SUMMARY
Human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) recapitulate numerous disease and drug response phenotypes, but cell immaturity may limit their accuracy and fidelity as a model system. Cell culture medium modification is a common method for enhancing maturation, yet prior studies have used complex media with little understanding of individual component contribution, which may compromise long-term hiPSC-CM viability. Here, we developed high-throughput methods to measure hiPSC-CM maturation, determined factors that enhanced viability, and then systematically assessed the contribution of individual maturation medium components. We developed a medium that is compatible with extended culture. We discovered that hiPSC-CM maturation can be sub-specified into electrophysiological/EC coupling, metabolism, and gene expression and that induction of these attributes is largely independent. In this work, we establish a defined baseline for future studies of cardiomyocyte maturation. Furthermore, we provide a selection of medium formulae, optimized for distinct applications and priorities, that promote measurable attributes of maturation.
Graphical abstract
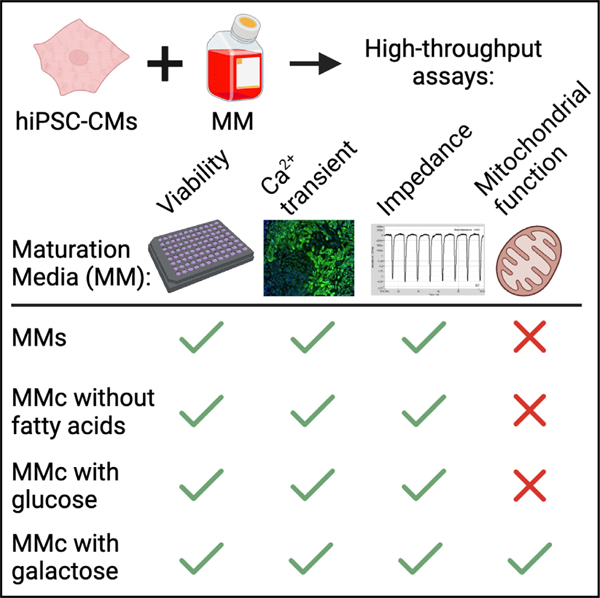
In brief
Fetterman et al. systematically evaluated individual medium component contribution to hiPSC-CM maturation and long-term viability using high-throughput methods. They developed an optimized maturation media and discovered that hiPSC-CM maturation can be sub-specified into functional, metabolic, and transcriptomic maturation. This study establishes a framework for screening novel promoters of maturation.
INTRODUCTION
Human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) are a powerful in vitro tool to screen therapeutics and to model patient-specific diseases and drug responses. hiPSC-CMs offer a minimally invasive source with unlimited supply compared to difficult-to-obtain and -maintain primary human CMs. However, a frequently cited limitation of hiPSC-CMs is their immaturity, as they have been suggested to more closely resemble fetal CMs. hiPSC-CMs differ from adult CMs structurally, electrophysiologically, mechanically, metabolically, and transcriptionally.1 Several methods have been described to induce more adult-like properties in hiPSC-CMs, including the use of long-term culture (80+ days),2,3 engineered growth surfaces that more closely resemble the consistency of the human heart,4 and mechanical and electrical stimulation.5,6 However, these methodologies are either slow or low throughput and not suitable for hiPSC-CM applications requiring large quantities of cells or that use multiple cell lines. An alternative approach is modification of the cell culture medium composition.1,5 Induction of maturation through biochemical cues, by adding components to the medium rather than culturing in specialized systems, allows for reproducible, high-yield generation of more mature hiPSC-CMs amendable to high-throughput applications for disease modeling and drug screening.
Previous studies of cell culture medium-based maturation have focused on biochemical cues involved in human cardiac maturation during the neonatal period, including metabolic substrates and hormones. At birth, the heart switches from glycolysis to more efficient fatty acid β-oxidation (FAO) to support greater postnatal energy demands.7 Because hiPSC-CMs primarily rely on glycolysis as their main energy source, researchers investigated methods to promote FAO to enhance maturation. One strategy is to add fatty acids such as palmitate and oleate, which are the most abundant fatty acids in the serum of infants, to the medium as oxidative metabolism substrates.8–10 Another strategy, used in combination with the addition of fatty acids, is to make glycolysis inefficient by reducing glucose concentration,11 removing glucose,12 or replacing glucose with galactose.8
In addition to metabolic substrates, thyroid hormones and glucocorticoids are essential for development and maturation of the fetal heart in vivo.13,14 Elevated levels of the thyroid hormone triiodothyronine (T3) occur concurrent with rapid maturation of the heart postnatally.14 Plasma concentrations of the endogenous glucocorticoid cortisol increase before birth and remain elevated immediately after birth.15 Insulin growth factor 1 (IGF1) and insulin support cardiac growth in the developing heart through interactions with the IGF1 receptor.14 Studies have shown that T3, the glucocorticoid dexamethasone, and IGF1/insulin induce maturation in hiPSC-CMs, and, consequently, these components are frequently included in maturation media.16–18
Additional components have been added to maturation media to support the energy demands and overall survival of more mature hiPSC-CMs. Ascorbic acid, an antioxidant, both enhances cardiac differentiation by increasing proliferation of cardiac progenitor cells and induces maturation.19,20 Taurine and L-carnitine are believed to enhance metabolism by supporting FAO through buffering the mitochondrial matrix and transporting fatty acids into the mitochondria, respectively.21 Combinations of creatine, sodium selenite, transferrin, and insulin have been used to maintain cell viability in serum-free culture.21,22 Existing maturation media formulations vary in complexity and together provide an extensive list of components that may contribute variably to promote maturation of hiPSC-CMs. However, these studies utilize formulations composed of many components, and the individual contribution of each component is rarely investigated, resulting in a lack of thorough investigation into the mechanism and necessity of each component for cell survival or maturation.
We established high-throughput techniques to study functional and metabolic maturation of hiPSC-CMs using individual medium components previously established in the literature to promote hiPSC-CM maturation and viability as a baseline. This in-depth analysis demonstrated that functional, metabolic, and transcriptional maturation of hiPSC-CMs are not inherently linked. We determined that certain composition modifications, such as replacement of galactose with glucose or removal of fatty acids, only affected mitochondrial function but did not change intracellular Ca2+ dynamics, impedance, or voltage-gated sodium (Nav) current parameters. Overall, our work develops and extensively characterizes multiple maturation medium formulations that range in complexity and creates a powerful platform for unbiased screening for novel medium formulations.
RESULTS
High-throughput assays detect changes in maturation status of hiPSC-CMs
To assess the effect of cell culture media on CM maturation, hiPSC-CMs were maintained in either glucose-based maintenance medium (RBAI-RPMI with BSA, ascorbic acid, and insulin) or galactose and fatty acid-based complete maturation medium (MMc; Figures 1A and 1B; Table S1). MMc includes components cited in the literature to promote maturation and viability. To determine whether culturing in MMc produced more mature hiPSC-CMs, we developed two high-throughput assays to assess maturation metrics: Ca2+ dynamics and microelectrode array (MEA)-based impedance recordings. After 9 days of MMc exposure, MMc-cultured hiPSC-CMs had a significantly shorter Ca2+ transient duration at 75% (CTD75) compared to RBAI-cultured hiPSC-CMs (MMc: 0.53 ± 0.01 vs. RBAI: 1.0 ± 0; Figures 1C and 1D). Maximum upstroke and downstroke velocity were also significantly faster after 9 days cultured in MMc (1.78 ± 0.10 and 1.14 ± 0.10) compared to the control RBAI (1.0 ± 0; Figures S1A and S1B). Although all time points between 9 and 21 days in MMc produced similar Ca2+ transient parameters, only 21 days in MMc produced a homogeneous response in all recorded wells (Figures 1D, S1A, S1B, S1E, and S1F). For the Ca2+ transient assay, hiPSC-CMs tolerated culture in galactose for 21 days, the latest time point we assessed in this study (Figures 1C, 1D, S1A, and S1B). After testing a narrowed window of 17–21 days, we determined that hiPSC-CMs need at least 19 days in MMc starting at day 16 for all wells to fully respond (Figures S1C–S1F). While day 16 hiPSC-CMs required 19 days of MMc exposure to achieve a uniform Ca2+ transient response, hiPSC-CMs cultured in MMc medium for only 12 days starting at day 20 or 10 days starting at day 24 produced an equivalent CTD75, indicating that older hiPSC-CMs respond faster to MMc (Figures S1G and S1H).
Figure 1. High-throughput assays detect changes in maturation status of hiPSC-CMs.
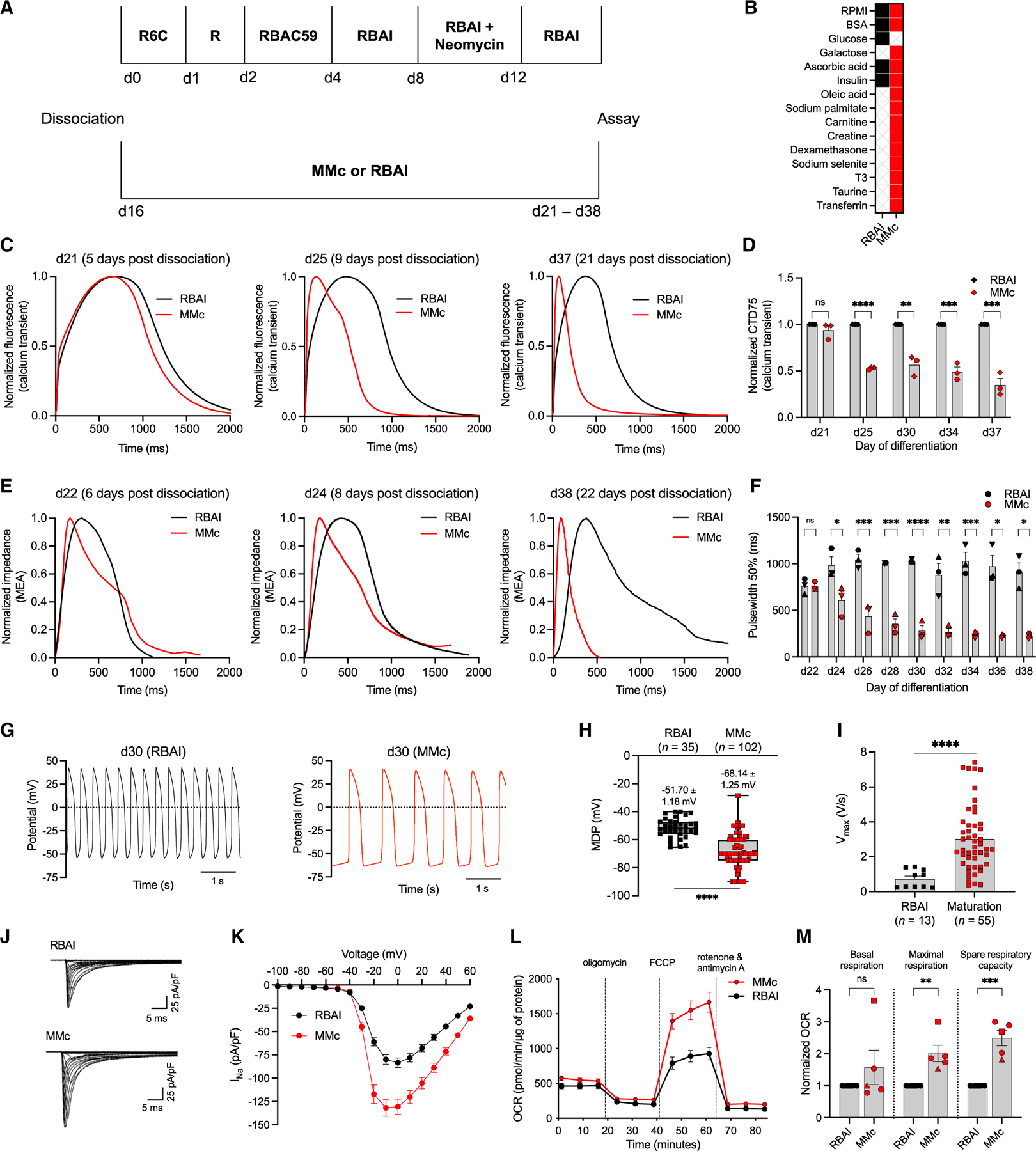
(A) Schematic of hiPSC-CM differentiation with RBAI or MMc starting at day 16 until day of assay.
(B) Components of RBAI and MMc.
(C) Representative spontaneous high-throughput Ca2+ transient recordings of hiPSC-CMs in RBAI or MMc.
(D) CTD75 of Ca2+ transient recordings (n = 3).
(E) Representative spontaneous high-throughput impedance recordings of hiPSC-CMs in RBAI or MMc.
(F) Pulse width at 50% of impedance recordings (n = 3).
(G) Representative spontaneous AP traces of hiPSC-CMs in RBAI or MMc using manual patchclamp.
(H) Maximum diastolic potentials (MDPs) of individual cells measured in spontaneously beating hiPSC-CMs (n > 3).
(I) Maximum upstroke velocities of individual cells measured in hiPSC-CMs paced at 1 Hz (n > 3).
(J) Representative recordings of Nav current (INav) normalized to cell capacitance.
(K) Mean INav current-voltage plot from automated patch-clamp experiments (n = 7).
(L) Mean oxygen consumption rate (OCR) of hiPSC-CMs cultured in MMc or RBAI (n = 5).
(M) Basal respiration, maximal respiration, and spare respiratory capacity of hiPSC-CMs cultured in MMc or RBAI (n = 5).
Each data point shape indicates a different hiPSC line. Data are presented as mean ± SEM. n = experimental replicates, unpaired Student’s t test, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.005, ****p ≤ 0.0001; ns, not significant. See also Figure S1.
Using impedance recordings as a proxy for contraction of hiPSC-CM monolayers, we determined that hiPSC-CMs cultured in MMc for at least 8 days starting at day 16 had a significantly shorter pulse width at 50% compared to those grown in RBAI (Figures 1E and 1F). Although not always significant, hiPSC-CMs cultured in MMc also tended to produce a faster maximum upstroke and downstroke velocity compared to RBAI at all time points (Figures S1I and S1J). For the impedance assay, hiPSC-CMs tolerated culture in galactose for 22 days, the latest time point we assessed in this study (Figures 1C, 1D, S1A, and S1B). Because the contractility response of hiPSC-CMs is consistent following at least 8 days of MMc exposure starting on either day 16 or day 20, we designated 12 days of MMc starting at day 20 as the optimal time frame to induce maturation based on Ca2+ transient and impedance recordings (Figure S1K).
Action potential (AP) and Nav current recordings revealed that culture in MMc resulted in hiPSC-CMs with more mature electrophysiological properties. Specifically, spontaneously beating hiPSC-CMs maintained in MMc exhibited a significantly more hyperpolarized maximum diastolic potential (MDP; −68.14 ± 1.25 mV) relative to RBAI (−51.70 ± 1.18 mV; Figures 1G and 1H). Paced hiPSC-CMs cultured in MMc had a significantly higher maximum upstroke velocity (Vmax) relative to those maintained in RBAI (MMc: 30.23 ± 2.76 mV/ms vs. RBAI: 7.33 ± 1.64 mV/ms), while the AP duration at 90% of repolarization (APD90) (MMc: 43.04 ± 4.39 ms vs. RBAI: 44.35 ± 3.41 ms) and peak amplitude (MMc: 94.21 ± 3.62 mV vs. RBAI: 89.67 ± 1.88 mV) remained unchanged (Figure 1I, S1L, and S1M). Nav currents were recorded by automated patch-clamp recording. Whole-cell recordings revealed that cells cultured in MMc exhibited greater peak current density compared to cells grown in RBAI (at 0 mV, MMc: −130.6 ± 8.17 pA/pF vs. RBAI: −80.73 ± 4.83 pA/pF; Figures 1J, 1K, and S1N). Cell capacitance did not differ between cells cultured in RBAI or MMc (MMc: 19.02 ± 0.44 vs. RBAI: 19.01 ± 0.49; Table S2). The calculated current voltage dependence of activation V½ was similar for both MMc- and RBAI-treated cells, and both curves exhibited similar slopes (k). Likewise, the voltage dependence of inactivation V1/2 for the NaV currents was similar between both treatments, and the inactivation curves also had similar slopes (Figure S1O; Table S2).
During mitochondrial stress tests, hiPSC-CMs cultured in MMc had more mature metabolic parameters compared to those cultured in RBAI, as indicated by increased oxygen consumption rates (OCRs). hiPSC-CMs maintained in MMc had the same basal respiration as RBAI-cultured cells (MMc: 1.57 ± 0.54 vs. RBAI: 1.0 ± 0) but greater maximal respiration (MMc: 2.01 ± 0.26 vs. RBAI: 1.0 ± 0) and spare respiratory capacity (MMc: 2.49 ± 0.24 vs. RBAI: 1.0 ± 0), indicating an enhanced mitochondrial oxidative capacity (Figures 1L and 1M).
Optimized candidate component contribution to viability and Ca2+ transient phenotype
To determine the necessity of candidate components, we evaluated hiPSC-CM viability and Ca2+ dynamics by performing single-addition experiments in a minimal formulation consisting of base components (MMb) and then single-subtraction experiments in MMc (Figure 2A). Due to the complexity of the experiments, we established an experimental design to identify contributing components through strict statistical analysis on three experimental replicates. The final optimized candidate component concentrations determined by viability closely match those used in other hiPSC-CM maturation studies, except for a lower galactose concentration (Figure S2). Based on these results, we lowered the concentration of galactose from 10 mM to 2 mM to perform inclusion and exclusion experiments for each component as well as in all remaining studies. Inclusion of sodium selenite (1.11 ± 0.01) or insulin (1.25 ± 0.08) to MMb significantly improved viability, while exclusion of sodium selenite (0.92 ± 0.03), transferrin (0.91 ± 0.01), insulin (0.91 ± 0.02), ascorbic acid (0.88 ± 0.04), or T3 (0.81 ± 0.03) from MMc resulted in significantly lower viability (Figures 2B, 2C, and S3I).
Figure 2. Optimized candidate component contribution to viability and Ca2+ transient phenotype.
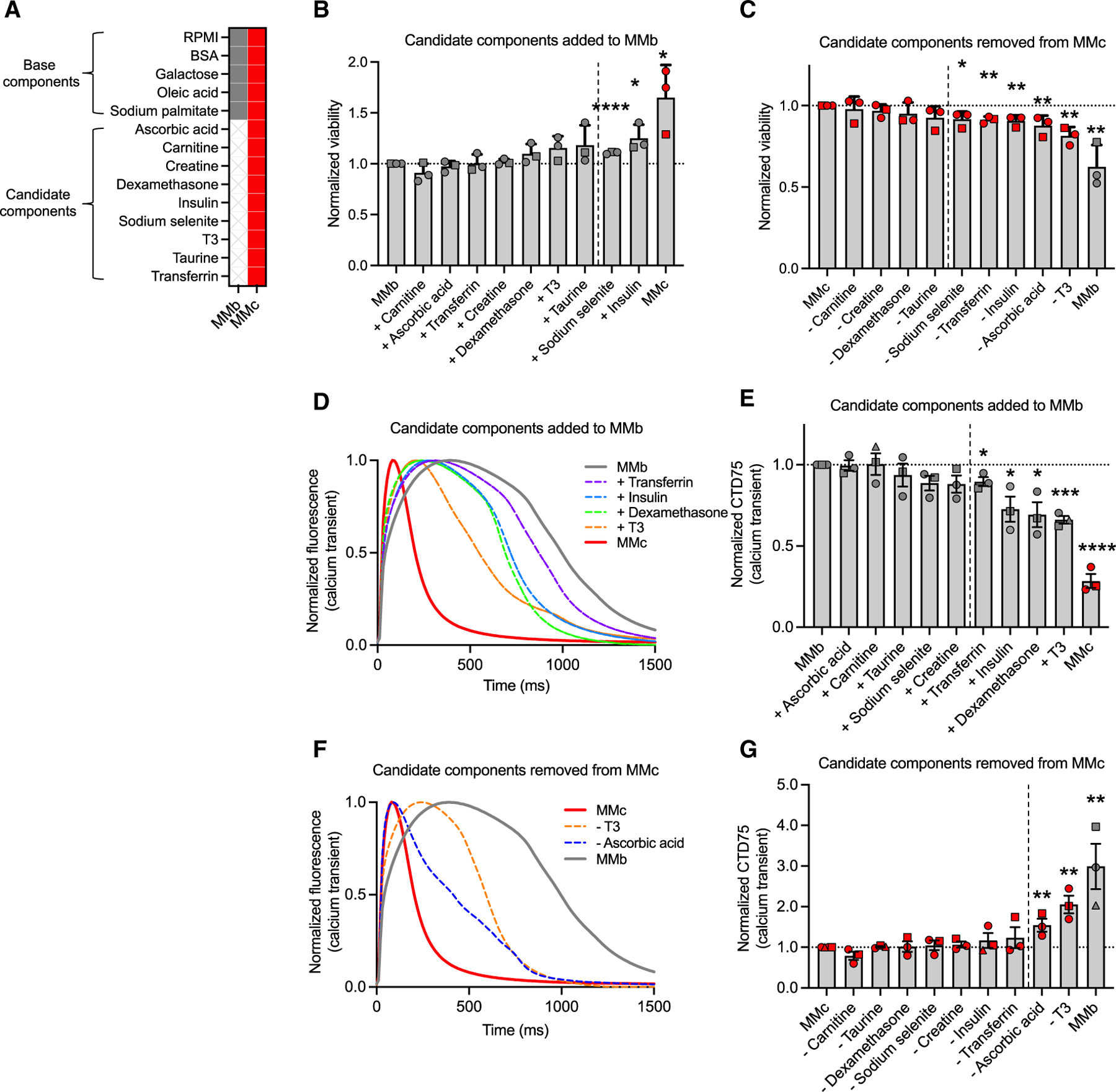
(A) MMb and MMc formulations.
(B) Additive approach to determine changes in hiPSC-CM viability when each candidate component is added to MMb one at a time from day 20 to day 33 (n = 3).
(C) Subtractive approach to determine changes in hiPSC-CM viability when each candidate component is removed from MMc one at a time from day 20 to day 33 (n = 3).
(D) Representative spontaneous hiPSC-CM Ca2+ transient recordings of candidate components that significantly shorten CTD75 when added one at time to MMb from day 20 to day 35.
(E) CTD75 measured when each candidate component is added to MMb (n = 3).
(F) Representative spontaneous hiPSC-CM Ca2+ transient recordings of candidate components that significantly increase CTD75 when removed one at a time from MMc.
(G) CTD75 measured when each candidate component is removed from MMc (n = 3).
Each data point shape indicates a different hiPSC line. Data are presented as mean ± SEM. n = experimental replicates, unpaired Student’s t test, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.005, ****p ≤ 0.0001. All components to the right of the dashed line have p ≤ 0.05. See also Figure S3.
When comparing Ca2+ transient parameters, MMc cultured hiPSC-CMs, compared to those grown in MMb, had a significantly shorter CTD75 (MMc: 0.28 ± 0.04 vs. MMb: 1 ± 0) as well as a faster upstroke (MMc: 2.28 ± 0.24 vs. MMb: 1 ± 0) and downstroke velocity (MMc: 2.32 ± 0.40 vs. MMb: 1 ± 0; Figures 2D, 2F, and S3A–S3D). Using the additive approach, no single candidate component included in MMb was able to produce Ca2+ transients equivalent to MMc. However, inclusion of transferrin (0.90 ± 0.03), insulin (0.73 ± 0.08), dexamethasone (0.69 ± 0.08), or T3 (0.66 ± 0.02) to MMb produced a shorter CTD75 compared to MMb only (1.0 ± 0; Figures 2D and 2E). The upstroke velocity was faster with inclusion of insulin (1.26 ± 0.09) relative to MMb only (1 ± 0; Figures S3A and S3B). Inclusion of sodium selenite (1.07 ± 0.01), ascorbic acid (1.16 ± 0.03), dexamethasone (1.30 ± 0.08), or insulin (1.32 ± 0.08) resulted in faster downstroke velocity compared to MMb only (1 ± 0; Figures S3C and S3D). Using the subtractive approach, we found that the exclusion of either ascorbic acid (1.54 ± 0.16) or T3 (2.05 ± 0.22) resulted in a longer CTD75 compared to MMc (1 ± 0; Figures 2F and 2G). Additionally, exclusion of ascorbic acid from MMc slowed both the upstroke and downstroke velocity (Figures S3E–S3H). While the additive and subtractive approaches were largely concordant regarding which candidate components promote a more mature Ca2+ transient phenotype, we observed that removing insulin or sodium selenite resulted in a faster upstroke velocity or a faster upstroke and downstroke velocity, respectively (Figures S3E–S3H and S3J).
Simple maturation medium reduces mitochondrial function compared to MMc
We next developed a simplified maturation medium (MMs) formulation by excluding the candidate components (carnitine, creatine, and taurine) that did not contribute to improving viability or promoting more mature Ca2+ transients (Figure 3A). We compared viability, Ca2+ transients, impedance, and metabolism between cells cultured in MMs and MMc. There was no difference in viability of hiPSC-CMs cultured in each medium (MMs: 1.00 ± 0.07 vs. MMc: 1 ± 0; Figure 3B). We observed a slight but significant improvement in Ca2+ transient parameters with MMs, indicated by a shorter CTD75 (MMs: 0.80 ± 0.04 vs. MMc: 1 ± 0) and by greater maximum upstroke (MMs: 1.19 ± 0.05 vs. MMc: 1 ± 0) and downstroke (MMs: 1.19 ± 0.04 vs. MMc: 1 ± 0) velocities (Figures 3C and 3D). For measured impedance, there was no difference in pulse width at 50% (MMs: 1.14 ± 0.30 vs. MMc: 1 ± 0), maximum upstroke velocity (MMs: 1.41 ± 0.30 vs. MMc: 1 ± 0), or maximum downstroke velocity (MMs: 1.24 ± 0.24 vs. MMc: 1 ± 0) between paced hiPSC-CMs grown in MMs or MMc (Figures 3E and 3F). Although there was no difference in basal respiration (MMs: 1.08 ± 0.08 vs. MMc: 1 ± 0), the maximal respiration (MMs: 0.79 ± 0.04 vs. MMc: 1 ± 0) and spare respiratory capacity (MMs: 0.67 ± 0.04 vs. MMc: 1 ± 0) were lower for cells grown in MMs (Figures 3G and 3H).
Figure 3. Simple maturation medium reduces mitochondrial function compared to MMc.
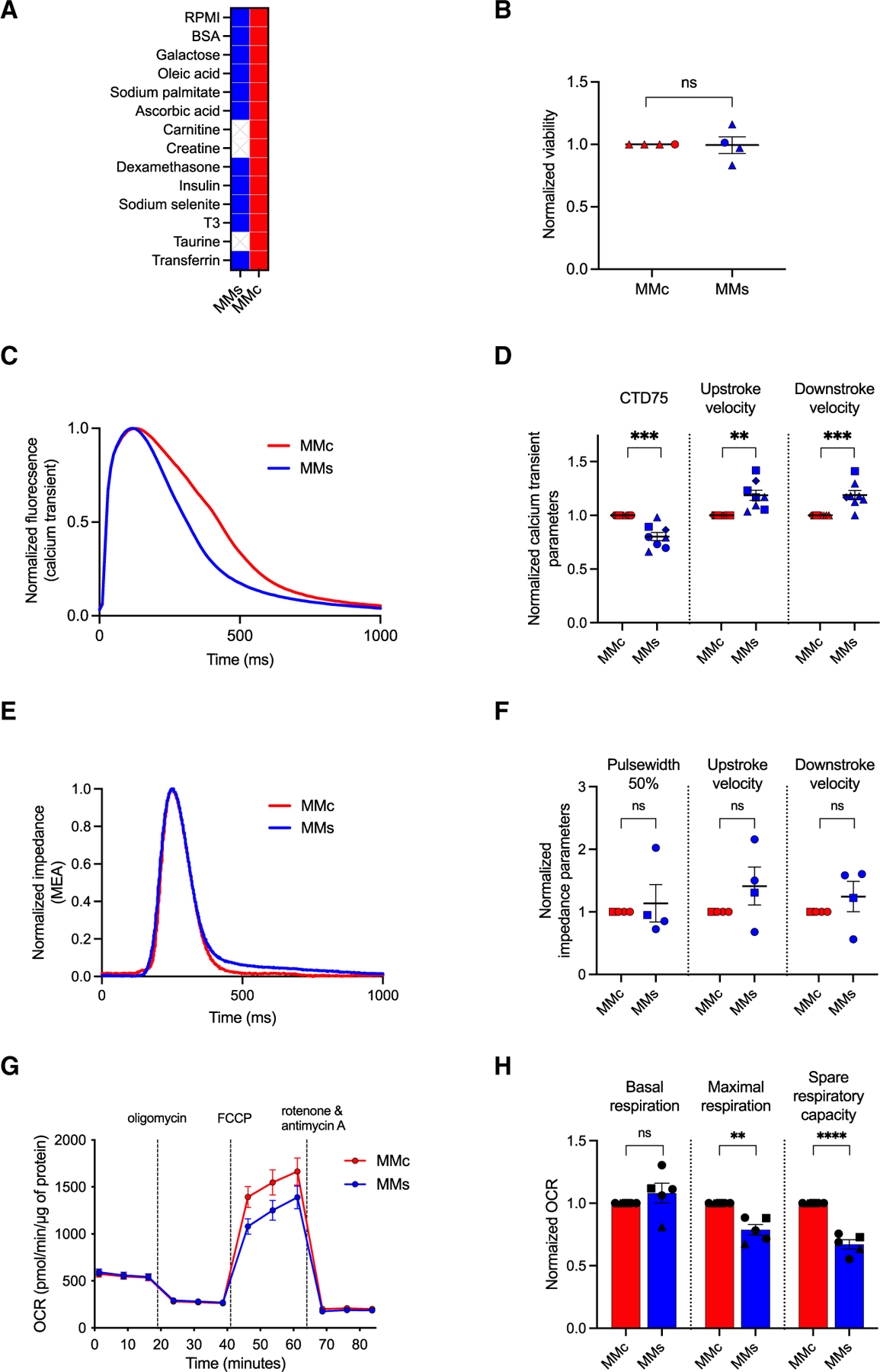
(A) MMs and MMc formulations.
(B) Viability of hiPSC-CMs cultured in MMs or MMc from day 20 to day 33 (n = 4).
(C) Representative spontaneous Ca2+ transient recordings of hiPSC-CMs cultured in MMs or MMc from day 20 to day 35.
(D) CTD75 measured when hiPSC-CMs are cultured in MMs or MMc (n = 8).
(E) Representative paced (0.8 Hz) impedance recordings of hiPSC-CMs cultured in MMs or MMc.
(F) Pulse width at 50% measured from impedance recordings of hiPSC-CMs cultured in MMs or MMc from day 20 to day 32 (n = 4).
(G) Mean OCR of hiPSC-CMs cultured in MMs or MMc from day 20 to day 33 (n = 5).
(H) Basal respiration, maximal respiration, and spare respiratory capacity of hiPSC-CMs cultured in MMs or MMc (n = 5).
Each data point shape indicates a different hiPSC line. Data are presented as mean ± SEM. n = biological replicates, unpaired Student’s t test, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.005, ****p ≤ 0.0001.
Galactose improves mitochondrial basal respiration
We next examined the effect of base components by first comparing glucose and galactose formulations of MMb, MMs, and MMc on hiPSC-CM viability and maturation (Figure 4A). Prior hiPSC-CM maturation media used galactose instead of glucose as an input to glycolysis. Metabolism of galactose through glycolysis generates no net ATP and, in turn, undergoes subsequent mitochondrial oxidative phosphorylation to generate additional energy.23 Therefore, replacing glucose with galactose is expected to shift hiPSC-CMs from glycolysis to an oxidative metabolism that can support the increased energy demands of more mature cells. We found that glucose and galactose formulations did not differ in cell viability, Ca2+ transient recordings, or impedance parameters (Figures 4B–4F and S4A–S4D). Of note, hiPSC-CMs grown in glucose containing MMb never reached the expected beat rate dictated by our pacing protocol, and therefore impedance parameters were not analyzed for this condition. hiPSC-CMs cultured in glucose- or galactose-containing MMs or MMc had no differences in peak Nav current density and the same V1/2 and k values for Nav current voltage-dependence of activation and inactivation (Figures S4E–S4G; Table S2). Mitochondrial stress tests revealed higher basal respiration when galactose replaced glucose in MMb (MMb-gal: 1.28 ± 0.15 vs. MMb-glu: 0.86 ± 0.09), MMs (MMs-gal: 1.08 ± 0.08 vs. MMs-glu: 0.55 ± 0.08), and MMc (MMc-gal: 1.0 ± 0 vs. MMc-glu: 0.57 ± 0.08; Figures 4G and 4H). However, substitution of glucose with galactose in MMs or MMc did not alter maximal respiration or spare respiratory capacity (Figures 4G and 4H).
Figure 4. Galactose improves mitochondrial basal respiration.
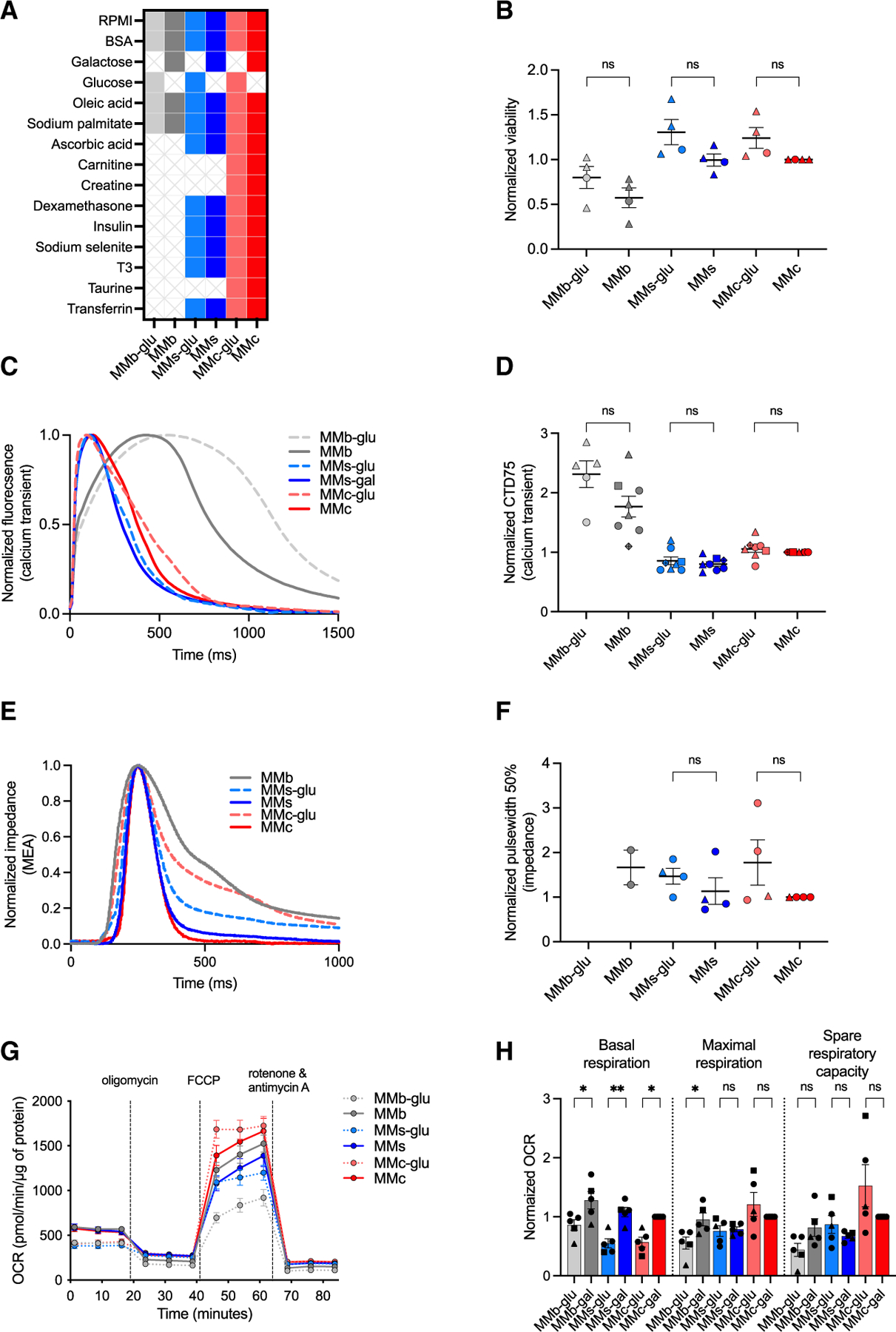
(A) Glucose and galactose formulations of MMb, MMs, and MMc.
(B) Viability of hiPSC-CMs cultured in glucose or galactose formulations of MMb, MMs, or MMc from day 20 to day 33 (n = 4).
(C) Representative spontaneous Ca2+ transient recordings of hiPSC-CMs cultured in glucose or galactose formulations of MMb, MMs, or MMc from day 20 to day 35.
(D) CTD75 of hiPSC-CMs cultured in glucose or galactose formulations of MMb, MMs, or MMc (n = 8).
(E) Representative paced (0.8 Hz) impedance recordings of hiPSC-CMs cultured in glucose or galactose formulations of MMb, MMs, or MMc from day 20 to day 32.
(F) Pulse width at 50% measured from impedance recordings of paced (0.8 Hz) hiPSC-CMs cultured in glucose or galactose formulations of MMb, MMs, or MMc (n = 4).
(G) Mean OCR of hiPSC-CMs cultured in glucose or galactose formulations of MMb, MMs, or MMc from day 20 to day 33 (n = 5).
(H) Basal respiration, maximal respiration, and spare respiratory capacity of hiPSC-CMs cultured in glucose or galactose formulations of MMb, MMs, or MMc (n = 5).
Each data point shape indicates a different hiPSC line. Data are presented as mean ± SEM. n = experimental replicates, unpaired Student’s t test, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.005, ****p ≤ 0.0001. See also Figure S4.
Fatty acids contribute to metabolism but not viability, Ca2+ dynamics, impedance, or Nav current
To evaluate the necessity of fatty acids (FAs), we tested MMs and MMc formulations with FA (MMs and MMc) and without FA (MMs-no FA and MMc-no FA; Figure 5A). Removing FAs from MMs or MMc did not change viability, Ca2+ dynamics, or impedance parameters (Figures 5B–5F and S5A–S5D). For Nav current recordings, we observed a lower peak density and different V1/2 for inactivation when FAs were removed from MMs. No difference was observed when FAs were removed from MMc (Figures S5E–S5I; Table S2). For metabolic function, we saw no change in measured parameters when we removed FAs from MMs. However, in the absence of FAs, basal respiration (MMc: 1.0 ± 0 vs. MMc-no FA: 0.75 ± 0.06), maximal respiration (MMc: 1.0 ± 0 vs. MMc-no FA: 0.71 ± 0.08), and spare respiratory capacity (MMc: 1.0 ± 0 vs. MMc-no FA: 0.70 ± 0.09) were significantly lower compared to hiPSC-CMs cultured in MMc (Figures 5G and 5H).
Figure 5. FA contribution to metabolism but not viability, Ca2+ transient, or impedance.
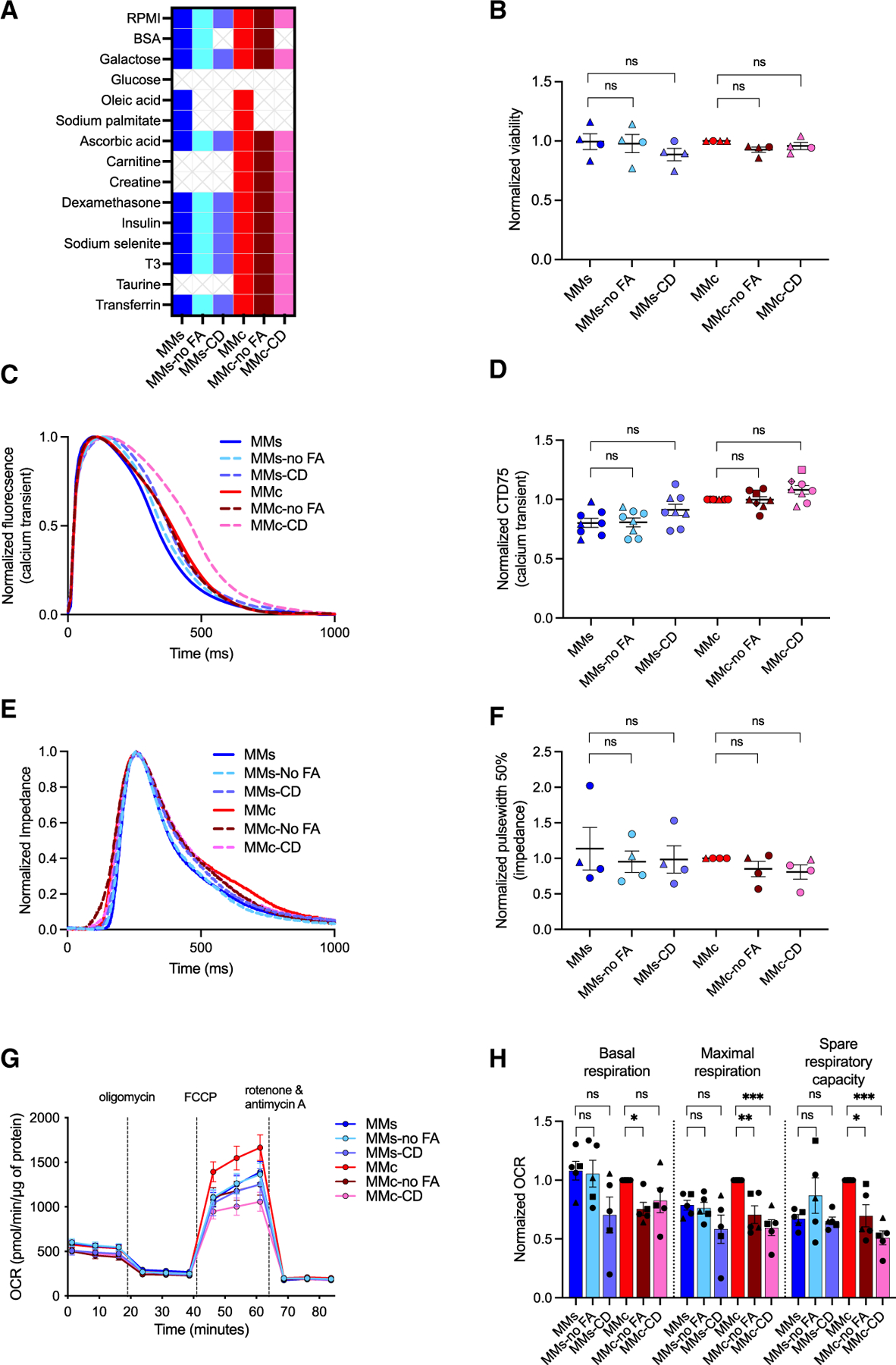
(A) MMb, MMs, and MMc formulations with FAs, without FAs (no FA), and chemically defined (CD).
(B) Viability of hiPSC-CMs cultured in MMb, MMs, or MMc formulations ± FA and CD from day 20 to day 33 (n = 4).
(C) Representative spontaneous Ca2+ transient recordings of hiPSC-CMs cultured in MMb, MMs, or MMc formulations ± FA and CD from day 20 to day 35.
(D) CTD75 of hiPSC-CMs cultured in MMb, MMs, or MMc formulations ± FA and CD (n = 8).
(E) Representative paced (0.8 Hz) impedance recordings of hiPSC-CMs cultured in MMb, MMs, or MMc formulations ± FA and CD from day 20 to day 32.
(F) Pulse width at 50% measured from impedance recordings of hiPSC-CMs cultured in MMb, MMs, or MMc formulations ± FA and CD (n = 4).
(G) OCR of hiPSC-CMs cultured in MMb, MMs, or MMc formulations ± FA and CD from day 20 to day 33 (n = 5).
(H) Basal respiration, maximal respiration, and spare respiratory capacity of hiPSC-CMs cultured in MMb, MMs, or MMc ± FA and CD (n = 5).
Each data point shape indicates a different hiPSC line. Data are presented as mean ± SEM. n = biological replicates, ANOVA, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.005, ****p ≤ 0.0001. See also Figure S5.
To evaluate whether FAs and BSA could be removed to make the formulation chemically defined (CD), we tested MMs and MMc formulations without FA and BSA (MMs-CD and MMc-CD; Figure 5A). We observed no change in viability or impedance parameters for cells grown in MMs and MMc compared to those grown in their respective CD formulations. Only cells cultured in the MMc-CD formulation had Ca2+ transients with lower maximum upstroke and downstroke velocities (Figures 5B–5F and S5A–S5D). We observed no change in peak Nav current density for hiPSC-CMs grown in MMs and MMc compared to those grown in MMs-CD or MMc-CD, respectively. Kinetic parameters of Nav channels remained the same, except for decreased V1/2 values of both Nav curent activation and inactivation when FAs and BSA were removed from MMs (Figures S5E–S5I; Table S2). For metabolic function, we found that basal respiration, maximal respiration, and spare respiratory capacity were not different between hiPSC-CMs grown in MMs and MMs-CD. Cells grown in MMc and MMc-CD exhibited comparable basal respiration, while maximal respiration (MMc: 1.0 ± 0 vs. MMc-CD: 0.60 ± 0.07) and spare respiratory capacity (MMc: 1.0 ± 0 vs. MMc-CD: 0.51 ± 0.06) were significantly higher for hiPSC-CMs grown in MMc (Figures 5G and 5H).
Energy substrate is a key driver of hiPSC-CM transcriptomics
To assess differences in gene expression, RNA sequencing (RNA-seq) was performed on day 33 hiPSC-CMs cultured in MMc, MMc-glu, MMc-no FA, MMc-CD, MMs, MMs-glu, MMs-no FA, MMs-CD, or RBAI for 2 weeks. Both hierarchical clustering of the top 100 expressed genes and principal-component analysis revealed that glucose- and galactose-based formulations grouped together, suggesting that energy substrate was an important determinant of clustering and resulted in the largest transcriptomic shift (Figures 6A and 6B). Differential gene expression analysis demonstrated a strong separation between glucose and galactose conditions and very little separation between various galactose conditions. For example, galactose-based MMc-cultured cells compared to glucose-based MMc-glu- and RBAI-grown cells had 4,347 and 4,847 differentially expressed genes, respectively (Table S3). Similarly, cells grown in MMs compared to MMs-glu and RBAI had 3,816 and 4,670 differentially expressed genes, respectively. MMc cultured cells had very few differentially expressed genes compared to cells cultured in other galactose-based formulations (MMc vs. MMs: 2 genes, MMc vs. MMc-no FA: 0 genes, and MMc vs. MMc-CD: 16 genes). hiPSC-CMs grown in MMc had differentially expressed genes involved in sarcomere structure, ion channels, and Ca2+ handling compared to cells cultured in RBAI and MMc-glu (Figure 6C; Table S3). These differences correlate with known features of maturation. For example, we observed a switch in sarcomeric genes from fetal (TNNI1 and MYH6) to adult (TNNI3 and MYH7) isoforms. We observed higher TNNI3 expression in hiPSC-CMs cultured in MMc compared to those grown in MMc-glu or RBAI. With respect to ion channel expression, hiPSC-CMs grown in MMc had lower HCN4 expression, no change in SCN5A expression, and higher CACNA1C, KCNQ1, and KCNJ2 expression compared to cells cultured with RBAI. We also observed higher expression of Ca2+ cycling genes, such as ATP2A2, PLN, and RYR2, in cells grown in MMc compared to RBAI and no change in expression of these genes in cells cultured in MMc compared to those grown in MMc-glu. Finally, many genes involved in glycolysis are downregulated, and many genes involved in FAO are upregulated, for MMc-cultured cells compared to RBAI- and MMc-glu-cultured hiPSC-CMs (Figure 6C; Table S3).
Figure 6. Energy substrate is key driver of hiPSC-CM transcriptomics.
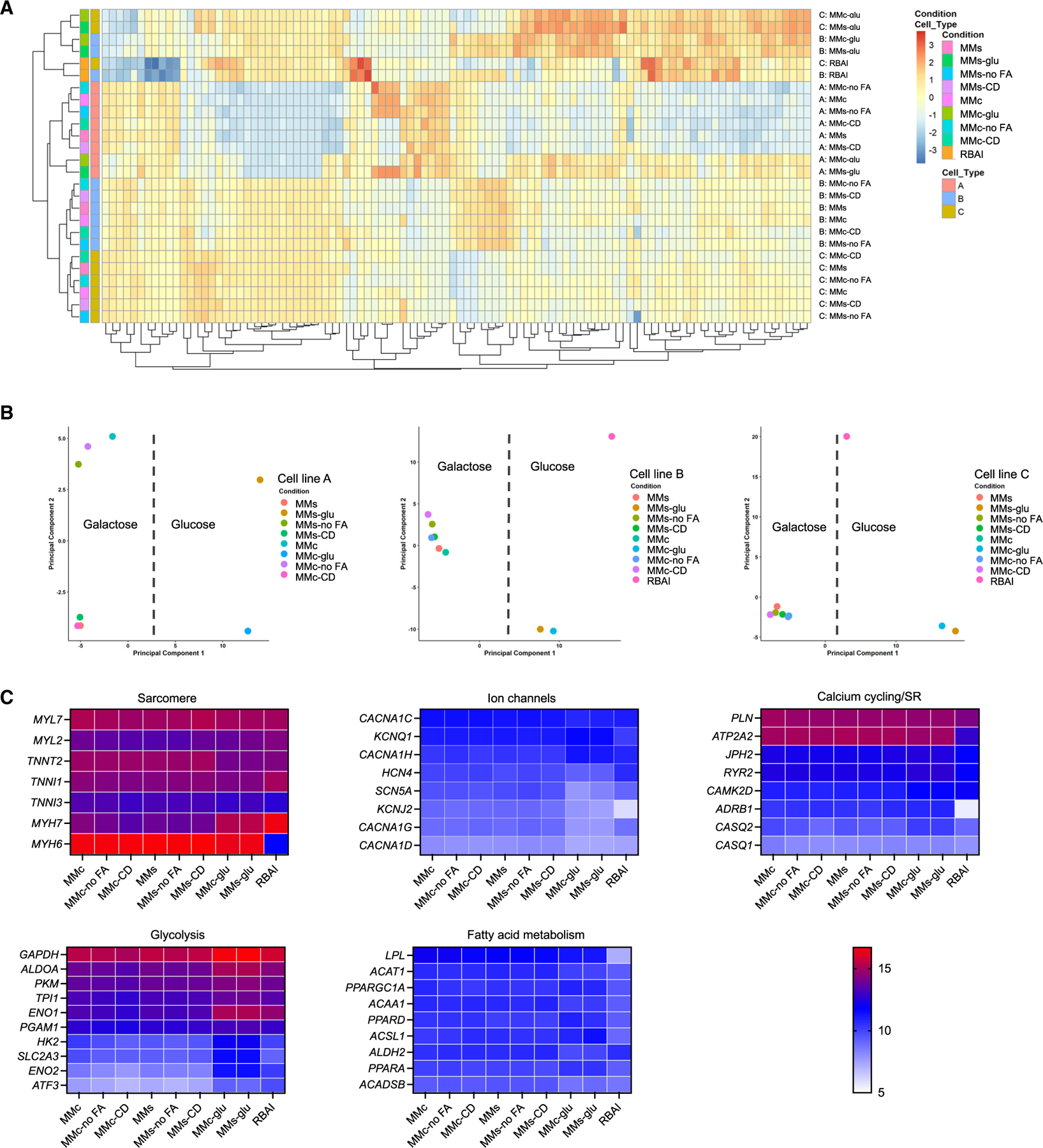
(A) Heatmap of top 100 expressed genes for hiPSC-CMs generated from 3 different hiPSC lines and cultured in 9 different medium formulations from day 20 to day 33.
(B) Principal-component analysis (PCA) plots of hiPSC-CMs generated from 3 different hiPSC lines and cultured in 9 different medium formulations.
(C) Heatmaps of genes related to metabolism and cardiac structure and function.
See also Table S3.
DISCUSSION
This study introduces a high-throughput platform to screen inducers of CM maturation and provides insight into the impact of individual components on specific aspects of maturation. By combining many well-established components that induce hiPSC-CM maturation, including replacing glucose with galactose and adding FAs, T3, and dexamethasone, we developed, for the first time, a concentration-optimized formulation, MMc, that improved hiPSC-CM maturation status functionally, metabolically, and transcriptionally. We further explored the individual roles of each component and demonstrated that certain components enhance functional maturation and others enhance metabolic maturation.
Previous medium-based maturation studies have often compared hiPSC-CMs cultured in a complex maturation medium to those cultured in a simple maintenance medium, making the effect of individual components on viability and maturation unclear. Although all components in this study have been included in maturation medium formulations in previous studies, the individual role of each component pertaining to maturation has only been examined for a few of them. One study found that ascorbic acid improved structural and functional maturation of hiPSC-CMs.19 Another study demonstrated that, when added to a maintenance medium, T3 improved morphological, molecular, Ca2+ transient, contractility, and metabolic parameters of hiPSC-CM maturation.16 We found that ascorbic acid and T3 produced more mature Ca2+ transients, which aligns with these two studies.16,19 Studies have shown that dexamethasone and IGF1, which activates the same receptor as insulin, did not improve the specific areas of hiPSC-CM maturation investigated. However, when each is combined with T3 or all three combined together, synergistic effects promoting maturation have been reported.17,18,24 For example, Birket et al. found that T3 alone, along with various combinations of T3 with dexamethasone and IGF1, positively modulated the resting membrane potential, as determined through a proxy fluorescence assay. They further demonstrated that combining all three components consistently outperformed single or dual combinations in regard to improved maturation of metabolic, AP, and contractility properties.17 Although the combinations of carnitine, creatine, and taurine11,12,21 as well as insulin, transferrin, and selenium12,21 have been included in multicomponent maturation medium formulations, the individual role of each was not explored. For the first time, we determined that dexamethasone, insulin, sodium selenite, and transferrin are individually important for Ca2+ dynamics, while carnitine, creatine, and taurine are dispensable. It is unsurprising that the removal of these 3 components identified as dispensable from the Ca2+ transient screen did not change the performance of hiPSC-CMs in subsequent Ca2+ dynamic, impedance, and Nav current recordings, since these measure closely related aspects of the AP and excitation-contraction (EC) coupling. This suggest that Ca2+ imaging is sensitive enough to detect components that induce maturation of hiPSC-CM AP and EC coupling-related functions and is an appropriate methodology to screen established and novel components of maturation in future studies.
When investigating metabolic parameters of hiPSC-CMs, we discovered that although carnitine, creatine, and taurine were dispensable for AP-related assays, they were not dispensable for mitochondrial function. These three components are often added to maturation media together and, although not individually examined and validated, it is hypothesized that they contribute to hiPSC-CM bioenergetics.11,12,21 Cells cultured in MMc had higher mitochondrial respiration parameters compared to cells cultured in MMc without carnitine, creatine, and taurine, which is indicative of a transition from a more immature glycolytic profile to a more mature and efficient oxidative metabolism. This correlates with the number of reports showing their importance in regulating cell energy demands. Taurine is a regulator of the electron transport chain. Schaffer et al. observed increased glycolysis, reduced oxidation of glucose and FAs, and decreased ATP production in mice with taurine-deficient hearts.25 This can explain why the absence of taurine resulted in a less oxidative metabolism as observed in our study. Within the creatine kinase pathway, creatine is responsible for shuttling ATP from the mitochondria to the cytosol and regulates this process based on the energy demands of the cell.26 Removing creatine from the medium potentially decreases the creatine pool available to shuttle ATP and makes the cell less equipped to handle cell stressors, which is an indicator of immaturity. Carnitine is required for long-chain FA transport from the cytosol into the mitochondria, where subsequent FAO occurs.27 The absence of carnitine from the medium would then result in a less mature metabolism due to reduced FAO and could explain the decrease we observed in mitochondrial function. Interestingly, carnitine palmitoyltransferase 1A (CPT1A), which encodes an enzyme for carnitine-dependent transport into the mitochondria, was one of only two genes differentially expressed in hiPSC-CMs cultured in MMs compared to MMc, indicating that the presence of carnitine in MMc might be influencing this gene regulation.28 With the knowledge that one or more of these components have the potential to increase ATP production within hiPSC-CMs, a future study can investigate the OCR of cells cultured in carnitine, creatine, or taurine to identify which of these three components are contributing to improved metabolic maturation.
In previous studies, an alternative energy substrate to the high glucose concentrations found in most basal media, such as low glucose, no glucose, or galactose, was part of complex maturation medium formulations being compared to a simple maintenance medium. When there was a direct comparison of the same formulation with two different energy substrates, only metabolic maturation was assessed leaving the impact on other aspects of maturation unknown. For example, Rana et al. demonstrated that energy substrate, specifically galactose, improved metabolic maturation.29 However, the effect of these metabolic substrates on other areas of maturation, such as structure or physiological function, was not reported. In addition to improved metabolic maturation, Correia et al. demonstrated improved transcriptional, structural, and functional maturation status of hiPSC-CMs grown in a galactose-based medium with FAs compared to a glucose-based medium without FAs, making it difficult to distinguish if the observed difference was due to galactose, FAs, or the combination of the two.8 Our study builds upon these studies by directly comparing galactose and glucose versions of three medium formulations to distinguish the effect of the energy substrate on functional and metabolic hiPSC-CM maturation. Our findings align with these studies in that galactose rather than glucose improves metabolic maturation status. Glucose and galactose versions of the same medium also had around 4,000 differentially expressed genes, indicating that energy substrate greatly influences the transcriptional profile of hiPSC-CMs. This suggests that, when the components contributing to more mature Ca2+ transients are included in the medium, replacing glucose with galactose enhances metabolic maturation and influences the transcriptome but does not further enhance Ca2+ transient, impedance, or Nav current maturation status.
We found that removing FAs from the formulations also decreased mitochondrial function while not impacting functional parameters. Only a handful of studies that include FAs have direct comparisons of medium formulations with and without FAs, and often both FAs and energy substrate are being altered at the same time. Of these studies, there are conflicting reports on whether FAs improve maturation. For example, Yang et al. used a glucose-based medium and compared adding palmitate, oleate, and linoleate to the same medium and demonstrated that these additions improved Ca2+ transient kinetics, contractile force, AP parameters, and mitochondrial function.9 This supports the concept that FAs, even in the presence of glucose, have a role in several aspects of hiPSC-CM maturation. Hu et al. also added FAs to a glucose-based medium but found no significant improvement in metabolic or structural maturation. However, when they added FAs to a formulation without glucose or galactose, metabolic, structural, contractile, and calcium transient parameters improved, indicating increased maturation.30 These results indicate that an alternative energy substrate to glucose is required for FAs to impact maturation. Our study supports the concept that the combination of FAs and galactose improves metabolic maturation; however, we did not see an improvement in other areas of maturation. When we removed FAs from MMc, we saw a decreased oxidative metabolism similar to the levels observed in cells cultured in MMs. But interestingly, no additional loss was observed with the removal of FAs from MMs suggesting that without carnitine, creatine, or taurine, FAs cannot be utilized as an oxidative substrate. This could explain the difference between our study and the findings of Rana et al., where they observed no change in metabolic maturation when adding FAs to a galactose- or glucose-based medium.29 Additionally, we did not see a difference in transcriptional profiles of hiPSC-CMs cultured with or without FAs, making them dispensable for gene regulation.
This leaves us with the question of how much further can we refine hiPSC-CM functional and metabolic maturation using a medium-based approach and whether certain aspects should be focused on depending on the applications. For example, for drug screening and safety testing, future studies should examine whether there is a differential drug response with varying levels of functional and metabolic maturity. If both are important, we recommend combining Ca2+ imaging and mitochondrial stress tests for future screens of inducers of maturation. These screens can include novel or established components targeting identified pathways important for maturation in both MMb to better understand their individual role as well as in MMc to determine whether there is an additive effect. For example, small molecules that have previously been identified to improve hiPSC-CM maturation and are regulators of metabolic pathways, the cell cycle, or developmental pathways can be screened.30–33 Additional medium alterations, such as increasing Ca2+ concentrations in the basal medium, which is less than one-third of physiological levels, could increase the maturation status of Ca2+ transients or contractility, as observed previously in increased contractile force of human engineered heart tissues.34
In summary, we found that ascorbic acid, dexamethasone, insulin, sodium selenite, T3, and transferrin contribute to viability and more mature Ca2+ transients and contractility. Galactose and FAs, along with a combination of carnitine, creatine, and taurine, contribute to improved metabolism. Hence, we recommend MMc as a balanced formulation for enhancing multiple facets of maturation and to use as a baseline in combination with the high-throughput strategies we established to screen for novel inducers of maturation.
Limitations of the study
In this study, we generated both a high-throughput system to investigate various facets of hiPSC-CM maturation and an optimized maturation medium using components established in the literature as a baseline. Therefore, we did not screen and identify novel inducers of maturation or viability in this study. Also, while we probed multiple aspects of maturation using specific assays to measure viability, Ca2+ transients, impedance, mitochondrial function, and electrophysiology, we did not further examine component effects on structures or pathways related to these assays, such as the presence of t-tubules, sarcomeric organization, exit from the cell cycle, or multinucleation. Additionally, previous studies have indicated that galactose could have toxic effects on CMs.35 In this study, most experiments were performed using hiPSC-CMs cultured in galactose for 2 weeks and, in the case of Ca2+ dynamic and impedance recordings, up to 3 weeks. Although we did not observe toxicity to hiPSC-CMs in these studies, potential toxic effects of extended culture in galactose could be examined in future studies. Last, both beneficial and detrimental effects of the maturation media could be masked due to the culture of hiPSC-CMs in monolayers rather than in a 3D tissue format. This high-throughput system will be a powerful tool to identify components contributing to maturation and to inform further in-depth mechanistic studies.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Paul Burridge (paul.burridge@northwestern.edu).
Materials availability
Plasmids used in this study have been deposited to Addgene (pAAVS1-Puro-TNNT2-Neo, Addgene#214013). hiPSC lines used in this study are available from the lead contact, Paul Burridge (paul.burridge@northwestern.edu), with a completed Materials Transfer Agreement.
Data and code availability
RNA-seq datasets generated in this study are available at the NCBI Gene Expression Omnibus (GEO) as accession number GSE244962.
The software and algorithms for data analyses used in this study are published and referenced.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Human induced pluripotent stem cell culture
The hiPSC lines used in this publication include 19c3 (M), 21c10 (F), 22c10 (M), 23c9 (M), 25c3 (M), 26c3 (M), GW124c9 (M), and GW132c2B (F).36,37 Protocols were approved by the Northwestern University Institutional Review Board and written informed consent was obtained from all volunteers. hiPSC lines were derived from isolated peripheral blood mononuclear cells of healthy volunteers using a Sendai virus (Invitrogen, A16518) reprogramming protocol and cultured in B8 as previously described.36 Once established, each line was transfected with an exogenous TNNT2 promoter-driven neomycin resistance cassette targeting the AAVS1 locus for cardiomyocyte selection during differentiation. Cells were maintained in B8 medium on 1:800 growth factor reduced Matrigel (Corning, 356234) diluted in DMEM (Corning, 10013CV), except for the first 24 h after passage when B8 was supplemented with 2 μM thiazovivin (B8T, LC Labs, T-9753).36,38,39 hiPSC lines at ~75% confluency were passaged in B8T at a ratio of 1:15 every 4 days using 0.5 mM EDTA (Corning, 46–034-Cl) diluted in DPBS−/− (Corning, 20–031-CV) to a final concentration of 0.5 μM for 6 min at RT. All established pluripotent lines and lines undergoing reprogramming were maintained at 37°C in Heracell VIOS 160i humidified incubators (Thermo) with 5% CO2 and 5% O2. All cultures were routinely tested for mycoplasma using a MycoAlert PLUS Kit (Lonza, LT07–710) and a Varioskan LUX (Thermo) plate reader.
Cardiac differentiation
hiPSC lines were differentiated into cardiomyocytes using a previously described RBAI protocol with slight modifications.40 Briefly, hiPSC lines were grown in B8 medium for 4 days until reaching ~75% confluency. To start the differentiation (day 0), B8 medium was changed to R6C which is RPMI 1640 (Corning, 10–040-CM) supplemented with 6 μM of glycogen synthase kinase 3-b inhibitor CHIR99021 (LC Labs, C-6556). On day 1, medium was changed to R which is RPMI 1640 (Corning, 10–040-CM) basal medium alone, and on day 2, medium was changed to RBA-C59 which is RPMI 1640 (Corning, 10–040-CM) supplemented with 2 mg/mL fatty acid-free bovine serum albumin (GenDEPOT, A0100), 200 μg/mL L-ascorbic acid 2-phosphate (Wako, 321–44823), and 2 μM Wnt-C59 (Biorbyt, orb181132). Medium was then changed every other day starting on day 4 with RBAI which is RPMI 1640 (Corning, 10–040-CM) supplemented with 2 mg/mL fatty acid-free bovine serum albumin, 200 μg/mL L-ascorbic acid 2-phosphate, and 5 μg/mL E. coli-derived recombinant human insulin (Gibco, A11382IJ). Contracting cells were observed starting at day 7 and were treated with 100 μg/mL of neomycin (Gibco, 10131027) from day 8 to day 12 to select for TNNT2+ cardiomyocytes. On day 16 of differentiation, hiPSC-CMs were dissociated using DPBS (Corning, 21–031-CV) for 20 min at 37°C, 5% CO2 followed by 1:200 Liberase TH (Roche, 5401151001) diluted in DPBS for 20 min at 37°C, 5% CO2. The collected cells were centrifuged at 300 × g for 5 min and filtered through a 100 μm cell strainer (Fisherbrand, 22–363-549). Cells were then plated in RBAI +10% Cosmic Calf Serum (Hyclone, SH3008703HI) for 2 days on 1:800 Matrigel-coated plates for each assay. Media was then replaced with RBAI or maturation medium which was changed every 2–3 days until cells were assayed. Cells were maintained at 37°C in Heracell VIOS 160i humidified incubators (Thermo) with 5% CO2 and atmospheric O2 during differentiation.
Maturation media formulation
Complete maturation media (MMc) basal medium is RPMI 1640 with no glucose (Corning, 10–043-CV). Sodium palmitate (Sigma, P9767) was complexed with fatty acid-free bovine serum albumin to enhance solubility. Sodium palmitate was dissolved in 50% ethanol at 52°C for 10 min and added to fatty acid-free bovine serum albumin at 37°C for 1 h with occasional mixing for a final concentration of 3 mM sodium palmitate and 120 mg/mL BSA. The following components were added to the basal medium: 2 mM galactose (Fisher, BP656–500), 50 μM oleic acid (water soluble, Sigma, O1383), 50 μM sodium palmitate complexed with 2 mg/mL fatty acid-free bovine serum albumin (GenDEPOT, A0100), 5 mM creatine phosphate (Dot Scientific, DSC70300–25), 5 mM taurine (Sigma T8691), 200 μg/mL L-ascorbic acid 2-phosphate, 20 mg/mL Oryza sativa-derived recombinant human transferrin (InVitria, 777TRF029), 15 ng/mL sodium selenite (Sigma, S5261), 2 mM L-carnitine hydrochloride (Sigma, C0283), 10 μg/mL E. coli-derived recombinant human insulin (Gibco, A11382IJ), 1 μM dexamethasone (Sigma, D4902), and 3,3′,5-Triiodo-L-thyronine sodium salt (T3, Sigma T6397). Base maturation media (MMb) and simple maturation media (MMs) are composed of 5 and 11 of the above components, respectively (Table S1). Additional derivatives of MMc and MMs, including fatty acid-free and chemically defined formulations can be found in Table S1.
Intracellular Ca2+ transient imaging
Intracellular Ca2+ transients were recorded using an IC200 Kinetic Image Cytometer (KIC, Vala Sciences) as previously described.41 Briefly, hiPSC-CMs were dissociated on d16 and plated at 60,000 cells per well in RBAI +10% Cosmic Calf Serum (Hyclone, SH3008703HI) on a Matrigel-coated 96-well black microplate (Greiner, 655090). Media was replaced with RBAI on d18. Starting on d20, medium in each well was changed to RBAI, MMc, MMc-glu, MMc-no FA, MMc-CD, MMs, MMs-glu, MMs-no FA, or MMs-CD every 3 days until day of assay (d35). On the day of imaging, cells were incubated at 37°C, 5% CO2 in Hanks’ Balanced Salt Solution (HBSS, Corning, 21–023-CV), 20 mM HEPES (Corning, 25–060-CI), 0.04% Pluronic F-127 (Sigma, P2443), 2.5 mM probenecid (Sigma, P8761), 2 drops per 10 mL NucBlue Live ReadyProbes Reagent (Invitrogen, R37605), and 2 μM Cal-520 a.m. (AAT Bioquest, 21130) for 1 h. The Ca2+ dye loading solution was then replaced with FluoroBrite DMEM (Gibco, A1896702) and 2.5 mM probenecid. Cells were imaged on an IC200 KIC at 37°C, 5% CO2 for a 10 s time series at an acquisition frequency of 99.75 Hz Ca2+ transients at the single cell level were analyzed for all wells using CyteSeer Analysis software (Vala Sciences). We used the average of all single cell Ca2+ transients per well for traces and trace quantification including maximum downstroke velocity, maximum upstroke velocity, peak value, Ca2+ transient duration at 75% (CTD75).
Impedance recordings
Impedance recordings were measured using a CardioExcyte96 (Nanion). Briefly, hiPSC-CMs were dissociated on d16 and plated at 100,000 cells per well in RBAI +10% Cosmic Calf Serum (Hyclone, SH3008703HI) on Matrigel-coated Cardioexcyte96 sensor plates with stimulation electrode (Nanion, 20–1003). Media was replaced with RBAI on d18. Starting on d20, medium in each well was changed to RBAI, MMc, MMc-glu, MMc-no FA, MMc-CD, MMs, MMs-glu, MMs-no FA, or MMs-CD every 3 days until day of assay (d32). Impedance was measured at 37°C, 5% CO2, and 80% humidity using CardioExcyte Control 96 software (Nanion). Spontaneous and then paced (0.8 Hz) recordings were captured using a 30 s sweep duration every 5 min for a minimum of 1 h. Impedance traces were analyzed using Data Control (Nanion) software and the beat rate (beat/min), pulsewidth at 50% (sec), upstroke velocity (ohm/s), and downstroke velocity (ohm/s) were calculated. All wells were analyzed unless their impedance value was less than 1 or wells had less than 45 beats per min when paced at 0.8 Hz.
Action potential recordings
hiPSC-CMs were dissociated on d20 and plated in RBAI +10% Cosmic Calf Serum (Hyclone, SH3008703HI) on Matrigel-coated 18 mm glass coverslips (Warner Instruments, 64–0384). Medium was replaced every three days with RBAI or MMc. Action potentials (APs) were measured on d30 hiPSC-CMs with manual patch clamp at 37°C from spontaneously beating hiPSC-CMs using the amphotericin-B perforated patch-clamp technique with a Multiclamp 700B amplifier and Clampex 10.3 software (Molecular Devices). Pipettes (resistance 3–3.5 MΩ) were pulled from thin wall borosilicate glass capillaries (WPI, 1B120F-4) using a horizontal microelectrode puller (Sutter instrument, P-1000). Signals were low-pass-filtered with a cutoff of 10 kHz and digitized at 10 kHz. Bath solution contained: 140 mM NaCl, 4.0 mM KCl, 1.0 mM CaCl2, 0.5 mM MgCl2, 10 mM glucose, 10 mM HEPES; pH 7.4 (NaOH). Pipettes were filled with solution containing: 125 mM K-gluconate, 20 mM KCl, 5.0 mM NaCl, 0.26 mM amphotericin-B, 5.0 mM HEPES; pH 7.2 (KOH). To overcome the spontaneous activity and depolarized state of hiPSC-CMs, an ohmic current ~4 pA/pF was continuously injected to maintain a stable resting potential at approximately −80 mV, except where mentioned otherwise. APs were evoked at 1 Hz by 4-ms, ~1.3× threshold current pulses through the patch pipette and were characterized by maximum AP amplitude (APA), AP duration at 90% of repolarization (APD90), and maximal upstroke velocity (Vmax). The maximal diastolic potential was analyzed during spontaneous activity. Potentials were corrected for the calculated liquid junction potential.
hiPSC-CM isolation for automated high-throughput electrophysiology
hiPSC-CMs were dissociated on d16 and plated in Matrigel-coated 6-well plates at a density of 4 x 106 cells per well in RBAI +10% Cosmic Calf Serum (Hyclone, SH3008703HI). Medium was changed on d18 to RBAI. Starting on d20, medium in each well was changed to RBAI, MMc, MMc-glu, MMc-no FA, MMc-CD, MMs, MMs-glu, MMs-no FA, or MMs-CD every 3 days until day of assay (d32). Only wells with beating (contracting) cells were utilized. The day of the experiment, each well was first washed with 1 mL DPBS (Corning, 21031CV). Solution was removed from the well and then incubated with 2 mL DPBS for 30 min at 37°C, 5% CO2. After this incubation, the solution was aspirated and replaced with 1 mL DPBS + liberase (50 units/ml, Liberase TH Research Grade, Sigma), and incubated for 25–27 min at 37°C, 5% CO2. hiPSC-CMs were then dissociated by gentle mechanical cell separation (trituration) using a 5 mL stripette. Two mL DPBS were added to cell suspension and cell number and viability were determined by automated cell counting (ViCell, Beckman Coulter). hiPSC-CMs were diluted to 0.5 x 106 per mL viable count with DPBS and placed on the electrophysiology platform cell container and allowed to recover for at least 30 min at 16°C, shaking at 250 RPM.
Automated high-throughput electrophysiology
High-throughput automated electrophysiology was successfully used to record and analyze voltage-gated sodium whole-cell currents from hiPSC-CMs in a 384-well plate format using the SyncroPatch 384 platform (Nanion Technologies) as previously described.42,43 Nav currents recordings were performed at 21°C. The external solution contained (in mM): 140 NaCl, 4 KCl, 2.0 CaCl2, 1 MgCl2, 10 HEPES, 5 glucose, pH 7.4. The internal solution consisted of (in mM): 110 CsF, 10 CsF, 10 NaCl, 20 EGTA, 10 HEPES, 2 mM MgATP, pH 7.2. Pulse generation and data collection were performed with PatchController384 V2.2 (Nanion Technologies). Whole-cell currents were filtered at 3 kHz and digitized at 10 kHz. The access resistance and apparent membrane capacitance were estimated using built-in protocols. Series resistance was compensated for 70% and leak and capacitance artifacts were subtracted using the P/4 method. Whole-cell current peak current amplitude and voltage-dependence of activation and inactivation were measured using a double pulse protocol from a holding potential of −120 mV.
Automated high-throughput electrophysiology data analysis
Data were analyzed and plotted using a combination of DataController384 V2.3 (Nanion Technologies), Excel (Microsoft), and GraphPad Prism 10 (GraphPad Software). The peak current was normalized for cell capacitance and plotted against step voltage to generate peak current density–voltage relationships. Whole-cell conductance (GNa) was calculated as GNa = I/(V – Erev), where I is the measured peak current, V, is the step voltage, and Erev is the calculated sodium reversal potential. GNa at each voltage step was normalized to the maximum conductance between −80 mV and 40 mV. To calculate voltage dependence of activation normalized GNa was plotted against voltage and fitted with the Boltzmann function G/Gmax= (1 + exp[(V – V1/2)/k])−1, where V1/2 indicates the voltage at half-maximal activation and k is a slope factor describing voltage sensitivity of the channel. Voltage dependence of steady-state inactivation was assessed by plotting currents generated by the −20 mV postpulse voltage step normalized to the maximum current against the 500 ms prepulse voltage step from −120 to 0 mV in 10-mV increments. Normalized currents were plotted against voltage and fitted with the Boltzmann function. Number of cells (n) is given in the figure legends. Statistical significance was established at p ≤ 0.05.
Viability
hiPSC-CMs were dissociated on d16 and plated at 100,000 cells per well in RBAI +10% Cosmic Calf Serum (Hyclone, SH3008703HI) on a Matrigel-coated 96-well black microplate (Greiner, 655090). Media was replaced with RBAI on d18. Starting on d20, medium in each well was changed to RBAI, MMc, MMc-glu, MMc-no FA, MMc-CD, MMs, MMs-glu, MMs-no FA, or MMs-CD every 3 days until day of assay (d33). Viability was assessed by diluting PrestoBlue (Invitrogen, A13262) 1:10 with DPBS−/− (Corning, 21–031-CV) and replacing the media in 96-well plates with 100 μL per well. The plate was then incubated for 2 h at 37°C, 5% CO2. Fluorescence (560 nm excitation, 590 nm emission) was then measured using a Varioskan LUX (Thermo Scientific) plate reader with ‘top read’ function.
Seahorse OCR/ECAR measurement
The Seahorse XF96 extracellular flux analyzer was used to assess cellular metabolism. On d16, hiPSC-CMs were dissociated and plated at 100,000 cells per well in RBAI +10% Cosmic Calf Serum (Hyclone, SH3008703HI) on a Matrigel-coated XF96 plate (Agilent, 101085–004). Media was replaced with RBAI on d18. Starting on d20, medium in each well was changed to RBAI, MMc, MMc-glu, MMc-no FA, MMc-CD, MMs, MMs-glu, MMs-no FA, or MMs-CD every 3 days until day of assay (d33). On day 34, RPMI with no glucose and no sodium bicarbonate (Made in house) was supplemented with 10 mM glucose, 2 M L-glutamine, and 1 mM sodium pyruvate (pH 7.4). The medium in the XF96 plate was replaced with this and the plate was incubated for 1 h in a 37°C, no-CO2 incubator. The wells of a hydrated sensor cartridge were then loaded with mitochondrial stress test inhibitors. A standard mitochondrial stress test was then conducted and the following inhibitors were injected during the protocol: oligomycin (1 μM), FCCP (2 μM), and antimycin A and rotenone (1 μM). The oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) values were normalized to total protein content. Basal respiration was determined by subtracting the OCR after antimycin/rotenone injection from the OCR at baseline. Spare respiratory capacity was calculated by subtracting the baseline OCR from the OCR after FCCP injection. Nonmitochondrial respiration was determined from the OCR value after antimycin/rotenone injection. Maximal respiration was determined by subtracting nonmitochondrial respiration from OCR after FCCP injection.
Protein concentration measurement
After OCR/ECAR measurements, wells were rinsed twice with DPBS and 100 μL of 1:5 radioimmunoprecipitation assay buffer (Thermo, 89900) in DPBS was added to each well. Cell lysates were transferred to a 96-well PCR plate and stored at −20°C. BCA protein assay (Thermo, 23227) was performed according to the manufacturer’s instructions. Briefly, the working reagent was made by mixing 50 parts of BCA reagent A and 1 part of BCA reagent B. In a 96-well plate, 200 μL of the working reagent and 25 μL of the cell lysates or serial dilutions of the standard (BSA) were added per well and mixed on a plate shaker for 30 s. After a 30 min incubation at 37°C, absorbance was read at a wavelength of 562 nm using a plate reader (Varioskan LUX, Thermo). To calculate protein concentration, absorbance values were fitted to the generated protein standard curve. Total protein mass per well of the XF96 plate was calculated by multiplying the protein concentration by the total volume of the lysate (100 μL).
RNA-sequencing
hiPSC-CMs were dissociated on d16 and plated in Matrigel-coated 6-well plates at a density of 4 x 106 cells per well in RBAI +10% Cosmic Calf Serum (Hyclone, SH3008703HI). Medium was changed on d18 to RBAI. Starting on d20, medium in each well was changed to RBAI, MMc, MMc-glu, MMc-no FA, MMc-CD, MMs, MMs-glu, MMs-no FA, or MMs-CD every 3 days. On d33, cell lysis was collected with 300 μL per well of TRIzol Reagent (Thermo Fisher, 15596026). RNA was then purified using Direct-zol RNA MicroPrep kit (Zymo, R2062) including on-column DNase digestion to remove genomic DNA. Sample purity was confirmed using a Thermo Scientific NanoDrop 8000 and samples were then shipped to Novogene on dry ice for library preparation and sequencing. Upon arrival, all samples underwent QC using electrophoresis, Nanodrop, and Agilent2100 to determine RNA Integrating Number (RIN). The eukaryotic mRNA library with poly A enrichment was prepared and resulted in the construction of 250–300 bp insert cDNA library. Paired-end (150 bp) RNA-sequencing on hiPSC-CMs was performed using the Illumina NovaSeq 6000 sequencing platform at Novogene.
Using SOAPnuke,44 raw fastq files were processed to trim/remove adaptors, low-quality reads, and N reads. Clean sequencing reads were mapped to the GRCh38 reference genome using STAR45 and counted using RSEM.46 Genes with less than 10 counts across all 26 samples were excluded from subsequent analysis. Following variance stabilizing transformation in DESeq2 R package,47 principal component analysis was performed to visualize the clustering of samples. Using the 250 most variable genes, unsupervised hierarchical clustering based on Euclidean distance was performed by the pheatmap R package. Differential expression analysis was performed using the DESeq2 R package47 accounting for the matched pair design, cell passage, and an interaction term assessing the gene expression change by cell passage. Results were shown by a volcano plot generated using the ggplot2 R package. Genes with adjusted p-value < 0.05 corrected for multiple testing using the Benjamini Hochberg method were considered as differentially expressed. Ingenuity Pathway Analysis (IPA) was performed to identify canonical pathways based on the differentially expressed genes.48 This was used to identify differences in pathways related to biological processes that were relevant between our samples.
QUANTIFICATION AND STATISTICAL METHODS
Data were analyzed in Excel and GraphPad Prism. Detailed statistical information is included in the corresponding figure legends. Data were presented as mean ± SEM. Data were checked for normal distribution and comparisons were conducted via an unpaired two-tailed Student’s t-test or one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test. Significant differences are defined as p < 0.05 (*), p < 0.01 (**), p < 0.005 (***), and p < 0.0001 (****). No statistical methods were used to predetermine sample size. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
|
| ||
| CytoTune-iPS 2.0 Sendai Reprogramming Kit | Invitrogen | Cat#A16518 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Thiazovivin | LC Labs | Cat#T-9753 |
| CHIR99021 | LC Labs | Cat#C-6556 |
| Wnt-C59 | Biorbyt | Cat#orb1881132 |
| l-ascorbic acid 2-phosphate trisodium salt | Wako | Cat#321-44823 |
| Fatty acid-free bovine serum albumin | GenDEPOT | Cat#A0100 |
| Recombinant human insulin | Gibco | Cat#A11382IJ |
| Neomycin | Gibco | Cat#10131027 |
| Liberase TH | Roche | Cat#5401151001 |
| Galactose | Fisher | Cat#BP656-500 |
| Sodium palmitate | Sigma | Cat#P9767 |
| Oleic acid | Sigma | Cat#O1383 |
| Creatine phosphate disodium salt tetrahydrate | Dot Scientific | Cat#DSC70300-25 |
| Taurine | Sigma | Cat#T8691 |
| Recombinant human transferrin | In Vitria | Cat#777TRF029 |
| Sodium selenite | Sigma | Cat#S5261 |
| L-carnitine hydrochloride | Sigma | Cat#C0283 |
| Dexamethasone | Sigma | Cat#C0283 |
| 3,3′,5-Triiodo-L-thyronine sodium salt | Sigma | Cat#T6397 |
| Pluronic F-127 | Sigma | Cat#P2443 |
| Probenecid | Sigma | Cat#P8761 |
|
| ||
| Critical commercial assays | ||
|
| ||
| MycoAlert PLUS Kit | Lonza | Cat#LT07-705 |
| PrestoBlue | Invitrogen | Cat#A13262 |
| Cal-520 a.m. | AAT Bioquest | Cat#21130 |
| Seahorse XF Cell Mito Stress Test Kit | Agilent Technologies | Cat#103015-100 |
| BCA protein assay | Thermo Scientific | Cat#23227 |
| Direct-zol RNA MicroPrep kit | Zymo | Cat#R2062 |
|
| ||
| Deposited data | ||
|
| ||
| Raw and analyzed data | This paper | GEO: GSE244962 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| 19c3 | Kuo et al.36 | N/A |
| 21c10 | Lyra-Leite et al.37 | N/A |
| 22c10 | Lyra-Leite et al.37 | N/A |
| 23c1 | Kuo et al.36 | N/A |
| 25c1 | This paper | N/A |
| 26c3 | This paper | N/A |
| GW124c9 | This paper | N/A |
| GW132c2B | This paper | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pAAVS1-Puro-TNNT2-Neo | This paper | RRID: Addgene_214013 |
|
| ||
| Software and algorithms | ||
|
| ||
| SOAPnuke | Chen et al.44 | https://github.com/BGI-flexlab/SOAPnuke |
| STAR | Dobin et al.45 | https://code.google.com/archive/p/rna-star/ |
| RSEM | Li et al.46 | http://deweylab.github.io/RSEM/ |
| DESeq2 R package | Love et al.47 | https://www.bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Excel (version 16.81) | Microsoft | https://www.microsoft.com/en-us/microsoft-365/ |
| GraphPad Prism (version 10.0.1) | GraphPad | https://www.graphpad.com/features/ |
| CyteSeer Analysis (version 2.9) | Vala Sciences | https://valasciences.com/ |
| CardioExcyte Control 96 (version 1.8) | Nanion Technologies | https://www.nanion.de/ |
| Data Controller 384 (version 1.6) | Nanion Technologies | https://www.nanion.de/ |
| Seahorse Wave (version 2.6) | Agilent Technologies | https://www.agilent.com/en/product/cell-analysis/real-time-cell-metabolic-analysis/xf-software |
Highlights.
High-throughput evaluation of hiPSC-CM viability and maturation
Identification of individual medium component contribution to viability and function
Functional and metabolic maturation are largely independent
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health R01 CA220002 and CA261898 (to P.W.B.), the Leducq Foundation (to P.W.B.), and S10-OD026867 (to A.L.G.). The graphical abstract was created with BioRender.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2024.114160.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Karbassi E, Fenix A, Marchiano S, Muraoka N, Nakamura K, Yang X, and Murry CE (2020). Cardiomyocyte maturation: advances in knowledge and implications for regenerative medicine. Nat. Rev. Cardiol 17, 341–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lundy SD, Zhu WZ, Regnier M, and Laflamme MA (2013). Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cell. Dev 22, 1991–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dai DF, Danoviz ME, Wiczer B, Laflamme MA, and Tian R (2017). Mitochondrial Maturation in Human Pluripotent Stem Cell Derived Cardiomyocytes. Stem Cell. Int 2017, 5153625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feaster TK, Cadar AG, Wang L, Williams CH, Chun YW, Hempel JE, Bloodworth N, Merryman WD, Lim CC, Wu JC, et al. (2015). Matrigel Mattress: A Method for the Generation of Single Contracting Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Circ. Res 117, 995–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo Y, and Pu WT (2020). Cardiomyocyte Maturation: New Phase in Development. Circ. Res 126, 1086–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlos-Oliveira M, Lozano-Juan F, Occhetta P, Visone R, and Rasponi M (2021). Current strategies of mechanical stimulation for maturation of cardiac microtissues. Biophys. Rev 13, 717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopaschuk GD, and Jaswal JS (2010). Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J. Cardiovasc. Pharmacol 56, 130–140. [DOI] [PubMed] [Google Scholar]
- 8.Correia C, Koshkin A, Duarte P, Hu D, Teixeira A, Domian I, Serra M, and Alves PM (2017). Distinct carbon sources affect structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Sci. Rep 7, 8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X, Rodriguez ML, Leonard A, Sun L, Fischer KA, Wang Y, Ritterhoff J, Zhao L, Kolwicz SC Jr., Pabon L, et al. (2019). Fatty Acids Enhance the Maturation of Cardiomyocytes Derived from Human Pluripotent Stem Cells. Stem Cell Rep 13, 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramachandra CJA, Mehta A, Wong P, Ja KPMM, Fritsche-Danielson R, Bhat RV, Hausenloy DJ, Kovalik JP, and Shim W (2018). Fatty acid metabolism driven mitochondrial bioenergetics promotes advanced developmental phenotypes in human induced pluripotent stem cell derived cardiomyocytes. Int. J. Cardiol 272, 288–297. [DOI] [PubMed] [Google Scholar]
- 11.Feyen DAM, McKeithan WL, Bruyneel AAN, Spiering S, Hörmann L, Ulmer B, Zhang H, Briganti F, Schweizer M, Hegyi B, et al. (2020). Metabolic Maturation Media Improve Physiological Function of Human iPSC-Derived Cardiomyocytes. Cell Rep 32, 107925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drawnel FM, Boccardo S, Prummer M, Delobel F, Graff A, Weber M, Gérard R, Badi L, Kam-Thong T, Bu L, et al. (2014). Disease modeling and phenotypic drug screening for diabetic cardiomyopathy using human induced pluripotent stem cells. Cell Rep 9, 810–821. [DOI] [PubMed] [Google Scholar]
- 13.Rog-Zielinska EA, Thomson A, Kenyon CJ, Brownstein DG, Moran CM, Szumska D, Michailidou Z, Richardson J, Owen E, Watt A, et al. (2013). Glucocorticoid receptor is required for foetal heart maturation. Hum. Mol. Genet 22, 3269–3282. [DOI] [PubMed] [Google Scholar]
- 14.Chattergoon NN (2019). Thyroid hormone signaling and consequences for cardiac development. J. Endocrinol 242, T145–T160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fowden AL, Li J, and Forhead AJ (1998). Glucocorticoids and the preparation for life after birth: are there long-term consequences of the life insurance? Proc. Nutr. Soc 57, 113–122. [DOI] [PubMed] [Google Scholar]
- 16.Yang X, Rodriguez M, Pabon L, Fischer KA, Reinecke H, Regnier M, Sniadecki NJ, Ruohola-Baker H, and Murry CE (2014). Tri-iodol-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells. J. Mol. Cell. Cardiol 72, 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birket MJ, Ribeiro MC, Kosmidis G, Ward D, Leitoguinho AR, van de Pol V, Dambrot C, Devalla HD, Davis RP, Mastroberardino PG, et al. (2015). Contractile Defect Caused by Mutation in MYBPC3 Revealed under Conditions Optimized for Human PSC-Cardiomyocyte Function. Cell Rep 13, 733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parikh SS, Blackwell DJ, Gomez-Hurtado N, Frisk M, Wang L, Kim K, Dahl CP, Fiane A, Tønnessen T, Kryshtal DO, et al. (2017). Thyroid and Glucocorticoid Hormones Promote Functional T-Tubule Development in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Circ. Res 121, 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao N, Liu Z, Chen Z, Wang J, Chen T, Zhao X, Ma Y, Qin L, Kang J, Wei B, et al. (2012). Ascorbic acid enhances the cardiac differentiation of induced pluripotent stem cells through promoting the proliferation of cardiac progenitor cells. Cell Res 22, 219–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, et al. (2014). Chemically defined generation of human cardiomyocytes. Nat. Methods 11, 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horikoshi Y, Yan Y, Terashvili M, Wells C, Horikoshi H, Fujita S, Bosnjak ZJ, and Bai X (2019). Fatty Acid-Treated Induced Pluripotent Stem Cell-Derived Human Cardiomyocytes Exhibit Adult Cardiomyocyte-Like Energy Metabolism Phenotypes. Cells 8, 1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu C, He JQ, Kamp TJ, Police S, Hao X, O’Sullivan C, Carpenter MK, Lebkowski J, and Gold JD (2006). Human embryonic stem cell-derived cardiomyocytes can be maintained in defined medium without serum. Stem Cell. Dev 15, 931–941. [DOI] [PubMed] [Google Scholar]
- 23.Batho CAP, Mills RJ, and Hudson JE (2020). Metabolic Regulation of Human Pluripotent Stem Cell-Derived Cardiomyocyte Maturation. Curr. Cardiol. Rep 22, 73. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Wada Y, Ballan N, Schmeckpeper J, Huang J, Rau CD, Wang Y, Gepstein L, and Knollmann BC (2021). Triiodothyronine and dexamethasone alter potassium channel expression and promote electrophysiological maturation of human-induced pluripotent stem cell-derived cardiomyocytes. J. Mol. Cell. Cardiol 161, 130–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaffer SW, Shimada-Takaura K, Jong CJ, Ito T, and Takahashi K (2016). Impaired energy metabolism of the taurine-deficient heart. Amino Acids 48, 549–558. [DOI] [PubMed] [Google Scholar]
- 26.Del Franco A, Ambrosio G, Baroncelli L, Pizzorusso T, Barison A, Olivotto I, Recchia FA, Lombardi CM, Metra M, Ferrari Chen YF, et al. (2022). Creatine deficiency and heart failure. Heart Fail. Rev 27, 1605–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verkerk AO, Knottnerus SJG, Portero V, Bleeker JC, Ferdinandusse S, Guan K, IJlst L, Visser G, Wanders RJA, Wijburg FA, et al. (2020). Electrophysiological Abnormalities in VLCAD Deficient hiPSC-Cardiomyocytes Do not Improve with Carnitine Supplementation. Front. Pharmacol 11, 616834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44, D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rana P, Anson B, Engle S, and Will Y (2012). Characterization of human-induced pluripotent stem cell-derived cardiomyocytes: bioenergetics and utilization in safety screening. Toxicol. Sci 130, 117–131. [DOI] [PubMed] [Google Scholar]
- 30.Hu D, Linders A, Yamak A, Correia C, Kijlstra JD, Garakani A, Xiao L, Milan DJ, van der Meer P, Serra M, et al. (2018). Metabolic Maturation of Human Pluripotent Stem Cell-Derived Cardiomyocytes by Inhibition of HIF1alpha and LDHA. Circ. Res 123, 1066–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chirico N, Kessler EL, Maas RGC, Fang J, Qin J, Dokter I, Daniels M, Šarić T, Neef K, Buikema JW, et al. (2022). Small molecule-mediated rapid maturation of human induced pluripotent stem cell-derived cardiomyocytes. Stem Cell Res. Ther 13, 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji S, Tu W, Huang C, Chen Z, Ren X, He B, Ding X, Chen Y, and Xie X (2022). The Aurora Kinase Inhibitor CYC116 Promotes the Maturation of Cardiomyocytes Derived from Human Pluripotent Stem Cells. Mol. Cell 45, 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garay BI, Givens S, Abreu P, Liu M, Yücel D, Baik J, Stanis N, Rothermel TM, Magli A, Abrahante JE, et al. (2022). Dual inhibition of MAPK and PI3K/AKT pathways enhances maturation of human iPSC-derived cardiomyocytes. Stem Cell Rep 17, 2005–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen S, Sewanan LR, Shao S, Halder SS, Stankey P, Li X, and Campbell SG (2022). Physiological calcium combined with electrical pacing accelerates maturation of human engineered heart tissue. Stem Cell Rep 17, 2037–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang SS, Zhang X, Ke ZZ, Wen XY, Li WD, Liu WB, Zhuang XD, and Liao LZ (2022). D-galactose-induced cardiac ageing: A review of model establishment and potential interventions. J. Cell Mol. Med 26, 5335–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuo HH, Gao X, DeKeyser JM, Fetterman KA, Pinheiro EA, Weddle CJ, Fonoudi H, Orman MV, Romero-Tejeda M, Jouni M, et al. (2020). Negligible-Cost and Weekend-Free Chemically Defined Human iPSC Culture. Stem Cell Rep 14, 256–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyra-Leite DM, Copley RR, Freeman PP, Pongpamorn P, Shah D, McKenna DE, Lenny B, Pinheiro EA, Weddle CJ, Gharib M, et al. (2023). Nutritional requirements of human induced pluripotent stem cells. Stem Cell Rep 18, 1371–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fonoudi H, Lyra-Leite DM, Javed HA, and Burridge PW (2020). Generating a Cost-Effective, Weekend-Free Chemically Defined Human Induced Pluripotent Stem Cell (hiPSC) Culture Medium. Curr. Protoc. Stem Cell Biol 53, e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyra-Leite DM, Fonoudi H, Gharib M, and Burridge PW (2021). An updated protocol for the cost-effective and weekend-free culture of human induced pluripotent stem cells. STAR Protoc 2, 100213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magdy T, Jouni M, Kuo HH, Weddle CJ, Lyra-Leite D, Fonoudi H, Romero-Tejeda M, Gharib M, Javed H, Fajardo G, et al. (2022). Identification of Drug Transporter Genomic Variants and Inhibitors That Protect Against Doxorubicin-Induced Cardiotoxicity. Circulation 145, 279–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blancard M, Fetterman KA, and Burridge PW (2022). Pharmacogenomic Screening of Drug Candidates using Patient-Specific hiPSC-Derived Cardiomyocyte High-Throughput Calcium Imaging. Methods Mol. Biol 2547, 241–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vanoye CG, Desai RR, Ji Z, Adusumilli S, Jairam N, Ghabra N, Joshi N, Fitch E, Helbig KL, McKnight D, et al. (2022). High-throughput evaluation of epilepsy-associated KCNQ2 variants reveals functional and pharmacological heterogeneity. JCI Insight 7, e156314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson CH, Potet F, Abramova TV, DeKeyser JM, Ghabra NF, Vanoye CG, Millichap JJ, and George AL (2023). Epilepsy-associated SCN2A (NaV1.2) variants exhibit diverse and complex functional properties. J. Gen. Physiol 155, e202313375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y, Chen Y, Shi C, Huang Z, Zhang Y, Li S, Li Y, Ye J, Yu C, Li Z, et al. (2018). SOAPnuke: a MapReduce acceleration-supported soft-ware for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience 7, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krämer A, Green J, Pollard J Jr., and Tugendreich S (2014). Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq datasets generated in this study are available at the NCBI Gene Expression Omnibus (GEO) as accession number GSE244962.
The software and algorithms for data analyses used in this study are published and referenced.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.


