Abstract
Purpose:
Minimal residual disease (MRD) detection can identify the recurrence in patients with colorectal cancer (CRC) following definitive treatment. We evaluated a plasma-only MRD assay to predict recurrence and survival in patients with metastatic CRC who underwent curative intent procedures (surgery and/or radiotherapy), with or without (neo)adjuvant chemotherapy. The primary objective of this study was to assess the correlation of postprocedure tumor cell–free DNA detection status with radiographic disease recurrence.
Experimental Design:
Preprocedure and postprocedure longitudinal samples were collected from 53 patients and analyzed with a multiomic MRD assay detecting circulating tumor DNA (ctDNA) from genomic and epigenomic signals. Preprocedure and postprocedure ctDNA detection correlated with recurrence-free and overall survival (OS).
Results:
From 52 patients, 230/233 samples were successfully analyzed. At the time of data cutoff, 36 (69.2%) patients recurred with median follow-up of 31 months. Detectable ctDNA was observed in 19/42 patients (45.2%) with ctDNA analyzed 3 weeks postprocedure. ctDNA detection 3 weeks postprocedure was associated with shorter median recurrence-free survival (RFS; HR, 5.27; 95% CI, 2.31–12.0; P < 0.0001) and OS (HR, 12.83; 95% CI, 3.6–45.9; P < 0.0001). Preprocedure ctDNA detection status was not associated with RFS but was associated with improved OS (HR, 4.65; 95% CI, 1.4–15.2; P = 0.0111). Undetectable ctDNA preprocedure had notable long-term OS, >90% 3 years postprocedure.
Conclusions:
In this cohort of oligometastatic CRC, detection of ctDNA preprocedure or postprocedure was associated with inferior outcomes even after accounting for known prognostic clinicopathologic variables. This suggests ctDNA may enhance current risk stratification methods helping the evaluation of novel treatments and surveillance strategies toward improving patient outcomes.
Translational Relevance
Oligometastatic colorectal cancer (CRC) is potentially curable, yet patient outcomes are suboptimal with a high rate of recurrence. Commonly used biomarkers, such as carcinoembryonic antigen, are not sufficient to accurately predict recurrence prior to radiographic detection. Early identification of minimal residual disease (MRD) through circulating tumor DNA (ctDNA) analysis might afford better risk stratification to identify patients who may or may not benefit from additional treatment. Here, we evaluate a plasma-only ctDNA-based MRD assay as a prognostic tool by analyzing precurative and postcurative intent procedure samples collected at structured time points. This assay demonstrated a high positive predictive value for recurrence with >6 months lead time when compared with the standard radiographic imaging. Postprocedure ctDNA-negative patients who developed recurrence demonstrated a longer overall survival than ctDNA-positive patients. In a multivariable analysis, ctDNA was the only clinical variable evaluated to demonstrate statistical association with recurrence. These data support the feasibility and potential clinical utility of ctDNA assays to predict patient outcomes following curative intent therapy in oligometastatic CRC.
Introduction
Colorectal cancer (CRC) is the third most common cancer and the second leading cause of cancer death in the world (1). With the advancement of surgical and radiotherapy techniques, approximately 20% of patients with recurrent or metastatic CRC (mCRC) undergo procedures with curative intent, e.g., definitive surgery and/or radiotherapy, aimed to render the patient with no evidence of disease radiographically (2). Approximately 30% of such patients undergoing curative intent procedures will be alive for 10 years postprocedure. The recurrence rate in this population is ∼70% (3), and it is not clear that adjuvant chemotherapy (ACT) improves outcomes after definitive surgery and/or radiotherapy (4–7).
Perioperative chemotherapy may be offered to patients undergoing lung or liver resection to treat potential micrometastatic disease beyond the resected metastasis (7). A meta-analysis found that perioperative chemotherapy improved disease-free survival (DFS), but not overall survival (OS) in 1,896 patients undergoing liver resection (8). Surveillance is recommended following a curative intent strategy with the goal of detecting recurrence early enough to offer additional curative intent therapy (9, 10). Despite this, the existing tools to predict recurrence risk are imprecise. Carcinoembryonic antigen (CEA) is a widely used serum biomarker to assess individual risk (11, 12). However, not all CRC cases produce CEA (13, 14), and CEA elevation is not sensitive or specific for the presence of micrometastatic disease (15). A more precise biomarker is needed to detect occult mCRC prior to clinical recurrence, as early detection and intervention may offer patients an opportunity for cure.
Circulating tumor DNA (ctDNA) analysis is an established tool for advanced-stage cancer to identify genomic drivers and resistance alterations for therapeutic decision-making without the need for tumor tissue (16–18). More recently, newer methods of ctDNA analysis have been developed for use in early-stage CRC to detect evidence of residual disease after curative intent treatment and to predict recurrence (19–26). Multiple studies have shown that detection of ctDNA following definitive treatment is prognostic and may provide a tool to identify patients at high-risk for recurrence with better sensitivity, specificity, and lead time than current surveillance modalities (27–31). Most of these studies utilized assays that require a priori analysis of tumor tissue to inform ctDNA detection in plasma. The need for prior tissue analysis may be logistically challenging in oligometastatic CRC, particularly among patients undergoing nonsurgical procedures or for those receiving neoadjuvant treatment.
The purpose of the present study was to evaluate a plasma-only ctDNA assay incorporating both genomic and epigenomic signatures to determine the ctDNA detection rate after a curative intent procedure in patients with mCRC and associated with longitudinal postprocedure ctDNA detection with radiographic disease recurrence.
Materials and Methods
Study population
Adult patients with mCRC planning to undergo curative intent surgery and/or radiotherapy were prospectively recruited at the University of California, San Francisco (UCSF) and Massachusetts General Hospital from February 2017 until May 2018. Patients were eligible if they had radiographic evidence of metastatic disease confined to the liver, lungs, and/or ovaries. Patients with a known second primary cancer in addition to CRC were excluded. This study retrospectively analyzed ctDNA. Physicians were not informed of the ctDNA results. The study protocol was approved by the respective Institutional Review Boards and was conducted in accordance with the Declaration of Helsinki. All patients provided written informed consent.
Patients received standard-of-care (SOC) treatment as determined by the treating multidisciplinary oncology teams. Protocol-mandated surveillance included cross-sectional imaging of the chest, abdomen, and pelvis beginning at 10 weeks postprocedure. This was repeated every 12 weeks (±4 weeks) in years 1 to 2 and every 24 weeks in years 3 to 5. CEA was assessed at the same intervals if elevated preprocedure. Elevated CEA was defined as >5 ng/mL based on a reference range [Abbott ARCHITECT i1000sr System (RRID:SCR_019328)]. Clinicopathological, treatment, and clinical outcome data were abstracted from the electronic health record.
Sample collection and ctDNA testing methodology
Peripheral blood was collected in three 10-mL Streck tubes preprocedure, approximately 3 and 10 weeks postprocedure, and then longitudinally every 12 to 24 weeks for up to 5 years (Supplementary Fig. S1). Research blood draws were intended to occur in conjunction with SOC follow-up visits, with concurrent CEA testing when indicated. The “primary” collection time point was drawn approximately 3 weeks postprocedure. Patients were included in the longitudinal analysis if they had two or more postprocedure samples. In this analysis, patients were categorized as ctDNA positive if they had one or more ctDNA positive postprocedure results and ctDNA negative if all postprocedure ctDNA results were negative.
De-identified samples were shipped to a single site (Guardant Health) for analysis. As per methods described previously, cell-free DNA (cfDNA) was extracted from plasma using the Qiagen circulating nucleic acid kit (32). Extracted cfDNA was analyzed with the Guardant Reveal assay (ref. 21; version L1.2) according to previously described methods (21). Briefly, cfDNA fragments were partitioned based on extent of methylation, enriched using a ∼500 kb panel that targets informative genomic and epigenomic regions, barcoded, and pooled for sequencing. Raw data from sequencing data files were analyzed with a proprietary bioinformatic variant classifier to identify common clonal driver mutations (SNVs and Indels) as well as differential methylation patterns consistent with CRC. The assay returned a binary result of “ctDNA detected” or “ctDNA not detected” based on predefined thresholds. The classifier is designed to exclude nontumor-derived variants (e.g., clonal hematopoiesis) without the need for additional sequencing of tissue samples or peripheral blood cells. For this study, ctDNA analysis was performed retrospectively and blinded to the clinical outcome data. Neither the treating physicians nor patients were informed of the results of the ctDNA analysis.
Statistical analysis
The primary objective of this study was to assess the 3-week postprocedure ctDNA detection rate in patients with oligometastatic CRC being treated with curative intent surgery and/or radiotherapy. Secondary objectives were to evaluate preprocedure and postprocedure ctDNA detection status and CEA elevation status with recurrence, including sensitivity, specificity, positive and negative predictive value, recurrence-free survival (RFS), and OS. RFS was measured from the day of the curative intent procedure to the first radiographic recurrence or death from CRC. Patients were censored at the date of last follow-up or non–CRC-related death. OS was measured from the day of the curative intent procedure to death from any cause; patients were censored at the date of last follow-up. The data cutoff date was January 15, 2022. The Kaplan–Meier method was used to estimate RFS and OS outcomes.
A multivariable analysis was performed with a Cox regression analysis to analyze the association of ctDNA detection and other clinicopathologic prognostic factors with RFS in patients available for analysis at a 3-week postprocedure (“primary”) time point. Clinicopathologic variables included gender, location of the primary tumor (ascending or descending colon), administration of predefinitive or postdefinitive chemotherapy, metastatic lesion size (measured radiographically preprocedure as the sum of the tumor diameter of all target lesions), and age. CEA elevation could not be evaluated in the multivariable analysis due to the high rate of missing CEA data as, per protocol, CEA was not routinely collected if it was not elevated at diagnosis—applicable to 30/52 (57.7%) patients included in this study. Thus, to supplement the multivariable analysis, a univariate Cox proportional hazards model was conducted to evaluate the relationship between RFS and CEA elevation status at 3 weeks postprocedure in patients with CEA available. A second univariate analysis was conducted to explore associations between preprocedure ctDNA detection and the following clinical variables: lesion size (maximum diameter), time between end of neoadjuvant chemotherapy treatment and curative intent procedure, metastatic disease site, preprocedure CEA, and Fong Clinical Risk Score (14) by chi-square test (for categorical variables) or Wilcoxon rank-sum test (for continuous variables). Statistical analyses were performed using R (version 4.0.5), Graphpad PRISM (version 9.0 for Windows, GraphPad Software), and SAS software package 9.4 (SAS Institute). Statistical significance was declared as P value <0.05 and no multiple testing adjustments were performed.
Data availability
All relevant data are provided within the article and the accompanying Supplementary Data. Because of Health Insurance Portability and Accountability Act (HIPAA) requirements, we are not consented to share individualized patient data, which contains potentially identifying or sensitive patient information. We are committed to collaborative data analysis, and more information and mechanisms for data access can be obtained by contacting the corresponding authors.
Results
This study enrolled 53 microsatellite stable patients between February 2017 and May 2018. Demographics and baseline clinicopathologic features are available in Table 1. Patients received surgical resection (88.7%), radiotherapy (9.4%), or both (1.9%) as their curative intent therapy. One patient was excluded after the procedure was deemed to be noncurative due to the presence of unresectable disease. Of the remaining 52 patients, 84.6% received neoadjuvant chemotherapy and 32.7% of patients received both neoadjuvant and adjuvant chemotherapy. No patients received only ACT. The median follow-up was 36.1 months (range 4.9–59.6 months). This was based on 51 patients as one patient was lost at follow up. At the time of data cut-off, recurrences were detected in 69.2% of patients (36/52) and 25% (13/52) were deceased.
Table 1.
Demographics, clinicopathologic, and procedure-related features of study participants.
| Clinicopathologic and procedure-related variables (N = 53) | |||||
|---|---|---|---|---|---|
| Age | Months between diagnosis and procedure, mean (SD) | 20.5 | 19.5 | ||
| Age at diagnosis, median (range) | 56 | (39–75) | Procedure type, n (%) | ||
| Gender, n (%) | Surgery | 47 | 89% | ||
| Female | 31 | 58% | Radiation | 5 | 9% |
| Male | 22 | 42% | Surgery and radiation | 1 | 2% |
| Race, n (%) | Curative intent procedure, n (%) | ||||
| White | 38 | 72% | Liver | 31 | 58% |
| Asian | 7 | 13% | Liver + colon | 13 | 25% |
| Other | 8 | 15% | Lung | 8 | 15% |
| Ethnicity, n (%) | Ovaries + colon | 1 | 2% | ||
| Hispanic/Latino | 8 | 15% | Prior treatment % | ||
| Other | 45 | 85% | Chemotherapy | 96% | |
| Primary site, n (%) | Relevant genomics, n (total n reported) | ||||
| Ascending colon | 16 | 30% | RAS mutated | 21 | 42 |
| Transverse colon | 2 | 4% | BRAF V600E | 0 | 28 |
| Descending colon | 22 | 41% | MSI-H detected | 0 | 48 |
| Rectum | 13 | 25% | Elevated CEA at baselinea | ||
| Differentiation, n (%) | N elevated (%) | 22 | 42% | ||
| Well/moderately | 48 | 91% | Median (ng/mL) | 9.6 | |
| Poor/undifferentiated | 4 | 7% | Range (ng/m L) | (5.2–531.1) | |
| Unknown | 1 | 2% | SD (ng/m L) | 110.7 | |
| M staging at diagnosis, n (%) | |||||
| MO (metachronous metastases) | 19 | 36% | |||
| M1 (synchronous metastases) | 34 | 64% | |||
CEA was not consistently collected in 58% of the patients (30/52) because it was not elevated at baseline.
Abbreviation: MSI‐H, microsatellite instability.
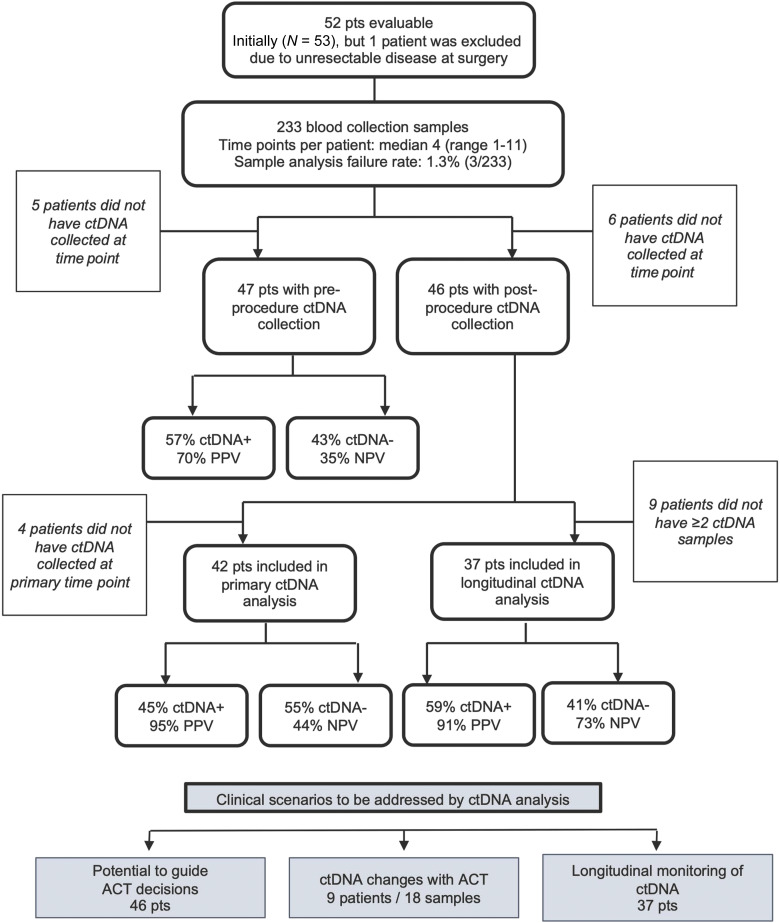
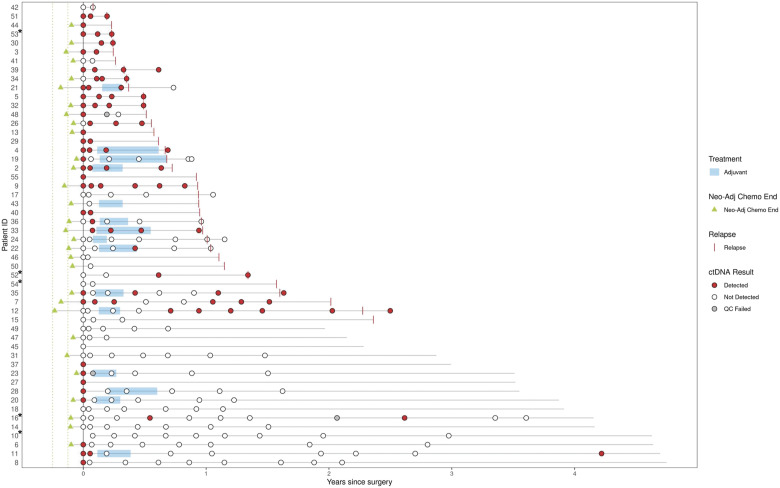
In total, 233 samples from the 52 eligible patients were collected and analyzed, including 47 preprocedure, 42 primary postprocedure (i.e., 3 weeks postprocedure), and 143 samples collected after the 3-week postprocedure time point (Fig. 1). Three samples (1.3%; including one primary and two longitudinal) failed ctDNA analysis. Fourteen patients had chemotherapy following their 3-week time point collection. Patient de-identified record IDs are listed alongside the respective analyses in Supplementary Table S1. A swimmer plot outlining the treatment course and ctDNA results are listed by patient ID in Fig. 2.
Figure 1.
CONSORT flow diagram. Diagram depicts patient enrollment, exclusions, the number of patients included in the primary and longitudinal analyses, and the number of ctDNA-positive vs. ctDNA-negative results. NPV, negative predictive value; PPV, positive predictive value.
Figure 2.
Swimmer plot describing the treatment course and ctDNA results by patient ID (N = 52). Time at 0 year denotes the time of curative intent procedure. Neoadjuvant course only shown for patients with treatment within 3 months of curative intent procedure. Dotted lines around neoadjuvant data points represent −1.5 and −3 months before procedure. Record IDs with an asterisk (*) indicate patients who received radiation as their curative intent procure. All other patients received curative intent surgery.
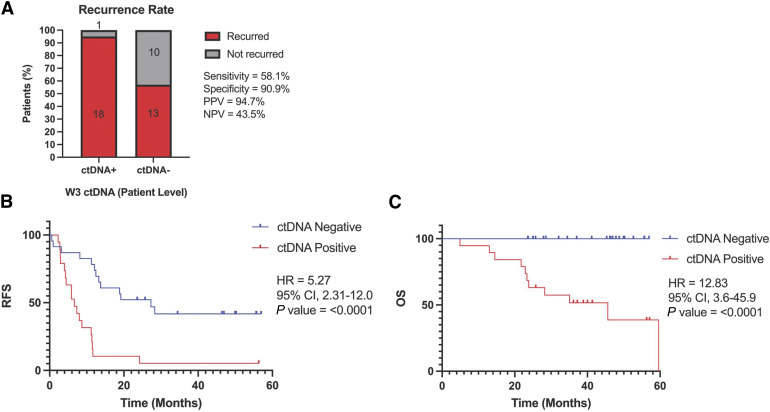
Primary postprocedure ctDNA status predicts RFS and OS
A total of 19 patients had ctDNA detected at the primary time point, of whom 18 subsequently developed recurrent disease (PPV 94.7%; Fig. 3A). In contrast, 13/23 patients with ctDNA not detected recurred (NPV 10/23; 43.5%). ctDNA detection was associated with significantly shorter RFS (median 6.6 vs. 27.3 months; P < 0.0001) and OS (median 45.6 months vs. not reached; P < 0.0001) compared to ctDNA-negative patients (Fig. 3B and C). The median follow up time for ctDNA-negative patients was 45.3 months (range 23.6–56.9 months).
Figure 3.
Primary postprocedure ctDNA status predicts RFS and OS. A, Recurrence rate stratified by ctDNA status. Ninety-five percent CI as follows: sensitivity (40.8%–73.6%), specificity (62.3%–99.5%), PPV (75.4%–99.7%), and NPV (25.6%–63.2%). Kaplan–Meier curves at the primary time point stratified by ctDNA status (n = 42) for (B) RFS and (C) OS. CI, confidence interval.
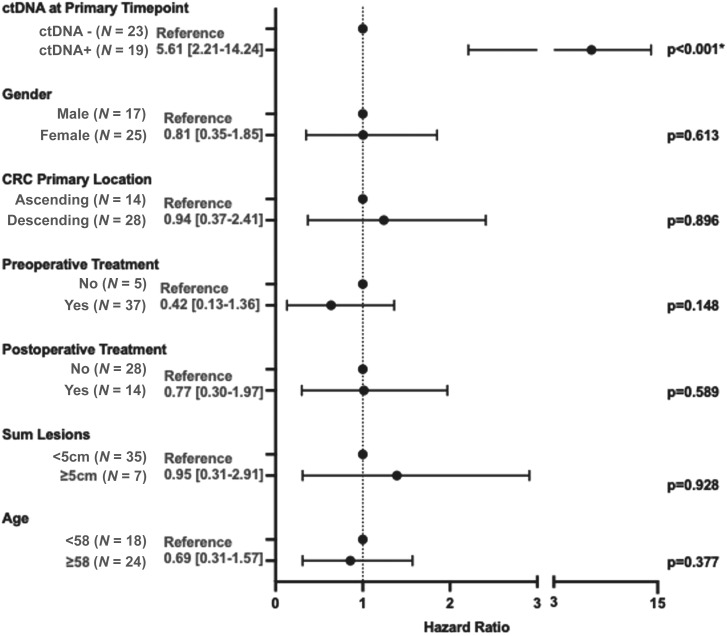
The sensitivity of ctDNA analysis at the primary time point was 58.1% (18/31) for any future recurrence (Fig. 3A) and 77.3% (17/22) for recurrences that happened within 1 year of the procedure. The specificity of ctDNA analysis at the primary time point was 90.9% (10/11). Patient ID 11 had ctDNA detected at the primary time point and did not recur. They appear to have cleared ctDNA with subsequent postprocedure chemotherapy (Fig. 2). Interestingly, all the 23 patients with undetectable ctDNA at the primary time point were still alive at the end of the study period after a median follow-up of 45.3 months, including all 13 who developed recurrent disease (median follow-up of 37.1 months; Fig. 3C). In contrast, only 42.1% (8/19) of ctDNA-positive patients were still alive at the end of the study period (median follow-up of 35.1 months). To evaluate the association of ctDNA and other clinicopathological factors with RFS, a multivariable analysis was conducted. ctDNA status at the primary time point was the only clinicopathological factor evaluated that was significantly associated with RFS (P < 0.001; Fig. 4). The demographics, clinicopathologic features, and procedure-related features of study participants in this primary time point analysis are outlined in Supplementary Table S2.
Figure 4.
Multivariable analysis of risk factors associated with RFS. ctDNA positivity is defined as ctDNA positive at the primary time point. In this study, “preoperative therapy” refers to chemotherapy within 3 months of procedure. Primary tumor types were binned into ascending (ascending or transverse tumor) or descending (descending, sigmoid, or rectal tumor).
Longitudinal ctDNA analysis improves sensitivity to detect relapse
Thirty-seven patients had at least two postprocedure samples (including the 3-week time point where available) and were included in the longitudinal analysis, 24 of whom had recurrence. The longitudinal analysis evaluated 174 total samples (median of 4 per patient, range 2–9), 30 (17.2%) of which were collected prior to ACT completion, and 144 of which were collected following all treatment. Among the 37 patients included in the longitudinal analysis, 16 were positive at the 3-week primary time point and 6 became positive at a later time point at a median of 7.0 months (range 5.1–13.9 months).
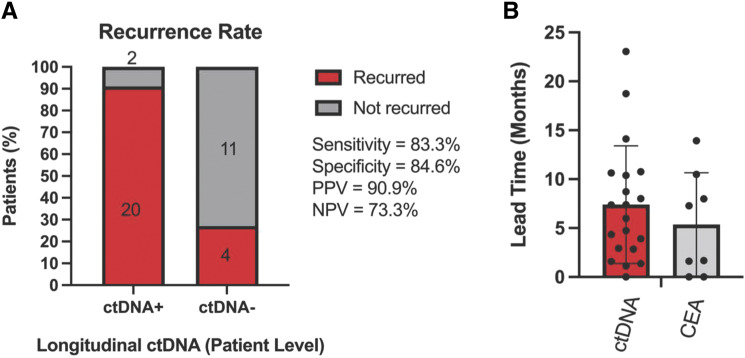
Sensitivity of longitudinal ctDNA analysis was 83.3% (20/24) with a median lead time of 6.7 months (mean of 7.4 months; range of 0.0–23.0 months; Fig. 5A and B). All 4 patients with ctDNA-negative results and recurrence were still alive at the study cut-off with a median follow-up of 46.6 months. Patient IDs 11 and 16 were ctDNA positive in the longitudinal analysis and did not recur, one of whom had only 5.8 months of follow-up from their last positive draw (specificity 84.6%, 11/13; Fig. 2). The longitudinal samples with CEA evaluated had a median lead time of 4.5 months (mean of 5.4 months; range of 0.0–13.9 months; N = 8), which was not statistically significant compared to ctDNA (unpaired t test; P = 0.0671; Fig. 5B). We also evaluated the specificity among samples collected after all treatment to remove the confounding effect of subsequent chemotherapy use and the possibility of ctDNA clearance. The post-treatment sample-level specificity was 95.7% (67/70, 95% CI, 88.1%–98.8%). There were 28 patients with a reported site of recurrence in this cohort. Recurrence in the peritoneal cavity, pelvic implants, or multiple sites was detected by ctDNA for all patients. Recurrence in the lymph node, liver, or lung was detected by ctDNA for 67% to 71% of patients.
Figure 5.
Longitudinal ctDNA analysis improves sensitivity to detect relapse. A, Patient-level ctDNA status 95% CI as follows: sensitivity (64.2%–93.3%), specificity (57.8%–97.3%), PPV (72.2%–98.4%), and NPV (48.1%–89.1%). B, Lead time comparison of ctDNA vs. CEA as measured by disease recurrence measured by time to relapse (months; n = 20; P < 0.0001).
Preprocedure ctDNA detection is associated with overall survival
Forty-seven patients had an available preprocedure ctDNA sample, i.e., a blood sample taken before curative intent therapy. Of these patients, 55% (26/47) received chemotherapy 3 months prior to their curative intent procedure and prior to sample collection completed a median of 37 days prior to curative intent procedure (range 20–1,986 days). ctDNA was detected in 57.4% (27/47) of patients prior to procedure. Among the subset of patients who received neoadjuvant chemotherapy within 3 months prior to the preprocedure sample collection (N = 26), the median time between the last dose of chemotherapy and sample collection was 5.2 weeks.
Preprocedure ctDNA status was not statistically associated with RFS (P = 0.3418); however, patients with undetectable ctDNA demonstrated significantly improved OS (median 59.6 months vs. not reached; P = 0.0111; Supplementary Fig. S2A and S2B). One patient was lost at follow-up and thus was included in the RFS but not the OS calculation. In univariate analyses, ctDNA positivity preprocedure was associated with sum total lesions (P = 0.007) but not with other clinicopathological features including preprocedure CEA (P = 0.092; Supplementary Table S3). The 3-year OS rate was 60.9% for patients with ctDNA detected preprocedure vs. 90.9% for patients with undetectable ctDNA preprocedure.
ctDNA dynamics associated with treatment
We analyzed 37 patients who had paired preprocedure and postprocedure ctDNA samples and 13 patients who had paired preadjuvant and postadjuvant chemotherapy samples to assess ctDNA dynamics (Supplementary Fig. S3A and S3B). Patients who cleared ctDNA with surgery and/or radiation and local therapy (3/19) trended toward a longer RFS than those who remained positive; however, this difference was not statistically significant (not reached for ctDNA+/ctDNA− vs. 7.7 months for ctDNA+/ctDNA+; P = 0.1069; Supplementary Fig. S3C). There were 4/13 patients with ctDNA detected preadjuvant chemotherapy. One patient cleared ctDNA postadjuvant chemotherapy and had a numerically longer RFS compared to patients without clearance (19.2 months vs. a median 8.7 months; Supplementary Fig. S3C).
CEA
In this population, 42% (22/52) of patients had CEA elevated prior to curative intent procedure. and there were 21 patients with available CEA at the 3-week postprocedure time point. CEA elevation was not associated with RFS at the postprocedure time point (P = 0.1140) but did associate with OS (P = 0.0064; Supplementary Fig. S4A–S4C). Among 32 patients with CEA evaluated in the longitudinal setting, sensitivity and specificity for recurrence were 13/17 (76.5%) and 4/15 (26.7%), respectively (Supplementary Fig. S4D). The PPV and NPV for recurrence were 13/24 (54.2%) and 4/8 (50.0%), respectively. The median lead time of all CEA elevated samples with recurrence was 1.6 months.
Discussion
Despite treatment with curative intent, a majority of patients with oligometastatic CRC will have recurrence and ultimately die of their disease. The selection of patients who derive greatest benefit from chemotherapy in addition to local procedures such as surgical resection and/or radiotherapy is challenging given that there is theoretical ability of chemotherapy to eradicate minimal residual disease (MRD) but with limited data available and no clear improvement in OS proven to date.
In this study, we found that ctDNA detection at 3 weeks postsurgery and/or radiotherapy was a significant predictor of RFS and OS. In contrast, the elevation of the commonly used tumor biomarker CEA (using a cutoff of 5 ng/mL) was predictive of OS but not RFS and was only available for 21 patients. Patients with detectable ctDNA had nearly five-fold greater risk of recurrence (approaching 100% in the absence of additional therapy) and more than 12-fold greater risk of death. Interestingly, while the sensitivity of the ctDNA assay at this single primary time point was modest (58.1%), patients with undetectable ctDNA experienced a longer median time to recurrence and all were still alive after more than 23 months of follow-up. Patients with undetectable ctDNA may have a lower volume of micrometastases compared to patients with detectable ctDNA which could take longer to become observable radiographically and progress to lethal cancer. It is also possible that patients with more indolent tumors have low ctDNA shedding which could also explain the more favorable outcomes in patients with undetectable ctDNA. We also note some differences in ctDNA detection by site of recurrence. However, there were too few extra-hepatic recurrences to draw conclusions based on site. We observed that for 4 patients in the longitudinal cohort who showed recurrences, three of the four recurrences were in the lungs, consistent with previous data but noting the small number of lung recurrences in this cohort. The observation that ctDNA status predicted OS among preprocedure samples (when disease is radiographically evident) lends support to this theory. However, given the high degree of heterogeneity in the treatment of patients, patterns of relapse, and relatively small sample size, further studies are necessary. Patients with more indolent CRC could theoretically have a lower benefit of chemotherapy compared to patients with more aggressive tumors. There was not enough data in this study to evaluate the efficacy of chemotherapy based on ctDNA detection status, and further studies to develop approaches that identify patients likely to benefit from both chemotherapy and curative intent procedures would be valuable. Toward this, the ongoing COSMOS-CRC-03 (NCT2072220055), OPTIMISE (NCT04680260), NCT05815082, and NCT05797077 trials will be valuable for understanding the role of ctDNA in predicting chemotherapy benefit in patients with oligometastatic CRC.
We utilized a plasma-only ctDNA assay to detect MRD. To date, most ctDNA-based MRD assays utilize tumor tissue to either design or inform the plasma-based assay. Tissue-based assays may have logistical challenges for patients with oligometastatic CRC in that tumor tissue may not be readily available for all patients, particularly those undergoing nonsurgical localized therapy approaches. In addition, there may be time delays and resourcing issues associated with acquiring and testing new tissue or retrieving archival tissue. While cross-study comparisons are challenging due to differences in study design, patient population, patterns of treatment and recurrence, and cohort maturity, the findings in this study are similar to other studies that have investigated the use of ctDNA for recurrence detection in patients with oligometastatic CRC undergoing locoregional treatment strategies. This study, like others, found a high risk for recurrence in patients with ctDNA detected following treatment. In a recent meta-analysis, the majority of studies evaluating post-treatment ctDNA detection found a statistically significant reduction in RFS among patients with detectable ctDNA, despite a large degree of interstudy heterogeneity (33). Hazard ratios ranged from 2.7 to 7.6 with a pooled RFS of 4.5 (95% CI, 3.4–5.1; n = 569). The two studies evaluating OS had striking similarity to our study, with excellent OS (>80% at 5 years) reported in patients with undetectable ctDNA postintervention (28,29). Pretreatment ctDNA detection was associated with inferior RFS and OS in most but not all studies.
Additionally, our study confirms findings from other studies that sensitivity for recurrence improves with longitudinal sampling. The timing of recurrence is variable due to heterogeneity in disease biology between patients (i.e., tumor growth rate and starting volume of residual disease post-treatment), and it may not be realistic for a single sample to identify all relapses, particularly those that occur more than a year from the time of sample collection. Even assays with extremely high analytic sensitivity will face limitations in clinical sensitivity. When ctDNA concentration is low, the likelihood of sampling tumor DNA in a clinically reasonable volume of blood is reduced. The anatomic site, size, and biology of clinically detected relapses may also impact ctDNA detection. Future interventional strategies may need to involve serial collection of ctDNA to maximize detection of high-risk patients. Postintervention strategies could involve giving adjuvant therapy only if and when ctDNA becomes detected (de-escalation strategy) and/or more frequent or intensive radiographic imaging if ctDNA is detected with the goal of detecting recurrence when locoregional techniques may still be curative. The optimal frequency and duration of longitudinal testing in patients with oligometastatic CRC is not yet established. The average lead time from ctDNA detection to radiographic progression/recurrence was 7.4 months which suggests testing every 6 months may be reasonable, or perhaps, more frequently in the first 1 to 2 years postprocedure when the risk of recurrence is highest (e.g., every 3 months). Frequent longitudinal sampling may reduce the risk of false negatives due to sampling error.
We also had 2 patients that had longitudinal samples that were initially positive turn to negative without intervention and who remained recurrence free. While it is difficult to conclusively explain these findings, we hypothesize the following points. (i) The assay is detecting a coexistent disease that is associated with differential methylation patterns and that overlaps with that of CRC. This would be an analytical true positive, but a clinical false positive for the intended use of recurrence detection. A published example of this involved a patient with hepatitis C who had ctDNA detected without recurrence (34). (ii) These are true positive samples in patients who did not have recurrence within the study follow-up period. This has the potential for stochastic sampling error leading to subsequent false negatives. (iii) This is an analytical false positive, i.e., a signal incorrectly identified as tumor derived by the assay algorithm. (iv) There was spontaneous clearance that occurred, as has been previously described. To the second point, in our longitudinal analysis, we demonstrated ctDNA lead times up to 23 months; patients with follow-up durations shorter than 23 months from the time of the sample collection are difficult to confidently assign as “recurrence-free.”
Assessment of ctDNA outperformed CEA, the standard-of-care biomarker for monitoring CRC. While easy to measure, CEA has limited sensitivity and is only reliable in the subset of patients who have initial CEA elevation. Indeed, in this study, CEA could not be evaluated in the multivariable analysis because 11 patients were not evaluable for CEA. Additionally, in evaluable patients, CEA level did not demonstrate a significant difference in RFS at the primary time point.
There were several limitations to this study. First, this was a modest cohort. More validation is needed with a larger cohort to demonstrate the efficacy of this assay clinically. Second, longitudinal samples were taken at providers’ discretion and were not taken at predefined cadence. Further studies with predefined sample time point numbers and timing would provide more information about longitudinal monitoring.
Conclusion
Despite the high degree of heterogeneity in patient disease burden and treatment patterns, our findings with a tissue-free assay are consistent with other studies demonstrating that post-treatment ctDNA detection is prognostic of both RFS and OS in patients with oligometastatic CRC, and pretreatment ctDNA detection status may also provide prognostic value. Future studies are necessary to understand the clinical utility of ctDNA testing in the oligometastatic population to optimize therapeutic strategies to improve outcomes while reducing toxicity.
Supplementary Material
Supplementary Figure S1. Schema of ctDNA sampling.
Supplementary Figure S2. Kaplan-Meier curves for patients with pre-procedure ctDNA stratified by ctDNA status.
Supplementary Figure S3. ctDNA dynamic changes for patients pre-and post-curative intent therapy (CI) (left) or adjuvant chemotherapy (ACT) (right).
Supplementary Figure S4. Survival outcomes and recurrence rates stratified by CEA status.
Supplementary Table S1. Patient deidentified record ID by analysis. Description of the patient record IDs used per analysis to be cross-referenced with the swimmer plot in Figure 2.
Supplementary Table S2. Demographics, clinicopathologic and procedure-related features table. Table depicts study participants in primary timepoint analysis.
Supplementary Table S3. Univariate analysis by pre-operative ctDNA status. Depicted are multiple clinicopathologic variables analyzed by ctDNA status.
Acknowledgments
We would like to thank Brandon Shih, Marin Pollak, Paige Steiding, Islam Baiev, Daniel Dix, Justin I. Odegaard, and Mingyang Cai for their assistance on this manuscript. Guardant Health provided funding for this study.
A.R. Parikh is also supported by a NIH K08 CA 273688.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors’ Disclosures
A.R. Parikh reports grants from Guardant during the conduct of the study, as well as equity in C2i Genomics, XGenomes, Cadex, Vionix, and Parithera. A.R. Parikh also reports being an advisor/consultant for Eli Lilly, Mirati, Pfizer, Inivata, Biofidelity, Checkmate Pharmaceuticals, FMI, Guardant, AbbVie, Bayer, Delcath, Taiho, CVS, Value Analytics Lab, Seagen, Saga, AstraZeneca, Scare Inc., Illumina, Taiho, Hookipa, Kahar Medical, Xilio Therapeutics, Sirtex, Takeda, and Science For America; fees from UpToDate; travel fees from Karkinos Healthcare; being on the DSMC for a Roche study and on the Steering Committee for Exilixis; and research funding to institution from PureTech, PMV Pharmaceuticals, Plexxicon, Takeda, BMS, Mirati, Novartis, Erasca, Genentech, Daiichi Sankyo, Syndax, Revolution Medicine, and Parthenon. J. Tsai reports other support from Guardant Health outside the submitted work, and is a shareholder with Guardant Health. T.A. Rich reports employment and stock holdings of Guardant Health. K.S. Price reports other support from Guardant Health during the conduct of the study, as well as other support from Guardant Health outside the submitted work. L. Zhang reports personal fees from Samay Inc. and Smith-Kettlewell Eye Research Institute outside the submitted work. E.E. Van Seventer reports other support from Blueprint Medicines outside the submitted work. V.M. Raymond reports other support from Guardant Health during the conduct of the study, as well as other support from Guardant Health outside the submitted work. R.B. Corcoran reports personal fees from Guardant and Natera, as well as personal fees and other support from nRichDx, Alterome Therapeutics, and Sidewinder Therapeutics outside the submitted work. K. Van Loon reports other support from Natera outside the submitted work. C.E. Atreya reports other support from Guardant Health during the conduct of the study, as well as other support from Inivata, Roche/GNE, Pfizer, Novartis, Merck, Bristol-Myers Squibb, Erasca, and Gossamer Bio outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
A.R. Parikh: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, methodology, writing–original draft, project administration, writing–review and editing. B.H. Chee: Resources, data curation, formal analysis, writing–review and editing. J. Tsai: Formal analysis, writing–original draft, project administration, writing–review and editing. T.A. Rich: Writing–original draft, writing–review and editing. K.S. Price: Formal analysis, writing–original draft. S.A. Patel: Formal analysis. L. Zhang: Formal analysis. F. Ibrahim: Data curation. M. Esquivel: Data curation. E.E. Van Seventer: Data curation. J.X. Jarnagin: Data curation. V.M. Raymond: Writing–review and editing. C.U. Corvera: Data curation. K. Hirose: Data curation. E.K. Nakakura: Data curation. R.B. Corcoran: Writing–review and editing. K. Van Loon: Writing–review and editing. C.E. Atreya: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2. Kopetz S, Chang GJ, Overman MJ, Eng C, Sargent DJ, Larson DW, et al. Improved survival in metastatic colorectal cancer is associated with adoption of hepatic resection and improved chemotherapy. J Clin Oncol 2009;27:3677–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shindoh J, Loyer EM, Kopetz S, Boonsirikamchai P, Maru DM, Chun YS, et al. Optimal morphologic response to preoperative chemotherapy: an alternate outcome end point before resection of hepatic colorectal metastases. J Clin Oncol 2012;30:4566–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med 2004;350:2335–42. [DOI] [PubMed] [Google Scholar]
- 5. Kawaguchi Y, Vauthey JN. The landmark series: randomized control trials examining perioperative chemotherapy and postoperative adjuvant chemotherapy for resectable colorectal liver metastasis. Ann Surg Oncol 2020;27:4263–70. [DOI] [PubMed] [Google Scholar]
- 6. Pan Z, Peng J, Lin J, Chen G, Wu X, Lu Z, et al. Is there a survival benefit from adjuvant chemotherapy for patients with liver oligometastases from colorectal cancer after curative resection? Cancer Commun 2018;38:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Benson AB, Venook AP, Al-Hawary MM, Arain MA, Chen YJ, Ciombor KK, et al. Colon cancer, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 2021;19:329–59. [DOI] [PubMed] [Google Scholar]
- 8. Wang ZM, Chen YY, Chen FF, Wang SY, Xiong B. Peri-operative chemotherapy for patients with resectable colorectal hepatic metastasis: a meta-analysis. Eur J Surg Oncol 2015;41:1197–203. [DOI] [PubMed] [Google Scholar]
- 9. Steele SR, Chang GJ, Hendren S, Weiser M, Irani J, Buie WD, et al. Practice guideline for the surveillance of patients after curative treatment of colon and rectal cancer. Dis Colon Rectum 2015;58:713–25. [DOI] [PubMed] [Google Scholar]
- 10. Lin J, Peng J, Zhao Y, Luo B, Zhao Y, Deng Y, et al. Early recurrence in patients undergoing curative resection of colorectal liver oligometastases: identification of its clinical characteristics, risk factors, and prognosis. J Cancer Res Clin Oncol 2018;144:359–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nicholson BD, Shinkins B, Pathiraja I, Roberts NW, James TJ, Mallett S, et al. Blood CEA levels for detecting recurrent colorectal cancer. Cochrane Database Syst Rev 2015;2015:CD011134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Benson AB, Venook AP, Al-Hawary MM, Arain MA, Chen YJ, Ciombor KK, et al. NCCN guidelines insights: rectal cancer, version 6.2020. J Natl Compr Canc Netw 2020;18:806–15. [DOI] [PubMed] [Google Scholar]
- 13. Nordlinger B, Guiguet M, Vaillant JC, Balladur P, Boudjema K, Bachellier P, et al. Surgical resection of colorectal carcinoma metastases to the liver. A prognostic scoring system to improve case selection, based on 1568 patients. Association Française de Chirurgie. Cancer 1996;77:1254–62. [PubMed] [Google Scholar]
- 14. Fong Y, Fortner J, Sun RL, Brennan MF, Blumgart LH. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg 1999;230:309–18; discussion 318–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sørbye H, Dahl O. Carcinoembryonic antigen surge in metastatic colorectal cancer patients responding to oxaliplatin combination chemotherapy: implications for tumor marker monitoring and guidelines. J Clin Oncol 2003;21:4466–7. [DOI] [PubMed] [Google Scholar]
- 16. Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reinert T, Henriksen TV, Christensen E, Sharma S, Salari R, Sethi H, et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol 2019;5:1124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tie J, Cohen JD, Wang Y, Christie M, Simons K, Lee M, et al. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol 2019;5:1710–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parikh AR, Seventer EEV, Siravegna G, Hartwig AV, Jaimovich A, He Y, et al. Minimal residual disease detection using a plasma-only circulating tumor DNA assay in patients with colorectal cancer. Clin Cancer Res 2021;27:5586–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chakrabarti S, Xie H, Urrutia R, Mahipal A. The promise of circulating tumor DNA (ctDNA) in the management of early-stage colon cancer: a critical review. Cancers (Basel) 2020;12:2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tarazona N, Gimeno-Valiente F, Gambardella V, Zuñiga S, Rentero-Garrido P, Huerta M, et al. Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized colon cancer. Ann Oncol 2019;30:1804–12. [DOI] [PubMed] [Google Scholar]
- 24. Kotani D, Oki E, Nakamura Y, Yukami H, Mishima S, Bando H, et al. Molecular residual disease and efficacy of adjuvant chemotherapy in patients with colorectal cancer. Nat Med 2023;29:127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tie J, Cohen JD, Lahouel K, Lo SN, Wang Y, Kosmider S, et al. Circulating tumor DNA analysis guiding adjuvant therapy in stage II colon cancer. N Engl J Med 2022;386:2261–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016;8:346ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Naidoo M, Gibbs P, Tie J. ctDNA and adjuvant therapy for colorectal cancer: time to re-invent our treatment paradigm. Cancers 2021;13:346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tie J, Wang Y, Cohen J, Li L, Hong W, Christie M, et al. Circulating tumor DNA dynamics and recurrence risk in patients undergoing curative intent resection of colorectal cancer liver metastases: a prospective cohort study. PLoS Med 2021;18:e1003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Loupakis F, Sharma S, Derouazi M, Murgioni S, Biason P, Rizzato MD, et al. Detection of molecular residual disease using personalized circulating tumor DNA assay in patients with colorectal cancer undergoing resection of metastases. JCO Precis Oncol 2021;5:PO.21.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Overman MJ, Vauthey JN, Aloia TA, Conrad C, Chun YS, Pereira AAL, et al. Circulating tumor DNA (ctDNA) utilizing a high-sensitivity panel to detect minimal residual disease post liver hepatectomy and predict disease recurrence. J Clin Oncol 2017;35(Suppl 15):3522. [Google Scholar]
- 31. Newhook TE, Overman MJ, Chun YS, Dasari A, Tzeng CWD, Cao HST, et al. Prospective study of perioperative circulating tumor DNA dynamics in patients undergoing hepatectomy for colorectal liver metastases. Ann Surg 2023;277:813–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One 2015;10:e0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Callesen LB, Takacova T, Hamfjord J, Würschmidt F, Oldhafer KJ, Brüning R, et al. Circulating DNA in patients undergoing loco-regional treatment of colorectal cancer metastases: a systematic review and meta-analysis. Ther Adv Med Oncol 2022;14:17588359221133171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lutfi A, Afghan MK, Swed B, Kasi PM. False-positive liquid biopsy assays secondary to overlapping aberrant methylation from non-cancer disease states. Case Rep Oncol 2023;16:1536–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Schema of ctDNA sampling.
Supplementary Figure S2. Kaplan-Meier curves for patients with pre-procedure ctDNA stratified by ctDNA status.
Supplementary Figure S3. ctDNA dynamic changes for patients pre-and post-curative intent therapy (CI) (left) or adjuvant chemotherapy (ACT) (right).
Supplementary Figure S4. Survival outcomes and recurrence rates stratified by CEA status.
Supplementary Table S1. Patient deidentified record ID by analysis. Description of the patient record IDs used per analysis to be cross-referenced with the swimmer plot in Figure 2.
Supplementary Table S2. Demographics, clinicopathologic and procedure-related features table. Table depicts study participants in primary timepoint analysis.
Supplementary Table S3. Univariate analysis by pre-operative ctDNA status. Depicted are multiple clinicopathologic variables analyzed by ctDNA status.
Data Availability Statement
All relevant data are provided within the article and the accompanying Supplementary Data. Because of Health Insurance Portability and Accountability Act (HIPAA) requirements, we are not consented to share individualized patient data, which contains potentially identifying or sensitive patient information. We are committed to collaborative data analysis, and more information and mechanisms for data access can be obtained by contacting the corresponding authors.