Abstract
Background
There have been studies on the role of sperm-associated antigen 6 (SPAG6) in cytoskeleton formation and growth cone stability, but it is also unknown how spag6 affect tumor growth and development. The aim of this study was to clarify the role of SPAG6 in pan-cancer, with some findings about thyroid carcinoma (THCA) validated through experiments.
Methods
We examined the role of SPAG6 in pan-cancer, with the data being collected from databases. Further analysis was conducted to assess its correlations with prognosis, gene heterogeneity, stemness, and tumor immunity. The interacting proteins of SPAG6 were also identified, and gene ontology enrichment analysis was performed to determine its biological function. We preliminarily confirmed the role of SPAG6 via in vitro experiments and immunofluorescence staining.
Results
This study found that SPAG6 expression was differentially expressed in cancers and at various tumor stages and grades. In stomach and esophageal carcinoma (STES), stomach adenocarcinoma (STAD), kidney renal clear cell carcinoma (KIRC), lung squamous cell carcinoma (LUSC), and adrenocortical carcinoma (ACC), SPAG6 expression was correlated with gender. SPAG6 expression was also found to be correlated with prognostic value, with low expression being associated with poor prognosis. Furthermore, SPAG6 expression was positively linked with immune-related cells in HNSC, chemokine receptors in LUSC, and immune checkpoint genes in THCA. Furthermore, SPAG6 overexpression suppressed the malignant phenotypes of THCA cells, manifested by slower proliferation and decreased migration. The different SPAG6 expression in THCA led to different malignant phenotypes, which are involved in the upregulation of DNA repair, MYC targets, peroxisome, and G2M checkpoint.
Conclusions
SPAG6 plays a significant role as an oncogene and can be used as a marker to predict the prognosis of cancer. SPAG6 influences both the tumor immune infiltration and microenvironment, making it a promising immunotherapeutic target for tumor therapy.
Keywords: Sperm-associated antigen 6 (SPAG6), pan-cancer, tumor immunity, immune infiltration, tumor microenvironment (TME)
Highlight box.
Key findings
• SPAG6 plays a significant role in pan-cancer, especially in thyroid carcinoma (THCA).
What is known and what is new?
• SPAG6 is a member of the cancer-testis antigen family.
• SPAG6 is differentially expressed in different cancers, with varying tumor stages and grades, and its expression is also linked to poor prognosis, indicating that it can be used as a marker to predict cancer prognosis.
What is the implication, and what should change now?
• SPAG6 expression is positively linked with immune-related cells, immunomodulators, and immune checkpoint genes, suggesting its potential as a target for cancer immunotherapy. Spag6 could act as a promising target for cancer treatment and prognostication, especially in THCA.
Introduction
The tumorigenesis and development of cancers involve complicated changes that result in varying downstream effects. Pan-cancer gene analysis is conducted to identify the similarities and differences among various types of cancer cells (1,2). Cancer-testis antigens (CTAs) have emerged as a promising class of proteins that can stimulate an anticancer immune response (3). CTAs are considered to be novel biomarker, as they are typically only expressed in immune-privileged sites. When expressed in somatic cells, the proteins encoded by these genes elicit both humoral and cell-mediated immune responses, making CTAs highly attractive targets for cancer immunotherapy (4).
Sperm-associated antigen 6 (SPAG6), belonging to the CTA family, is essential for microtubule binding and plays a critical role in cytoskeleton formation, growth cone stability, and cilia motility (5-8). Initially identified in human testicular tissue, SPAG6 governs sperm flagella motility and germ cell maturation (9). Recent investigations have unveiled the association of SPAG6 with tumorigenesis and progression (10-14). Nonetheless, limited studies have examined the precise mechanisms underlying SPAG6’s involvement in cancer and immunity.
The presence of tumor-associated immunogenic proteins poses challenges in developing antigen-specific cancer treatments. Despite the efficacy of tumor immunotherapy, a significant proportion of patients do not benefit. Programmed death 1 (PD-1) is a prominent immune checkpoint receptor (15,16). Dendritic cells (DCs) play a crucial role in antigen presentation and immune response promotion (17). Cancers evade immune recognition through various mechanisms (18). Previous research has linked SPAG6 to immunodeficiency, which included reduced CD8 cytotoxicity, decreased CD8 T-cell interferon-γ (IFNγ) secretion, and impaired antibody production (19). However, the role of SPAG6 in thyroid carcinoma (THCA) and other cancers remains unclear.
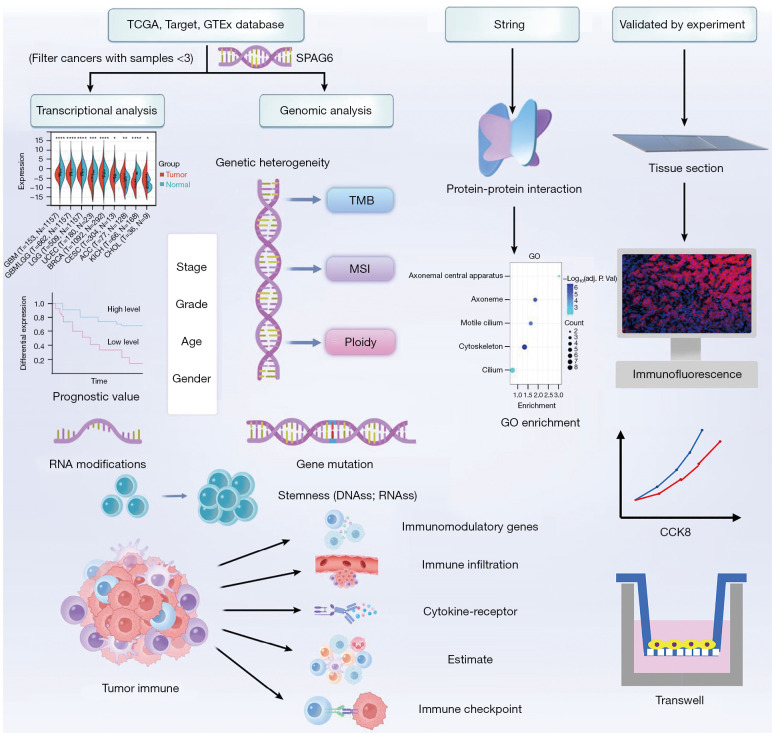
In this study, we aimed to clarify the relationship between SPAG6 and multiple cancers by integrating data from multiple databases using polyomics methods. We analyzed the expression, prognosis, gene heterogeneity, and tumor microenvironment (TME) of SPAG6 and further examined the association between SPAG6 and immunotherapy across different types of cancers (Figure 1). The results of this study provide a comprehensive understanding of the role of SPAG6 in various types of cancer, and serve as a valuable reference for further research. We present this article in accordance with the MDAR reporting checklist (available at https://gs.amegroups.com/article/view/10.21037/gs-24-157/rc).
Figure 1.
Flowchart of the study. TCGA, The Cancer Genome Atlas; GTEx, Genotype-Tissue Expression; TMB, tumor mutation burden; MSI, microsatellite instability; GO, Gene Ontology; CCK8, Cell Counting Kit 8.
Methods
Data collection
Data for 34 cancers were downloaded from The Cancer Genome Atlas (TCGA), Therapeutically Applicable Research to Generate Effective Treatment (TARGET), and the Genotype-Tissue Expression (GTEx) databases via the University of California Santa Cruz (UCSC) browser (https://xenabrowser.net/); meanwhile, single-nucleotide variation data were obtained from the Genomic Data Commons (GDC) portal (https://portal.gdc.cancer.gov/). A list of tumor abbreviations can be found in the Table S1.
SPAG6 expression, pathology, and clinical trait analysis
The expression level of SPAG6 was analyzed using Posit software, which further integrated information from the databases and a previous study (20). Pearson’s correlation method was used to calculate the association between SPAG6 expression and other factors.
Survival analysis
Kaplan-Meier survival analysis was conducted to assess the disease progression outcome in patients with high or low expression of SPAG6. The parameters considered for survival analysis included overall survival (OS), progression-free interval (PFI), disease-free survival (DFS), and disease-specific survival (DSS). The results were visualized using the “survival” R package (The R Foundation of Statistical Computing), and the Cox proportional hazards regression model was calculated using the coxph function.
Genetic heterogeneity analysis
For the data obtained from TCGA pan-cancer database, Pearson’s correlation was used to determine the correlation between SPAG6 and tumor mutational burden (TMB), microsatellite instability (MSI), tumor ploidy, and tumor purity.
Mutation analysis
R software was used to analyze the expression level of SPAG6, with the “maftools” R package being employed to determine the protein structure domains.
Cancer stemness analysis
After collection of pan-cancer data, the expression of SPAG6 was obtained across various cancer types. Subsequently, messenger RNA (mRNA) expression-based stemness scores (RNAss) and DNA methylation-based stemness scores (DNAss) were obtained (21).
TME and immune-related factor analysis
The “ESTIMATE” R package was used to calculate immune score, stromal score, and ESTIMATE score. The “IOBR” R package was used to estimate SPAG6 gene expression data (22). The Tumor Immune Estimation Resource (TIMER; cistrome.shinyapps.io/timer) platform was used to determine the correlation of SPAG6 expression with immune infiltration factors (23). Pearson correlation coefficients were calculated using the corr.test function in the “psych” R package for tumor scores and immunomodulators (24).
Protein-protein interaction network construction and enrichment analysis
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://string-db.org/) was accessed for hub targets and to conduct Gene Ontology (GO) enrichment analysis using parameters “medium” (≥0.4) and “Homo sapiens”. Results were visualized using Bioinformatics (http://www.bioinformatics.com.cn/). Adjusted P values were calculated using the false discovery rate (FDR) algorithm for result filtering. Gene set enrichment analysis (GSEA) was conducted with GSEA software (version 3.0) from the GSEA website (http://software.broadinstitute.org/gsea/index.jsp). The SPAG6 high-expression group (≥50%) and low-expression group (<50%) were defined. Gene analysis was conducted using h.all.v7.4. symbols.gmt in GSEA software with a minimum gene set of 5 and a maximum gene set of 5,000.
Immunohistochemical staining
The tissues samples were obtained from the patients with liver hepatocellular carcinoma (LIHC), cholangiocarcinoma (CHOL), ovarian serous cystadenocarcinoma (OV), uterine corpus endometrial carcinoma (UCEC), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), or THCA from pathology department, Shandong Provincial ENT Hospital, between September 2021 to February 2024. Formalin-fixed tissue sections from various cancers were fully embedded and cut into 10 µm sections for immunohistochemical staining. After dewaxing and antigen retrieval, the sections were blocked with PBT-1 for 60 minutes at room temperature. The sections were then incubated overnight with a primary antibody against spag6 (anti-SPAG6, 1:200, ab155653, Abcam, Cambridge, MA, USA) at 4 °C. Subsequently, a secondary antibody conjugated with Alexa Fluor 546 (1:1,000, Invitrogen, Carlsbad, CA, USA) and DAPI (1:1,000, D9542, Sigma-Aldrich, St. Louis, MO, USA) were incubated for 1 hour at room temperature. The sections were washed with buffer at each step. The final staining was visualized using a laser scanning confocal microscope (Leica SP8, Leica, Wetzlar, Germany).
Cell transfection
PEIpro (Polyplus) was applied for conducting cell transfection, which lasted 48 hours as per its standard guide. B-CPAP, and KTC-1 cells (Stem Cell Bank, Chinese Academy of Sciences; American Type Culture Collection) were transfected with SPAG6-overexpressed plasmid or empty vector as a negative control by GeneChem.
Cell Counting Kit 8 assay (CCK8)
B-CPAP, and KTC-1 cells were processed as required and seeded into 96-well plates at a density of [3–5]×103 per well. After treatment for 0, 24, 48, 72 and 96 h, 10 µL CCK8 (BioSharp, Anhui, China) was added to each well, and absorbance was measured at 450 nm with a spectrophotometer after incubation for 2 h.
Transwell migration assays
Cell migration was assessed using an 8-µm Transwell chamber (BioSharp, Anhui, China). Serum-free 1640 medium (200 µL) containing 4×104 cells were seeded in the upper chamber. 500 µL 1640 medium containing 20% FBS was added to the lower chamber. After incubation for 24 h, the cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Migrated cells were photographed in four random fields using an inverted light microscope (Olympus, Tokyo, Japan). The experiments were repeated three times independently. Data were quantified via the ImageJ software.
Statistical analysis
Data were log2 transformed. The Wilcoxon rank-sum and signed-rank tests were used to compare two groups, while the Kruskal-Wallis test was used to compare multiple groups, and the logrank test was used for survival analysis. Significance was defined as a P value <0.05. Pearson correlation coefficient was used to analyze the correlation of SPAG6 expression with other factors.
Ethical statement
The studies involving human samples were approved by the Shandong Provincial ENT Hospital Ethical Committee (No. 2024-019-01). Written informed consent to participate in this study was provided by the participants. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Clinical landscape of SPAG6 expression in pan-cancer
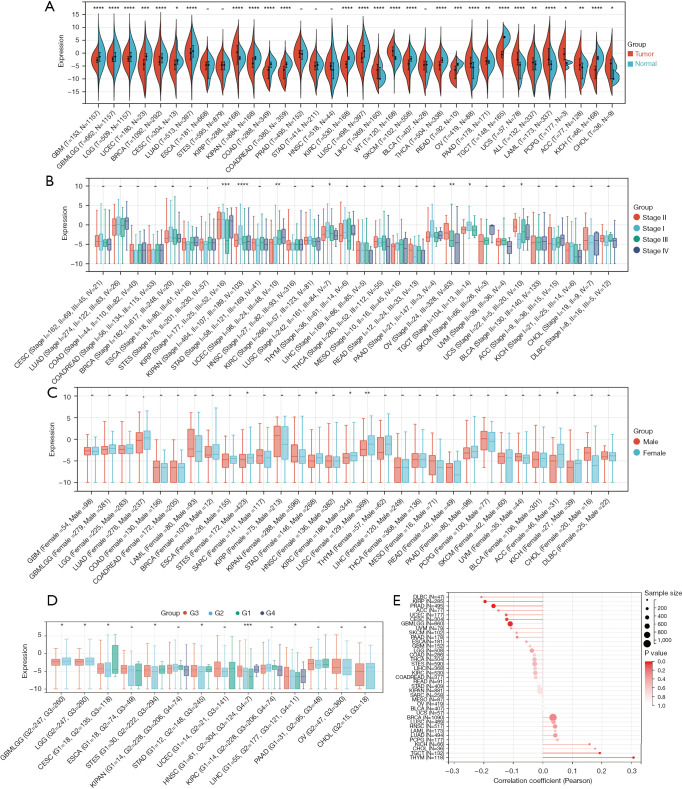
The analysis revealed that SPAG6 expression was upregulated in 11 cancers [kidney renal papillary cell carcinoma (KIRP), LIHC, high-risk Wilms tumor (WT), OV, pancreatic adenocarcinoma (PAAD), uterine carcinosarcoma (UCS), acute lymphoblastic leukemia (ALL), acute myeloid leukemia (LAML), pheochromocytoma and paraganglioma (PCPG), adrenocortical carcinoma (ACC), and CHOL] and downregulated in 17 cancers [glioblastoma multiforme (GBM), glioma (GBMLGG), brain lower-grade glioma (LGG), UCEC, breast invasive carcinoma (BRCA), CESC, lung adenocarcinoma (LUAD), pankidney cohort (KIPAN), colon adenocarcinoma (COAD), colon adenocarcinoma/rectum adenocarcinoma esophageal carcinoma (COADREAD), kidney renal clear cell carcinoma (KIRC), lung squamous cell carcinoma (LUSC), skin cutaneous melanoma (SKCM), THCA, rectum adenocarcinoma (READ), testicular germ cell tumors (TGCT), and kidney chromophobe (KICH)] (Figure 2A). Tumor stage showed a correlation with SPAG6 expression in seven cancers (KIRP, KIPAN, UCEC, LUSC, OV, TGCT, and UCS) (Figure 2B). Gender-based differences in SPAG6 expression were observed in five cancers, with higher expression in male patients in the stomach and esophageal carcinoma (STES), stomach adenocarcinoma (STAD), KIRC, and LUSC and higher expression in female patients in ACC (Figure 2C). SPAG6 expression also varied with tumor grade in seven cancers [GBMLGG, LGG, CESC, STES, STAD, head and neck squamous cell carcinoma (HNSC), and LIHC] (Figure 2D). Age-based differences in SPAG6 expression were found in six cancers, with a positive association in thymoma (THYM) and TGCT and a negative association in GBMLGG, CESC, KIRP, and prostate adenocarcinoma (PRAD) (Figure 2E).
Figure 2.
Expression of SPAG6 in the clinical data. (A) The differential expression of SPAG6 in pan-cancer (red) and normal tissues (blue) downloaded from different databases. (B) The correlation between tumor stage and SPAG6 expression. Stage I = blue, stage II = red, stage III = green, and stage IV = purple. (C) The correlation between patients’ gender and SPAG6 expression. Male = red and female = blue. (D) The correlation between patients’ grade and SPAG6 expression. Grade 1 = green, grade 2 = blue, grade 3 = red, and grade 4 = purple. (E) The correlation between patients’ age and SPAG6 expression. –, no statistical significance; *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001. G, grade.
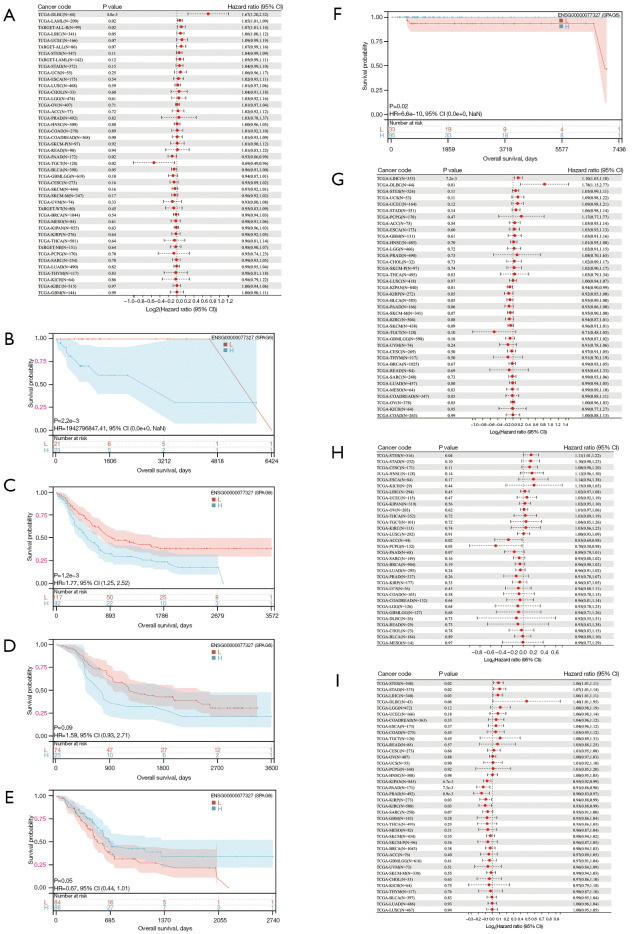
Prognostic value of SPAG6 across cancers
Cox regression analysis revealed a correlation between SPAG6 expression and five cancer types. High SPAG6 expression was associated with poor prognosis in LAML, ALL, and lymphoid neoplasm diffuse large b-cell lymphoma (DLBC), while low expression was associated with poor prognosis in PAAD and TGCT (Figure 3A). Kaplan-Meier curves further confirmed the impact of SPAG6 expression on osteosarcoma (OS), with high expression associated with poor prognosis in LAML, ALL, and DLBC (Figure 3B-3D) and low expression associated with poor prognosis in PAAD and TGCT (Figure 3E,3F). Additionally, SPAG6 expression correlated with DSS in LIHC, DLBC, KIRP, KIPAN, and bladder urothelial carcinoma (BLCA). High SPAG6 expression correlated with poor prognosis in LIHC and DLBC, while low expression correlated with poor prognosis in KIRP, KIPAN, and BLCA (Figure 3G). The analysis also indicated a relationship between SPAG6 expression and DFI in three cancers (Figure 3H), with high expression correlating with poor prognosis in STES and low expression correlating with poor prognosis in PCPG and ACC. SPAG6 expression correlated with PFI in eight cancers (Figure 3I), with high expression correlating with poor prognosis in STES, STAD, and LIHC and low expression correlating with poor prognosis in KIRP, KIPAN, PRAD, KIRC, and PAAD.
Figure 3.
Prognostic value of SPAG6 across cancers. (A) The correlation between OS and SPAG6 expression. (B-F) Kaplan-Meier analysis of significant outcomes in OS, namely DLBC, LAML, ALL, PAAD, and TGCT. (G) The correlation between DSS and SPAG6 expression. (H) The correlation between DFI and SPAG6 expression. (I) The correlation between PFI and SPAG6 expression. HR, hazard ratio; CI, confidence interval; NaN, not a number; L, low; H, high; OS, overall survival; DLBC, lymphoid neoplasm diffuse large b-cell lymphoma; LAML, acute myeloid leukemia; ALL, acute lymphoblastic leukemia; PAAD, pancreatic adenocarcinoma; TGCT, testicular germ cell tumor; DSS, disease-specific survival; DFI, disease-free interval; PFI, progression-free interval.
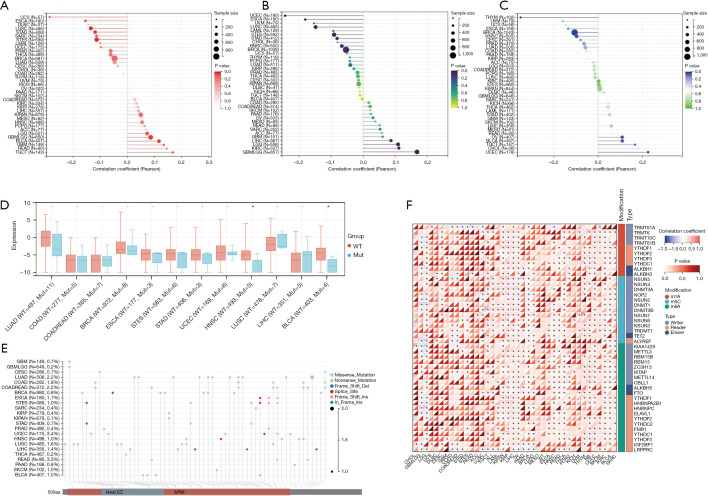
Correlation between SPAG6 expression and genetic heterogeneity
Our analysis revealed a correlation between SPAG6 expression and TMB (Figure 4A). Positive correlations between SPAG6 expression and TMB were observed in GBMLGG, TGCT, and BLCA, while negative correlations were found in esophageal carcinoma (ESCA), STES, STAD, LUSC, and UCS. Similarly, the correlation between SPAG6 expression and MSI was analyzed (Figure 4B). Positive relationships were observed in GBMLGG, LGG, and KIRC, while negative relationships were found in ESCA, STES, UCEC, and LUSC. Furthermore, the correlation between SPAG6 expression and tumor ploidy was analyzed (Figure 4C). Positive associations were found in UCEC, OV, TGCT, and BLCA, while negative associations were observed in BRCA, HNSC, and THYM.
Figure 4.
The expression of SPAG6 was associated with heterogeneity, gene mutation, and RNA modifications. (A-C) The lollipop plots indicate that SPAG6 was associated with genetic heterogeneity, namely TMB (A), MSI (B), and tumor ploidy (C). (D) The correlation between SPAG6 and gene mutation. Wild type = red and mutation = blue. (E) The landscape of the single-nucleotide variants of SPAG6. (F) The correlation between RNA modifications and SPAG6 expression. –, no statistical significance; *, P<0.05. WT, wild type; Mut, mutant; TMB, tumor mutational burden; MSI, microsatellite instability.
Correlation between SPAG6 expression and gene mutation
The box plot (Figure 4D) revealed a striking difference in SPAG6 mutation between HNSC and BLCA. To gain a deeper understanding of SPAG6 gene mutations, we analyzed level 4 samples from TCGA and obtained the mutation status of SPAG6 protein’s structural domain in various cancer types (Figure 4E).
Relationship of SPAG6 expression and RNA modifications
We examined the association between 41 cancer types and three RNA regulators (m1A, m5C, and m6A) (Figure 4F). The heatmap depicted methyltransferases to be “writers”, demethylases to be “erasers”, and binding proteins to be “readers”. The results revealed a negative correlation between SPAG6 and most RNA regulators in LGG and GBMLGG, while a positive correlation was observed in KIPAN, KIRP, LIHC, OV, neuroblastoma (NB), UCEC, KIRC, and BLCA for most RNA modifications.
Analysis of SPAG6 expression and TME
The TME is a complex milieu comprising diverse cell types that can either facilitate or impede tumor growth (1,25). The diagram depicted a significant correlation between SPAG6 expression and tumor purity in 16 cancer types (Figure 5A). In KIPAN, high SPAG6 expression was associated with high tumor purity, while the opposite trend was observed in the remaining 15 cancer types, including GBM, GBMLGG, LGG, LUAD, COAD, COADREAD, ESCA, STES, STAD, PRAD, LUSC, THCA, READ, BLCA, and DLBC.
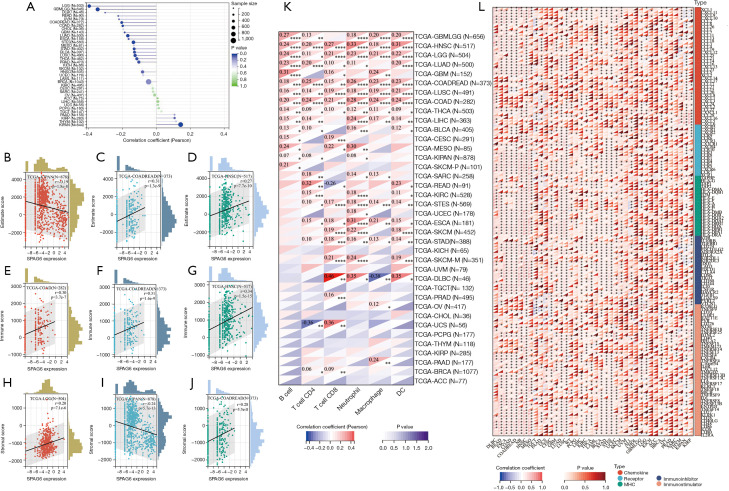
Figure 5.
The association between SPAG6 and tumor-related immunity in the tumor microenvironment. The correlation between tumor purity and SPAG6 expression. (A-D) The correlation between SPAG6 and infiltration as calculated by ESTIMATE score. (E-G) The correlation between SPAG6 and infiltration as calculated by immune score. (H-J) The correlation between SPAG6 and infiltration as calculated by stromal score. (K) The correlation between immune-related cells and SPAG6 expression. (L) The correlation between immunomodulators and SPAG6 expression. *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001. DC, dendritic cell; MHC, major histocompatibility complex.
We then investigated the association between SPAG6 activity and immune infiltration by calculating immune score, stromal score, and ESTIMATE score in pan-cancer. In the majority of the 44 cancers analyzed, all 3 scores exhibited a positive association with SPAG6 expression. Interestingly, SPAG6 expression was positively associated with the 3 scores in 13 cancers, including COAD, COADREAD, ESCA, GBM, HNSC, LGG, LUAD, LUSC, READ, SKCM, SKCM-M, STAD, and STES, but was negatively associated in KIPAN and OV. The cancers with the lowest P value in each score for ESTIMATE score, immune score, and stromal score are presented in Figure 5B-5D, Figure 5E-5G, and Figure 5H-5J, respectively.
Furthermore, the results showed a significant positive correlation between immune-related cells and almost cancers, except for DLBC and UCS. Figure 5K shows that all six immune-related cells were positively associated with SPAG6 expression in HNSC, LGG, COADREAD, LUSC, COAD, and THCA. These results were consistent with those of the three tumor immune scores.
The result also showed that SPAG6 expression and was positively correlated with immunomodulators in various cancers. In HNSC, it showed positive associations with most chemokines, but it was negatively correlated to nearly a quarter of the chemokines in KIPAN (Figure 5L). In LUSC, SPAG6 expression was positively correlated with almost all types of chemokine receptors, with no negative correlation observed. We also found a clear positive association between SPAG6 expression and immune inhibitors in HNSC, LUAD, and STES but a negative association with PDCD1LG2 in THYM. Additionally, a positive association was found between SPAG6 expression and immunostimulators in HNSC and a negative association with TGCT. Notably, SPAG6 expression was positively correlated with all detected major histocompatibility complex (MHC) molecules in HNSC. In contrast, SPAG6 expression in ALL demonstrated a negative correlation with several types of MHC.
The role of SPAG6 in cancer stemness
Cancer stem cells, characterized by self-renewal abilities, contribute critically to tumor initiation, progression, and metastasis (26). SPAG6 expression exhibited negative associations with DNAss and RNAss in cancers, except for LGG, PRAD, LIHC, and TGCT, where a positive correlation was observed with DNAss (Figure 6A). Additionally, LIHC, OV, PCPG, and BLCA showed a positive correlation with SPAG6 expression and RNAss (Figure 6B).
Figure 6.
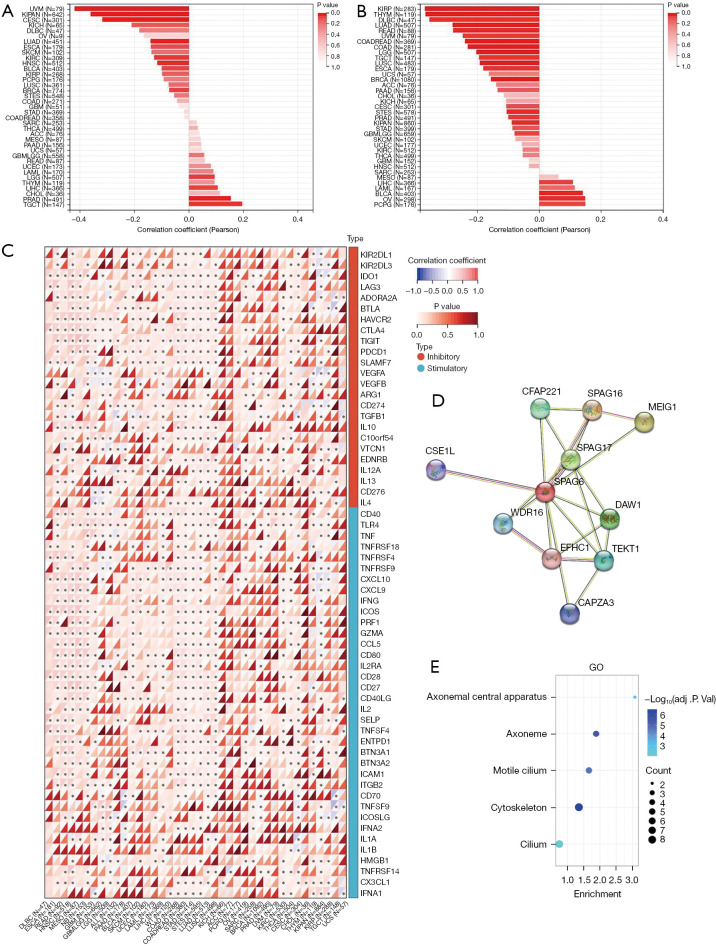
Correlation of SPAG6 expression as tumor stemness and immune checkpoint genes and the enrichment analysis its interacting targets. (A,B) The association between SPAG6 expression and tumor stemness, including DNAss (A) and RNAss (B). (C) The association between SPAG6 expression and immune checkpoints in pan-cancer. (D) The protein-protein interaction network of SPAG6. (E) The Gene Ontology enrichment analysis of SPAG6 and its interacting targets. *, P<0.05. GO, Gene Ontology.
Correlation between SPAG6 expression and immune checkpoint genes
The analysis included 60 immune checkpoint genes (Figure 6C), and SPAG6 expression showed a strong positive correlation with several genes in multiple cancers, such as READ, NB, COAD, COADREAD, STAD, STES, LUAD, KIRC, and THCA. Conversely, negative associations were observed with genes such as GBMLGG, LGG, KIRP, and KIRP. This suggests that SPAG6 has a significant role as a potential target for immunotherapy in various cancers. However, negative immunological associations were found with certain immune inhibitory genes (e.g., VEGFA, CD274) and immune stimulatory genes (e.g., TNFRSF4, CXCL10), indicating that elevated SPAG6 expression levels might hinder the efficacy of therapy in tumors.
GO enrichment analysis based on the protein-protein interaction network
The results indicate that the top 10 proteins with the strongest interactions with SPAG6 were CFAP221, SPAG16, MEIG1, SPAG17, DAW1, TEKT1, CAPZA3, EFHC1, WDR16, and CSE1L (Figure 6D). The top five most enriched biological processes (BPs) were the axonemal central apparatus, axoneme, motile cilium, cytoskeleton, and cilium (Figure 6E).
SPAG6 overexpression suppressed proliferation, migration, and invasion of THCA cell lines
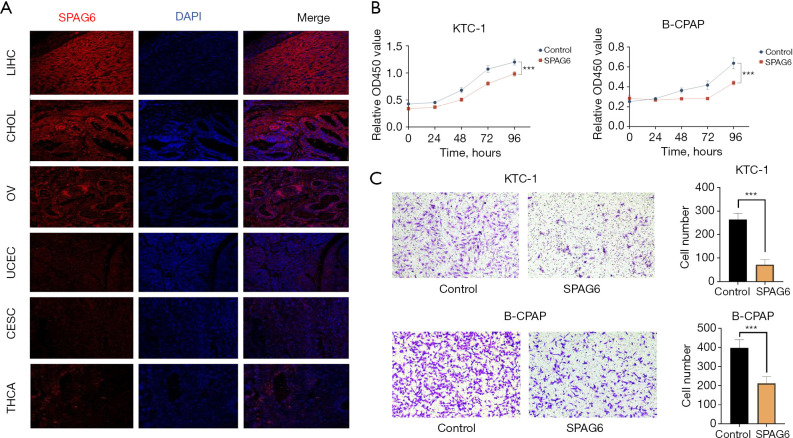
Immunofluorescence staining demonstrated high SPAG6 expression in LIHC, CHOL, and OV tumor samples and a low expression in UCEC, CESC, and THCA (Figure 7A). There exist distinct pathological subtypes within THCA, each characterized by unique etiopathogenic and clinical perspectives. Papillary THCA arises from the follicular thyroid cells, representing the predominant subtype, constituting over 80% of THCA cases. B-CPAP cells and KTC-1 cells are two common types of cell lines for studying THCA. Notably, KTC-1 cells are utilized in research involving human thyroid cancer, while B-CPAP cells also serve as a model for studying human thyroid cancer, especially for papillary THCA. Then, further investigation focused on the potential role of SPAG6 in THCA. Overexpression of SPAG6 in THCA cell lines resulted in decreased proliferation and migration ability, as observed in the CCK8 assay and Transwell migration assay, respectively (Figure 7B,7C). These experimental results validated the findings from the bioinformatics analysis.
Figure 7.
Experimental validation of SPAG6 expression. (A) Multiple immunofluorescence staining of SPAG6 in pan-cancer tissue sections. Views of cancers, including high expression (LIHC, CHOL, and OV) and low expression (UCEC, CESC, and THCA) under microscopy (200×). SPAG6 = red, cellular nuclei = blue (DAPI). (B) Cell Counting Kit 8 assay of SPAG6. (C) Transwell migration assay of SPAG6 (10× magnification). The cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Migrated cells were photographed with a microscope. ***, P<0.001. LIHC, liver hepatocellular carcinoma; CHOL, cholangiocarcinoma; OV, ovarian serous cystadenocarcinoma; UCEC, uterine corpus endometrial carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; THCA, thyroid carcinoma; DAPI, 4,6-diamidino-2-phenyiindole 2 hci.
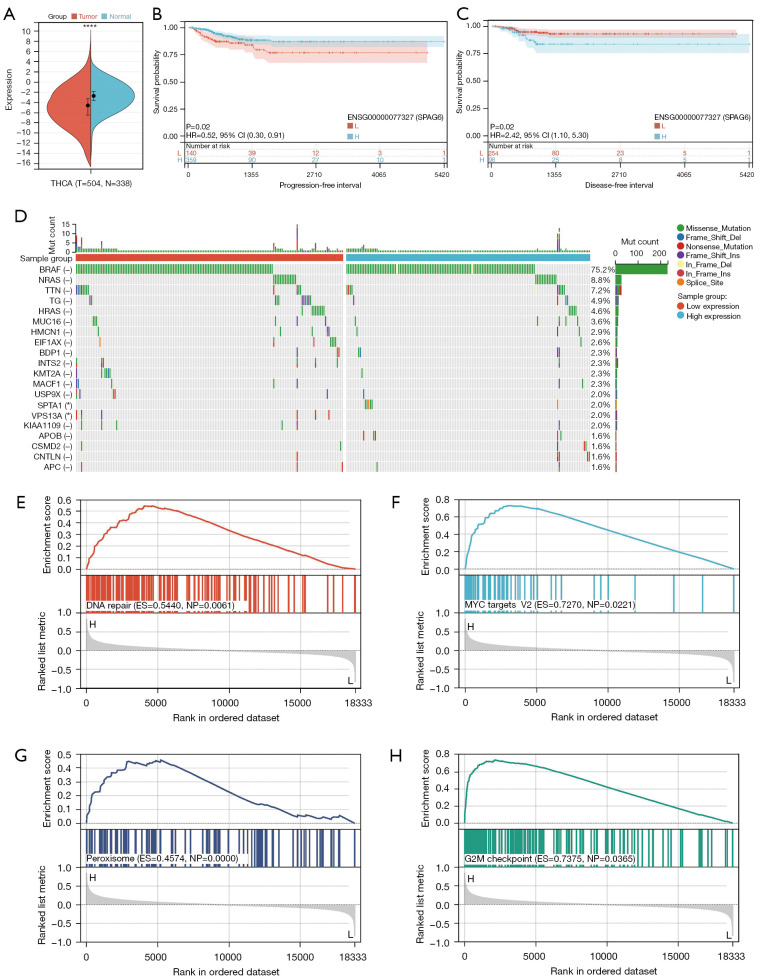
The expression of SPAG6 was lower in tumor samples compared to normal samples. The prognostic value of SPAG6 in THCA varied (Figure 8A), with high expression being associated with high PFI and low expression being associated with high DFI (Figure 8B,8C). Low SPAG6 expression indicated a lower risk of THCA and tumor recurrence but a higher risk of death from nonneoplastic causes. Mutational analysis revealed frequent BRAF (–) gene mutations in both the SPAG6 high- and low-expression groups, with distinct mutations in the SPTA1 gene for the high expression group and the VPS13A gene for the low expression group (Figure 8D). GSEA revealed distinct signaling pathways associated with high and low SPAG6 expression, including DNA repair, MYC targets, peroxisome, and G2M checkpoint, which could explain the observed clinical outcomes (Figure 8E-8H).
Figure 8.
The effect of SPAG6 overexpression on the THCA cell lines. (A) Differential expression of SPAG6 in normal and tumor tissues. (B,C) Prognostic value of SPAG6 in THCA. (D) Mutated genes in high and low SPAG6 expression group. (E-H) GSEA of SPAG6. –, no statistical significance; *, P<0.05; ****, P<0.0001. THCA, thyroid carcinoma; HR, hazard ratio; CI, confidence interval; GSEA, gene set enrichment analysis.
Discussion
In this comprehensive study, we analyzed SPAG6 expression across 34 different cancers using multiple databases. Our findings support previous research (5,6,27), indicating that SPAG6 is upregulated in 11 cancers and downregulated in 17 cancers. In this study, we focused on the high incidence of thyroid cancer and conducted in vitro experiments to preliminarily confirm the possible role of spag6 in cancer. The expression of SPAG6 was downregulated in THCA and was associated with prognosis. The differential expression of SPAG6 regulates especially proliferation and metastasis in THCA. In thyroid cancer with SPAG6 expression, its expression is associated with immune cell infiltration. Additionally, the expression of immune checkpoints shows a strong correlation with SPAG6. When SPAG6 expression is low, the risk of THCA tumor recurrence is lower, but the risk of death from non-tumor causes is relatively higher. Therefore, for the treatment and prognosis evaluation of THCA, SPAG6 expression may be an important target. The relationships found between the expression of SPAG6 and the cells of the immune system may be of interest from a clinical point of view. Immunotherapy currently has limited clinical application in advanced thyroid cancer. The use of pembrolizumab (28), a PD-1 inhibitor, and the combination of the CTLA-4 inhibitor ipilimumab with the PD-1 inhibitor nivolumab have been tested (29). In all cases clinical efficacy has been poor. The results of the present study open a new potential avenue of action against thyroid cancer through the immune system. A study has suggested that gene fusion can lead to upregulation of SPAG6, as observed in leukemia samples (30). Additionally, silencing of SPAG6 expression has been linked to apoptosis and differentiation of leukemia cells through the PI3K–AKT signaling pathway (31). These results provide preliminary insight into the role of SPAG6 in cancer initiation and progression.
Our study suggests that SPAG6 expression was associated with certain clinical characteristics of malignant tumors. Additionally, the combination of SPAG6 with other markers has been proposed as a promising biomarker for early breast cancer diagnosis via a liquid biopsy approach (32). Our results also show gender-specific differences, with high SPAG6-expression cancers being more prevalent in women (e.g., ACC) and low SPAG6-expression cancers being more common in men (e.g., GBM, GBMLGG, and LUSC). The Human Protein Atlas indicates SPAG6 expression in various tissues, including the brain, respiratory system, male tissues, and female tissues, although the level of expression varies between males and females, providing insights into gender-specific cancer diagnosis.
SPAG6 expression is associated with prognosis in various cancers. High SPAG6 expression is linked to better survival in LAML and ALL, while low expression is associated with poor prognosis in TGCT. These findings suggest SPAG6 as a potential prognostic biomarker. In hematologic cells, SPAG6 regulates proliferation and apoptosis and can be targeted therapeutically through the P53 pathway (33). Our study is the first to examine SPAG6 in THCA and to report that it is involved in DNA repair, MYC targets, peroxisome, and G2M checkpoint, but further experimental validation is needed to clarify the mechanism underlying this relationship.
SPAG6 expression is also associated with important biomarkers such as TMB, MSI, and tumor ploidy, which are crucial in predicting the response to immunotherapy (34). The expression of SPAG6 can serve as a guide for immunotherapy in different types of cancer. Additionally, our study revealed a link between SPAG6 expression and genetic heterogeneity, including DNA mutations and RNA modifications, particularly in BLCA. Contrary to previous findings, we observed that lower SPAG6 expression was associated with higher stemness in tumors, indicating its facilitative role in cancer development (35). These findings highlight the significance of SPAG6 in cancer progression and provide insights for potential therapeutic approaches.
In addition, immunotherapy has been shown to provide durable responses in some clinical patients, but only a small percentage of individuals respond to the treatment. A study has shown that increasing immunogenicity by modifying the TME can resolve these issues (36). Our study found a positive association between SPAG6 expression and immune score, stromal score, and ESTIMATE score in various cancers. The presence and function of immune cells in the TME are crucial for antitumor immunity, and TME heterogeneity affects treatment response and clinical outcomes (37-39). Our investigation confirmed the positive relationship between cancer and SPAG6 expression, as observed in the study regarding B cells and DCs (11). The correlation between SPAG6 expression and various immunomodulators, including chemokines, receptors, MHCs, immune inhibitors, and immunostimulators, was also evaluated. SPAG6 expression is closely linked to immune cell infiltration in the TME, impacting cancer immunotherapy and the need to shift the TME from a tolerogenic state to an immunogenic one. The study supports the role of SPAG6 in tumor immunotherapy by revealing its relationship with immune checkpoint genes. SPAG6 expression was positively associated with a successful response in most cancers, indicating its potential as an immune checkpoint. If the role of SPAG6 in the initiation and progression of THCA and its relationship with prognosis can be demonstrated, it could be considered a promising target for new drugs against this tumor. These results show that a quest for greater precision in molecular analysis could be the key to greater precision in personalized medicine.
However, the effects of immunotherapy are complex and are influenced by multiple and specific factors in each cancer. This is a limitation of our study and requires further in vivo experiments and clinical observations for the role of SPAG6 in tumorigenesis and progression to be fully understood. Despite these limitations, our findings provide a foundation for future studies on SPAG6 in the development and treatment of cancers, especially THCA.
Conclusions
In conclusion, this study found that the expression level of SPAG6 was differentially expressed in various cancers at different tumor stages and grades, with gender-associated expression observed in certain cancers. The variation in SPAG6 expression between progression and survival suggests its prognostic value in cancer. The results indicate that SPAG6 affects immune infiltration, regulates the TME, and plays a role in the development of cancers. Additionally, SPAG6 was positively correlated with a successful tumor therapy and may be a potential immune checkpoint.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was funded by Taishan Scholars Program of Shandong Province (No. ts20130913), National Natural Science Foundation of China (No. 82171150), and the National Natural Science Foundation of Shandong Province (No. ZR2020MH179).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The studies involving human samples were approved by the Shandong Provincial ENT Hospital Ethical Committee (No. 2024-019-01). Written informed consent to participate in this study was provided by the participants. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://gs.amegroups.com/article/view/10.21037/gs-24-157/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://gs.amegroups.com/article/view/10.21037/gs-24-157/coif). The authors have no conflicts of interest to declare.
Data Sharing Statement
Available at https://gs.amegroups.com/article/view/10.21037/gs-24-157/dss
References
- 1.Du T, Gao J, Li P, et al. Pyroptosis, metabolism, and tumor immune microenvironment. Clin Transl Med 2021;11:e492. 10.1002/ctm2.492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Priestley P, Baber J, Lolkema MP, et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature 2019;575:210-6. 10.1038/s41586-019-1689-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siliņa K, Zayakin P, Kalniņa Z, et al. Sperm-associated antigens as targets for cancer immunotherapy: expression pattern and humoral immune response in cancer patients. J Immunother 2011;34:28-44. 10.1097/CJI.0b013e3181fb64fa [DOI] [PubMed] [Google Scholar]
- 4.Yang P, Meng M, Zhou Q. Oncogenic cancer/testis antigens are a hallmarker of cancer and a sensible target for cancer immunotherapy. Biochim Biophys Acta Rev Cancer 2021;1876:188558. 10.1016/j.bbcan.2021.188558 [DOI] [PubMed] [Google Scholar]
- 5.Kulkarni P, Shiraishi T, Rajagopalan K, et al. Cancer/testis antigens and urological malignancies. Nat Rev Urol 2012;9:386-96. 10.1038/nrurol.2012.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng DF, Wang Q, Wang JP, et al. The Emerging Role of Sperm-Associated Antigen 6 Gene in the Microtubule Function of Cells and Cancer. Mol Ther Oncolytics 2019;15:101-7. 10.1016/j.omto.2019.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Xu L, Sun G, et al. Spag6 Mutant Mice Have Defects in Development and Function of Spiral Ganglion Neurons, Apoptosis, and Higher Sensitivity to Paclitaxel. Sci Rep 2017;7:8638. 10.1038/s41598-017-08739-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Xu L, Li J, et al. Otitis media in sperm-associated antigen 6 (Spag6)-deficient mice. PLoS One 2014;9:e112879. 10.1371/journal.pone.0112879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sapiro R, Kostetskii I, Olds-Clarke P, et al. Male infertility, impaired sperm motility, and hydrocephalus in mice deficient in sperm-associated antigen 6. Mol Cell Biol 2002;22:6298-305. 10.1128/MCB.22.17.6298-6305.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bao Z, Zhu R, Fan H, et al. Aberrant expression of SPAG6 and NM23 predicts poor prognosis of human osteosarcoma. Front Genet 2022;13:1012548. 10.3389/fgene.2022.1012548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Q, Yan Y, Shi S, et al. DNMT3b-mediated SPAG6 promoter hypermethylation affects lung squamous cell carcinoma development through the JAK/STAT pathway. Am J Transl Res 2022;14:6964-77. [PMC free article] [PubMed] [Google Scholar]
- 12.Altenberger C, Heller G, Ziegler B, et al. SPAG6 and L1TD1 are transcriptionally regulated by DNA methylation in non-small cell lung cancers. Mol Cancer 2017;16:1. 10.1186/s12943-016-0568-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang R, Zhu H, Yuan Y, et al. SPAG6 promotes cell proliferation and inhibits apoptosis through the PTEN/PI3K/AKT pathway in Burkitt lymphoma. Oncol Rep 2020;44:2021-30. 10.3892/or.2020.7776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao B, Yin J, Ding L, et al. SPAG6 regulates cell proliferation and apoptosis via TGF-β/Smad signal pathway in adult B-cell acute lymphoblastic leukemia. Int J Hematol 2024;119:119-29. 10.1007/s12185-023-03684-x [DOI] [PubMed] [Google Scholar]
- 15.Yi M, Zheng X, Niu M, et al. Combination strategies with PD-1/PD-L1 blockade: current advances and future directions. Mol Cancer 2022;21:28. 10.1186/s12943-021-01489-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Q, Jiang L, Li SC, et al. Small molecule inhibitors targeting the PD-1/PD-L1 signaling pathway. Acta Pharmacol Sin 2021;42:1-9. 10.1038/s41401-020-0366-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bialek-Waldmann JK, Domning S, Esser R, et al. Induced dendritic cells co-expressing GM-CSF/IFN-α/tWT1 priming T and B cells and automated manufacturing to boost GvL. Mol Ther Methods Clin Dev 2021;21:621-41. 10.1016/j.omtm.2021.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jhunjhunwala S, Hammer C, Delamarre L. Antigen presentation in cancer: insights into tumour immunogenicity and immune evasion. Nat Rev Cancer 2021;21:298-312. 10.1038/s41568-021-00339-z [DOI] [PubMed] [Google Scholar]
- 19.Cooley LF, El Shikh ME, Li W, et al. Impaired immunological synapse in sperm associated antigen 6 (SPAG6) deficient mice. Sci Rep 2016;6:25840. 10.1038/srep25840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Lichtenberg T, Hoadley KA, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018;173:400-416.e11. 10.1016/j.cell.2018.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malta TM, Sokolov A, Gentles AJ, et al. Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell 2018;173:338-354.e15. 10.1016/j.cell.2018.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng D, Ye Z, Shen R, et al. IOBR: Multi-Omics Immuno-Oncology Biological Research to Decode Tumor Microenvironment and Signatures. Front Immunol 2021;12:687975. 10.3389/fimmu.2021.687975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. 10.1158/0008-5472.CAN-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thorsson V, Gibbs DL, Brown SD, et al. The Immune Landscape of Cancer. Immunity 2019;51:411-2. 10.1016/j.immuni.2019.08.004 [DOI] [PubMed] [Google Scholar]
- 25.Elhanani O, Ben-Uri R, Keren L. Spatial profiling technologies illuminate the tumor microenvironment. Cancer Cell 2023;41:404-20. 10.1016/j.ccell.2023.01.010 [DOI] [PubMed] [Google Scholar]
- 26.Chen P, Hsu WH, Han J, et al. Cancer Stemness Meets Immunity: From Mechanism to Therapy. Cell Rep 2021;34:108597. 10.1016/j.celrep.2020.108597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603-7. 10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehnert JM, Varga A, Brose MS, et al. Safety and antitumor activity of the anti-PD-1 antibody pembrolizumab in patients with advanced, PD-L1-positive papillary or follicular thyroid cancer. BMC Cancer 2019;19:196. 10.1186/s12885-019-5380-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorch J, Barletta J, NehsM, et al. A phase II study of nivolumab (N) plus ipilimumab (I) in radioidine refractory differentiated thyroid cancer (RAIR DTC) with exploratory cohorts in anaplastic (ATC) and medullary thyroid cancer (MTC). J Clin Oncol 2020;38:abstr 6513.
- 30.Mulaw MA, Krause A, Deshpande AJ, et al. CALM/AF10-positive leukemias show upregulation of genes involved in chromatin assembly and DNA repair processes and of genes adjacent to the breakpoint at 10p12. Leukemia 2012;26:1012-9. 10.1038/leu.2011.307 [DOI] [PubMed] [Google Scholar]
- 31.Jiang M, Chen Y, Deng L, et al. Upregulation of SPAG6 in Myelodysplastic Syndrome: Knockdown Inhibits Cell Proliferation via AKT/FOXO Signaling Pathway. DNA Cell Biol 2019;38:476-84. 10.1089/dna.2018.4521 [DOI] [PubMed] [Google Scholar]
- 32.Mijnes J, Tiedemann J, Eschenbruch J, et al. SNiPER: a novel hypermethylation biomarker panel for liquid biopsy based early breast cancer detection. Oncotarget 2019;10:6494-508. 10.18632/oncotarget.27303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang B, Wang L, Luo X, et al. SPAG6 silencing inhibits the growth of the malignant myeloid cell lines SKM-1 and K562 via activating p53 and caspase activation-dependent apoptosis. Int J Oncol 2015;46:649-56. 10.3892/ijo.2014.2768 [DOI] [PubMed] [Google Scholar]
- 34.Addeo A, Friedlaender A, Banna GL, et al. TMB or not TMB as a biomarker: That is the question. Crit Rev Oncol Hematol 2021;163:103374. 10.1016/j.critrevonc.2021.103374 [DOI] [PubMed] [Google Scholar]
- 35.Friedmann-Morvinski D, Verma IM. Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO Rep 2014;15:244-53. 10.1002/embr.201338254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murciano-Goroff YR, Warner AB, Wolchok JD. The future of cancer immunotherapy: microenvironment-targeting combinations. Cell Res 2020;30:507-19. 10.1038/s41422-020-0337-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujii SI, Shimizu K. Immune Networks and Therapeutic Targeting of iNKT Cells in Cancer. Trends Immunol 2019;40:984-97. 10.1016/j.it.2019.09.008 [DOI] [PubMed] [Google Scholar]
- 38.Efimova I, Catanzaro E, Van der Meeren L, et al. Vaccination with early ferroptotic cancer cells induces efficient antitumor immunity. J Immunother Cancer 2020;8:e001369. 10.1136/jitc-2020-001369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 2013;501:346-54. 10.1038/nature12626 [DOI] [PubMed] [Google Scholar]