Abstract
Apoptosis of infected cells is an important host defense mechanism, and many viruses have exploited antiapoptotic proteins that interfere with crucial cellular pathways. Viral FLICE inhibitory proteins (vFLIPs) are encoded by rhadinoviruses like herpesvirus saimiri, the related Kaposi's sarcoma-associated herpesvirus-human herpesvirus 8 (KSHV/HHV8), and the poxvirus responsible for molluscum contagiosum. The vFLIPs can block the interaction of the death receptor-adapter complex with the cellular effector FLICE (caspase-8), and this prevents the initiation of the downstream caspase cascade. KSHV/HHV8 vFLIP overexpression can confer resistance to T-cell-mediated apoptosis and acts as a tumor progression factor in a murine B-cell lymphoma model. To analyze the function of herpesvirus vFLIPs in the genetic background of the virus and in a model for viral pathogenesis, we deleted the vFLIP gene (open reading frame 71) from the genome of herpesvirus saimiri strain C488. The viral deletion mutant was viable and replicated like the wild-type virus. An antiapoptotic effect could be attributed to the vFLIP gene, but we also show that the vFLIP gene of herpesvirus saimiri is dispensable for viral transformation of T cells in vitro and for pathogenicity in cottontop tamarins in vivo.
Viruses utilize various strategies that relate to apoptosis of host cells or attacking effector cells of the immune system. Since the induction of programmed cell death in cells infected with various pathogens is an important and common host defense mechanism, many viruses have evolved proteins to evade this protective mechanism. On the other hand, apoptosis is used by some viruses to promote the release of progeny from infected cells (47).
Open reading frame 71 (ORF71) of the oncogenic herpesvirus saimiri (saimirine herpesvirus 2) encodes a putative antiapoptotic protein that is homologous to a family of cellular and viral inhibitory proteins which interfere with apoptosis signaled through the death receptor Fas-CD95 and the tumor necrosis factor receptor 1 (TNFR-1) (5, 30, 31, 54).
The binding of a specific ligand to death receptors expressed on eukaryotic cells induces multimerization of the receptor complex, and this clustering recruits adapter molecules like the FADD (Fas-associated death domain) protein or the TNFR-associated death domain protein via interactions between the death domain of the receptor and the death domain of the adapter. The formation of the death receptor-adapter complex recruits the upstream caspase-8 (FLICE) by interaction between the death effector domain (DED) of the adapter protein and the DED of the caspase. Together they form the death-inducing signaling complex, and caspase-8 is activated by proteolytic autocleavage, which initiates the downstream caspase cascade and results in apoptosis (3, 24).
Viral FLICE inhibitory proteins (vFLIPs) are found in the poxvirus responsible for molluscum contagiosum and in most of the Gammaherpesviridae of the genus Rhadinovirus, namely the Kaposi's sarcoma (KS)-associated herpesvirus-human herpesvirus 8 (KSHV/HHV8), rhesus rhadinovirus (RRV) (2, 50), herpesvirus saimiri, equine herpesvirus 2 (30), and bovine herpesvirus 4 (55). The vFLIPs contain two DEDs and have been shown to block the interaction of a death receptor-adapter complex like Fas-FADD with the cellular effector FLICE (caspase-8) and prevent its autoactivation (5, 30, 54). A cellular homolog to the vFLIPs has also been identified, although it occurs in two forms: the short cellular form, cFLIP(S), contains two DEDs and acts at the same level as the vFLIPs by preventing FADD-FLICE interaction; the long form, cFLIP(L), resembles the caspase-8 structure with two DEDs plus a caspase domain. However, the active site of the protease domain is mutated. cFLIP(L) can also interact with the autoactivation step of caspase-8 (31).
Overexpression of KSHV/HHV8 vFLIP by retroviral transduction of the murine B cell line A20 has been shown to confer resistance against death receptor-mediated apoptosis. It promotes clonal outgrowth in the presence of death stimuli, and caspase activation in the vFLIP-A20 transductants is inhibited. When transferred into syngeneic or semiallogeneic immune competent mice, the vFLIP-transduced B cells induced significantly more and faster-growing B-cell tumors than the mock-transduced line. This difference was not evident in immune response-compromised mice, suggesting that vFLIP is a tumor progression factor that can offer protection from T-cell-mediated apoptosis (15). Similarly, the cFLIP has been shown to prevent tumor rejection in a different mouse model, also presumably by escape from cytotoxic T-cell-mediated apoptosis, and lead to tumor progression (40). Additional evidence points to a modulation of the NF-κB pathway by the interaction of vFLIPs with signaling proteins (8). However, all these observations are from in vitro overexpression of recombinant proteins (5, 30) or transplantation of tumor cell transfectants into mice (15).
Studying the vFLIP function of herpesvirus saimiri provides (i) a permissive cell culture system that allows the construction of recombinant viruses, (ii) in vitro lymphocyte transformation assays in human and simian T cells, and (iii) a meaningful and stringent animal model for pathogenesis in common marmosets or cottontop tamarins. A previous study argues for the function of the herpesvirus saimiri vFLIP gene, since it demonstrates an antiapoptotic effect during the lytic infection of owl monkey kidney (OMK) cells (54). Although protection from apoptosis was evident in infected cells, the data is weakened by the fact that herpesvirus saimiri contains at least one other antiapoptotic protein, a viral Bcl-2 homolog that is functional in in vitro models (11, 43). Thus, vFLIP function has been proven neither in the normal genetic background of the virus nor in a model for viral pathogenesis.
Therefore, we generated a vFLIP deletion mutant by removing the vFLIP gene (ORF71) from the genome of herpesvirus saimiri strain C488. The C488ΔFLIP deletion mutant was replication competent, and we demonstrate that the antiapoptotic effect is lost after deletion of vFLIP. However, we also show that the vFLIP gene of herpesvirus saimiri is dispensable for the viral transformation of T cells in vitro and for pathogenicity in cottontop tamarins in vivo.
MATERIALS AND METHODS
Cell culture and virus propagation.
OMK cells (ATCC CRL1556), cultivated in Dulbecco's modified Eagle medium supplemented with glutamine (350 μg/ml), gentamicin (100 μg/ml), and 10% heat-inactivated fetal calf serum, were used for the propagation of herpesvirus saimiri. Virus stocks were generated by the infection of confluent OMK cells seeded in 175-cm2 tissue culture flasks at a low multiplicity of infection. When lysis was complete, supernatants were cleared from the cellular debris by centrifugation at 2000 × g for 15 min, and cell-free supernatants were stored at −80°C. Peripheral blood mononuclear cells (PBMC) of Callithrix jacchus and human umbilical cord blood lymphocytes (CBL) were isolated by density gradient centrifugation. The cells were cultivated in lymphocyte growth medium (LGM) (45% RPMI 1640 medium, 45% Panserin [Pansystems, Aidenbuch, Germany], 10% fetal calf serum [Pansystems], gentamicin [100 μg/ml], and glutamine [350 μg/ml]). LGM for human CBL was supplemented with 100 U of recombinant human interleukin-2 (IL-2) (aldesleukin; ProleukinR; Chiron, Ratingen, Germany) per ml or 20 U of human IL-2 (Roche Diagnostics, Mannheim-Penzberg, Germany) per ml.
Construction of the viral deletion mutant C488ΔFLIP.
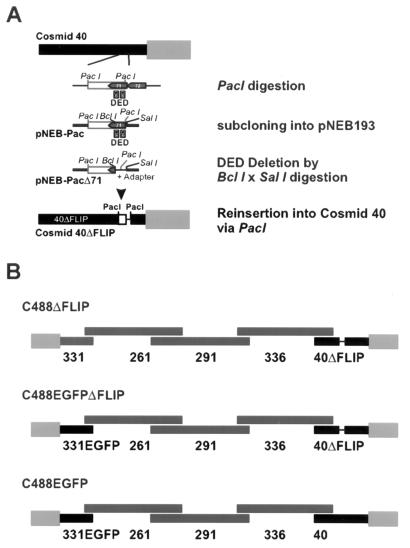
ORF71 was deleted from the herpesvirus saimiri strain C488 genome by a cosmid-based approach. All cloning procedures were performed by standard methods. A PacI fragment including most of ORF71 was subcloned from cosmid 40 into pNEB193 (New England Biolabs). Most of the ORF71 gene, including the two DEDs, was deleted by digestion with BclI-SalI. A double-stranded oligonucleotide adapter was designed from the oligonucleotides 5′-GATCGTTTAAACGTTAATTAATCGA-3′ and 5′-TCGATCGATTAATTAACGTTTAAAC-3′. This adapter contained an internal PacI site (bold), as well as BclI-and SalI-compatible 5′ overhangs (underlined) at the ends. It was inserted into the BclI-SalI-digested pNEB193-Pac to replace the deleted ORF71 segment (Fig. 1A). The altered PacI fragment encompassing the deleted ORF71 gene segment was reinserted into the PacI-digested cosmid 40, resulting in cosmid 40ΔFLIP. The correct insertion was verified by sequencing. Recombinant virus was generated by liposome-mediated cotransfection of a set of overlapping cosmids, including cosmid 40ΔFLIP, into permissive OMK cells (Fig. 1B). The cosmids were linearized before transfection by restriction with NotI; this also removed the pWE15 cloning vector, since two NotI sites flank the BamHI site that was used to clone the viral DNA. Virus-containing supernatant from completely lysed cultures was harvested by centrifugation, and the pelleted virions were lysed in 100 μl of PCR buffer containing 100 μg of proteinase K (Roche Diagnostics) per ml and 0.5% Tween 20 for 1 h at 56°C; then the proteinase K was heat inactivated for 15 min at 95°C. An aliquot of 2 to 4 μl was used for PCR analysis.
FIG. 1.
Construction of the recombinant viruses herpesvirus saimiri C488ΔFLIP, C488EGFP, and C488ΔFLIP-EGFP. The vFLIP encoding ORF71 of herpesvirus saimiri C488 was deleted from the right terminal cosmid 40. (A) A PacI fragment was subcloned into vector pNEB193, and most of the ORF71 gene was deleted by digestion with BclI and SalI and insertion of an adapter containing a PacI site. Then the altered PacI fragment from pNEB193 was reinserted into PacI-digested cosmid 40 to generate the cosmid 40ΔFLIP. (B) Recombinant virus was generated by the cotransfection of overlapping linearized cosmids 331 or 331EGFP, 261, 291, 336, and 40 or 40ΔFLIP.
Construction of C488EGFP recombinant.
The enhanced green fluorescent protein (EGFP) gene was PCR amplified from plasmid pEGFP-C1 (Clontech). PCR mixtures (50 μl each) contained 2 μl of template DNA, 0.2 μM each deoxynucleoside triphosphates (dNTP), 10 μM each primer, and 5 U of Pfu polymerase (Stratagene) in 1× Pfu reaction buffer. The primers 5′-GGGCGCGCCGAATGCAGTGAAAAAAATGC-3′ and 5′-GGCGCGCCATTAATAGTAATCAATTACG-3′ were used. After a 1-min initial denaturation step at 95°C, 25 cycles of 10 s at 95°C, 20 s at 55°C, and 3 min at 70°C were performed in an MJ Research PTC-200 thermal cycler, followed by a 4-min final extension step at 70°C. The PCR fragment was then subcloned into the single SwaI site of cosmid 331 located just upstream of the DHFR gene (ORF2) in a noncoding region. The construct was analyzed by sequencing, and expression of the EGFP was verified by transfection into OMK cells and fluorescence microscopy.
DNA sequence analysis.
Nucleotide sequences were determined with an ABI 377A automated sequencer (Applied Biosystems) using the Dye-Deoxy Terminator Sequencing kit according to the manufacturer's instructions (Perkin-Elmer). DNA sequence evaluation was done with XBAP software (10).
Virus stocks and replication studies.
For virus titration, OMK cells were grown in 48-well plates (Nalgene Nunc International, Roskilde, Denmark) and infected with serial ten-fold dilutions (10−3 to 10−7) of herpesvirus saimiri C488, C488ΔFlip, and the respective EGFP variants in 400 μl of Dulbecco's modified Eagle medium with supplements. A dilution step was defined as positive if the cells in at least 4 of 8 wells were completely lysed. The virus replication kinetics was determined by the infection of OMK cells (3 × 105 cells seeded in a 25-cm2 flask 2 days before infection) with 104 tissue culture infectious particles (TCIP) in 10 ml of medium. The titers of virus-containing supernatant taken on subsequent days were determined by limiting dilution as described above.
In vitro transformation of lymphocytes.
Lymphocytes were expanded after isolation for 2 to 3 days by stimulation with 0.5 to 1 μg of phytohemagglutinin A (Murex, Großburgwedel, Germany) per ml. Between 3 × 106 and 5 × 106 cells were infected with 1 ml of herpesvirus saimiri C488 or mutant virus C488ΔFLIP containing supernatants (titer, >106 TCIP/ml) and cultivated in LGM as described above. The transformation of the resulting T-cell lines was assessed microscopically and by the observation of accelerated growth.
Experimental infection of common marmosets.
In vivo oncogenicity of the herpesvirus saimiri C488 recombinants was assayed by experimental infection of Saguinus oedipus tamarins. The study was approved by the Institutional Animal Care and Use Committee and was performed according to governmental regulations with purpose-bred, healthy, adult cottontop tamarins at the Biomedical Primate Research Center (Rijswijk, The Netherlands). Two animals were infected with the experimental virus C488ΔFLIP, and only one animal was infected with the wild-type control virus C488. Since cottontop tamarins are an endangered and protected species, larger numbers of animals could not be justified from an ethical point of view. Two animals per experimental virus are considered necessary to obtain a meaningful result in a primate experiment, and this number is usually used in published studies. Since the wild-type virus C488 is highly pathogenic and generally causes lymphoma in all infected animals (C. jacchus or S. oedipus) (17, 35, 36), only one animal was allowed to obtain control tissues and cell lines. The animals were each intravenously injected with 1 ml of cell-free virus containing supernatant from infected OMK cultures containing 106 TCIP of virus. They were housed in separate cages and received a standard monkey diet and drinking water ad libitum. Blood samples were taken prior to infection, at weekly intervals, and before necropsy to expand T lymphoma cells and reisolate virus by cocultivation on OMK cells. The animals were euthanized as soon as illness became evident. Tissues were fixed in formalin and stained with hematoxylin and eosin and for immunohistochemistry additionally with antibodies specific for CD3 (Dako, Hamburg, Germany) and CD20 (Dako).
Stable growing transformed T-cell lines were obtained from PBMC, and different organs (thymus, spleen, liver, kidney, and lymph nodes) were obtained from each diseased animal. Briefly, tissue obtained at necropsy was cut into pieces with a sterile scalpel blade, and single-cell suspensions were obtained by passage through 70-μm nylon sieves (Falcon Cell Strainer, Becton Dickinson, Heidelberg, Germany). Cells were cultivated in LGM without IL-2.
Detection of viral DNA.
The status of viral DNA in the transformed cell lines was analyzed by PCR and Southern blotting. PCR analysis was carried out in 25-μl reaction mixtures, each containing 2 μl of template DNA, 0.2 μM each dNTP, 10 μM each primer, and 2.5 U of AmpliTaq in 1× AmpliTaq buffer (Perkin-Elmer). PCR conditions were as follows: a 5-min denaturation at 95°C; 29 cycles of 30 s at 95°C, 30 s at 56°C, and 1 min at 70°C; a 4-min extension at 70°C; and a 4°C hold. The following primer pairs specific for the respective ORFs were used for the analysis: StpC/Tip (TR1 5′-GTAGTAAACTAAGAGCAAAGCAAGC-3′ and TR2 5′-GTACAAGCTGTTCAAGTTTGTTAGC-3′), ORF3 (5′-CACAACACTGGTATGTACCAATG-3′ and 5′-CTGTGGAGGTAATGCAGATAC-3′), ORF75 (5′-TGGCTGCTAACAGGCATGG-3′ and 5′-AGCACGTTGCCCGAGATTG-3′), and ORF71/FLIP (5′-GGCGCGCCTCGAAATTCTGTAAATGGAC-3′ and 5′-ACAGAAAGAGACACAAGAG-3′).
DNA for Southern blotting was prepared by the addition of 400 μl of extraction buffer (100 mM NaCl, 10 mM Tris-HCl, 25 mM EDTA, 0.5% sodium dodecyl sulfate; proteinase K was added to a final concentration of 1 mg/ml) to 5 × 107 cells. After an overnight incubation at 56°C, the solution was digested with 40 μl of RNase A (5 mg/ml) at 37°C for 30 min. The DNA was extracted with buffer-saturated phenol:chloroform:isoamyl-alcohol (25:24:1), and then a one-half volume of ammonium acetate (7.5 M) was added to the aqueous phase, and the DNA was precipitated with 2 volumes of ethanol. After centrifugation, the pellet was washed with 70% (vol/vol) ethanol, dried briefly, and resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA). A total of 20 μg of cellular DNA was digested with SstI or PstI. The DNA fragments were size fractionated by electrophoresis through a 1% agarose gel. The DNA was then transferred to a nylon membrane (Hybond N; Amersham) and hybridized with a 32P-labeled DNA fragment. This 1.9-kb probe specific for ORF71 and -72 was amplified by PCR from cosmid 40 (primers, 5′-TGCGTTAGACAAATATCCC-3′ and 5′-CTAAAAATGCAGCATCGTCACC-3′; conditions were as above). The DNA fragment was purified from a 1% agarose gel (Qiaquick Gel Extraction kit; Qiagen, Hilden, Germany), and random labeling with [α-32P]dATP was performed (20).
RNA and cDNA analysis.
Total cellular RNA was prepared by the acidic phenol extraction method (9). Five micrograms of RNA was treated with RNase-free DNase I (Roche Diagnostics) in 1× DNAse I buffer for 30 min, followed by heat inactivation at 70°C for 10 min. Then, first-strand cDNA was synthesized with Superscript II reverse transcriptase (Gibco BRL): the RNA was incubated with 500 ng of random hexamer primers for 10 min at 70°C. After a short incubation on ice, 8 μl of the reaction mixture (4 μl of 5× first-strand buffer, 2 μl of 0.1 M dithiothreitol, 1 μl of 10 mM dNTPs, and 1 μl of Superscript II) was added, and the synthesis of the cDNA was started (10 min at 25°C and 50 min at 37°C). The enzyme was heat inactivated at 70°C for 15 min. RNA complementary to the cDNA was removed by the addition of 1 U of RNase H (MBI Fermentas) and incubation for 20 min at 37°C. An identical sample was prepared in parallel where the reverse transcriptase was omitted from the reaction mixture (as a control sample). A total of 2 μl of the reaction mixture was used for reverse transcription (RT)-PCR analysis. PCR conditions were as follows: a 5-min initial denaturation at 95°C; 39 cycles of 20 s at 95°C, 30 s at 52°C, and 1 min at 70°C; a 4-min final extension at 70°C; and a 4°C hold. Primers utilized were specific for the ORF71/FLIP (as above) and ß-actin (5′-CGGGAAATCGTGCGTGACAT-3′ and 5′-GAACTTTGGGGGATGCTCGC-3′).
Flow cytometry.
Transformed human and simian T cells were analyzed by flow cytometry with antibodies for B- and T-cell surface epitopes on a FACS-Calibur flow cytometer (Becton Dickinson). The directly labeled monoclonal antibodies (MAbs) (Cy-Chrome or phycoerythrin conjugated) were specific against CD2 (RPA-2.10; PharMingen, Heidelberg, Germany), CD3ɛ (SP34; PharMingen), CD3 (Leu-4; Becton Dickinson), CD4 (Leu-3a SK3; Becton Dickinson), CD8 (RPA-T8; PharMingen), CD20 (Leu16 L27; Becton Dickinson), HLA-DR (L243; Becton Dickinson), CD80 (L307.4; PharMingen), and CD86 (IT2.2; PharMingen). Directly labeled isotype-matched control MAbs were used (Becton Dickinson and PharMingen). CD95 surface expression was detected with a MAb directed to CD95 (DX2; PharMingen) and a secondary fluorescein isothiocyanate-labeled goat anti-mouse immunoglobin G F(ab′)2 fragment (Dianova, Hamburg, Germany).
Immunofluorescence.
A total of 104 OMK cells were seeded in four-well chamber slides (Nalgene Nunc International) and infected with the EGFP variants of C488 and C488ΔFLIP. After 24 h, the cells were overinfected with recombinant adenovirus expressing the soluble Fas ligand (Ad-FasL) (7), or 50 μM menadione was added and the cells were incubated for 4 or 8 h at 37°C and fixed with 4% paraformaldehyde (30 min at room temperature). To visualize nuclei and DNA, the cells were stained by the addition of 1 μg of Hoechst-33342 dye (Sigma) per ml. The apoptotic cells were viewed under a Zeiss Axiovert fluorescence microscope at a magnification of ×630.
Induction of apoptosis and determination of cell death.
A total of 104 OMK cells were grown in flat-bottomed 96-well plates (Nalgene Nunc International) overnight and infected with 5 × 104 to 10 × 104 TCIP of the recombinant viruses C488EGFP and C488ΔFLIP-EGFP (multiplicity of infection, 5 to 10). After 24 h the cells were superinfected with recombinant adenovirus (Ad) Ad-FasL (0.5 × 106 PFU) or the β-GAL gene (Ad-Z) as a control (GenVec, Inc., Gaithersburg, Md.). After incubation for 4 h at 37°C, cell death was quantified by an enzyme-linked immunosorbent assay (ELISA) for the detection of histones bound to fragmented DNA released into the cytoplasm of apoptotic cells (Cell Death Detection kit; Roche Molecular Biochemicals, Mannheim, Germany). Cytoplasmatic extracts were normalized to their total protein content that was determined by the BCA-Assay (Pierce, Inc., Rockford, Ill.). Specific protection from apoptosis was calculated as follows: 10,000 × (optical density/protein [μg/ml]). Uninfected cells treated with Ad-FasL were assigned a value of 100% apoptosis.
RESULTS
Construction of recombinant viruses.
The herpesvirus saimiri ORF71 encoding the vFLIP was deleted by a cosmid-based approach that allows the construction of recombinant viruses without contaminating wild-type virus. Previously, we had subcloned the genome of herpesvirus saimiri into overlapping cosmids (18). The right-terminal cosmid 40 was selected for the deletion of ORF71. In the genome of herpesvirus saimiri strain C488, the methionine start codon of ORF71 is located directly adjacent to the ochre stop codon terminating the ORF72 encoding the vCyclin. The ORF71 contains a PacI restriction site 72 bp downstream of the first ATG codon. A second PacI site is located at a distance of 2.15 kb in the noncoding region between ORF70 (encoding a thymidylate synthase) and ORF71. This 2.15-kb PacI restriction fragment encompassing most of vFLIP was subcloned from cosmid 40 into plasmid pNEB193 to generate pNEB193-Pac. The region including the two DEDs of vFLIP was removed by restriction of pNEB193-Pac with BclI and SalI and replaced by an oligonucleotide adapter containing a PacI site. The modified PacI fragment was then reinserted into cosmid 40 to generate cosmid 40ΔFLIP (Fig. 1A). The correct insertion was verified by restriction mapping and sequencing. A human cytomegalovirus-promoter driven EGFP expression cassette was inserted into a noncoding region of cosmid 331 to generate cosmid 331EGFP. Recombinant viruses were then constructed by cotransfection of linearized cosmids into OMK cells. The recombinant virus C488ΔFLIP was generated from cosmids 331, 261, 291, 336, and 40ΔFLIP. We also designed an EGFP-expressing recombinant vFLIP deletion virus and a corresponding control virus. The recombinant EGFP-expressing vFLIP deletion virus C488ΔFLIP-EGFP was generated from cosmids 331EGFP, 261, 291, 336, and 40ΔFLIP; the recombinant EGFP-expressing control virus C488EGFP was generated from cosmids 331EGFP, 261, 291, 336, and 40 (Fig. 1B).
Replication of recombinant viruses.
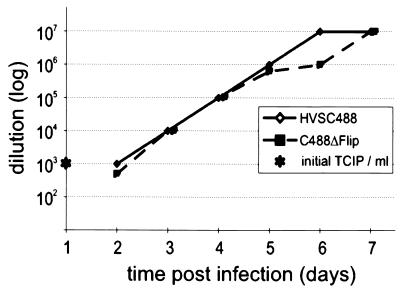
Recombinant viruses were obtained from cosmid cotransfection, demonstrating that the vFLIP gene of herpesvirus saimiri is not essential for lytic replication in OMK cells. No differences in plaque size or morphology in OMK cells were notable (data not shown). Lytic viral replication of C488ΔFLIP on OMK cells was compared to that of C488. Endpoint titers of both viruses were in the range of 106 to 107 TCIP per ml of supernatant from several independent cultures. The kinetics of viral replication was then compared by infection of OMK cultures with 104 TCIP and by titration of supernatant taken from successive days. There was no apparent difference between the viruses, lysis was complete after 7 days, and similar endpoint titers of 107 were observed (Fig. 2).
FIG. 2.
Replication of the recombinant viruses. OMK cells were infected with culture supernatants containing 104 infectious particles (determined by limiting dilution) of the EGFP variants of herpesvirus saimiri C488 and C488ΔFLIP. Virus titers were determined from supernatant taken on subsequent days until the cells were completely lysed by cytopathic effect.
Transformation of human and simian T cells in vitro.
Herpesvirus saimiri is able to transform human and simian T cells to permanent antigen-independent growth in vitro (6). Human CBL (13 donors) and PBMC from C. jacchus (5 donors) and S. oedipus (10 donors) were infected with C488 and C488ΔFLIP in parallel, and the proliferation of the cells was compared to that in uninfected controls. After 4 to 6 weeks of culture, the control cells had stopped growing. Both the deletion mutant virus C488ΔFLIP as well as the wild-type virus C488 transformed the T cells. The data are summarized in Table 1. The transformed T cell lines were analyzed by PCR and Southern blot analysis, and the specific viral genotype present in the cells was confirmed. Proliferation tests performed with the established T-cell lines showed no detectable difference (data not shown). Thus, the vFLIP is not essential for replication and T-cell transformation in vitro.
TABLE 1.
Transformation by recombinant viruses in vitro and pathogenesis in infected cottontop tamarins
| Virus strain or mutant | Species and/or cell type | In vitro transformation experiments (no. positive/total)a | Pathogenesis (survival time in days)b |
|---|---|---|---|
| C488 | Human CBL | 8/13 | NA |
| C. jacchus PBMC | 9/9 | ND | |
| S. oedipus PBMC | 10/10 | 15 | |
| C488ΔFLIP | Human CBL | 7/13 | NA |
| C. jacchus PBMC | 9/9 | ND | |
| S. oedipus PBMC | 10/10 | 14, 15 |
Performed with human CBL (13 donors) and PBMC from C. jacchus (5 donors) and S. oedipus (10 donors). Parallel cultures of cells were infected with herpesvirus saimiri C488 or C488ΔFLIP.
NA, not applicable; ND, not done.
Pathogenicity in cottontop tamarins.
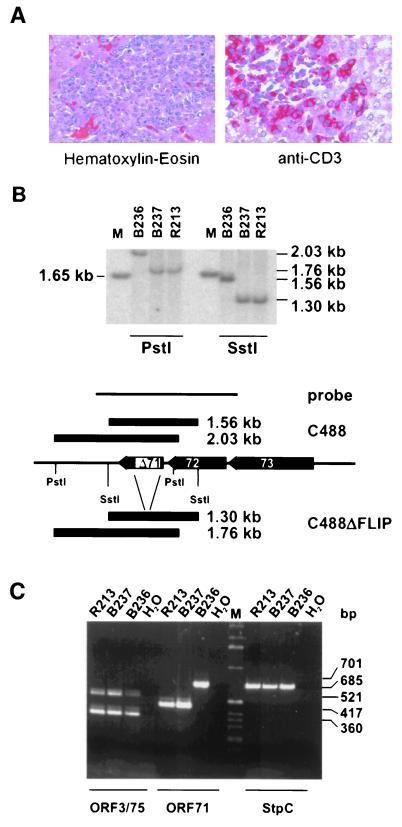
One putative function of vFLIP is the escape of transformed cells from immune surveillance by interference with cytotoxic T-cell-mediated apoptosis, which is difficult to study in vitro. Herpesvirus saimiri C488 is able to induce T-cell lymphoma in vivo in several species of New World primates. In vitro transformation is linked to pathogenicity in vivo for most published viral deletion mutants. However, there are reports where efficient in vitro transformation was observed but where the recombinant virus turned out to be apathogenic or less pathogenic than wild-type virus in vivo (16, 28, 38). The relevance of the vFLIP gene for the development of T-cell lymphoma was studied in cottontop tamarins (S. oedipus). Two animals were infected intravenously with the deletion mutant virus C488ΔFLIP (animals B237 and R213) and one was infected with the wild-type virus C488 carrying an intact vFLIP (animal B236). All animals developed disease between day 14 and 15 and were euthanized (Table 1). Necropsy was performed, and macroscopic findings were consistent with the typical lymphoproliferative disease induced by herpesvirus saimiri. Enlarged lymph nodes and spleen and infiltrations in various organs were recognizable. Immunohistochemistry revealed infiltrations of CD3+ blast-like lymphoid cells in various tissues of all three animals (Fig. 3A). Infiltration of virus-transformed T cells was widespread in the different organs or tissues, and the morphology of infiltrations was similar in the wild-type- and deletion virus-infected animals. Continuously growing T-cell lines from thymus, spleen, liver, kidney, and lymph nodes were established. The cultures grew stably over an observation period of more than 10 months, and there was no difference detectable in the proliferation of the cell lines transformed by the recombinant or the wild-type viruses. DNA from the transformed T-cell cultures established from the infected animals was used to confirm the presence of the specific viral genome by Southern blotting and PCR (Fig. 3B and C). Thus, the vFLIP gene is also dispensable for the capacity of herpesvirus saimiri to induce T-cell lymphoma in vivo.
FIG. 3.
Detection of recombinant herpesvirus saimiri in tumor cells. (A) Liver tissue sections from an C488ΔFLIP-infected animal that show infiltration of portal tracts by large blast-like lymphoid cells (left). Immunohistochemistry reveals expression of the CD3 T-cell antigen in these cells (right). (B) DNA was recovered from T-cell lines established from tumorous spleens of infected animals. Total cellular DNA was digested with SstI or PstI and analyzed by Southern blotting for the presence of the ORF71 deletion in the cell lines; the marker is a 1-kb ladder (Gibco BRL Life Technologies, Karlsruhe, Germany). Hybridization was done with the probe indicated. B236, C488 infected; R213 and B237, C488ΔFLIP infected. (C) PCR was performed with extracts from corresponding transformed T cells. StpC and ORF75 and -3 were not affected by the deletion, whereas the ORF71 deletion was confirmed in the recombinant viral genomes.
Expression of vFLIP and phenotype of transformed cells.
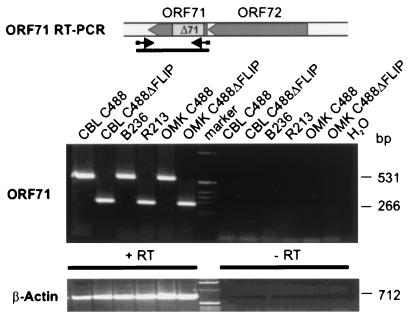
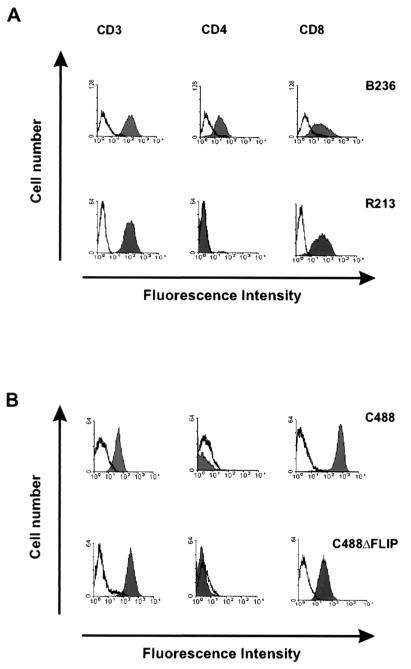
The transcription of the mRNA encoding vFLIP was confirmed by RT-PCR from infected OMK cells, S. oedipus ex vivo-cultured T-cell lines, and transformed human CBL-derived T-cell lines. Oligonucleotides flanking the deleted region were chosen that generated PCR amplification products of 266 bp from C488ΔFLIP and 531 bp from C488. Contamination of the reactions resulting from genomic viral DNA was ruled out by DNase I treatment and by analysis of an identically treated parallel sample in which the reverse transcriptase was omitted from the first-strand cDNA synthesis reaction. Fragments of the expected sizes were amplified and confirmed the transcription of herpesvirus saimiri vFLIP in C488-transformed and -infected cells and the transcript that includes the deletion in C488ΔFLIP-infected cells (Fig. 4). The amplified region also encloses the 3′ region of the vCyclin gene. However, by using several different sets of oligonucleotide primers, no amplification products corresponding to transcripts originating at the amino terminus of ORF73 that would be spliced to the vFLIP or vCyclin gene were detected (data not shown). In addition, the surface phenotype of the transformed cells was determined. We analyzed the transformed human CBL-derived and the simian tumor-derived T-cell lines from all animals and could not detect significant and consistent differences in the expression of cell surface markers. An example shows CD3, CD4, and CD8 expression on C488 or the C488ΔFLIP deletion virus-transformed cells (Fig. 5). At least four different lymphoid cell lines were established from each of the three animals; they showed no significant variation in growth in cell culture, and all were CD3+ T cells, with some variation in CD4 and CD8 expression also within cell lines established from the same animal. Presumably, during cultivation an initially polyclonal culture becomes nonhomogenous due to the outgrowth of subclones. All cells expressed surface markers which are typically found on mature activated T cells, like CD2, CD3, CD4 and/or CD8, HLA-DR, CD80, or CD86, but not B-cell markers like CD20 (data not shown).
FIG. 4.
Expression of vFLIP in different cell lines. Total RNA was used for an ORF71-specific RT-PCR. The middle panel shows the transcripts detected from infected OMKs or transformed human and simian T cells. The bottom panel shows the corresponding RNA as detected by RT-PCR for β-actin. +RT, RT-PCR from first-strand cDNA; −RT, PCR from parallel control reaction, where Superscript reverse transcriptase was omitted. The locations of primers are indicated.
FIG. 5.
Surface phenotype of transformed simian and human T cells. A typical example of the expression of the T-cell markers CD3, CD4, and CD8 is shown for the spleen-derived T-cell lines B236 (herpesvirus saimiri C488 infected) and R213 (C488ΔFLIP infected) (A) and for T-cell lines derived from human CBL transformed by C488 or C488ΔFLIP (B). The histograms show fluorescence intensity in logarithmic scale on the x axis and cell numbers in linear scale on the y axis. Open graphs represent negative isotype controls, and solid graphs represent specific staining.
Protection from apoptosis induced by FasL.
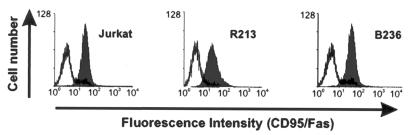
The surface expression of the death receptor Fas-CD95 in the different simian and human cell lines was studied by fluorescence-activated cell sorter analysis. The results of an experiment with transformed T cells obtained from the thymus of animals B236 (C488) and R213 (C488ΔFLIP) are shown in Fig. 6, and Jurkat cells served as a positive control. The CD95 surface marker was detectable on both wild-type virus C488 or C488ΔFLIP transformed cells (Fig. 6).
FIG. 6.
Expression of the Fas/CD95 on transformed T cells. Expression of CD95 on the surface of Jurkat cells (positive control), C488ΔFLIP-transformed T cells (S. oedipus animal R213), and herpesvirus saimiri C488-transformed T cells (S. oedipus animal B236) derived from infected animals. The open graph represents the negative isotype control, and the solid graph represents the specific staining of Fas/CD95.
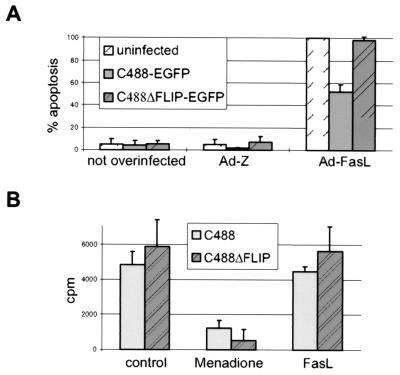
Apoptosis can be detected by morphological criteria. It is characterized by membrane blebbing, nuclear fragmentation, and chromatin condensation that can be observed microscopically. We used the EGFP-expressing variants of wild-type (C488EGFP) and deletion (C488ΔFLIP-EGFP) viruses to discriminate infected from uninfected cells. The herpesvirus saimiri-infected cells are easily identified by the green autofluorescence of EGFP when examined with a fluorescein isothiocyanate-compatible filter set. The FasL-induced apoptosis was detectable in the OMK cells by nuclear fragmentation that was visualized by the DNA binding dye Hoechst-33342. A smaller proportion of green C488-infected cells seemed to show nuclear fragmentation than the C488ΔFLIP-EGFP-infected cells; however, an exact quantification was difficult due to concurrent lytic replication of the virus (data not shown). The antiapoptotic effect of herpesvirus saimiri was then analyzed in a system that quantifies apoptotic events. Endogenous endonucleases are activated in apoptotic cells and cleave the DNA into small oligonucleosomes containing histone H1. These nucleosomal DNA fragments leak through the impaired nuclear membranes into the cytoplasm and are detected in cytoplasmatic extracts by the Cell Death Detection ELISA system. When apoptosis was induced by superinfection with a recombinant adenovirus expressing soluble FasL, an antiapoptotic effect was detectable in C488EGFP-infected OMK cells and not in C488ΔFLIP-EGFP-infected cells. The results of four independent experiments are shown in Fig. 7A. In contrast to the observations of OMK cells, the proliferation of transformed T cells (S. oedipus) was not affected by treatment with up to 1 μg soluble FasL per ml. For comparison, Jurkat T cells are highly susceptible to 20-fold-lower doses of soluble FasL. Thus, a protective effect was not detectable, presumably due to overall resistance of the transformed T cells. Only the Fas-independent proapoptotic substance menadione induced a decrease in proliferation in both cells transformed with the wild-type virus C488 and those transformed with the deletion mutant virus C488ΔFLIP (Fig. 7B).
FIG. 7.
Protection from apoptosis by herpesvirus saimiri. (A) Infection of the permissive cell line OMK with C488 and C488ΔFLIP. Cells were infected with herpesvirus saimiri and superinfected with recombinant Ads. Four hours after superinfection with FasL- or lacZ-expressing adenovirus (0.5 × 106 PFU), the resulting apoptotic effect was assayed by the Cell Death Detection ELISA. The results are mean values from four normalized experiments. (Left) negative control (cells not superinfected); (middle) control cells superinfected with the control Ad-Z; (right) cells superinfected with Ad-FasL. The protective effect from apoptosis provided by herpesvirus saimiri C488 was no longer detectable in the C488ΔFLIP infected cells. (B) Proliferation of C488- and C488ΔFLIP-transformed T-cell lines from S. oedipus tamarins. Proliferation was assayed by [3H]thymidine incorporation. Cells were studied for Fas-independent cell death by incubation with 50 μM menadione for 8 h, for susceptibility to Fas-mediated apoptosis with 1 μg of soluble FasL (cross-linked with anti-FLAG M2 antibody) per ml or by study of control cells in the presence of medium alone. Mean values from three transformed cell lines and two experiments are shown.
DISCUSSION
Herpesvirus saimiri is the prototype of the gamma-2 herpesviruses or Rhadinoviridae (1). It is apathogenic in the persistently infected natural host, the squirrel monkey (Saimiri sciureus), but causes rapidly progressive T-cell leukemia and lymphoma in several species of New World primates (22). Strains of the highly oncogenic subgroup C are capable of transforming primary human T cells to permanent antigen-independent growth (6), while subgroup A and B strains do not transform human cells. The herpesvirus saimiri genome (M-DNA) has a size of about 155 to 160 kb; it consists of a 113-kb AT-rich unique region (L-DNA) that is flanked by about 20 to 25 GC-rich, tandem repeats of 1.4 kb (H-DNA). This principal genome structure is shared by all rhadinoviruses except equine herpesvirus 2. Within the L-DNA region, herpesvirus saimiri has at least 76 ORFs plus genes for small nuclear URNAs, termed HSURs. The H-DNA terminal repeats of herpesvirus saimiri do not encode any known viral proteins (1). At the very left end of the L-DNA, all herpesvirus saimiri subgroups encode the saimiri transformation-associated proteins (STP-A, -B, or -C), a family of weakly conserved oncoproteins that are essential for the transformation of T cells and pathogenicity (12, 16, 33). The subgroup C strains additionally encode the tyrosine kinase-interacting protein, TIP, in this region, which is also essential for transformation (17). The HSURs are encoded in the region adjacent to the transforming genes; they are expressed in transformed cells but are dispensable for transformation in vitro (18). In addition to conserved gammaherpesvirus genes, herpesvirus saimiri shares several ORFs carrying sequestered cellular genes with the other rhadinoviruses KSHV/HHV8 and RRV, namely the vFLIP gene (ORF71), a vCyclin gene (ORF72), and a viral G protein-coupled receptor gene (ORF74, IL-8 receptor) (44). Recombinant herpesvirus saimiri has further been used to study the putative transforming protein K1 of KSHV/HHV8 (38) and is a useful model for gammaherpesvirus oncogenesis (32). RRVs are the closest relatives of KSHV/HHV8 (2, 50). Like herpesvirus saimiri, the RRVs do not seem to be tumorigenic in the natural host (39), and a lytic cell culture system in primary rhesus fibroblasts would allow the construction of recombinant viruses (13). However, no RRV-associated tumor or in vitro transformation model has been described so far.
Epidemiological data as well as the consistent detection of viral DNA in diseased tissue suggest that KSHV/HHV8 is most likely the major cofactor for the development of both classical KS and KS in immune response-compromised patients. In addition, KSHV/HHV8 is linked to several rare B-cell lymphoproliferative diseases, like primary effusion lymphoma (PEL) and multicentric Castleman's disease (27, 41). Although several reports hint at the possibility for the propagation of KSHV/HHV8 in cells derived from human endothelium or kidney (23, 42, 46), there is no accessible experimental model for the generation of recombinant viruses. Furthermore, there exist neither in vitro transformation models that involve the complete KSHV/HHV8 virus nor animal models for pathogenesis, e.g., for the development of KS- or PEL-like disease after viral infection.
Interestingly, the vFLIP- and vCyclin-encoding mRNA is found latently transcribed in KS in situ (51) and in KS and PEL by Northern blotting (14, 48, 49, 52, 53). The presence of a latent bicistronic mRNA encoding vFLIP and vCyclin, as well as a latent tricistronic mRNA encoding vFLIP, vCyclin, and the latency-associated nuclear antigen LANA in diseased tissue from KS and MCD hint at a role in progression or maintenance of the disease state by the inhibition of tumor cell apoptosis. vFLIP may also be required to counteract putative proapoptotic functions of the vCyclin gene under specific circumstances where high levels of cyclin-dependent kinase 6 are present in infected cells (45). Similarly, the KSHV/HHV8 LANA has been shown to exert an antiapoptotic function by inhibition of p53 transcriptional activity and thus promote cell survival (25). In contrast to the cultures established from KSHV/HHV8-associated PEL, where a small proportion of cells always produce viral particles and where the lytic cycle can be induced by stimulation with phorbol esters or sodium butyrate, the herpesvirus saimiri-transformed human T cells harbor the viral episomes in a latent state, and no infectious virus is produced even upon stimulation. We demonstrate that the ORF encoding vFLIP is transcribed in simian and human T cells transformed by herpesvirus saimiri (Fig. 4). However, vFLIP protein expression has not been published for KSHV/HHV8-associated tumors, and the protein was not detectable from our herpesvirus saimiri-transformed cell lines or other infected cells by immunoblot analysis with specific rabbit antisera (data not shown).
The antiapoptotic effect observed in the C488 wild-type virus-infected OMK cells was no longer detectable in the C488ΔFLIP deletion virus-infected cells (Fig. 7B). However, the fact that the herpesvirus saimiri vFLIP gene deletion reverses the protective effect of herpesvirus saimiri C488 in OMK cells does not rule out a relevant antiapoptotic function of the vBcl2 homolog. It may be that a missing balance to a potentially proapoptotic vCyclin masks vBcl2 effects. However, this could only be studied effectively if vBcl2 deletion viruses were available as well.
On one hand, induction of apoptosis by herpesvirus proteins, including homologs of cellular regulators of apoptosis, has been postulated as a way for more-efficient release of progeny virions from the infected cell and for the elimination of immune cells attacking the virus host cell. An example for herpesvirus proapoptotic proteins is the induction of apoptosis in mononuclear cells and bovine B-lymphoma BL-3 cells after binding of bovine herpesvirus 1 glycoprotein D (29). Herpes simplex virus type 1 (HSV-1) induces and blocks apoptosis at multiple steps during infection and protects cells from exogenous inducers in a cell type-dependent manner (26), and ICP27-deleted HSV-1 induces apoptosis in epithelial cells (4). HSV-2 induces apoptosis of macrophages in a Fas- and TNFR-independent manner (21). On the other hand, premature host cell destruction may be induced by the activation of cellular signaling and apoptotic pathways. Such signals could result from upregulation of surface receptor molecules, cytokine secretion by infected cells, increased production of death receptor ligands, or from endogenous events, e.g., those related to the p53 or NF-κB pathways. vFLIP genes could interfere with some of these events, and antiapoptotic gene expression in lytically infected cells could result in a more-efficient production of progeny virions. Although we speculated that a vFLIP deletion virus would be impaired during lytic replication, no significant difference in virus endpoint titer or viral replication kinetics was detectable (Fig. 2). Thus, the herpesvirus saimiri vFLIP does not increase viral particle yield under the conditions of lytic tissue culture in OMK cells, the standard cell line for propagation of herpesvirus saimiri (19).
Inhibition of death receptor-mediated apoptosis is a common mechanism exploited by several DNA viruses to protect infected cells from immune system attack by cytotoxic T cells. Adenovirus E3-14.5 and E3-10.4 proteins are inhibitors of death receptor internalization. The vFLIP proteins interfere with death receptor signal transduction, and cowpox virus CrmA, baculovirus p35, and Ad E3-14.7 proteins have been identified as downstream inhibitors of cellular caspases (reviewed by Roulston and colleagues [47]). Several studies suggest that the herpesvirus and cellular FLIPs act as putative tumor progression factors in vitro and in tumor models based on the introduction of FLIP-transduced cell lines in mice; they provide evidence that tumor progression is in fact mediated by resistance to T-cell-induced death receptor-mediated apoptosis (15, 40). Our data obtained with the C488ΔFLIP deletion mutant cannot support the view that the herpesvirus saimiri vFLIP is a cofactor for oncogenesis. There was no difference in the efficiency of viral transformation in vitro; nor was there a significantly different incubation period of herpesvirus saimiri disease in cottontop tamarins (Table 1). There may be small differences in incubation time or disease progression that might be detectable by titration of the pathogenic viruses in the highly susceptible tamarins.
We also could not detect a significant difference in proliferation between T cells transformed by herpesvirus saimiri C488 or C488ΔFLIP (Fig. 7B). The postulated protective effect of vFLIP may be too discrete to be detected in transformed T cells, since herpesvirus saimiri-transformed human and simian T cells are rather resistant to Fas-mediated apoptosis (37). However, this resistance is not due to lack of death receptor expression, since significant levels of Fas-CD95 are detectable on the transformed T cells (Fig. 6). Moreover, a balanced expression and/or interaction of Fas-FasL may promote T-cell growth, resulting in a net growth of the culture, along with some apoptosis in fewer cells than those replenished by expansion (34).
The ORF carrying the vFLIP gene of herpesvirus saimiri is transcribed in virus-infected and -transformed cells, and the antiapoptotic effect found after infection of OMK cells by herpesvirus saimiri C488 is reversed by deletion of the vFLIP gene. Thus, an antiapoptotic effect can be attributed to the vFLIP gene expression in virus-infected cells. However, in the herpesvirus saimiri system which allows testing of rhadinovirus transformation and pathogenesis, the deletion of the vFLIP gene did not affect transformation or oncogenicity. This may have implications for KSHV/HHV8-associated disease, since the vFLIP gene is also latently transcribed in cells persistently infected by KSHV/HHV8.
In principle, the vFLIPs may have evolved to counteract T-cell-mediated apoptosis of persistently infected cells in the natural host or to compensate for proapoptotic signals provided by other viral proteins like envelope glycoproteins or vCyclin. The latter is an attractive hypothesis: the growth-promoting vCyclin is expressed from the same bi- or polycistronic message that also encodes vFLIP, and the vCyclin of KSHV/HHV8 has been shown to promote apoptosis in transfected cells that have entered S phase (45). High levels of the vFLIP and vCyclin message are detectable in advanced lesions of KS (51). In this model, vFLIP would not offer protection from externally induced apoptosis but would balance proapoptotic stimuli associated with the growth promoting functions of the virus itself. Although the similar domain structure and conserved genomic context, as well as in vitro data obtained from overexpression studies in transfected cell lines, suggest an analogous function, it still may be that the vFLIPs of KSHV/HHV8 and herpesvirus saimiri play different roles in their respective target tissues in vivo, and the vFLIP of KSHV may be essential for the transformation by KSHV, in contrast to herpesvirus saimiri. However, rhadinoviruses usually cause either minor or no relevant pathology in their natural host. Since we do not find herpesvirus saimiri vFLIP relevant to viral transformation or pathogenesis, we speculate that vFLIP expression may result in improved survival of acutely or persistently infected or transformed cells and may consequently enlarge or maintain the cellular reservoir for viruses in the persistently infected natural host.
ACKNOWLEDGMENTS
We thank Martina Göen and Monika Schmidt for their excellent technical assistance and Helmut Fickenscher and Jürg Tschopp for reagents.
This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 466, Lymphoproliferation und Immundefizienz.
REFERENCES
- 1.Albrecht J C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander L, Denekamp L, Knapp A, Auerbach M R, Damania B, Desrosiers R C. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J Virol. 2000;74:3388–3398. doi: 10.1128/jvi.74.7.3388-3398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashkenazi A, Dixit V M. Death receptors: signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 4.Aubert M, O'Toole J, Blaho J A. Induction and prevention of apoptosis in human HEp-2 cells by herpes simplex virus type 1. J Virol. 1999;73:10359–10370. doi: 10.1128/jvi.73.12.10359-10370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertin J, Armstrong R C, Ottilie S, Martin D A, Wang Y, Banks S, Wang G H, Senkevich T G, Alnemri E S, Moss B, Lenardo M J, Tomaselli K J, Cohen J I. Death effector domain-containing herpesvirus and poxvirus proteins inhibit both Fas- and TNFR1-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:1172–1176. doi: 10.1073/pnas.94.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biesinger B, Müller-Fleckenstein I, Simmer B, Lang G, Wittmann S, Platzer E, Desrosiers R C, Fleckenstein B. Stable growth transformation of human T lymphocytes by herpesvirus saimiri. Proc Natl Acad Sci USA. 1992;89:3116–3119. doi: 10.1073/pnas.89.7.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruder J T, Appiah A, Kirkman III W M, Chen P, Tian J, Reddy D, Brough D E, Lizonova A, Kovesdi I. Improved production of adenovirus vectors expressing apoptotic transgenes. Hum Gene Ther. 2000;11:139–149. doi: 10.1089/10430340050016229. [DOI] [PubMed] [Google Scholar]
- 8.Chaudhary P M, Jasmin A, Eby M T, Hood L. Modulation of the NF-kappa B pathway by virally encoded death effector domains-containing proteins. Oncogene. 1999;18:5738–5746. doi: 10.1038/sj.onc.1202976. [DOI] [PubMed] [Google Scholar]
- 9.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 10.Dear S, Staden R. A sequence assembly and editing program for efficient management of large projects. Nucleic Acids Res. 1991;19:3907–3911. doi: 10.1093/nar/19.14.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derfuss T, Fickenscher H, Kraft M S, Henning G, Lengenfelder D, Fleckenstein B, Meinl E. Antiapoptotic activity of the herpesvirus saimiri-encoded Bcl-2 homolog: stabilization of mitochondria and inhibition of caspase-3-like activity. J Virol. 1998;72:5897–5904. doi: 10.1128/jvi.72.7.5897-5904.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desrosiers R C, Burghoff R L, Bakker A, Kamine J. Construction of replication-competent herpesvirus saimiri deletion mutants. J Virol. 1984;49:343–348. doi: 10.1128/jvi.49.2.343-348.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desrosiers R C, Sasseville V G, Czajak S C, Zhang X, Mansfield K G, Kaur A, Johnson R P, Lackner A A, Jung J U. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J Virol. 1997;71:9764–9769. doi: 10.1128/jvi.71.12.9764-9769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Djerbi M, Screpanti V, Catrina A I, Bogen B, Biberfeld P, Grandien A. The inhibitor of death receptor signaling, FLICE-inhibitory protein defines a new class of tumor progression factors. J Exp Med. 1999;190:1025–1032. doi: 10.1084/jem.190.7.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duboise M, Guo J, Czajak S, Lee H, Veazey R, Desrosiers R C, Jung J U. A role for herpesvirus saimiri orf14 in transformation and persistent infection. J Virol. 1998;72:6770–6776. doi: 10.1128/jvi.72.8.6770-6776.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duboise S M, Guo J, Czajak S, Desrosiers R C, Jung J U. STP and Tip are essential for herpesvirus saimiri oncogenicity. J Virol. 1998;72:1308–1313. doi: 10.1128/jvi.72.2.1308-1313.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ensser A, Pfinder A, Müller-Fleckenstein I, Fleckenstein B. The URNA genes of herpesvirus saimiri (strain C488) are dispensable for transformation of human T cells in vitro. J Virol. 1999;73:10551–10555. doi: 10.1128/jvi.73.12.10551-10555.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falk L A. Biology of herpesvirus saimiri and herpesvirus ateles. In: Klein G, editor. Viral oncology. New York, N.Y: Raven Press; 1980. pp. 813–833. [Google Scholar]
- 20.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 21.Fleck M, Mountz J D, Hsu H C, Wu J, Edwards III C K, Kern E R. Herpes simplex virus type 2 infection induced apoptosis in peritoneal macrophages independent of Fas and tumor necrosis factor-receptor signaling. Viral Immunol. 1999;12:263–275. doi: 10.1089/vim.1999.12.263. [DOI] [PubMed] [Google Scholar]
- 22.Fleckenstein B, Desrosiers R C. Herpesvirus saimiri and herpesvirus ateles. In: Roizman B, editor. The herpesviruses. Vol. 1. New York, N.Y: Plenum Press; 1982. pp. 253–332. [Google Scholar]
- 23.Flore O, Rafii S, Ely S, O'Leary J J, Hyjek E M, Cesarman E. Transformation of primary human endothelial cells by Kaposi's sarcoma-associated herpesvirus. Nature. 1998;394:588–592. doi: 10.1038/29093. [DOI] [PubMed] [Google Scholar]
- 24.French L E, Tschopp J. Inhibition of death receptor signaling by FLICE-inhibitory protein as a mechanism for immune escape of tumors. J Exp Med. 1999;190:891–894. doi: 10.1084/jem.190.7.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friborg J, Jr, Kong W, Hottiger M O, Nabel G J. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- 26.Galvan V, Roizman B. Herpes simplex virus 1 induces and blocks apoptosis at multiple steps during infection and protects cells from exogenous inducers in a cell-type-dependent manner. Proc Natl Acad Sci USA. 1998;95:3931–3936. doi: 10.1073/pnas.95.7.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganem D. Human herpesvirus 8 and its role in the genesis of Kaposi's sarcoma. Curr Clin Top Infect Dis. 1998;18:237–251. [PubMed] [Google Scholar]
- 28.Guo J, Williams K, Duboise S M, Alexander L, Veazey R, Jung J U. Substitution of ras for the herpesvirus saimiri STP oncogene in lymphocyte transformation. J Virol. 1998;72:3698–3704. doi: 10.1128/jvi.72.5.3698-3704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanon E, Keil G, Drunen Littel-van den Hurk S, Griebel P, Vanderplasschen A, Rijsewijk F A, Babiuk L, Pastoret P P. Bovine herpesvirus 1-induced apoptotic cell death: role of glycoprotein D. Virology. 1999;257:191–197. doi: 10.1006/viro.1999.9620. [DOI] [PubMed] [Google Scholar]
- 30.Hu S, Vincenz C, Buller M, Dixit V M. A novel family of viral death effector domain-containing molecules that inhibit both CD-95- and tumor necrosis factor receptor-1-induced apoptosis. J Biol Chem. 1997;272:9621–9624. doi: 10.1074/jbc.272.15.9621. [DOI] [PubMed] [Google Scholar]
- 31.Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, Bodmer J L, Schroter M, Burns K, Mattmann C, Rimoldi D, French L E, Tschopp J. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 32.Jung J U, Choi J K, Ensser A, Biesinger B. Herpesvirus saimiri as a model for gammaherpesvirus oncogenesis. Semin Cancer Biol. 1999;9:231–239. doi: 10.1006/scbi.1998.0115. [DOI] [PubMed] [Google Scholar]
- 33.Jung J U, Trimble J J, King N W, Biesinger B, Fleckenstein B W, Desrosiers R C. Identification of transforming genes of subgroup A and C strains of herpesvirus saimiri. Proc Natl Acad Sci USA. 1991;88:7051–7055. doi: 10.1073/pnas.88.16.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kennedy N J, Kataoka T, Tschopp J, Budd R C. Caspase activation is required for T cell proliferation. J Exp Med. 1999;190:1891–1896. doi: 10.1084/jem.190.12.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knappe A, Hiller C, Niphuis H, Fossiez F, Thurau M, Wittmann S, Kuhn E M, Lebecque S, Banchereau J, Rosenwirth B, Fleckenstein B, Heeney J, Fickenscher H. The interleukin-17 gene of herpesvirus saimiri. J Virol. 1998;72:5797–5801. doi: 10.1128/jvi.72.7.5797-5801.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knappe A, Thurau M, Niphuis H, Hiller C, Wittmann S, Kuhn E M, Rosenwirth B, Fleckenstein B, Heeney J, Fickenscher H. T-cell lymphoma caused by herpesvirus saimiri C488 independently of ie14/vsag, a viral gene with superantigen homology. J Virol. 1998;72:3469–3471. doi: 10.1128/jvi.72.4.3469-3471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kraft M S, Henning G, Fickenscher H, Lengenfelder D, Tschopp J, Fleckenstein B, Meinl E. Herpesvirus saimiri transforms human T-cell clones to stable growth without inducing resistance to apoptosis. J Virol. 1998;72:3138–3145. doi: 10.1128/jvi.72.4.3138-3145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee H, Veazey R, Williams K, Li M, Guo J, Neipel F, Fleckenstein B, Lackner A, Desrosiers R C, Jung J U. Deregulation of cell growth by the K1 gene of Kaposi's sarcoma-associated herpesvirus. Nat Med. 1998;4:435–440. doi: 10.1038/nm0498-435. [DOI] [PubMed] [Google Scholar]
- 39.Mansfield K G, Westmoreland S V, DeBakker C D, Czajak S, Lackner A A, Desrosiers R C. Experimental infection of rhesus and pig-tailed macaques with macaque rhadinoviruses. J Virol. 1999;73:10320–10328. doi: 10.1128/jvi.73.12.10320-10328.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medema J P, de Jong J, van Hall T, Melief C J, Offringa R. Immune escape of tumors in vivo by expression of cellular FLICE-inhibitory protein. J Exp Med. 1999;190:1033–1038. doi: 10.1084/jem.190.7.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore P S, Chang Y. Antiviral activity of tumor-suppressor pathways: clues from molecular piracy by KSHV. Trends Genet. 1998;14:144–150. doi: 10.1016/s0168-9525(98)01408-5. [DOI] [PubMed] [Google Scholar]
- 42.Moses A V, Fish K N, Ruhl R, Smith P P, Strussenberg J G, Zhu L, Chandran B, Nelson J A. Long-term infection and transformation of dermal microvascular endothelial cells by human herpesvirus 8. J Virol. 1999;73:6892–6902. doi: 10.1128/jvi.73.8.6892-6902.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nava V E, Cheng E H Y, Veliuona M, Zou S, Clem R J, Mayer M L, Hardwick J M. Herpesvirus saimiri encodes a functional homolog of the human bcl-2 oncogene. J Virol. 1997;71:4118–4122. doi: 10.1128/jvi.71.5.4118-4122.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neipel F, Albrecht J C, Fleckenstein B. Human herpesvirus 8—the first human rhadinovirus. J Natl Cancer Inst Monogr. 1998;23:73–77. doi: 10.1093/oxfordjournals.jncimonographs.a024178. [DOI] [PubMed] [Google Scholar]
- 45.Ojala P M, Tiainen M, Salven P, Veikkola T, Castanos V E, Sarid R, Biberfeld P, Makela T P. Kaposi's sarcoma-associated herpesvirus-encoded v-cyclin triggers apoptosis in cells with high levels of cyclin-dependent kinase 6. Cancer Res. 1999;59:4984–4989. [PubMed] [Google Scholar]
- 46.Renne R, Blackbourn D, Whitby D, Levy J, Ganem D. Limited transmission of Kaposi's sarcoma-associated herpesvirus in cultured cells. J Virol. 1998;72:5182–5188. doi: 10.1128/jvi.72.6.5182-5188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roulston A, Marcellus R C, Branton P E. Viruses and apoptosis. Annu Rev Microbiol. 1999;53:577–628. doi: 10.1146/annurev.micro.53.1.577. [DOI] [PubMed] [Google Scholar]
- 48.Sarid R, Flore O, Bohenzky R A, Chang Y, Moore P S. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarid R, Wiezorek J S, Moore P S, Chang Y. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J Virol. 1999;73:1438–1446. doi: 10.1128/jvi.73.2.1438-1446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Searles R P, Bergquam E P, Axthelm M K, Wong S W. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73:3040–3053. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stürzl M, Hohenadl C, Zietz C, Castanos V E, Wunderlich A, Ascherl G, Biberfeld P, Monini P, Browning P J, Ensoli B. Expression of K13/v-FLIP gene of human herpesvirus 8 and apoptosis in Kaposi's sarcoma spindle cells. J Natl Cancer Inst. 1999;91:1725–1733. doi: 10.1093/jnci/91.20.1725. [DOI] [PubMed] [Google Scholar]
- 52.Sun R, Lin S F, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J Virol. 1999;73:2232–2242. doi: 10.1128/jvi.73.3.2232-2242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Talbot S J, Weiss R A, Kellam P, Boshoff C. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology. 1999;257:84–94. doi: 10.1006/viro.1999.9672. [DOI] [PubMed] [Google Scholar]
- 54.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer J L, Schroter M, Scaffidi C, Krammer P H, Peter M E, Tschopp J. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 55.Wang G H, Bertin J, Wang Y, Martin D A, Wang J, Tomaselli K J, Armstrong R C, Cohen J I. Bovine herpesvirus 4 BORFE2 protein inhibits Fas- and tumor necrosis factor receptor 1-induced apoptosis and contains death effector domains shared with other gamma-2 herpesviruses. J Virol. 1997;71:8928–8932. doi: 10.1128/jvi.71.11.8928-8932.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]