Abstract
Although microwave ablation (MWA) is an important curative therapy in colorectal cancer liver metastasis, recurrence still occurs clinically. Our previous studies have shown that the expression of programmed cell death 1 ligand 1 (PD‐L1) is upregulated following MWA, suggesting that MWA combined with anti‐PD‐L1 treatment can serve as a promising clinical therapeutic strategy against cancer. Using MWA‐treated preclinical mice models, MWA combined with αPD‐L1 treatment decreased tumor growth and prolonged overall survival (OS). Furthermore, through flow cytometry and single‐cell RNA sequencing analysis, we determined that the MWA plus αPD‐L1 therapy significantly suppressed CD8+ T cell exhaustion and enhanced their effector function. A significant increase in γ‐interferon (IFN‐γ) stimulated transcription factors, specifically Irf8, was observed. This enhancement facilitated the polarization of tumor‐associated macrophages (TAM1s and TAM2s) through the nuclear factor‐κB/JAK‐STAT1 signaling pathway. Furthermore, the combination therapy stimulated the production of CXC motif chemokine ligand (CXCL9) by TAM1s and tumor cells, potentially increasing the chemotaxis of CD8 T cells and Th1 cells. Knocking out Cxcl9 in MC38 tumor cells or using CXCL9 blockade enhanced tumor growth of untreated tumors and shortened OS. Taken together, our study showed that blocking the IFN‐γ‐Cxcl9‐CD8+ T axis promoted tumor progression and discovered a potential involvement of IRF8‐regulated TAMs in preventing T cell exhaustion. Collectively, we identified that the combination of MWA with anti‐PD‐L1 treatment holds promise as a therapeutic strategy to rejuvenate the immune response against tumors. This merits further exploration in clinical studies.
Keywords: Cxcl9, immunotherapy, IRF8, microwave ablation, PD‐L1
Microwave ablation plus α programmed death ligand 1 therapy may mediate the polarization of tumor‐associated macrophages (TAM1s) in the tumor microenvironment, thereby slowing down tumor growth, which are at least partially related to the nuclear factor‐κB/JAK‐STAT1 signaling pathway. Furthermore, blocking the γ‐interferon‐Cxcl9‐CD8+ T axis promoted tumor progression and discovered a potential involvement of IRF8‐regulated TAMs in preventing T cell exhaustion.
1. INTRODUCTION
The local management of tumors has been emphasized by accumulating evidence, showcasing the crucial role of radiofrequency ablation (RFA) and microwave ablation (MWA), particularly in colorectal cancer liver metastasis (CRLM). 1 , 2 However, in most cases, even if the tumor is completely resected by MWA, clinical recurrence still occurs due to unclear tumor boundaries and the absence of micrometastases, which suggests that the tumor‐promoting immune microenvironment cannot be ignored. Simultaneously, it will be important to determine which cell populations mediate the immunosuppressive microenvironment after MWA treatment and their underlying mechanism. Furthermore, new therapeutic strategies effectively combined with MWA are urgently needed.
Numerous preclinical and clinical investigations have shown that thermal ablation promotes the infiltration of tumor cells and myeloid cells by upregulating programmed death ligand 1 (PD‐L1), thereby inducing immunosuppression of residual tumor. 3 Previous studies have shown that PD‐1 inhibitors combined with RFA or MWA synergistically enhance T cell responses and induce tumor rejection in animal models. 4 , 5 Abundance of tumor‐associated macrophage 1 (TAM1) within the tumor microenvironment (TME) in preclinical models is linked to tumor regression and is deemed essential for the efficacy of T cell‐based immunotherapies. 6 , 7 Tumor‐associated macrophage 1 normally yields interferon gamma‐induced chemokine (IFN‐γ), CXC motif chemokine ligand (CXCL) 9, and CXCL10, which have proinflammatory effects. 8 To the best of our knowledge, the responses of myeloid and tumor cells after thermal ablation have been seldom evaluated, and their mechanisms of action in combination therapy remain unclear. 9 , 10
The purpose of this research was to analyze the impact of MWA combined with αPD‐L1 therapy on cytokines and chemokines involved in the interaction between tumor‐infiltrating lymphocytes and tumor cells.
2. MATERIALS AND METHODS
2.1. Mice and mouse cell lines
Male C57BL/6j or BALB/c mice (6–8 weeks old) were purchased from Cavens Laboratory Animal and maintained in a dedicated pathogen‐free facility. All animal experiments were carried out in accordance with protocols allowed by the Ethics Committee of The Third Affiliated Hospital of Soochow University. The mouse colon cancer cell lines CT26 and MC38 were obtained from the Chinese Academy of Sciences, Shanghai Institutes for Biological Sciences. The MC38 Cxcl9−/− tumor cell was provided by Haixing Bioscien. The MC38 and CT26 cells were cultured in DMEM (Gibco, Thermo Fisher Scientific) supplemented with 10% (v/v) FBS (Gibco, Thermo Fisher Scientific), 100 U/mL penicillin, and 100 mg/mL streptomycin. Cells were incubated at 37°C with 5% CO2.
2.2. Animal models and in vivo treatment
For tumor inoculation, MC38 (1 × 106) and CT26 (1 × 106) in 100 μL PBS were inoculated subcutaneously into both sides of C57BL/6j and BALB/c mice. Seven days after inoculation, mice were randomly assigned to the Control group, MWA group, αPD‐L1 group, or MWA combined with αPD‐L1 group. Simultaneously, MWA was carried out only on right‐sided tumors only on day 7 after inoculation. The approach to performing MWA and measuring subcutaneous tumors aligns with the methodology established by Chen et al. 11 For subcutaneous tumors, mice were euthanized at 24 h after three combined therapies (day 15 after MWA) for flowing cytometric, single cell RNA sequencing (scRNA‐seq) and quantitative real‐time PCR (qRT‐PCR).
Mice with established subcutaneous tumor were treated with 200 μg αPD‐L1 (Clone 10F.9G2; BioXcell) or αCXCL9 (Clone MIG‐2F5.5; BioXcell) in 100 μL PBS i.p. four times every 3 days starting from day 1 after MWA. To deplete CD8+ T cells, mice were intraperitoneally injected with 200 μg αCD8 (Clone 2.43; BioXcell) in 100 μL PBS every 3 days for four times starting 1 day before MWA. For the blockage of IFN‐γ, αIFN‐γ Abs (Clone XMG1.2; BioXcell) were administered with αPD‐L1 treatment.
2.3. Cxcl9 gene knockout in MC38 tumor cells
CRISPR‐Cas9‐mediated Cxcl9 gene ablation was undertaken using the CRISPR‐Cas9 ribonucleoprotein complexes. These complexes contained expression cassettes for hSpCas9 and chimeric guide RNA. To target exons 1 to 4 of the Cxcl9 gene, two guide RNA sequences, CAACGGGGCTGGCCAAATGCTGG and GGTCTCGAAAGCTACGTGGGAGG, were selected using the https://portals.broadinstitute.org/gpp/public/analysis‐tools/sgrna‐design website. The plasmid containing the guide RNA sequence was electrotransfected into cells using the Neon Transfection System (Thermo Fisher Scientific) according to the manufacturer's instructions. Plasmids from eight to 10 single colonies were isolated and sequenced through Sanger sequencing (GENEWIZ). Clones with mutations in both alleles were selected for subsequent studies. The generated cell lines using this strategy include gene (+/+), gene (+/−), gene (+/−A), gene (+/−B), and gene (−/−) cells.
2.4. Flow cytometry
Tumors excised from the tumor‐bearing mice were minced and digested with Liberase TL (REF 05401020001; Roche) and DNase I (REF 10104159001; Roche) for 30 min at 37°C. Subsequently, the resulting cell suspension was filtered through a 100 μm nylon filter (BD Biosciences) and reconstituted in Hank's media supplemented with 1% FCS for subsequent analysis. Anti‐mouse Abs including CD45 (Clone I3/2.3), Ghost (Cell Signaling Technology), CD3 (Clone 17A2), CD4 (Clone GK1.5), CD8 (Clone 53–6.7), NK1.1 (Clone PK136), FOXP3 (Clone MF‐14), CD11b (Clone M1/70), CD11c (Clone HL3), F4/80 (Clone BM8), MHCII (Clone M5/114.15.2), CD103 (Clone M290), PD‐L1 (Clone 10F.9G2), CD206 (Clone C068C2), CXCL9 (Clone MIG‐2F5.5), CD31 (Clone MEC13.3), and CD140b (Clone APB5) were used for cell staining and analysis. To assess intracellular cytokine levels, cellular stimulation was initiated by treating the cells with PMA (50 ng/mL; Sigma‐Aldrich), ionomycin (1 mg/mL; Sigma‐Aldrich), and monensin (GolgiStop; BD Biosciences) at 37°C for 5 h. Poststimulation, surface marker Abs were used for cell staining, followed by fixation and permeabilization as per the manufacturer's protocol outlined in the Invitrogen Fixing/Permeabilization Solution kit. The fixed cells were further stained using Abs targeting IFN‐γ (Clone XMG1.2), tumor necrosis factor‐α (TNF‐α; Clone MP6‐XT22), and granzyme B (GZMB; Clone GB11). Analysis was carried out utilizing a BD FACSAria II flow cytometer, with data processed through FlowJo software.
2.5. Quantitative real‐time PCR analyzing IFN‐γ‐stimulated MC38 cells both in vivo and in vitro
In 6‐well plates, MC38 tumor cells were seeded at a density of 0.5 × 105 cells/well. In vitro experiments were undertaken by subjecting the cells to either medium alone or the specified concentration of IFN‐γ (10 ng/mL) at 37°C for 24 h.
For tumor cell sorting, single cells prepared as described above were stained with Abs against CD45 (Clone 30‐F11), CD140b (Clone APB5), and CD31 (Clone 390) followed by FACS to obtain tumor cells. After in vivo sorting or in vitro stimulation, MC38 tumor cells were assessed by qRT‐PCR.
Quantitative real‐time PCR was carried out for mRNA analysis. Total RNA extraction from cells was undertaken using TRIzol (Thermo Fisher Scientific). Subsequently, the PrimeScript qRT‐PCR kit (Takara) was utilized to synthesize first‐strand cDNA. Target gene expression levels were assessed using specific primers detailed in Table 1.
TABLE 1.
Primers involved in quantitative real‐time PCR.
| Gene | Primer | Sequence (5′‐3′) |
|---|---|---|
| GAPDH | FP | GCACCGTCAAGGCTGAGAAC |
| RP | ATGGTGGTGAAGACGCCAGT | |
| CD274 | FP | GCTCCAAAGGACTTGTACGTG |
| RP | TGATCTGAAGGGCAGCATTTC | |
| Cxcl9 | FP | GGAGTTCGAGGAACCCTAGTG |
| RP | GGGATTTGTAGTGGATCGTGC | |
| Cxcl10 | FP | CCAAGTGCTGCCGTCATTTTC |
| RP | GGCTCGCAGGGATGATTTCAA |
Abbreviations: FP, forward primer; RP, reverse primer.
2.6. Quality control, normalization, integration, reduction, and clustering of scRNA‐seq data analysis
As our previous research, we utilized Cellranger 7.0.0 in combination with the mm10 reference genome to align and quantify mouse transcriptome data. 11 , 12 To remove artificial doublets, we applied the DoubletFinder package. The individual data was converted into a Seurat object. Subsequently, we determined the proportion of artificial nearest neighbors (referred to as pANN) for each genuine cell by calculating the PC distance matrix. The final predictions of doublets were generated by subjecting the pANN values to a threshold (set at 0.075), which was set according to the expected number of doublets. The default parameters used for this thresholding included PCs within the range of 1–20, pN at 0.25, and pK at 0.01. Further analyses involved the selection of cells meeting specific criteria, including the “singlet” status, nFeature RNA count less than 6000, nCount RNA less than 60,000, a mitochondrial gene percentage less than 5%, and a ribosomal gene percentage below 40%.
The application of the FindVariableFeatures function allowed for normalizing the data, resulting in the identification of the top 2000 variable features using default parameters. The RunPCA function was used to execute dimensionality reduction. To eliminate the batch effect present in our data, we utilized the Harmony package. In order to visualize the cells in a reduced‐dimensional space, the RunUMAP algorithm was subsequently implemented, using the top 30 principal components. Meaningful annotations were then assigned to the obtained clusters by renaming them according to marker genes mentioned in relevant published reports. The Seurat and ggplot2 packages were used to create visual representations.
2.7. Trajectory analysis
Within the scope of our study, the gene identification process using Monocle2 entailed the following essential steps: genes had to meet the criteria of being expressed in more than 10 subsets of cells and exhibiting an average expression level greater than 0.5. Subsequently, we used the DDRTree method to estimate pseudotime, which enabled the assignment of temporal values to individual cells, thus adding a temporal dimension for further analysis. To visually elucidate and interpret the developmental trajectories of cells, we leveraged the Plot_cell_trajectory function.
Additionally, we used the velocyto package, a tool capable of estimating RNA velocities of single cells by distinguishing between unspliced and spliced mRNAs in standard scRNA‐seq protocols. Specifically, we utilized the velocyto run10x method to estimate unspliced and spliced mRNAs. For the purpose of visualization, we turned to the velocyto.R package. These techniques collectively allowed us to gain insights into the temporal aspects of gene expression and the dynamics of cellular development in our study.
2.8. Gene set enrichment analysis
In our analysis, we gathered gene sets from the Molecular Signatures Database (MSigDB) and a previous article, 13 and we utilized the AddModuleScore function to compute the signature scores for these gene sets. Subsequently, we harnessed the ggplot2 package for creating visual representations of our results. This approach enabled us to assess the functional enrichment and associations of specific gene sets in our dataset.
2.9. SCENIC analysis
We undertook a transcriptional regulatory network analysis on subclusters extracted from the integrated scRNA‐seq dataset. Initially, the coexpression network was computed using GRNBoost2, available in the Python package pySCENIC. Subsequently, regulons were discerned utilizing the R package RcisTarget. The activity of each regulon was quantified for every cell through the R package AUCell. For dimensionality reduction of regulon activity, we adopted t‐distributed stochastic neighbor embedding (t‐SNE), executed by the runSCENIC_3_scoreCells function in the SCENIC R package. Visualization of the area under the curve values for the regulons was facilitated using the DoHeatmap function within Seurat.
2.10. Cell–cell interaction analysis
In our analysis, the initial step involved converting the normalized data into a CellChat object. We focused on utilizing the Secreted Signaling module for the analysis of cell–cell communication. To do this, we subset the expression data specifically for signaling genes, using the default parameters. Subsequently, we used the mergeCellChat function to integrate data from different experimental conditions, facilitating a comprehensive analysis. To assess differences in cell–cell interactions, we applied the netVisual_diffInteraction function. The netAnalysis_signalingRole_heatmap function was applied to visualize selected outgoing or incoming signaling.
2.11. Statistical analyses
Statistical analysis was carried out by using the two‐tailed unpaired t‐test, one‐way ANOVA, two‐way ANOVA, Kruskal–Wallis multiple comparison test, and the log‐rank test or log‐rank survival analysis.
3. RESULTS
3.1. Blockade of PD‐L1 following MWA shows synergistic antitumor effects
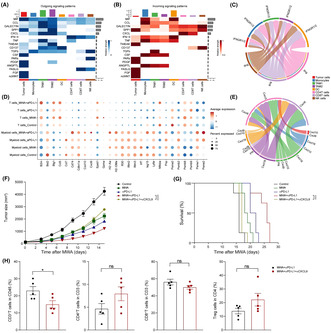
Using the aforementioned mouse model, we undertook combined treatment with αPD‐L1 Abs following MWA (Figure 1A). Compared with the control group, we observed that both standalone MWA and αPD‐L1 treatments resulted in a decelerated growth of the contralateral tumor (Figure 1B,D). Furthermore, treatment with either MWA or αPD‐L1 led to an extension survival time of mice (Figure 1C,E). Remarkably, αPD‐L1 therapy showed a remarkable efficacy when combined with MWA, as evidenced by significant differences in the rate of tumor growth and substantially prolonged survival compared with other treatment groups (Figure 1C,E). Together, the data above collectively suggest that the combination of MWA with αPD‐L1 demonstrates synergistic antitumor effects, distinguishing them from the outcomes observed with singular MWA or αPD‐L1 treatments.
FIGURE 1.
Microwave ablation (MWA) combined with α programmed death ligand 1 (αPD‐L1) therapy inhibits non‐MWA tumor growth and prolongs survival time. (A) Schematic diagram illustrating the investigation of the combined therapy involving MWA and PD‐L1 inhibition. MC38 or CT26 cells (1 × 106) were bilaterally subcutaneously injected into male C57BL/6 and BALB/c mice, respectively. MWA was carried out from day 7 after tumor inoculation. Day 1 after MWA, αPD‐L1 Ab (200 μg/100 μL) was administered (i.p.) with established subcutaneous tumor four times every 3 days. (B) Mean tumor growth and (C) Kaplan–Meier survival curves for C57BL/6 mice bearing MC38 tumors after treatment (n = 10). (D) Mean tumor growth and (E) Kaplan–Meier survival curves for BALB/c mice bearing CT26 tumors after treatment (n = 5). Two‐way ANOVA multiple comparison test and the log‐rank test were applied, and the mean ± SEM is shown. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
3.2. Microwave ablation combined with αPD‐L1 therapy increases proportion of infiltrated lymphocytes
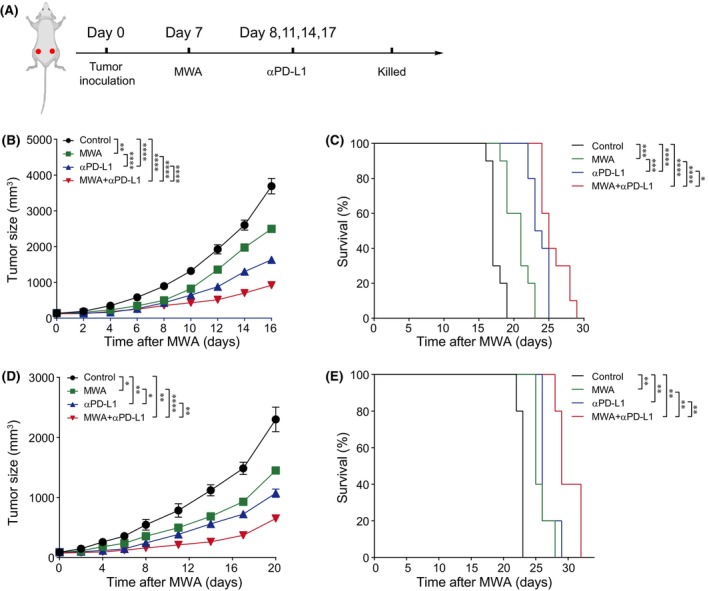
To investigate the synergistic anticancer mechanism of MWA combined with αPD‐L1 therapy, we undertook a comprehensive scRNA‐seq analysis of non‐MWA‐treated tumor tissues under the four aforementioned conditions. Compared to the control group, we observed that both MWA alone and combination therapy showed a decreased proportion of tumor cells and a higher proportion of immune cells, such as myeloid cells and T cells (Figure 2A,B). Furthermore, compared to the control group among subsets of T cells, MWA combined with αPD‐L1 therapy showed a notably increased proportion of effector CD8+ T cells, while the proportions of exhausted and cycling CD8+ T cells were decreased (Figure 2C,D). Importantly, the percentages of natural killer (NK) cells, stromal cells, and regulatory T cells (Tregs) showed almost no change (Figure 2B,D). Furthermore, in comparison to other groups, MWA plus αPD‐L1 therapy showed a notable increase in the percentages of CD3+ T cells and CD8+ T cells (Figures 2E,G and S1), which related to the scRNA‐seq analysis results. Additionally, it is worth noting that there were no substantial alterations in the proportions of NK cells or Tregs among the four groups (Figure 2F,H). Consequently, we hypothesized that αPD‐L1 enhances the MWA‐elicited antitumor immunity which may be associated with infiltrating T cells, particularly CD8+ T cells.
FIGURE 2.
Microwave ablation (MWA) combined with α programmed death ligand 1 (αPD‐L1) therapy induces CD8+ T cell accumulation in the tumor microenvironment of non‐MWA tumors. C57BL/6 mice were subcutaneously engrafted with 1 × 106 MC38 cells. From day 7 after tumor inoculation, MWA was performed. αPD‐L1 was administered through intraperitoneal injection to mice every 3 days for a total of four times,staring from day 1 after MWA. On day 15 post‐MWA, tumor samples were collected for flow cytometry and single cell RNA sequencing analysis. (A) UMAP analysis showing tumor cells, stromal cells, myeloid cells, T cells and natural killer (NK) cells present in MC38 tumors colored by cluster. (B) Bar plot displaying frequencies of cells related to (A). (C) UMAP analysis showing that T cells were classified into five subclusters. (D) Bar plot illustrating the distribution of T cell subsets with different groups. (E–H) Representative flow cytometry plots of tumor‐infiltrating lymphocytes (TILs) among four groups (n = 5). (E) Absolute cell numbers infiltrating CD3+ TILs. (F) Absolute cell numbers of infiltrating CD4+ TILs. (G) Absolute cell numbers of infiltrating CD8+ TILs. (H) Absolute cell numbers of infiltrating regulatory T cells (Tregs). One‐way ANOVA multiple comparison test was performed, and mean ± SEM is shown. *p < 0.05. ns, not significant.
3.3. Microwave ablation combined with αPD‐L1 therapy suppressed CD8 + T cell exhaustion and enhanced their effector function
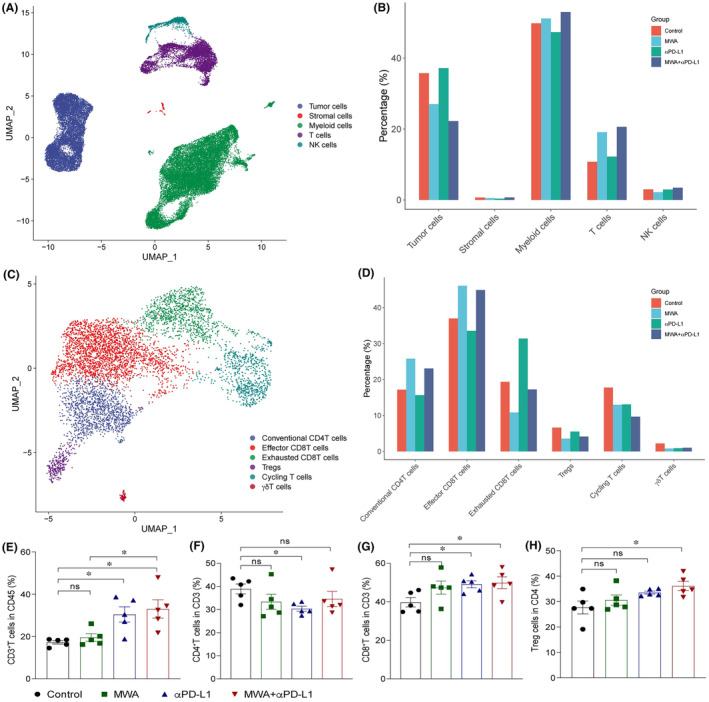
To comprehensively investigate functional changes, we undertook scRNA‐seq analysis of CD8+ T cell subpopulations, primarily subdividing them into effector CD8+ T cells, exhausted CD8+ T cells, and cycling CD8+ T cells (Figure 3A). Simultaneously, we utilized the monocle3 and velocyto.R 14 packages to construct developmental trajectories for these three CD8+ T cell subpopulations. The results revealed that effector CD8+ T cells further differentiate into cycling CD8+ T cells and exhausted CD8+ T cells (Figure 3B,C). Furthermore, compared with the control group, MWA alone upregulated the expression of chemokines associated with T cell recruitment and circulation (Cxcr3 and Cxcr4), as well as the molecule related effector function (Tnf). In contrast, αPD‐L1 alone upregulated the expression of immune checkpoint molecules (Pdcd1, Lag3, Havcr2, Ctla4), T‐cell receptor‐related molecules (Zap70, Nfact1), and the chemokine factor associated with T cell residence (Cxcr6). Within these gene expressions, MWA combined with αPD‐L1 therapy displayed a synergistic enhancing effect on molecules like Ifng and Gzmb, which are pertinent to T cell effector functions (Figure 3D). Subsequently, we undertook SCENIC 15 analysis to investigate the transcriptional regulatory networks of distinct CD8+ T cell subpopulations. Microwave ablation plus αPD‐L1 therapy promoted the regulatory activity of transcription factors E2F and the ETS family in effector CD8+ T cells (Figure 3E). Additionally, MWA plus αPD‐L1 therapy has a significant increase in the proportion of IFN‐γ+CD8+ T, TNF‐α+CD8+ T and GZMB+CD8+ T cells compared to the control group (Figure 3F–I). Assessing the role of CD8+ T cells in the context of MWA plus αPD‐L1 therapy, we used anti‐CD8 to deplete CD8+ T cells in the TME. The results revealed that tumor growth and overall survival (OS) among the four groups of mice carrying tumors tended to align (Figure 3J,K). Taken together, this study has highlighted the pivotal role of CD8+ T cells in the antitumor immune response of MWA plus αPD‐L1 therapy by using scRNA‐seq and flow cytometry.
FIGURE 3.
Combination of microwave ablation (MWA) with α programmed death ligand 1 (αPD‐L1) therapy reduces exhausted CD8+ T cells and increases the effector CD8+ T cell population in MC38 tumors. Tumor samples were analyzed by flow cytometry and scRNA‐seq on day 15 after MWA. (A) UMAP analysis showed that CD8+TILs were classified into effector CD8+T cell, exhausted CD8+T cells and cycling CD8+T cells. (B) Pseudotime analysis showing the developmental trajectory of CD8+ tumor‐infiltrating lymphocytes (TILs) in MC38 tumors. According to different gene markers, CD8+ TILs were classified into effector (Ccl5, Pdcd1, Ifng), exhausted (Havcr2, Lag3, Pdcd1), and cycling (Mki67) subpopulations. (C) RNA velocities analysis showed the developmental trajectory of CD8+ TILs within MC38 tumors. (D) Dot plot illustrating the expression of specific genes across various treatment groups. (E) Heatmap showing the area under curve of regulons' scores among distinct CD8+ T cell subpopulations. (F) Representative flow cytometry plots and absolute numbers of polyfunctional CD8+ T cells measured by co‐staining for (G) intracellular granzyme B (GZMB), (H) γ‐interferon (IFN‐γ), and (I) tumor necrosis factor‐α (TNF‐α), after ex vivo stimulation with PMA and ionomycin. (J, K) For depletion experiments, anti‐CD8 was injected i.p. into mice on day 1 prior to MWA. (J) The average growth of tumors and (K) survival curves using the Kaplan–Meier method were analyzed for mice hosting MC38 tumors posttreatment. (F–I) One‐way ANOVA and (J) two‐way ANOVA multiple comparison test and (K) the log‐rank test were carried out, and data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. ns, not significant.
3.4. Microwave ablation combined with αPD‐L1 therapy enhances infiltrations and functions of monocytes and macrophages
To obtain a profound understanding of how MWA plus αPD‐L1 therapy affects other cellular components of the TME, we undertook a subgroup analysis of myeloid cells. Analysis of scRNA‐seq data uncovered the diversity of myeloid cells within the tumor (Figure 4A). Specifically, there was an increase in the proportion of TAM1 and a decrease in TAM2 (Figure 4B). Next, we undertook a pseudotime analysis to assess the impact of MWA in combination with αPD‐L1 on monocytes and macrophages. Consistent with previous findings, 16 , 17 it appeared that these cells differentiate from monocytes into TAM1 and TAM2, and the combination therapy inhibited the differentiation of monocytes into TAM2 (Figure 4C). In addition, the percentage of TAM1 was increased, while TAM2 percentage was decreased after combination therapy by flow cytometry (Figure 4D–G). Furthermore, gene set enrichment analysis results revealed enhanced IFN‐γ signaling in monocytes, TAM1, TAM2, and cycling macrophages in the combination therapy (Figure 4H). Consecutively, we observed a significant upregulation of the IFN downstream target gene Cxcl9 in monocytes, TAM1, TAM2, and circulating macrophages of the MWA combined with αPD‐L1 treatment compared to the control group (Figure 4I). Next, SCENIC analysis revealed a significant upregulation in the regulatory activity of transcription factors in the IFN‐γ signaling pathway (Stat1, lrf1/5/7/8) and nuclear factor‐κB (NF‐κB) signaling pathway (Rel, Relb, and Nfkb1) in the combination therapy (Figure 4J). Collectively, these data indicated that MWA in combination with αPD‐L1 therapy can activate the NF‐κB and JAK‐STAT1 signaling pathways, thus promoting the polarization of TAM1 and TAM2 macrophages.
FIGURE 4.
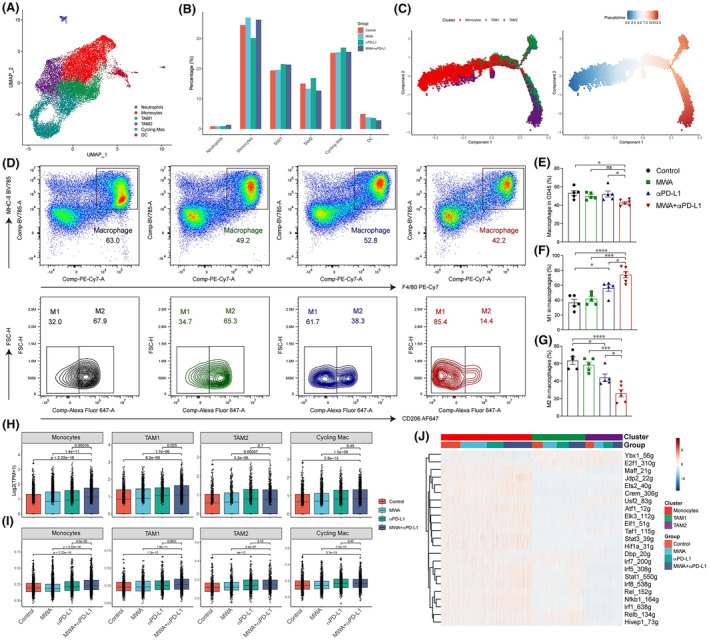
Microwave ablation (MWA) combined with α programmed death ligand 1 (αPD‐L1) therapy promotes the infiltration and functions of macrophages. Tumor samples were analyzed by flow cytometry and single cell RNA sequencing on day 15 after MWA. (A) UMAP analysis revealed the classification of myeloid cells. (B) Bar plot showing the proportions of myeloid cells in (A). (C) Pseudotime analysis showing the developmental trajectory of monocytes, tumor‐associated macrophages 1 (TAM1), and TAM2 in MC38 tumors. TAM1 and TAM2 are discerned through their positive expression of Cd68 and Mrc1 (CD206) genes, respectively. (D) Representative flow cytometry plots are displayed. (E) Macrophages are quantified in terms of absolute cell numbers. (F) Absolute cell numbers of TAM1. (G) Absolute cell numbers of TAM2. (H) Boxplot showing expression of Cxcl9 in monocytes, TAM1, TAM2, and cycling macrophages in different treatment groups. (I) Boxplot showing scores of γ‐interferon (IFN‐γ) signaling in monocytes, TAM1, TAM2, and cycling macrophages in different treatment groups. (J) Heatmap showing the area under the curve of regulons' scores among monocytes, TAM1, and TAM2 in different treatment groups. One‐way ANOVA and Kruskal–Wallis multiple comparison tests were performed, and data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
3.5. Microwave ablation combined with αPD‐L1 therapy enhances antigen presentation role of dendritic cells
Dendritic cells are the primary cells responsible for antigen presentation and are also the main cell type expressing PD‐L1. Subsequently, we undertook a subpopulation analysis of DCs, primarily categorizing them into four subgroups (Figure 5A). Notably, CD274 is primarily expressed in migratory DCs (Figure 5B). Through statistical analysis of various DC subgroups, we found almost no differences among them (Figure 5C). Following combination therapy, the proportion of DC1 cells was increased slightly, while DC2 cells remained largely unaffected by flow cytometry (Figure 5D–G). Furthermore, SCENIC analysis showed that the regulatory activity of IFN‐γ‐related transcription factors Stat1 and lrf1/8 in the combination therapy group was significantly enhanced (Figure 5H). Additionally, there was an enhancement in the expression of MHC‐I molecules in the combination therapy group (Figure 5I). Collectively, IFN‐γ from CD8+ T cells likely plays an essential role in the antigen presentation function of DCs in MWA plus αPD‐L1 therapy, thereby promoting the generation of an antitumor immune response.
FIGURE 5.
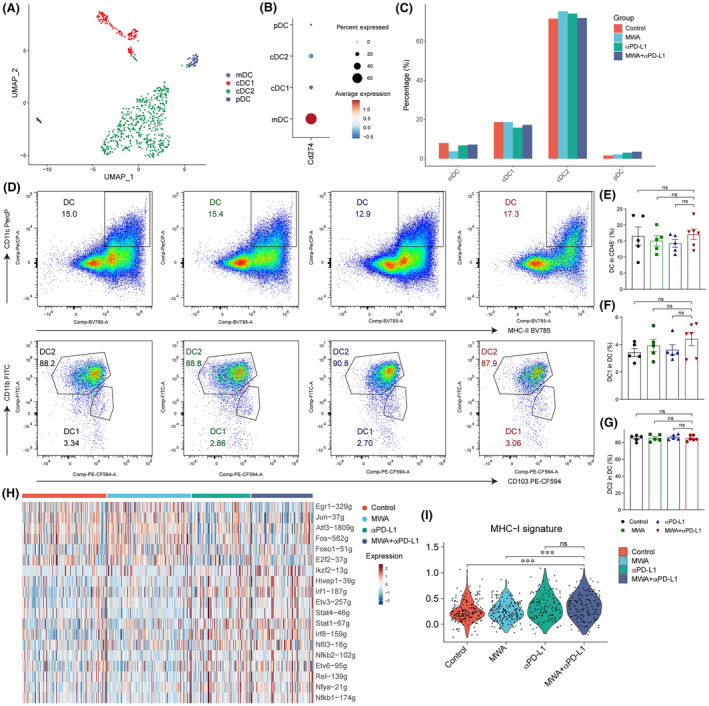
Antigen presentation role of dendritic cells (DCs) was promoted in microwave ablation (MWA) plus α programmed death ligand 1 (αPD‐L1) therapy. Tumor samples were analyzed by flow cytometry and single cell RNA sequencing on day 15 after MWA. (A) The UMAP analysis revealed the classification of tumor‐infiltrating DCs into migratory DC (mDC) (Ccr7), DC1 (Clea9a, Xcr1), DC2 (Cd209a), and plasmacytoid DC (pDCs) (Siglech). (B) Dot plot showing Cd274 expression in different treatment groups. (C) Bar plot depicting the distribution of DCs among various treatment groups. (D) Representative flow cytometry plots. (E) Absolute cell numbers of DCs. (F) Absolute cell numbers of DC1. (G) Absolute cell numbers of DC2. (H) Heatmap showing the area under the curve of regulons' scores among DC cells from various treatment methods. (I) Violin plot showing the scores of MHC‐I molecules in different treatment groups. One‐way ANOVA and Kruskal–Wallis multiple comparison test were performed, and data are shown as mean ± SEM. ns, not significant.
3.6. Microwave ablation combined with αPD‐L1 therapy alters interactions among different cellular populations within TME
In order to obtain a deeper understanding of the variances in cellular interactions, we utilized the CellChat package for the construction of a network that represents communication among cells. 18 Initially, the results revealed that tumor cells and myeloid cells are the primary sources of signaling molecules such as transforming growth factor‐β, CXCL, and CCL (Figure 6A), while CD4+ T cells and CD8+ T cells serve as the main providers of IFN‐γ and FAS ligand signals (Figure 6B). Subsequently, we observed that IFN‐γ originating from T cells plays a crucial immunomodulatory role on various immune cells within the TME (Figure 6C). In addition to genes associated with CXCL (such as Cxcl9 and Cxcl10), the combination therapy upregulated MHC‐II‐related molecules (such as Cd74 and H2‐Aa) and CCL5 (Figure 6D). In addition to the chemokines Cxcl9 and Cxcl10 downstream of the IFN‐γ signaling pathway, the chemokine Cxcl16 also plays a significant role in promoting cell recruitment (Figure 6E).
FIGURE 6.
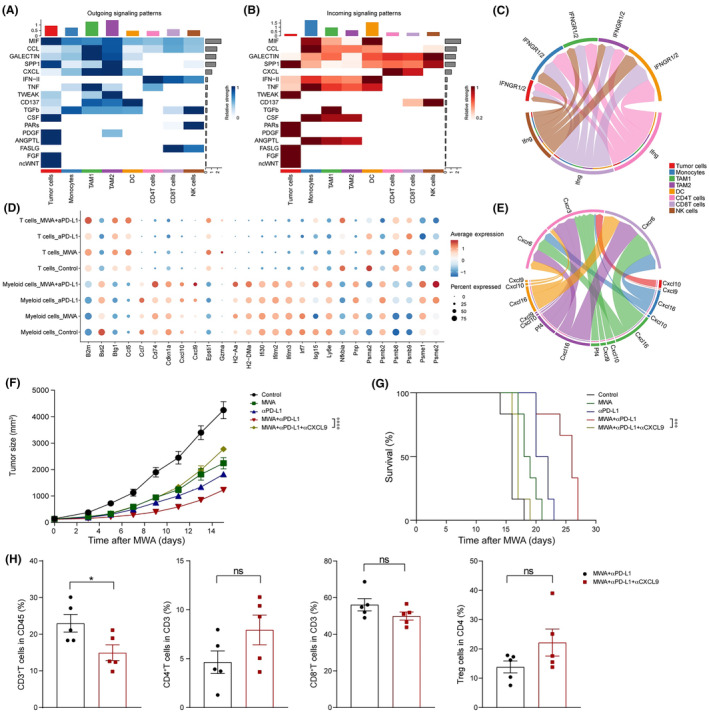
Interactions among different cellular populations were altered by microwave ablation (MWA) plus α programmed death ligand 1 (αPD‐L1) therapy. Tumor samples were analyzed by flow cytometry and single cell RNA sequencing analysis on day 15 after MWA. For the blockage of CXC motif chemokine ligand (CXCL9), αCXCL9 Abs (200 μg/100 μL) were given, consistent with treatment of αPD‐L1. (A, B) Heatmap showed the outgoing and incoming signaling pathways among various cellular populations, respectively. (C) The γ‐interferon (IFN‐γ) signaling pathway among different cellular populations. (D) Dot plots showing the expression levels of distinct chemokines in T cells or myeloid cells found in MC38 tumors among various treatment groups. (E) The CXCL signaling pathway among different cellular populations. (F) Representative mean tumor growth and (G) Kaplan–Meier survival curves of C57BL/6J mice (n = 5 mice per group) engrafted s.c. with MC38 cells and treated with MWA, αPD‐L1, MWA + αPD‐L1, or MWA + αPD‐L1 + αCXCL9. (H) Absolute cell numbers of CD3+ tumor‐infiltrating lymphocytes (TILs), CD4+ TILs, CD8+ TILs, and regulatory T cells (Tregs). Two‐way ANOVA multiple comparison test and log‐rank test were applied, and data are presented as mean ± SEM. NK, natural killer; TAM, tumor‐associated macrophage.
Furthermore, the results indicated that in the absence of CXCL9, both the tumor‐suppressive effect of αPD‐L1 and the antitumor effect induced by MWA were rendered ineffective (Figure 6F,G and S2). When αCXCL9 Ab was added, the percentages of CD3+ T cells in tumor tissue obviously decreased (Figure 6H), which suggests that MWA plus αPD‐L1 resulted in the tumor regression which requires Cxcl9 to facilitate antitumor immune responses through T cells.
3.7. Microwave ablation combined with αPD‐L1 therapy promotes the immunogenicity of tumor cells through IFN‐γ signaling
Through our previous research, we have discovered that tumor cells often upregulate the expression of PD‐L1 as a means to evade immune surveillance. Consequently, we undertook a further investigation into tumor cells and categorized them into two distinct types based on their functional characteristics: proliferative tumor cells and inflammatory tumor cells (Figure S3A). Proliferative tumor cells show heightened proliferative signaling pathways, whereas inflammatory tumor cells manifest more active inflammatory‐related signals (Figure S3B). Furthermore, compared to the other three groups, the expression of the genes Cd274, Stat1, Cxcl9, and Cxcl10 was upregulated in inflammatory tumor cells of the combination therapy group (Figure S3C), indicating a regulatory role of IFN‐γ on tumor cells. To validate the impact of IFN‐γ on tumor cells, we stimulated MC38 tumor cells with IFN‐γ in vitro and undertook qRT‐PCR analysis. We observed an obvious upregulation in the expression levels of the genes Cxcl9, Cxcl10, and Cd274 on MC38 tumor cells following IFN‐γ stimulation (Figure S3D). Furthermore, we isolated tumor cells from the tumor samples of all four treatment groups and undertook qRT‐PCR validation. The results indicated a significant upregulation in the expression of Cxcl9, Cxcl10, and Cd274 on tumor cells of the combination therapy group (Figure S3E). Moreover, the flow cytometry analysis corroborated these results, particularly in regard to the expression of CXCL9 on tumor cells (Figure S3F). Simultaneously, knockout of Cxcl9 in MC38 tumor cells promotes contralateral tumor progression after MWA plus anti‐PD‐L1 treatment, with accelerated tumor growth and shortened survival time, suggesting the immunoregulatory role of Cxcl9 in tumor cells (Figure S3G,H). In addition to IFN‐γ signaling, Cxcl9 is also upregulated by type I IFNs. To investigate this further, we utilized αIFN‐γ Abs targeting the IFN‐γ receptor (IFNGR) in conjunction with MWA and αPD‐L1 treatment. Upon blocking the IFNGR, the tumor growth rate accelerated, and the OS was shortened under the combined treatment of MWA plus αPD‐L1 (Figure S4A,B). In the context of MWA plus αPD‐L1 therapy, inhibition of IFN‐γ signaling resulted in elevated proportions of TAM2s (Figure S4C,D) and decreased expression levels of CXCL9 in both TAM1 and TAM2 cells (Figure S4E). Although the proportion of DCs remained unaffected following IFN‐γ blockade, a significant decrease was observed in the expression of CXCL9 specifically within DC2s (Figure S4F–H). Taken together, MWA plus αPD‐L1 therapy enhances tumor cell immunogenicity through IFN‐γ signaling, resulting in the upregulation of Cxcl9.
4. DISCUSSION
Thermal ablation induces “immunogenic cell death” and promotes antitumor immunity, providing opportunities for immunotherapy. 19 , 20 Our preclinical studies on mouse models supported MWA combined with immune checkpoint inhibitors not only had the potential to enhance CD8+ T cell infiltration in a distinct manner but also induced PD‐L1 expression in CD11b+ myeloid cells. 4 , 21 Herein, through the utilization of the mouse models of colon carcinoma (MC38 and CT26), we effectively illustrate that the combination of MWA alongside αPD‐L1 elicits a diverse immune reaction.
The upregulation of CXCL9 and CXCL10 was not only stimulated by IFN‐γ, but also by type‐I IFN signaling. 22 In our study, we showed that CXCL9 was translationally activated in tumor cells and TAMs after MWA through the IFN‐γ‐mediated signal transducer and activator of transcription 1 (STAT1) signaling pathway instead of type‐I IFN signaling, inducing T cell accumulation. Consistent with this, interrupting the IFN‐γ‐CXCL9‐CD8+ T axis by CD8+ T depletion, tumor‐intrinsic Cxcl9 KO or inhibition of IFN‐γ signaling significantly promote tumor progression. In addition, CCL5, which is a type of chemokine inherent to tumors, facilitates the movement of T cells within tumors and collaborates with CXCL9. 23 Tumors that showed diminished expression of CCL5 and CXCL9 became unresponsive to the treatment. 24 Overall, solid tumors commonly exhibit infiltration of CD8+ T cells, which is closely linked to the overexpression of CCL5 and CXCL9.
In our study, we found MWA combined with αPD‐L1 therapy not only enhanced their effector function, but also suppressed CD8+ T cell exhaustion. Specifically, in our investigation, we found that the application of MWA plus αPD‐L1 therapy yielded a decrease in the proportion of TAM2s. Furthermore, recent studies have revealed that continual interactions between TAM2s and CD8+ T cells impede the migration of T cells toward the tumor site. 25 Notably, the therapeutic outcomes were primarily attributed to a shift of TAMs toward the TAM1 phenotype. The accumulation of TAM1s resulted in an increased influx of T cells and augmented the tumor‐specific T helper 1 response through mechanisms involving direct cytotoxicity, as well as the generation of inflammatory cytokines and chemokines. 26
Interferon regulatory factor 5 (IRF5) and IRF8, as an essential transcription factor for TAM1 differentiation, 27 were obviously upregulated in MWA plus αPD‐L1 therapy. Interferon regulatory factor 8 is essential for the execution of DC1‐related functional processes, such as cross‐presentation. 28 Moreover, in patients with clear cell carcinoma, TAMs deficient in IRF8 show impaired proliferation and reduced expression of chemokine genes. Among these genes, their peculiar target, Cxcl9, assumes a vital function in drawing CTLs to interact with TAMs within the TME. In contrast, we found that MWA plus αPD‐L1 treatment improved survival by expressing higher levels of the IRF8‐TAM1 gene signature compared to massive CD8+ T cell infiltration. The reason for this difference is unclear but may be related to the use of different tumor tissues.
In summary, MWA plus αPD‐L1 therapy may mediate the polarization of TAM1s in TME, thereby slowing down tumor growth, which is at least partially related to the NF‐κB/JAK‐STAT1 signaling pathway. Furthermore, MWA plus αPD‐L1 treatment promoted MC38 tumor cells and TAMs to secrete CXCL9, which not only enhanced CD8+ T cell effector function, but also suppressed their exhaustion. Furthermore, exploring the potential involvement of IRF8‐regulated TAMs in preventing T cell exhaustion during MWA in combination with αPD‐L1 therapy can offer intriguing insights for innovative therapeutic strategies. Thus, this investigation yields novel understandings regarding the antitumor impact of MWA plus αPD‐L1 therapy by elucidating its underlying mechanism.
AUTHOR CONTRIBUTIONS
Ningning He: Data curation; formal analysis; investigation; methodology; validation; visualization; writing – original draft; writing – review and editing. Hao Huang: Data curation; formal analysis; investigation; methodology; validation; visualization; writing – original draft. Shaoxian Wu: Data curation; formal analysis; writing – original draft. Weipeng Ji: Data curation; investigation; writing – original draft. Yicheng Tai: Investigation; methodology; writing – original draft. Ruicheng Gao: Investigation; methodology; writing – original draft. Yingting Liu: Formal analysis; investigation; writing – original draft. Yungang Liu: Formal analysis; investigation; writing – original draft. Lujun Chen: Formal analysis; methodology; writing – original draft. Dawei Zhu: Conceptualization; funding acquisition; project administration; supervision; writing – original draft; writing – review and editing. Xiao Zheng: Conceptualization; funding acquisition; project administration; supervision; writing – original draft; writing – review and editing. Jingting Jiang: Conceptualization; funding acquisition; project administration; resources; supervision; writing – original draft; writing – review and editing.
FUNDING INFORMATION
National Natural Science Foundation of China (32270955, 82303164, 82202994), the Natural Science Foundation of Jiangsu Province (BK20211065), Postgraduate Research and Practice Innovation Program of Jiangsu Province (SJCX23_2031), Key R&D Project of Science and Technology Department of Jiangsu Province (BE2022721), the Science and Technology Support Plan (Social Development Project) of Changzhou (CE20235058).
CONFLICT OF INTEREST STATEMENT
The authors declares no conflict of interest.
ETHICS STATEMENTS
Approval of the research protocol by an institutional reviewer board: N/A.
Informed consent: N/A.
Registry and the registration no. of the study/trial: N/A.
Animal studies: The animal use in this study was carried out in compliance with the guidelines of the Ethics Committee of The Third Affiliated Hospital of Soochow University. All efforts were made to minimize suffering of mice.
Supporting information
Figure S1.
Figure S2.
Figure S3.
Figure S4.
ACKNOWLEDGMENTS
None.
He N, Huang H, Wu S, et al. Microwave ablation combined with PD‐L1 blockade synergistically promotes Cxcl9‐mediated antitumor immunity. Cancer Sci. 2024;115:2196‐2208. doi: 10.1111/cas.16182
Contributor Information
Dawei Zhu, Email: davysky@163.com.
Xiao Zheng, Email: zhengxiao@suda.edu.cn.
Jingting Jiang, Email: jiangjingting@suda.edu.cn.
REFERENCES
- 1. Solbiati L, Ahmed M, Cova L, Ierace T, Brioschi M, Goldberg SN. Small liver colorectal metastases treated with percutaneous radiofrequency ablation: local response rate and long‐term survival with up to 10‐year follow‐up. Radiology. 2012;265(3):958‐968. [DOI] [PubMed] [Google Scholar]
- 2. Popovic P, Surlan‐Popovic K, Lukic S, Mijailovic M, Jankovic S, Kuhelj D. Percutaneous imaging‐guided radiofrequency ablation of small renal cell carcinoma: techniques and outcomes of 24 treatment sessions in 18 consecutive patients. J BUON. 2011;16:127‐132. [PubMed] [Google Scholar]
- 3. Yu M, Pan H, Che N, et al. Microwave ablation of primary breast cancer inhibits metastatic progression in model mice via activation of natural killer cells. Cell Mol Immunol. 2021;18:2153‐2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shi L, Chen L, Wu C, et al. PD‐1 blockade boosts radiofrequency ablation‐elicited adaptive immune responses against tumor. Clin Cancer Res. 2016;22:1173‐1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang SLT, Chen Y, Liu J, et al. Microwave ablation combined with anti‐PD‐1 therapy enhances systemic antitumor immunity in a multitumor murine model of Hepa1‐6. Int J Hyperth. 2022;39:278‐286. [DOI] [PubMed] [Google Scholar]
- 6. House IGSP, Lai J, Chen AXY, et al. Macrophage‐derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin Cancer Res. 2020;26:487‐504. [DOI] [PubMed] [Google Scholar]
- 7. de Masson A, Darbord D, Dobos G, et al. Macrophage‐derived CXCL9 and CXCL11, T‐cell skin homing and disease control in mogamulizumab‐treated CTCL patients. Eur J Cancer. 2021;156:S19‐S20. [DOI] [PubMed] [Google Scholar]
- 8. Murray P, Allen J, Biswas S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schneider T, Sevko A, Heussel CP, et al. Serum inflammatory factors and circulating immunosuppressive cells are predictive markers for efficacy of radiofrequency ablation in non‐small‐cell lung cancer. Clin Exp Immunol. 2015;180:467‐474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu X, Zhang W, Xu Y, et al. Targeting PI3Kgamma/AKT pathway remodels LC3‐associated phagocytosis induced immunosuppression after radiofrequency ablation. Adv Sci (Weinh). 2022;9:e2102182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen Y, Huang H, Li Y, et al. TIGIT blockade exerts synergistic effects on microwave ablation against cancer. Front Immunol. 2022;13:832230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shao D, Chen Y, Huang H, et al. LAG3 blockade coordinates with microwave ablation to promote CD8(+) T cell‐mediated anti‐tumor immunity. J Transl Med. 2022;20:433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dixon KO, Tabaka M, Schramm MA, et al. TIM‐3 restrains anti‐tumour immunity by regulating inflammasome activation. Nature. 2021;595:101‐106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. La Manno G, Soldatov R, Zeisel A, et al. RNA velocity of single cells. Nature. 2018;560:494‐498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aibar S, Gonzalez‐Blas CB, Moerman T, et al. SCENIC: single‐cell regulatory network inference and clustering. Nat Methods. 2017;14:1083‐1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schetters STT, Rodriguez E, Kruijssen LJW, et al. Monocyte‐derived APCs are central to the response of PD1 checkpoint blockade and provide a therapeutic target for combination therapy. J Immunother Cancer. 2020;8:e000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Genin M, Clement F, Fattaccioli A, Raes M, Michiels C. M1 and M2 macrophages derived from THP‐1 cells differentially modulate the response of cancer cells to etoposide. BMC Cancer. 2015;15:577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jin S, Guerrero‐Juarez CF, Zhang L, et al. Inference and analysis of cell‐cell communication using CellChat. Nat Commun. 2021;12:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheng AL, Hsu C, Chan SL, Choo SP, Kudo M. Challenges of combination therapy with immune checkpoint inhibitors for hepatocellular carcinoma. J Hepatol. 2020;72:307‐319. [DOI] [PubMed] [Google Scholar]
- 20. Zhou CLY, Li J, Song B, et al. A phase 1/2 multicenter randomized trial of local ablation plus toripalimab versus toripalimab alone for previously treated unresectable hepatocellular carcinoma. Clin Cancer Res. 2023;29:2816‐2825. [DOI] [PubMed] [Google Scholar]
- 21. Shi L, Wang J, Ding N, et al. Inflammation induced by incomplete radiofrequency ablation accelerates tumor progression and hinders PD‐1 immunotherapy. Nat Commun. 2019;10:5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mikucki ME, Fisher DT, Matsuzaki J, et al. Non‐redundant requirement for CXCR3 signalling during tumoricidal T‐cell trafficking across tumour vascular checkpoints. Nat Commun. 2015;6:7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Spranger S, Dai D, Horton B, Gajewski TF. Tumor‐residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell. 2017;31:711‐723.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dangaj D, Bruand M, Grimm AJ, et al. Cooperation between constitutive and inducible chemokines enables T cell engraftment and immune attack in solid tumors. Cancer Cell. 2019;35:885‐900.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu G, Zhang R, Geng S, et al. Myeloid cell‐derived inducible nitric oxide synthase suppresses M1 macrophage polarization. Nat Commun. 2015;6:6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bergamaschi C, Pandit H, Nagy BA, et al. Heterodimeric IL‐15 delays tumor growth and promotes intratumoral CTL and dendritic cell accumulation by a cytokine network involving XCL1, IFN‐gamma, CXCL9 and CXCL10. J Immunother Cancer. 2020;8:e000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krausgruber T, Blazek K, Smallie T, et al. IRF5 promotes inflammatory macrophage polarization and Th1/Th17 response. Nat Immunol. 2011;12:231‐238. [DOI] [PubMed] [Google Scholar]
- 28. Nixon BG, Kuo F, Ji L, et al. Tumor‐associated macrophages expressing the transcription factor IRF8 promote T cell exhaustion in cancer. Immunity. 2022;55:2044‐2058.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.
Figure S2.
Figure S3.
Figure S4.


