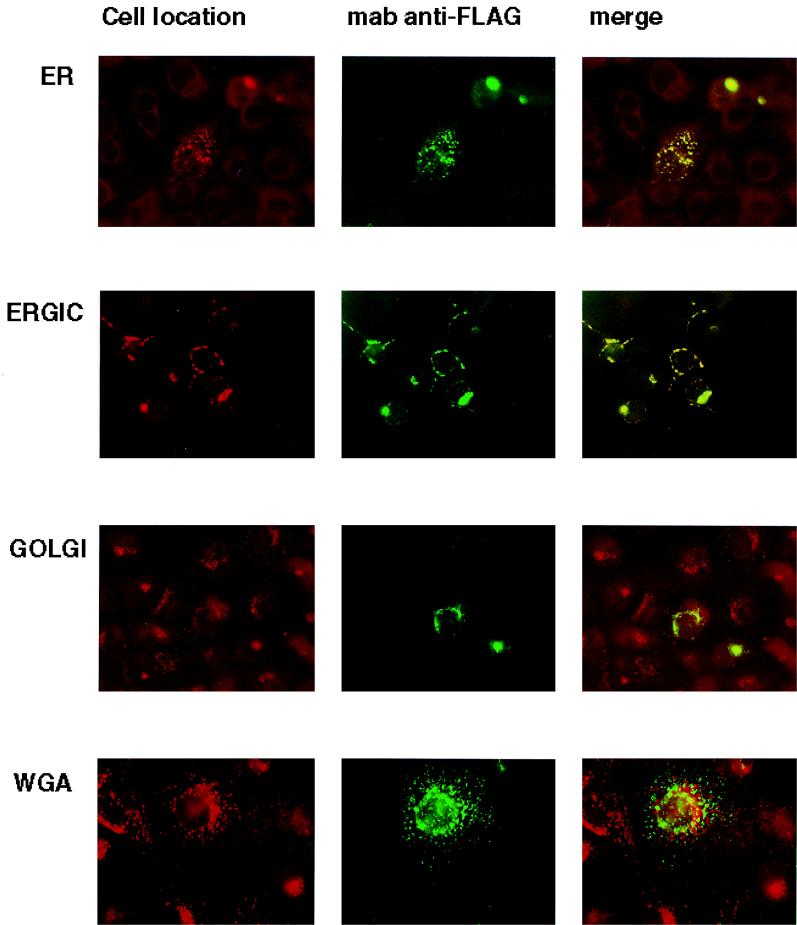
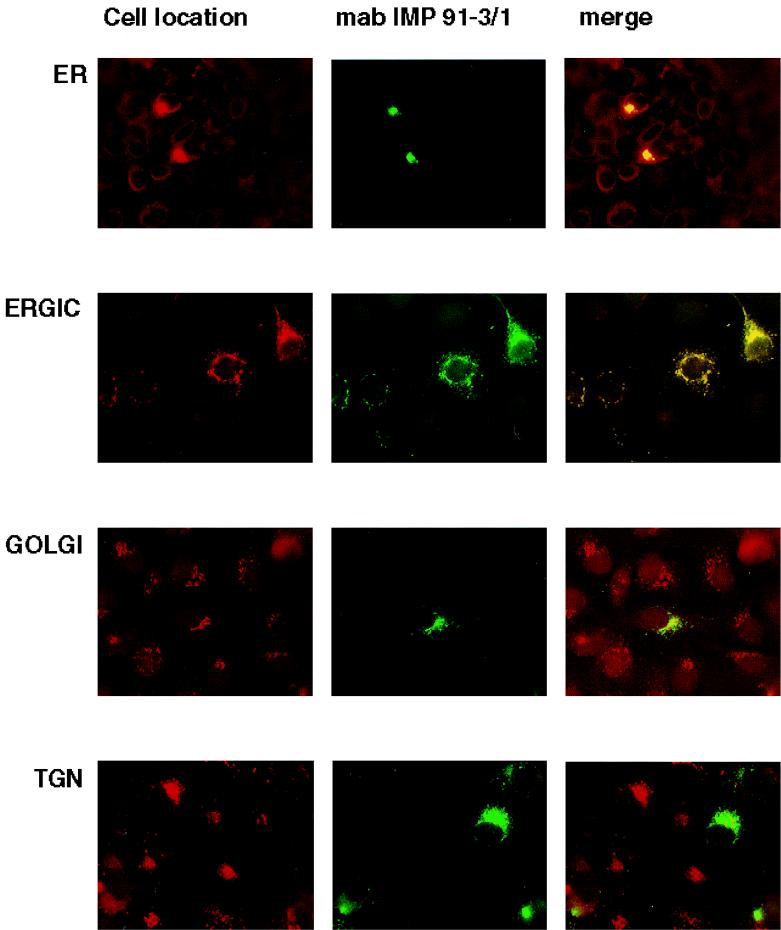
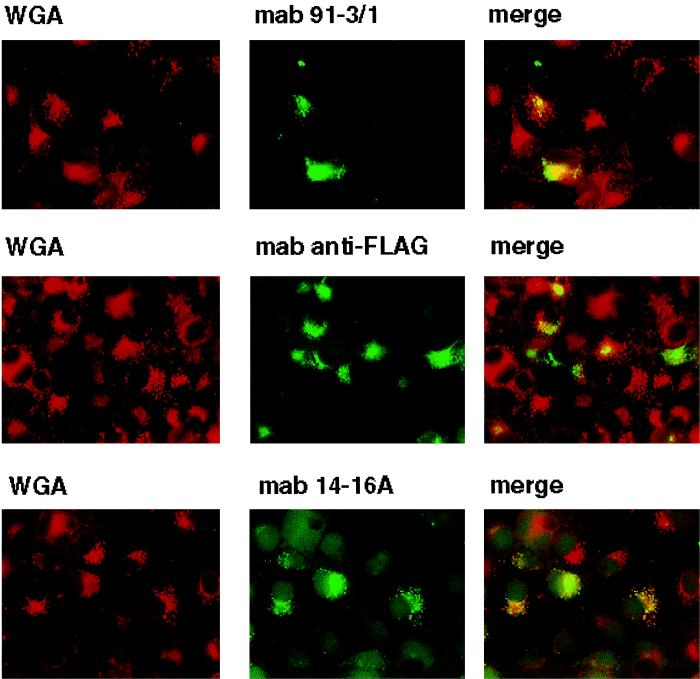
We investigated the intracellular trafficking of gM and gN and the gM-gN complex in an attempt to define the cellular compartment in which complex formation occurred. We initially defined the intracellular trafficking of gN and gM when expressed in cells transfected with a plasmid for only one of the glycoproteins. The distribution of UL73FLAG(gN) was confined to the ER and the ERGIC with no obvious colocalization with markers of the Golgi or TGN (Fig. 6). Similarly, when gM was expressed by itself, the protein colocalized only with markers of the ER and ERGIC and not of the Golgi or TGN (Fig. 7). In contrast to these results, when the expression plasmids for gNFLAG and gM were cotransfected into Cos-7 cells, reactivity for both gM and gNFLAG could be detected in the major compartments of the secretory system (data not shown). Cellular trafficking of the complex was next analyzed using antibody 14-16A. The complex could be shown to colocalize with markers for the TGN with minimal overlap of the signal with markers for the Golgi (Fig. 8) (data not shown). Together these results suggest that complex formation was required for transport of both gN and gM to distal compartments of the secretory system and that complex formation was required for assembly of the antibody binding site defined by antibody 14-16A. Furthermore, the finding that the molecular mass of gN increased from an apparent 18 kDa when expressed alone to an apparent 42 to 45 kDa when coexpressed with gM was consistent with its intracellular transport and posttranslational modification, most likely carbohydrate addition. Lastly, the specificity of MAb 14-16A for the form of gN expressed in the complex composed of gN and gM in the virion and its restricted reactivity for gN found in distal compartments of the secretory system suggest that this determinant was specific for a fully modified form of gN.
FIG. 6.
The product of the UL73 open reading frame, gN, localizes in the ER and ERGIC in transfected cells. Cos-7 cells were grown on glass coverslips and transfected with the plasmid for gNFLAG. The coverslips were then processed for imaging as described in Materials and Methods. The intracellular location of gN was determined by comparing the signal from the FLAG-specific antibody M2 (MAb anti-FLAG) with those of antibodies or lectins specific for markers of cellular components of the secretory system. The cellular markers and associated antibodies were as follows: ER, anticalreticulin; ERGIC, anti-p115(TAP); Golgi, anti-GM130; and TGN, WGA. The cellular markers were developed with Texas red and M2 antibody binding by fluorescein isothiocyanate-conjugated anti-mouse IgG. Yellow indicates colocalization of the signal.
FIG. 7.
The product of the UL100 open reading frame, gM, localizes in the ER and ERGIC in transfected cells. Cos-7 cells were grown on glass coverslips and transfected with a plasmid for gM. The coverslips were processed for imaging as described in Materials and Methods. The intracellular location of gM was determined by comparing the signal from the anti-gM antibody IMP 91-3/1 with those of antibodies or lectins specific for markers of the cellular secretory system. The cellular markers and associated antibodies were as follows: ER, anticalreticulin; ERGIC, anti-p115(TAP); Golgi, anti-GM130; and TGN, WGA. The antibodies reactive with the cellular markers were developed with Texas red and anti-gM MAb binding by fluorescein isothiocyanate-conjugated anti-mouse IgG. Yellow indicates colocalization of the signal.
FIG. 8.
The gM-gN complex formation allows transport of the gM and gN protein to distal compartments of the cellular secretory pathway. Cos-7 cells grown on coverslips were cotransfected with plasmids for gM and gNFLAG and processed as described in Materials and Methods. The coverslips were fixed and then reacted with antibodies against gM (MAb IMP 91-3/1), FLAG (MAb M2), or gN (MAb 14-16A). The TGN was detected with Texas red-conjugated WGA, which was added for the final 10 min of incubation with the fluorochrome-conjugated second antibody. Images were collected and digitized as described in Materials and Methods. A yellow signal indicates colocalization of the signal from the antibodies and colocalization of the two proteins. Note that the signal from MAb 14-16A is restricted to the TGN.
Antibody recognition of the gM-gN complex following natural infection.
In the final set of experiments we investigated the immunogenicity of the gM-gN complex following natural infection. 293T cells were transfected with plasmids expressing gM and/or gN, and indirect immunofluorescence was carried out using 34 human sera which were seropositive for HCMV, as determined by a commercially available assay of HCMV-specific antibody reactivity, and 5 negative control sera. Whereas 6 and 32% of the sera were found positive for gN and gM, respectively, 62% were positive for cells expressing both proteins (Table 1). Thus, the gM-gN complex of HCMV appeared to be antigenic in humans and was a target of antiviral antibody responses following natural infection. The fact that antibody 14-16A neutralized infectious virus very efficiently suggested that antibodies reactive with the gM-gN complexes which are present in human sera could also have virus-neutralizing activity.
TABLE 1.
Reactivity of IgG antibodies against gM, gN, and gM-gN complex in sera from HCMV-seropositive and -seronegative donorsa
| Category of sera | No. of reactive samples/no. tested (%) forb:
|
||
|---|---|---|---|
| gN | gM | gM-gN | |
| HCMV seropositive | 2/34 (6) | 11/34 (32) | 21/34 (62) |
| HCMV seronegative | 0/5 (0) | 0/5 (0) | 0/5 (0) |
293T cells were transfected with expression plasmids for gN (UL73) or gM (UL100) or were cotransfected with both plasmids as described in Materials and Methods. After 48 h, the cells were fixed in acetone and prepared for immunofluorescence assay.
Antibody reactivity was detected by a fluorescein isothiocyanate-conjugated goat anti-human IgG.
DISCUSSION
Our results have demonstrated that HCMV open reading frames UL100 and UL73 encode the gM and gN homologs of HCMV and that these proteins form a disulfide-linked complex in virus-infected cells and in extracellular virions. These results were consistent with previous descriptions of the gM-gN complex in both alpha- and gammaherpesviruses and indicated that the gM-gN protein complex is conserved in alpha-, beta-, and gammaherpesviruses. In contrast to descriptions of the gM-gN complex of EBV, we could demonstrate that a significant amount of HCMV gM and gN was present as a disulfide-linked complex in the virion and the infected cell (29). This finding was in agreement with those of studies of other herpesviruses, such as PRV and BHV-1, which have indicated that the gM-gN complex is disulfide linked (19, 32). Thus, the importance of disulfide bonds in maintaining the gM-gN complex in herpesviruses remains incompletely defined. Interestingly, an appreciable amount of gM and gN in the HCMV virion was also present in monomeric form, suggesting that covalent disulfide bonding was not absolutely required for the interaction between these proteins. However, formation of gM dimers, as has been seen in other herpesviruses, seems not to occur in HCMV (39). Furthermore, the noncovalent interaction between gM and gN was sufficient to permit intracellular transport and folding of gN, as evidenced by the recognition of the higher-molecular-weight gN monomers by MAb 14-16A. These results suggest that gM and gN form a complex through both covalent and noncovalent interactions and raise the possibility that the stoichiometry of this complex could be considerably more complex than a dimer containing a single molecule of gM and gN.
Genes encoding gM and gN homologs appeared to be conserved in all herpesviruses studied thus far, suggesting an important functional role of this complex. Yet several reports have demonstrated that the gM-gN complex is nonessential for virus replication in alphaherpesviruses in vitro (2, 12, 19, 38). More recent studies have shown that PRV gM deletion mutant viruses have reduced entry into permissive cells, a phenotype which has been linked to decreased penetration but not decreased attachment (19). However, this phenotype could not be definitively assigned to gM because the second component of the PRV gM-gN glycoprotein complex, gN, was also dependent on the expression of gM for its intracellular transport and incorporation into the virion. A PRV mutant virus in which the gN has been deleted has been reported to be infectious, but its phenotype has not been as extensively characterized as that of the gM-null virus (19). In the case of EBV, it is unclear whether gM and gN are virion proteins, but preliminary results have suggested that EBV gN may be required for the infection of B lymphocytes (L. Hutt-Fletcher, personal communication). Thus, the function of the gM-gN complex might only be revealed by in vivo experiments. In fact, a PRV mutant which contained a deletion in the gM open reading frame and which failed to express the gM-gN complex demonstrated an attenuated phenotype in the natural host (11). Yet this mutant exhibited spread within the central nervous systems of mice similar to that of the wild-type virus (37). Not surprisingly, the function of the envelope gM-gN complex in HCMV infection is not known, but previous reports have indicated that this protein complex could bind heparin and suggested that this complex could have a possible role in virion attachment (23). Earlier studies have clearly shown that the gM-gN complex is abundantly expressed on the surface of infectious virions and that in the presence of exogenous complement, MAb 14-16A could very efficiently neutralize virus infectivity (5). Thus, the function of the gM-gN complex in alpha and gammaherpesvirus replication remains undefined, although recent studies have indicated that in contrast to these groups of herpesviruses, the glycoprotein complex containing gM is essential for HCMV replication in vitro (15).
Previous studies by Gretch and colleagues described three disulfide-linked complexes within the envelope of HCMV: gCI, gCII, and gCIII (14). Reports from this and other laboratories indicated that the gCI was composed of homodimers of gB and that gCIII was composed of gH, gL, and gO (8, 16–18, 31). The composition of gCII has remained undetermined, although it was suggested that at least one component of this complex was gM (26). The findings in the present study suggest that the gM-gN complex was the HCMV gCII glycoprotein complex. The migration of the several forms of the gCII complex in SDS-PAGE in both the presence and absence of reducing agents was similar to the migration of different forms of the gM-gN complex which are described in this study (23, 25, 26). Of note was that several of the previous studies failed to use solubilization and gel conditions which could allow separation of the various forms of the gM-gN complex but used heat to denature the immune complexes. Heating to 100°C has been shown to convert gM-gN complexes into insoluble aggregates, most of which fail to enter the resolving gel under the conventional conditions of SDS-PAGE (M. Mach and W. Britt, personal observations). As a result, the initial description of some of the more slowly migrating components of the gCII complex could represent an in vitro artifact consisting of denatured aggregates of this complex. The use of MAb 14-16A to identify native gM-gN complexes in virions has suggested that within the mature particle, gM and gN formed a more limited number of complexes, of which one was disulfide bonded. Finally, preliminary studies by Kari and Gehrz using MAb 14-16A suggested that this MAb was reactive with the protein complex described by these investigators as gCII (B. Kari, personal communication). Thus, taken together, our data suggest that the gCII complex of HCMV is composed of gM and gN.
Similar to what was noted in previous descriptions of the gM of HSV, HCMV, and equine herpesvirus type 1, the HCMV gM-gN complex was highly hydrophobic and aggregated readily when heated (3, 38). This biochemical property of the gM-gN complex delayed the characterization of this protein complex in other herpesviruses and, in the case of HCMV, initially led us to believe that this protein complex represented a minor component of the virion envelope (5). By utilizing a denaturing SDS-PAGE system which included urea and solubilization of immune complexes in 8 M urea at room temperature, it was possible to separate the protein components of the larger oligomers and aggregates of the gM-gN complex. Virions solubilized under these conditions allowed us to demonstrate that the gM-gN complex was a relatively abundant component of the envelope of purified virions, a finding which was also consistent with a significant level of expression of the protein complex in infected cells. Analysis of virus-infected cells using MAb 14-16A and immune electron microscopy also indicated that the gM-gN complex was abundantly expressed on the surface of virions and infected cells (Britt, unpublished observations). In contrast to these findings, it has been suggested that the EBV gM-gN complex can be solubilized as well at 37°C as when the immunoprecipitated protein is heated to 100°C (29). It should be noted that these investigators qualified their interpretation of these findings by suggesting that under the specific experimental conditions used to study EBV gM-gN, it was not possible to accurately quantify the amounts of soluble and insoluble material (29). The gM of all herpesviruses thus far studied is a type III glycoprotein which contains multiple, hydrophobic, membrane-spanning domains. These hydrophobic domains likely contribute to the relative insolubility of the gM-gN protein complex and the tendency towards aggregation following denaturation. The lack of reactivity of MAb 14-16A for gN which was present in heat-aggregated forms of the gM-gN complex further argues that MAb 14-16A is specific for a conformational epitope present on mature, native forms of gN.
The structure of the carbohydrate modifications on HCMV gN remains to be determined. Most likely the molecule carries a limited number of N-linked and a large number of O-linked sugars, as indicated by (i) the small decrease in size after digestion with endoglycosidase F; (ii) the findings of Kari and Gehrz, who reported a large amount of O-linked sugar modification of one of the proteins of the gCII complex (21); and (iii) the presence of O-linked carbohydrates on gN molecules from other herpesviruses (20, 29). It is particularly interesting that this relatively small protein with a predicted mass of 18 kDa contained well more than 30 kDa of carbohydrate modifications, a characteristic consistent with the observation that over one-third of the primary amino acid sequence was either serine or threonine residues. The large amount of carbohydrate modifications present on HCMV gN was unique among herpesvirus gN and greatly exceeded the quantity of carbohydrate present on other herpesvirus gN molecules. The gNs of PRV and EBV appear to contain approximately 7 to 8 kDa of carbohydrate modifications, amounts similar to the predicted level of carbohydrate modifications of gN molecules from other herpesviruses (20, 29). Interestingly, gNs from other herpesviruses have been reported to be nonglycosylated proteins (32). This difference between the structure of gN of HCMV and the gNs of other herpesviruses was not reflected in the posttranslational modifications of HCMV gM. The gM of PRV has been proposed to contain a single N-linked sugar modification (12). Our findings also suggest that HCMV gM contains a single N-linked carbohydrate modification. The exact composition of the carbohydrate modifications on other herpesvirus gM molecules is unclear because of the lack of reagents for characterization of these proteins; however, the gM of EBV has been suggested to contain two predicted N-linked glycosylation sites, of which at least one is used (29). Thus, we can only speculate on the importance of the extensive carbohydrate, likely O-linked, modifications of the gN molecule of HCMV to the function of the HCMV gM-gN complex. However, this finding could suggest a unique role for this glycoprotein complex in the tissue and cell tropism of HCMV.
Complex formation between gM and gN likely occurred in the ER, based on results from our imaging studies. When expressed by themselves, both gM and gN remained localized to the ER and ERGIC. Coexpression of gN and gM resulted in formation of a gM-gN complex that was distributed throughout compartments of the cellular secretory pathway, as evidenced by a signal overlap from antibodies directed against gN and gM. Interestingly, MAb 14-16A recognized only a mature form of gN, perhaps a form that had undergone terminal carbohydrate modifications. Although biochemical evidence supporting this claim could not be obtained because of the insensitivity of the mature gN to available endoglycosidases, findings from imaging studies of cells transfected with plasmids for both gM and gN provided convincing evidence for this interpretation. Significant overlap was observed from signals from MAb 14-16A and markers for the TGN, and to a lesser extent, for the Golgi. No overlap was observed with markers for the ER or ERGIC, indicating that although gM-gN complexes were present in these compartments, the epitope recognized by MAb 14-16A was not expressed by gM-gN complexes localized to these compartments. Biochemical evidence supporting this interpretation was provided by the finding that MAb 14-16A recognized a form of gN within virus-infected cells which appeared to be identical to the gN present in extracellular virions. Together with the findings that the epitope defined by MAb 14-16A was expressed in the envelope of infectious virions, these results indicate that maturation of the gM-gN complex occurred in the distal compartments of the secretory system. In agreement with this interpretation is the recent finding that MAb 14-16A was reactive with gN (previously designated gp65) localized to a cytoplasmic compartment which is believed to be a site of virion assembly in HCMV-infected cells (42). Thus, the demonstration that a mature form of gN complexed with gM was localized exclusively to this compartment in HCMV-infected cells was consistent with the hypothesis that this cytoplasmic compartment was indeed a site of virion assembly and did not merely represent a cytoplasmic accumulation of virion structural proteins in various states of maturation.
Thus far, gB and gH have been identified as major targets for the neutralizing humoral immune response in human sera. When recombinant gB and gH were used in preadsorption experiments, between 0 and 98% (gB) and 0 and 58% (gH) of the total neutralizing capacity could be removed from human sera (7, 35, 36, 45). Thus, additional antigens must contribute to the induction of neutralizing antibodies. In the present study, more than 60% of human convalescent-phase sera contained antibodies reactive with the gM-gN complex. The contribution of antibodies directed against the individual glycoproteins to this response is unknown. The seropositivity rate of approximately 30% for gM is probably accurate, since the isolated expression of gM as a recombinant protein results in a polypeptide which carries terminal modifications and likely adopts a near-native structure. Therefore, this protein could be detected by gM-specific antibodies produced following natural infection. The antigen recognized by the additional 30% of sera when assayed against the gM-gN complex is unclear. The likeliest possibility is that the processing and transport of gN when complexed to gM expose antibody binding sites on gN which are not expressed in the absence of gM. Since gN lacks significant posttranslational modifications and likely native conformation when expressed by itself, the seroreactivity rate of 6% in this serum panel also likely reflects the requirements of terminal carbohydrate modification and native folding for recognition of gN. Antibodies specific for epitopes unique to the complex could also contribute to the increased rate of seroreactivity for the gM-gN complex. In any case, the gM-gN complex was identified in this study as an additional major, antigen for the humoral immune response against HCMV. The potential importance of this response has been suggested by previous findings, which indicated that the majority of infected newborns lack detectable antibodies against the gCII (gM-gN) complex, whereas most adult convalescent-phase sera contain gCII-specific antibodies (22).
In summary, we have identified the UL73 gene product of HCMV as gN. We were able to demonstrate that the protein product of the UL73 gene is processed and transported authentically only in the presence of the gM protein and that it forms a disulfide-linked complex with gM. This complex represents a major constituent of HCMV virions and is highly immunogenic during natural infection. Future experiments will be directed towards defining the functional and immunological properties of the gM-gN complex.
ACKNOWLEDGMENTS
This work was supported by grants from the Deutsche Forschungsgemeinschaft (MA 929/4-2), the Wilhelm Sander-Stiftung, and the National Institute of Allergy and Infectious Diseases, National Institutes of Health (AI35602).
REFERENCES
- 1.Alford C A, Britt W J. Cytomegalovirus. In: Fields B N, Knipe D M, editors. Virology. New York, N.Y: Raven Press, Ltd.; 1990. pp. 1981–2010. [Google Scholar]
- 2.Baines J D, Roizman B. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J Virol. 1991;65:938–944. doi: 10.1128/jvi.65.2.938-944.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baines J D, Roizman B. The UL10 gene of herpes simplex virus 1 encodes a novel viral glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J Virol. 1993;67:1441–1452. doi: 10.1128/jvi.67.3.1441-1452.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Britt W J. Neutralizing antibodies detect a disulfide-linked glycoprotein complex within the envelope of human cytomegalovirus. Virology. 1984;135:369–378. doi: 10.1016/0042-6822(84)90193-4. [DOI] [PubMed] [Google Scholar]
- 5.Britt W J, Auger D. Identification of a 65 000 dalton virion envelope protein of human cytomegalovirus. Virus Res. 1985;4:31–36. doi: 10.1016/0168-1702(85)90018-8. [DOI] [PubMed] [Google Scholar]
- 6.Britt W J, Auger D. Synthesis and processing of the envelope gp55–116 complex of human cytomegalovirus. J Virol. 1986;58:185–191. doi: 10.1128/jvi.58.1.185-191.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britt W J, Vugler L, Butfiloski E J, Stephens E B. Cell surface expression of human cytomegalovirus (HCMV) gp55–116 (gB): use of HCMV-recombinant vaccinia virus-infected cells in analysis of the human neutralizing antibody response. J Virol. 1990;64:1079–1085. doi: 10.1128/jvi.64.3.1079-1085.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Britt W J, Vugler L G. Oligomerization of the human cytomegalovirus major envelope glycoprotein complex gB (gp55–116) J Virol. 1992;66:6747–6754. doi: 10.1128/jvi.66.11.6747-6754.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cha T A, Tom E, Kemble G W, Duke G M, Mocarski E S, Spaete R R. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C A, Kouzarides T, Martignetti J A, et al. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 11.Dijkstra J M, Gerdts V, Klupp B G, Mettenleiter T C. Deletion of glycoprotein gM of pseudorabies virus results in attenuation for the natural host. J Gen Virol. 1997;78:2147–2151. doi: 10.1099/0022-1317-78-9-2147. [DOI] [PubMed] [Google Scholar]
- 12.Dijkstra J M, Visser N, Mettenleiter T C, Klupp B G. Identification and characterization of pseudorabies virus glycoprotein gM as a nonessential virion component. J Virol. 1996;70:5684–5688. doi: 10.1128/jvi.70.8.5684-5688.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellinger S, Mach M, Korn K, Jahn G. Cleavage and purification of prokaryotically expressed HIV gag and env fusion proteins for detection of HIV antibodies in the ELISA. Virology. 1991;180:811–813. doi: 10.1016/0042-6822(91)90097-u. [DOI] [PubMed] [Google Scholar]
- 14.Gretch D R, Kari B, Rasmussen L, Gehrz R C, Stinski M F. Identification and characterization of three distinct families of glycoprotein complexes in the envelopes of human cytomegalovirus. J Virol. 1988;62:875–881. doi: 10.1128/jvi.62.3.875-881.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobom U, Brune W, Messerle M, Hahn G, Koszinowski U. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J Virol. 2000;74:7720–7729. doi: 10.1128/jvi.74.17.7720-7729.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber M T, Compton T. Characterization of a novel third member of the human cytomegalovirus glycoprotein H-glycoprotein L complex. J Virol. 1997;71:5391–5398. doi: 10.1128/jvi.71.7.5391-5398.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huber M T, Compton T. The human cytomegalovirus UL74 gene encodes the third component of the glycoprotein H-glycoprotein L-containing envelope complex. J Virol. 1998;72:8191–8197. doi: 10.1128/jvi.72.10.8191-8197.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber M T, Compton T. Intracellular formation and processing of the heterotrimeric gH-gL-gO (gCIII) glycoprotein envelope complex of human cytomegalovirus. J Virol. 1999;73:3886–3892. doi: 10.1128/jvi.73.5.3886-3892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jons A, Dijkstra J M, Mettenleiter T C. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J Virol. 1998;72:550–557. doi: 10.1128/jvi.72.1.550-557.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jons A, Granzow H, Kuchling R, Mettenleiter T C. The UL49.5 gene of pseudorabies virus codes for an O-glycosylated structural protein of the viral envelope. J Virol. 1996;70:1237–1241. doi: 10.1128/jvi.70.2.1237-1241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kari B, Gehrz R. Isolation and characterization of a human cytomegalovirus glycoprotein containing a high content of O-linked oligosaccharides. Arch Virol. 1988;98:171–188. doi: 10.1007/BF01322167. [DOI] [PubMed] [Google Scholar]
- 22.Kari B, Gehrz R. Analysis of human antibody responses to human cytomegalovirus envelope glycoproteins found in two families of disulfide linked glycoprotein complexes designated gC-I and gC-II. Arch Virol. 1990;114:213–228. doi: 10.1007/BF01310750. [DOI] [PubMed] [Google Scholar]
- 23.Kari B, Gehrz R. A human cytomegalovirus glycoprotein complex designated gC-II is a major heparin-binding component of the envelope. J Virol. 1992;66:1761–1764. doi: 10.1128/jvi.66.3.1761-1764.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kari B, Gehrz R. Structure, composition and heparin binding properties of a human cytomegalovirus glycoprotein complex designated gC-II. J Gen Virol. 1993;74:255–264. doi: 10.1099/0022-1317-74-2-255. [DOI] [PubMed] [Google Scholar]
- 25.Kari B, Goertz R, Gehrz R. Characterization of cytomegalovirus glycoproteins in a family of complexes designated gC-II with murine monoclonal antibodies. Arch Virol. 1990;112:55–65. doi: 10.1007/BF01348985. [DOI] [PubMed] [Google Scholar]
- 26.Kari B, Li W, Cooper J, Goertz R, Radeke B. The human cytomegalovirus UL100 gene encodes the gC-II glycoproteins recognized by group 2 monoclonal antibodies. J Gen Virol. 1994;75:3081–3086. doi: 10.1099/0022-1317-75-11-3081. [DOI] [PubMed] [Google Scholar]
- 27.Kari B, Radeke R, Gehrz R. Processing of human cytomegalovirus envelope glycoproteins in and egress of cytomegalovirus from human astrocytoma cells. J Gen Virol. 1992;73:253–260. doi: 10.1099/0022-1317-73-2-253. [DOI] [PubMed] [Google Scholar]
- 28.Klupp B G, Nixdorf R, Mettenleiter T C. Pseudorabies virus glycoprotein M inhibits membrane fusion. J Virol. 2000;74:6760–6768. doi: 10.1128/jvi.74.15.6760-6768.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lake C M, Molesworth S J, Hutt-Fletcher L M. The Epstein-Barr virus (EBV) gN homolog BLRF1 encodes a 15-kilodalton glycoprotein that cannot be authentically processed unless it is coexpressed with the EBV gM homolog BBRF3. J Virol. 1998;72:5559–5564. doi: 10.1128/jvi.72.7.5559-5564.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehner R, Meyer H, Mach M. Identification and characterization of a human cytomegalovirus gene coding for a membrane protein that is conserved among human herpesviruses. J Virol. 1989;63:3792–3800. doi: 10.1128/jvi.63.9.3792-3800.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L, Nelson J A, Britt W J. Glycoprotein H-related complexes of human cytomegalovirus: identification of a third protein in the gCIII complex. J Virol. 1997;71:3090–3097. doi: 10.1128/jvi.71.4.3090-3097.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang X, Chow B, Raggo C, Babiuk L A. Bovine herpesvirus 1 UL49.5 homolog gene encodes a novel viral envelope protein that forms a disulfide-linked complex with a second virion structural protein. J Virol. 1996;70:1448–1454. doi: 10.1128/jvi.70.3.1448-1454.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang X, Tang M, Manns B, Babiuk L A, Zamb T J. Identification and deletion mutagenesis of the bovine herpesvirus 1 dUTPase gene and a gene homologous to herpes simplex virus UL49.5. Virology. 1993;195:42–50. doi: 10.1006/viro.1993.1344. [DOI] [PubMed] [Google Scholar]
- 34.MacLean C A, Robertson L M, Jamieson F E. Characterization of the UL10 gene product of herpes simplex virus type 1 and investigation of its role in vivo. J Gen Virol. 1993;74:975–983. doi: 10.1099/0022-1317-74-6-975. [DOI] [PubMed] [Google Scholar]
- 35.Marshall G S, Rabalais G P, Stout G G, Waldeyer S L. Antibodies to recombinant-derived glycoprotein B after natural human cytomegalovirus infection correlate with neutralizing activity. J Infect Dis. 1992;165:381–384. doi: 10.1093/infdis/165.2.381. [DOI] [PubMed] [Google Scholar]
- 36.Marshall G S, Stout G G, Knights M E, Rabalais G P, Ashley R, Miller H, Rossier E. Ontogeny of glycoprotein gB-specific antibody and neutralizing activity during natural cytomegalovirus infection. J Med Virol. 1994;43:77–83. doi: 10.1002/jmv.1890430115. [DOI] [PubMed] [Google Scholar]
- 37.Masse M J, Jons A, Dijkstra J M, Mettenleiter T C, Flamand A. Glycoproteins gM and gN of pseudorabies virus are dispensable for viral penetration and propagation in the nervous systems of adult mice. J Virol. 1999;73:10503–10507. doi: 10.1128/jvi.73.12.10503-10507.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osterrieder N, Neubauer A, Brandmuller C, Braun B, Kaaden O R, Baines J D. The equine herpesvirus 1 glycoprotein gp21/22a, the herpes simplex virus type 1 gM homolog, is involved in virus penetration and cell-to-cell spread of virions. J Virol. 1996;70:4110–4115. doi: 10.1128/jvi.70.6.4110-4115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osterrieder N, Neubauer A, Fakler B, Brandmuller C, Seyboldt C, Kaaden O R, Baines J D. Synthesis and processing of the equine herpesvirus 1 glycoprotein M. Virology. 1997;232:230–239. doi: 10.1006/viro.1997.8561. [DOI] [PubMed] [Google Scholar]
- 40.Pilling A, Davison A J, Telford E A, Meredith D M. The equine herpesvirus type 1 glycoprotein homologous to herpes simplex virus type 1 glycoprotein M is a major constituent of the virus particle. J Gen Virol. 1994;75:439–442. doi: 10.1099/0022-1317-75-2-439. [DOI] [PubMed] [Google Scholar]
- 41.Ross J, Williams M, Cohen J I. Disruption of the varicella-zoster virus dUTPase and the adjacent ORF9A gene results in impaired growth and reduced syncytia formation in vitro. Virology. 1997;234:186–195. doi: 10.1006/viro.1997.8652. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez V, Greis K D, Sztul E, Britt W J. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J Virol. 2000;74:975–986. doi: 10.1128/jvi.74.2.975-986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schoppel K, Hassfurther E, Britt W J, Ohlin M, Borrebaeck C A, Mach M. Antibodies specific for the antigenic domain 1 (AD-1) of glycoprotein B (gpUL55) of human cytomegalovirus bind to different substructures. Virology. 1996;216:133–145. doi: 10.1006/viro.1996.0040. [DOI] [PubMed] [Google Scholar]
- 44.Talbot P, Almeida J D. Human cytomegalovirus: purification of enveloped virions and dense bodies. J Gen Virol. 1977;36:345–349. doi: 10.1099/0022-1317-36-2-345. [DOI] [PubMed] [Google Scholar]
- 45.Urban M, Klein M, Britt W J, Hassfurther E, Mach M. Glycoprotein H of human cytomegalovirus is a major antigen for the neutralizing humoral immune response. J Gen Virol. 1996;77:1537–1547. doi: 10.1099/0022-1317-77-7-1537. [DOI] [PubMed] [Google Scholar]
- 46.Wu S X, Zhu X P, Letchworth G J. Bovine herpesvirus 1 glycoprotein M forms a disulfide-linked heterodimer with the UL49.5 protein. J Virol. 1998;72:3029–3036. doi: 10.1128/jvi.72.4.3029-3036.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]