Abstract
INTRODUCTION
Brain‐derived extracellular vesicles (BEVs) in blood allows for minimally‐invasive investigations of central nervous system (CNS) ‐specific markers of age‐related neurodegenerative diseases (NDDs). Polymer‐based EV‐ and immunoprecipitation (IP)‐based BEV‐enrichment protocols from blood have gained popularity. We systematically investigated protocol consistency across studies, and determined CNS‐specificity of proteins associated with these protocols.
METHODS
NDD articles investigating BEVs in blood using polymer‐based and/or IP‐based BEV enrichment protocols were systematically identified, and protocols compared. Proteins used for BEV‐enrichment and/or post‐enrichment were assessed for CNS‐ and brain‐cell‐type‐specificity, extracellular domains (ECD+), and presence in EV‐databases.
RESULTS
A total of 82.1% of studies used polymer‐based (ExoQuick) EV‐enrichment, and 92.3% used L1CAM for IP‐based BEV‐enrichment. Centrifugation times differed across studies. A total of 26.8% of 82 proteins systematically identified were CNS‐specific: 50% ECD+, 77.3% were listed in EV‐databases.
CONCLUSIONS
We identified protocol steps requiring standardization, and recommend additional CNS‐specific proteins that can be used for BEV‐enrichment or as BEV‐biomarkers.
Highlights
Across NDDs, we identified protocols commonly used for EV/BEV enrichment from blood.
We identified protocol steps showing variability that require harmonization.
We assessed CNS‐specificity of proteins used for BEV‐enrichment or found in BEV cargo.
CNS‐specific EV proteins with ECD+ or without were identified.
We recommend evaluation of blood‐BEV enrichment using these additional ECD+ proteins.
Keywords: age‐related neurodegenerative diseases, Alzheimer's disease, biofluid based biomarkers, blood, brain‐derived extracellular vesicles, exosomes, immunoprecipitation‐based enrichment, microvesicles, novel biomarkers, protocol variability
1. INTRODUCTION
The number of individuals living with neurodegenerative conditions has more than doubled from 1990 to 2016, owing to population aging and growth. 1 , 2 By 2050, more than 150 million are expected to live with neurodegenerative diseases of aging (NDDs), with Alzheimer's disease (AD) being the most prevalent. 1 NDDs have a long pre‐symptomatic stage during which neuropathological changes occur prior to symptom onset. 3 , 4 While neuroimaging and cerebrospinal fluid (CSF) biomarkers have been established for AD diagnosis, 5 they are expensive and/or invasive and available only in specialized centers. Major efforts are being devoted to the development of reliable early diagnostic biomarkers to facilitate identification of at‐risk persons prior to symptom‐onset. Availability of minimally‐invasive blood biomarkers for early diagnosis and/or investigating disease mechanisms and potentially novel therapeutic targets would be of great value.
While blood collection is easy to perform, measurement of brain‐derived markers from blood poses a challenge due to the complex composition of blood itself, and the relatively low quantities of molecules released from the brain into the peripheral circulation. 6 Technical and analytical advances over the past decade are, however, starting to enable specific and sensitive measurements of a handful of NDD‐biomarkers in blood, with most being relevant to AD (e.g., Aβ, and specific tau phosphorylated forms). 7 , 8 , 9 , 10 , 11 Despite this advancement, many of the current blood NDD biomarkers are not brain‐specific, but instead also expressed at high levels in peripheral tissues (e.g., Aβ expression in red blood cells). 12 This renders the interpretation of blood‐based measurements challenging. Approaches that allow for the enrichment of brain‐ and brain‐cell‐specific biomarkers from blood are thus being actively sought after.
On this front, extracellular vesicles (EVs) in blood comprise a promising minimally‐invasive biomarker source for many diseases, including cancer and NDDs. 13 , 14 Released by all cells in the body, 15 EVs are lipid‐delimited nanoparticles of different intracellular origins, with cell‐cell communication being a main function. 16 EVs contain molecules (e.g., proteins) that mirror the parental cell content and expression level, thereby providing a snapshot of the homeostatic status of their cell of origin. Given their small size, EVs can diffuse into biological fluids (e.g., blood, CSF (see review 17 ), and bidirectionally cross the blood‐brain barrier. 18 Their ability to diffuse from CNS to blood, from where they can be isolated, make them an attractive resource for novel biomarker discovery and mechanistic insights into brain disease.
Enrichment of both EVs and brain‐derived EVs (BEVs) from blood is not without its challenges given the high non‐EV content of blood (e.g., serum/plasma proteins, liposomes). 19 Presently, several methods for enrichment of BEVs from blood exist, each with its advantages and disadvantages. 14 , 20 Of these, polymer‐based precipitation followed by immunocapture with specific antibodies, commonly known as the immunoprecipitation (IP)‐based BEV enrichment method, is widely used in NDD research. 21 While Fiandaca et al. 22 using the L1CAM antibody pioneered this method for enrichment of neuron‐derived EVs (NEVs), others have since adapted/optimized it for enrichment of other brain‐cell‐type‐derived EVs using other antibodies (e.g., glial). 21 , 23 , 24 , 25 , 26 , 27 Given the growing use of IP‐based BEVs enriched from blood in NDD research, the objectives for this study were to: (1) systematically assess protocols for IP‐based BEV enrichment from blood, and (2) assess CNS‐specificity and extracellular accessibility of proteins used for BEV enrichment and/or detected in BEV enriched isolates. Note that this study was conducted by the Alzheimer's Association International Society to Advance Alzheimer's Research and Treatment: Biofluid Based Biomarkers Professional Interest Area—Exosome Working Group (ISTAART‐BBB‐PIA‐EWG).
2. METHODS
2.1. Literature search
A comprehensive PubMed review on BEVs enriched from blood in AD had identified 26 articles published up to October 2019. 21 We extended this search to March 22, 2022, using six searches (Figure S1). In addition to AD, we comprehensively reviewed the literature on BEVs enriched from blood in (a) other NDDs, namely, Parkinson's disease, vascular dementia, frontotemporal dementia, and Lewy body dementia, and (b) Down syndrome—given that these individuals represent the largest population at genetic‐risk for AD 28 (searches in Figure S1). Given that the study of BEVs enriched from blood is an emerging field in NDD research, we conducted an additional 18 searches using less stringent keywords to avoid missing relevant studies (Figure S1).
Overall, only original research articles published in English and employing an IP‐based BEV enrichment method from human blood were included. Additional articles were identified by scanning the reference lists of included articles. Articles excluded: reviews/case‐reports; those not investigating BEVs in NDDs; those investigating BEVs in biofluids other than blood; non‐human samples; and cell culture.
2.2. Data extraction
Data extracted from articles that met inclusion/eligibility criteria were as follows: cohort demographics (number of participants, sex distribution); last‐author demographics and publication date; blood‐portion (plasma or serum) used for BEV‐enrichment; protocol preparing plasma/serum for downstream EV and/or BEV work; EV‐enrichment protocol details; IP‐protein used for BEV‐enrichment and other protocol details; EV and BEV validation methods; and generating a list of proteins used for BEV‐enrichment and/or investigated post‐enrichment. For articles that provided partial or no protocol details, the published protocol(s) referenced by these articles was used to gather these details.
RESEARCH IN CONTEXT
Systematic review: Authors systematically assessed PubMed for articles on age‐related neurodegenerative diseases (NDDs) related to polymer‐based extracellular vesicle (EV) and/or immunoprecipitation (IP)‐based brain‐derived EV (BEV) enrichment protocols from blood. Fifty‐four keyword combinations were used for the search. The relevant studies were appropriately cited.
Interpretation: Variability across IP‐based BEV enrichment protocols was identified, all of which would benefit from harmonization. Authors identified central nervous system (CNS)‐specific proteins with extracellular domains (ECD) that can potentially be used for IP‐based BEV enrichment and EV cargo proteins that could be used as BEV biomarkers.
Future directions: While we demonstrate that commonly used IP‐proteins for BEV isolation from blood are CNS‐enriched, we recommend the evaluation of additional proteins that appear to be more brain‐specific; contain ECDs; and are present in EV databases. Harmonization of BEV enrichment protocols is crucial, given the need for CNS‐specific blood biomarkers for NDDs.
2.3. Characterization of proteins in our list
Proteins in our list (list generation described in Section 2.2) were converted to their corresponding UniProt 29 gene names to allow mapping to public databases. To determine CNS‐specificity, “RNA consensus tissue gene data” from The Human Protein Atlas (v21.0 and Ensembl v103.38; proteinatlas.org, 30 ) was utilized. It includes gene expression, corresponding to normalized expression values (nTPM), from 61 regions: 12 CNS and 49 peripheral regions (listed in Figure S2). CNS‐specificity of each gene was calculated by comparing its expression levels in the 12 CNS regions with those in the 49 peripheral regions. Genes demonstrating greater than or equal to four‐fold higher average in CNS than peripheral regions, and showing a statistically significant p‐value < 0.05 between the two groups (Mann‐Whitney U‐test) were defined as “CNS‐specific”. Extracellular domain‐containing (ECD) proteins, as potential targets for IP‐based BEV‐enrichment, were identified from the UniProt human database (v2022_01). 29 EV‐specificity was identified using Exocarta (exocarta.org) and Vesiclepedia (microvesicles.org) databases. To determine brain‐cell‐type‐specificity, a single‐cell RNAseq dataset (accession GSE67835, 31 ) reporting gene expression in six brain‐cell‐types in humans was downloaded from the NCBI GEO repository. 32 The six brain‐cell‐types were: neurons, oligodendrocyte progenitor cells (OPC), oligodendrocytes, astrocytes, microglia, and brain endothelial cells. For each identified CNS‐specific gene, the cell‐type‐specific gene expression value was extracted from the GSE67835 dataset and normalized to the average expression among the six cell‐types. If a gene had a normalized value greater than or equal to four ‐fold higher for a particular cell‐type, it was tagged as specific to that cell‐type, else it was tagged as present in multiple cell types.
3. RESULTS
3.1. Literature search and identification of IP‐based BEV articles
Our search workflow identified 39 articles investigating BEVs enriched from blood using IP‐based methods (Figure 1A, Table S1_IP‐protocols), of which 35 (89.7%) performed ELISA‐based assays following enrichment. Most articles (64.1%) investigated AD or Parkinson disease (25.6%) (Table S2_Participant‐Information). Publication dates of articles ranged from 2014 to 2021, with most (28.2%) published in 2020 (Figure 1B), and 55.1% reporting a last‐author in the United States (Figure 1C).
FIGURE 1.
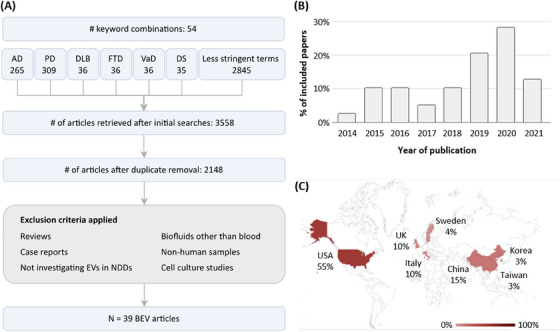
Literature search. (A) Flowchart of the article selection process. (B) Distribution of publication dates of the 39 BEV articles. (C) Geographic distribution of last‐authors’ affiliations of the 39 IP‐based BEV enrichment articles. AD, Alzheimer's disease; BEV, brain‐derived extracellular vesicles; DLB, Lewy body dementia; DS, Down syndrome; EV, extracellular vesicles; FTD, frontotemporal dementia; PD, Parkinson's disease; VaD, vascular dementia.
3.2. Assessment of sample preparation, EV, and IP‐based BEV enrichment protocols
Figure 2 provides a graphical breakdown of protocols used for sample preparation, and EV and IP‐based BEV enrichment, with additional details provided in Table S1_IP‐protocols.
FIGURE 2.
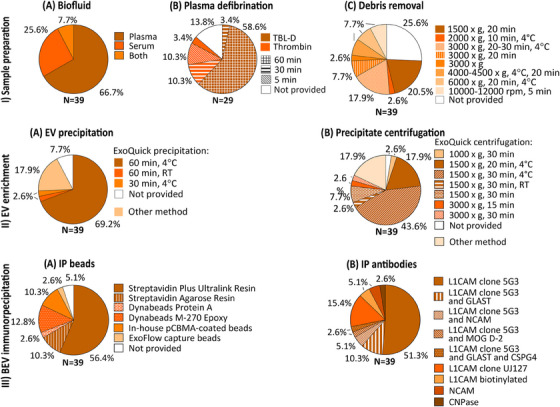
Sample preparation, EV and IP‐based BEV enrichment protocols used. (I) Sample preparation: (A) biofluid used; (B) plasma defibrination — reagent used and incubation time; and (C) centrifugation for debris removal — speed, time, and temperature. (II) EV enrichment: (A) method used for EV precipitation; for ExoQuick‐based precipitation: incubation time and temperature; (B) centrifugation for EV precipitation — speed, time, and temperature. (III) BEV immunoprecipitation (IP): (A) IP beads used; (B) antibodies used for IP. BEV, brain‐derived extracellular vesicles; EV, extracellular vesicles; IP, immunoprecipitation; TBL‐D, thromboplastin‐D.
3.2.1. Sample preparation for downstream enrichment of BEVs
The initial biofluid (plasma or serum) volume ranged from 250 to 500 μL across the 39 studies, with 26 (66.7%) using plasma only, 10 (25.6%) serum only, and 3 (7.7%) both (Figure 2‐IA). Of the 29 studies using plasma, 25 (86.2%) reported a defibrination‐step using thromboplastin‐D (N = 18, 62.1%, 100–200 μL) or thrombin (N = 7, 24.1%, 2.5–5 μL), while the remaining 4 did not mention this step (Figure 2‐IB). All studies using thromboplastin‐D incubated for 60 min at room temperature (RT), with the exception of one study (30 minutes, RT) (Figure 2‐IB). For thrombin, while an equal number of studies (N = 3 each) incubated for 5 or 30 minutes at RT (Figure 2‐IB), one study lacked specifics. Following incubation, all studies using thromboplastin‐D, and four studies using thrombin added protease and/or phosphatase inhibitor cocktails diluted in Dulbecco's phosphate buffered saline (DPBS, 150–495 μL). While a defibrination step is not needed for serum, five such studies also added DPBS (150–500 μL).
All 39 studies performed a centrifugation step to remove fibrinogen clot (relevant for plasma EV studies) and/or cell debris (Figure 2‐IC). Centrifugation conditions (speed, time) varied between studies (Figure 2‐IC). A speed of 3000 × g, 20–30 minutes (N = 12 studies, 11 plasma) was most used, followed by 1500 × g, 20 minutes (N = 9 studies, 8 plasma). Centrifugation temperature was not reported by the majority of studies (N = 24, 61.5%), but when reported (N = 15, 38.5%), it was 4°C (Figure 2‐IC).
3.2.2. EV enrichment
Thirty‐two (82.1%) studies performed polymer‐based EV precipitation on defibrinated plasma or serum using ExoQuick (Figure 2‐IIA). Of these, 28 studies incubated with ExoQuick for 60 minutes (temp: N = 27, 4°C or on ice, N = 1, RT), 1 for 30 minutes (at 4°C), and 3 did not report incubation conditions (Figure 2‐IIA). For precipitation of EVs following incubation, most ExoQuick studies (N = 28, 71.8%) centrifuged at 1500 × g for 20–30 minutes, with 24/28 performing this step at 4°C (Figure 2‐IIB). Precipitated EVs were resuspended in solution containing protease/phosphatase inhibitor cocktails under continuous rotation. Of the seven (17.9%) studies not using ExoQuick, one used a combination of EV precipitation and size exclusion chromatography, three used sequential spins for EV enrichment, and three others did not perform EV enrichment.
3.2.3. IP‐based BEV‐enrichment from blood
All 39 studies performed IP‐based BEV‐enrichment. The majority of studies used anti‐L1CAM products (36/39, 92.3%), albeit different types, to enrich NEVs (Figure 2‐IIIB). Of these, eight studies (22.2%) used an additional sample to enrich NEVs or other brain‐cell‐type‐derived EVs with another antibody: NCAM (for NEVs), MOG (for oligodendrocyte‐derived EVs), GLAST (for astrocyte‐derived EVs), or CSPG4 (for CSPG4‐cell‐derived EVs). Resultant BEV preparations were incubated with magnetic or resin beads. Twenty‐two (56.4%) of the studies used Streptavidin Plus UltraLink Resin beads (Figure 2‐IIIA) for immunoprecipitation.
3.3. EV and BEV characterization by size, shape, and cellular origin
Twenty‐four (61.5%) studies reported assessing biophysical properties of EVs and/or BEVs. Techniques most used were nanoparticle tracking analysis (NTA) and transmission electron microscopy (TEM), employed by 18 (46.2%) and 16 (41.0%) studies, respectively (see Table S1_IP‐protocols). Reported EV sizes ranged between 78 and 126 nm which is consistent with the known size ranges of small EVs or exosomes. Of the studies conducting NTA, few reported particle concentrations ranging from 191.1 particles/mL 22 to >10^9 particles/mL, 24 , 33 while the majority did not.
Thirty‐three (84.6%) studies reported assessing the presence of EV‐specific markers, with tetraspanin protein marker CD81 (N = 24, 61.5%) and the intracellular marker Alix (N = 10, 25.6%) being most used. Other EV‐markers used for validation were CD63 (N = 6, 15.4%), CD9 (N = 6, 15.4%), TSG101 (N = 5, 12.8%), and Hsp90 (N = 1, 2.6%).
The neuronal origin of BEVs was validated by 12 different markers: L1CAM, NCAM, NfL, neuronal‐specific (NS) enolase, synaptophysin, Tau‐1, MAP‐2, NeuN, Enolase‐2, MAPT, GRIA1, and PLP1. Of these, L1CAM (N = 16, 41.0%), followed by NCAM (N = 5, 12.8%) and NfL (N = 4, 10.3%), were most used. Glutamine synthetase (GluSyn) was mostly used to validate astrocyte‐derived EVs (AEVs). Studies using MOG or CNP‐ase as an IP antibody, validated the presence of these proteins post IP (e.g., using enzyme‐linked immunosorbent assay [ELISA]). Fourteen studies (35.9%) did not mention any brain cell marker or provided a reference paper for its validation.
3.4. CNS‐specificity, ECD status, and presence in public EV databases of proteins in our list
We generated a list of 117 proteins that were used for BEV enrichment and/or investigated post‐enrichment by the 39 articles, and these corresponded to 87 unique genes (Table 1). Protein products of five of these genes are known EV markers, namely, CD81, CD9, CD63, PDCD6IP (or Alix), and TSG101. CNS versus peripheral gene expression levels analysis on the remaining 82 genes identified 22 (26.8%) to be CNS‐specific (Figure 3A). Of these, 11 had ECDs (MOG, SYT1, SYP, SLC1A3, NRXN2, STX1A, NCAM1, L1CAM, NLGN1, GRIA4, SYT2), and 11 did not (GFAP, SNAP25, GAP43, NEFL, UCHL1, MAPT, ENO2, SNCA, OMG, NRGN, SYN1). Relative expression levels across the 12 CNS and 49 peripheral regions of the 17 CNS‐specific genes whose protein products are known to be present in EVs are shown in Figure 3B–D. Relative expression levels across the CNS and peripheral regions of all 87 genes are shown in Figure S3.
TABLE 1.
List of 87 unique genes
| No. | Protein name | Gene name |
|---|---|---|
| 1 | p‐gp/ABCB1 | ABCB1 |
| 2 | pAkt(Ser473), Akt (panel) | AKT1 |
| 3 | Aβ40, Aβ42, sAPPa, sAPPb | APP |
| 4 | BACE‐1 | BACE1 |
| 5 | pBADSer136 | BAD |
| 6 | Butyrylcholinesterase | BCHE |
| 7 | BDNF | BDNF |
| 8 | C1 complement complex – C1q | C1QA |
| 9 | Complement C3b, Complement C3d | C3 |
| 10 | Complement fragment C4b | C4B |
| 11 | Terminal complement complex C5b‐C9 | C5 |
| 12 | CD46 | CD46 |
| 13 | DAF (CD55) | CD55 |
| 14 | CD59 | CD59 |
| 15 | CD63 | CD63 |
| 16 | CD81 | CD81 |
| 17 | CD9 | CD9 |
| 18 | Complement B, Complement factor B–Bb, Factor B | CFB |
| 19 | Complement factor D | CFD |
| 20 | Factor I | CFI |
| 21 | Clusterin | CLU |
| 22 | CR1 | CR1 |
| 23 | CRP | CRP |
| 24 | Cystatin C | CST3 |
| 25 | Cathepsin D | CTSD |
| 26 | NS‐enolase | ENO2 |
| 27 | FGF‐13 | FGF13 |
| 28 | FGF‐2 | FGF2 |
| 29 | GAP43 | GAP43 |
| 30 | GDNF | GDNF |
| 31 | GFAP | GFAP |
| 32 | GluSyn | GLUL |
| 33 | AMPA4 | GRIA4 |
| 34 | PGRN | GRN |
| 35 | pGSK‐3B(Ser9), GSK‐3β | GSK3B |
| 36 | Gelsolin | GSN |
| 37 | HGF | HGF |
| 38 | HSF1 | HSF1 |
| 39 | HSP70 | HSPA1A |
| 40 | ICAM1 | ICAM1 |
| 41 | IGF‐1 | IGF1 |
| 42 | p‐IGF‐1R | IGF1R |
| 43 | IL‐1β | IL1B |
| 44 | IL‐6 | IL6 |
| 45 | pIR | INSR |
| 46 |
P‐serine 312‐IRS‐1, p‐panTyr‐IRS‐1, Total IRS‐1, p‐panTyr‐IRS‐1, Ser616‐IRS‐1, pIRS1(Ser636) |
IRS1 |
| 47 | L1CAM | L1CAM |
| 48 | LAMP‐1 | LAMP1 |
| 49 | LRP6 | LRP6 |
| 50 | Erk1/2 | MAPK1 |
| 51 | P38 MAPK | MAPK11 |
| 52 | JNK | MAPK8 |
| 53 |
PTau‐T181, PTau‐S202, PTau‐S231, PTau‐S396, PTau‐T205, T‐tau, Tau N‐123, Full length tau |
MAPT |
| 54 | MBL | MBL2 |
| 55 | MMP‐9 | MMP9 |
| 56 | MOG | MOG |
| 57 | mTOR, pmTORSer2448 | MTOR |
| 58 | NCAM‐1 | NCAM1 |
| 59 | NFL | NEFL |
| 60 | NLGN1 | NLGN1 |
| 61 | NPTX2 | NPTX2 |
| 62 | Neurogranin | NRGN |
| 63 | NRXN2alfa | NRXN2 |
| 64 | OMG | OMG |
| 65 | Alix | PDCD6IP |
| 66 | G‐secretase | PSENEN |
| 67 | pPTEN(Ser380) | PTEN |
| 68 | REST | REST |
| 69 | p70S6K(T389), pS6Ser235/Ser236 | RPS6KB1 |
| 70 | Syntenin‐1 | SDCBP |
| 71 | GLAST | SLC1A3 |
| 72 | Glut‐1 | SLC2A1 |
| 73 | LAT‐1 | SLC7A5 |
| 74 | SNAP‐25 | SNAP25 |
| 75 | Alpha‐synuclein | SNCA |
| 76 | Syntaxin 1 | STX1A |
| 77 | Synapsin 1 | SYN1 |
| 78 | Synaptopodin | SYNPO |
| 79 | Synaptophysin | SYP |
| 80 | Synaptotagmin‐1 | SYT1 |
| 81 | Synaptotagmin‐2 | SYT2 |
| 82 | TDP‐43 | TARDBP |
| 83 | TNF‐α | TNF |
| 84 | TSG101 | TSG101 |
| 85 | Ubiquitin | UBB |
| 86 | UCH‐L1 | UCHL1 |
| 87 | VAMP‐2 | VAMP2 |
FIGURE 3.
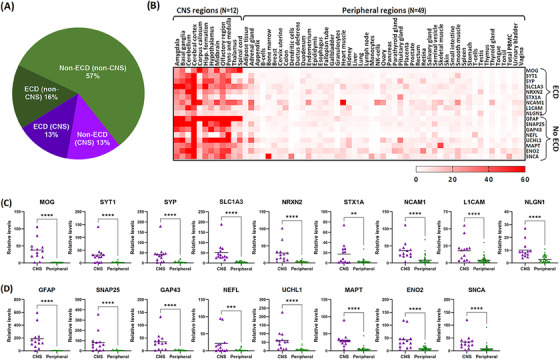
Characterization of proteins in our list. (A) Pie chart showing the distribution of CNS‐specific and ECD‐containing proteins in our list. Relative gene expression levels across brain (N = 12) and peripheral (N = 49) regions of 17 proteins identified as CNS‐specific and known to be present in EVs presented as both (B) a heatmap and (C–D) scatter plots. Also indicated are their ECD status, with (C) showing ECD‐containing proteins and (D) showing non‐ECD‐containing proteins. The heatmap color scale indicates normalized expression values (nTPM) ranging from 0 to 60. Asterisks in the scatter plots represent statistically significant differences of p < 0.0001 (****) or p < 0.01 (**) from Mann‐Whitney U‐test analysis. CNS, central nervous system; ECD, extracellular domains; EV, extracellular vesicles.
3.4.1. Brain cell‐type‐specificity of proteins in our list identified as CNS‐specific
Of the 22 CNS‐specific genes identified, the majority (N = 15, 68.2%) were found to be neuron‐specific, with the remaining being oligodendrocyte‐ or astrocyte‐specific, or were present in multiple cell types (Figure 4).
FIGURE 4.
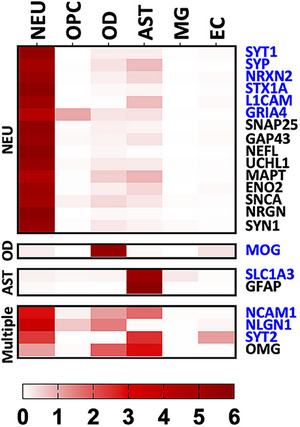
Brain cell‐type heatmap. The heatmap shows the relative gene expression from a single‐cell RNAseq dataset (accession GSE67835, 32 ) for each identified CNS‐specific protein. Expression in six cell‐types is shown: NEU, OPC, OD, AST, MG, and EC. The heatmap color scale indicates normalized expression values (relative to average of the six cell types) ranging from 0 to 6. Proteins showing normalized values greater than or equal to four were tagged as specific to that cell‐type, else they were tagged as present in multiple cell types. Protein names in blue indicate extracellular domain‐containing proteins. AST, astrocytes; CNS, central nervous system; EC, endothelial cells; MG, microglia; NEU, neurons; OD, oligodendrocytes cells; OPC, oligodendrocyte progenitor.
4. DISCUSSION
Given the increasing usage of BEVs enriched from blood in NDD research, a main goal of the ISTAART‐BBB‐PIA‐EWG was to investigate BEV characterization and enrichment methods from plasma and/or serum—a topic often heavily debated by the scientific community. Below, we summarize (a) commonly used experimental choices made across NDD studies for EV and BEV characterization and enrichment, and (b) offer ISTAART‐BBB‐PIA‐EWG's recommendations for CNS‐specific alternatives to L1CAM for IP‐based NEV enrichment from blood.
4.1. Commonly used experimental choices across NDD studies
4.1.1. Biophysical characterization of EVs based on size and shape
Different subgroups of EVs (e.g., exosomes, microvesicles) exist, and these can be distinguished to some extent by their biophysical characteristics, for example, exosomes exhibit diameters between 30 and 150 nm. 34 We found that techniques most used by included studies to assess the biophysical properties (size and shape) of EVs were NTA and TEM, respectively, albeit findings were not consistently reported across all studies. This may be because the Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018, 35 ) guidelines recommend several different methodologies for proper EV characterization, and the combination of methods used remain at the discretion of the investigator. However, to ensure consistency across studies where isolated BEVs are being assessed, we recommend that size range verification should be consistently reported in all future EV studies in order to differentiate smaller subpopulations of EVs from other microvesicles subpopulations or apoptotic bodies. 34 , 36 , ,
4.1.2. Choice of plasma over serum as starting material
We found that the majority (66.7%) of studies used plasma rather than serum as a starting material to precipitate EVs. This may be due to some of the obvious advantages of plasma, such as (i) the larger volume obtained from a fixed volume of blood, and (ii) no clotting time delay, 37 given that rapid processing of blood post‐collection is deemed crucial to avoid increasing EV instability with longer (>30 minutes) incubation periods. 38 , 39 Plasma is also considered the most physiological milieu to study blood EVs, taking into account that the number of EVs is higher in serum due to clot‐induced platelet vesiculation. 38 Recently, comparisons of fresh versus frozen plasma found no significant difference in protein content of enriched EVs, which may further boost the use of biobanked plasma for EV research. 40
Choice of starting material (i.e., plasma or serum) may, however, impact markers selected for EV validation. 41 While we found tetraspanin CD81 to be the most reported (N = 24 studies) EV marker, recently, Karimi et al. 41 demonstrated that CD81‐positive EVs constituted the rarest subpopulation in plasma and serum. Instead, CD9‐positive EVs comprised the majority, with considerable enrichment for CD9‐ and CD63‐positive EVs observed in plasma and serum, respectively. 41 While it is possible that release of EVs from activated platelets in plasma may account for the abundance of CD81‐positive EVs, an earlier study by Heijnen et al. 42 had reported platelet‐released EVs to be selectively CD63‐positive. The distinction between platelet‐rich and platelet‐poor plasma was not well described across most articles assessed in the current study, with only two articles from the same group 43 , 44 mentioning the use of platelet‐free plasma. It is evident that blood (serum and plasma) contains subpopulations of EVs that carry various tetraspanins. The contribution of platelet released EVs requires further investigation. Moreover, the presence of different subpopulations of EVs in platelet‐rich and platelet‐poor plasma that co‐express L1CAM or similar CNS‐related proteins also requires further investigation.
4.1.3. Choice of thromboplastin‐D over thrombin for defibrination
We found that thromboplastin‐D, and not thrombin, was used by the majority (62.1%) of plasma studies for defibrination. The protease thromboplastin‐D converts prothrombin to thrombin during the clotting of blood, while the enzyme thrombin facilitates blood clotting by converting fibrinogen to fibrin. Relative to untreated plasma, pre‐treatment with thromboplastin‐D was reported to (i) remove clouding factors and prevent aggregation of ExoQuick enriched EVs, 45 and (ii) not introduce contaminants, when using human recombinant thromboplastin and enriching for EVs using ultracentrifugation, 40 though use of rabbit thromboplastin introduced exogenous tau contaminants. 24 Thrombin pre‐treatment, unlike that observed with rabbit thromboplastin, lacked exogenous tau contaminants. 24 Pre‐treatment with thrombin, however, significantly lowered Exoquick EV yield compared to untreated plasma, 46 with the authors hypothesizing that the induced clotting entrapped a significant number of EVs, thereby leading to an underestimation. 46
4.1.4. Choice of ExoQuick over other approaches for EV enrichment
We found that polymer‐based EV precipitation/enrichment on cleared plasma or serum using ExoQuick was used by most (N = 32, 82.1%) studies. The observation that 68.8% of the ExoQuick studies followed manufacturer recommendations for incubation time and temperature (60 minutes, 4°C), as well as centrifugation speed and temperature (1500 × g, 4°C) for precipitation, points to a majority consensus for these parameters. However, (a) the majority of studies (N = 19, 59.4%) doubled the manufacturer's recommendation for ExoQuick for a given volume of plasma or serum, and (b) centrifugation time for precipitation lacked consensus. The impact of these protocol changes to ExoQuick‐based EV enrichment requires further investigation.
On the choice of ExoQuick itself for precipitation of EVs from blood, Serrano‐Pertierra and colleagues 47 reported that enrichment of plasma‐derived EV using ExoQuick was more efficient compared to ultracentrifugation and the Invitrogen kit. Moreover, a recent study, assessing EVs enriched using five commonly used methods, namely, precipitation (ExoQuick ULTRA), membrane affinity (exoEasy Maxi Kit), size‐exclusion chromatography (qEVoriginal), iodixanol gradient (OptiPrep), and phosphatidylserine affinity (MagCapture), reported that ExoQuick was a better method for plasma than for conditioned cell media. 48 The study highlighted the importance of selecting an EV enrichment protocol suited to the sample type (e.g., plasma, serum, cell media). 48 ExoQuick was also found to enrich the most EV‐proteins from low plasma volumes (e.g., 250 μL), an important consideration for biomarker studies and clinical trials that lack access to large biosample volumes. However, earlier versions of the ExoQuick kit precipitated both EVs and non‐EV‐particles (mostly lipoproteins), 48 , 49 a drawback that may be currently mitigated by the ExoQuick‐LP kit, which contains a lipoprotein pre‐clearing reagent. 48 It should also be noted that the addition of the immunocapture step with a cell‐specific antibody further helps with clearing lipoprotein contaminants. 50 Overall, ExoQuick is considered an easy to perform, fast, reproducible, scalable, and relatively low‐cost EV enrichment method. 48
4.1.5. Choice of L1CAM for BEV enrichment
L1CAM or CD171 is a transmembrane protein, known to be widely expressed in neurons, 51 and currently serves as the gold standard for enriching NEVs from blood. Numerous studies have demonstrated that NEVs contain Aβ and tau species, relevant to neurodegeneration. 22 , 26 , 52 , 53 Additional work has shown that NEVs and AEVs contain complement proteins, which can accurately predict conversion of MCI to AD. 54 , 55 , 56 Overall, employing cargo analysis of L1CAM‐positive BEVs has expanded the field of plasma biomarkers in early AD diagnosis exponentially. However, the specificity of L1CAM itself constitutes the main source of controversy. In 2021, Norman and colleagues 57 reported that L1CAM was not associated with plasma and/or CSF enriched EVs, thereby suggesting that prior BEV biomarker studies were measuring soluble cleaved versions of L1CAM that were not associated with the CNS. Finally, the authors advocated against the use of L1CAM as a marker for enrichment of NEVs. 57 In contrast to the studies investigated in our current work, Norman et al. 57 had used size exclusion chromatography (SEC) to elute their EVs, with L1CAM signal still observable, albeit at lower signal strengths, in fractions where tetraspanin signals (i.e., protein markers of EVs) were also present (e.g., fractions 9–12). Given that EVs are heterogeneous and can range in different sizes, it is possible that L1CAM may be associated with smaller EVs rather than larger EVs. This was not addressed by the authors as the size profile of the EVs eluted in their fractions were not thoroughly investigated. Moreover, given that both soluble and membrane‐bound L1CAM can exist together in the same fraction, additional studies robustly differentiating between the two forms are needed. Nonetheless, these findings have sparked a much‐needed discussion about the validation and harmonization of protocols in the EV field and require further investigation to counter the numerous studies that utilize L1CAM (e.g., 58 , 59 , 60 , N = 36 articles indicated in Table S1_IP‐protocols). We found that L1CAM continues to serve as the most used CNS‐specific marker for NEV enrichment from blood.
4.2. Recommendations for CNS‐specific alternatives to L1CAM for NEV enrichment from blood
The issue of BEV‐specificity and purity arises due to blood containing an admixture of EVs from multiple tissues/cell types. In the current study, we used publicly available RNAseq and EV‐databases, to categorize proteins in our list as CNS‐ and brain‐cell‐type‐specific. We found high expression in 12 CNS compared to 49 peripheral regions for (a) L1CAM, and (b) proteins used for IP‐based AEV enrichment, such as GLAST (or SLC1A3) and GFAP. In contrast, CD81 and CD63, well characterized EV markers, demonstrated a wide distribution across both CNS and peripheral regions. Interrogation of the CNS‐specific proteins in our list for ECD presence allowed for the identification of potential alternate markers for IP‐based BEV enrichment of neuronal EVs. Specifically, we put forth SYT1, SYP, NRXN2, GRIA4, as potential novel neuronal markers for BEV enrichment using IP‐based capture methods. Our findings also suggest that while the proteins NCAM1 and NLGN1 demonstrate a high CNS‐specificity, they may not be associated with a specific brain‐cell type. Our analyses further revealed candidate proteins with a CNS origin yet lacking an ECD. These are likely EV cargo proteins that could be used during secondary validation methods. Collectively, the candidate proteins identified here have a wide range of biological functions that may be relevant to neurodegeneration but require further investigation. Our future work includes validating candidate markers using ultra‐sensitive bioassays and proteomic tools for CNS‐specific confirmation and novel BEV cargo identification.
4.3. CONCLUSIONS
Harmonization of protocols are essential steps for blood‐enriched BEV biomarker work in AD and other NDDs and is advocated by the ISTAART‐BBB‐PIA‐EWG. For IP‐based BEV enrichment protocols, we have identified steps with obvious variability across studies that require harmonization. We also noted that most studies use L1CAM for enrichment of a specific subpopulation of NEVs. Using RNA‐seq databases, we put forth SYT1, SYP, NRXN2, GRIA4, as potential novel neuronal alternative markers to L1CAM. Moreover, these additional CNS‐specific ECD‐containing proteins identified in our study can potentially be used for both IP‐based BEV enrichment and as BEV biomarkers. Similarly, CNS‐specific cargo proteins can potentially be used for BEV biomarkers, however they require further investigation and must be validated experimentally. Future investigations include systematically assessing the consistency of BEV content across different studies, and establishing multi‐institutional collaborations for biomarker validation of CNS‐specific ECD containing alternatives to L1CAM.
AUTHOR CONTRIBUTIONS
AmanPreet Badhwar led the study group. AmanPreet Badhwar, Arsalan S. Haqqani, and Charisse N. Winston designed the study. Systematic assessment of protocols were performed by AmanPreet Badhwar, Yael Hirschberg, Natalia Valle‐Tamayo, M. Florencia Iulita, Aurélie Ledreux and Charisse N. Winston. CNS‐specificity assessments were performed by Arsalan S. Haqqani and AmanPreet Badhwar. All authors wrote, edited, and approved the final manuscript. All authors meet the ICMJE criteria for authorship.
CONFLICT OF INTEREST STATEMENT
All authors declare that they have no competing interests/conflicts. MFI is now a full‐time employee of Altoida Inc. and may hold stock options in the company. Author disclosures are available in the supporting information.
CONSENT STATEMENT
We confirm that consent was not necessary for this work.
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
ACKNOWLEDGMENTS
This manuscript was facilitated by the Alzheimer's Association International Society to Advance Alzheimer's Research and Treatment (ISTAART), through the Exosome Working Group of the Biofluid Based Biomarkers Professional Interest Area (ISTAART‐BBB‐PIA‐EWG). The views and opinions expressed in this publication represent those of the authors and do not necessarily reflect those of the BBB‐PIA membership, ISTAART, or the Alzheimer's Association. The authors thank the ISTAART‐BBB‐PIA for their help in initiating this work. This work was supported by the Fonds de Recherche Québec—Santé (FRQS) [Chercheur boursiers Junior 1, 2020‐2024], Fonds de soutien à la recherche pour les neurosciences du vieillissement from the Fondation Courtois (AmanPreet Badhwar); and NIH/NIA R01 grants AG058252, AG073979, AG079303 to Robert A. Rissman and K99AG070390‐01 to Charisse N. Winston; Flemish Institute for Technological Research (VITO, Belgium) (Yael Hirschberg); NIH RF1 AG070153 and R01 AG071228 to Aurélie Ledreux; NIH R21‐AG067755, P20‐AG068077, and UNM Grand Challenges Initiative to Rawan M. Tarawneh.
COLLABORATORS—BBB PIA Exosome Working Group (EWG), Established October 2021
Alberto Lleo. ALleo@santpau.cat, Amanpreet Badhwar, amanpreet.badhwar@umontreal.ca, Charisse Winston, chwinston@health.ucsd.edu, Chi Udeh‐Momoh, c.udeh@imperial.ac.uk, Henrik Zetterberg, henrik.zetterberg@clinchem.gu.se, Luc Buee, luc.buee@inserm.fr, luc.buee2@gmail.com, Maria Florencia Iulita, MIulita@santpau.cat, Michelle M. Mielke, mielke.michelle@mayo.edu, Morvane Colin, morvane.colin@inserm.fr, Paul Slowey, pds@4saliva.com, Rawan Tarawneh, rawan.altarawneh@osumc.edu, Robert Rissman, rrissman@health.ucsd.edu, Samuel Gandy, samgandy@gmail.com, Sylvain Lehmann, sylvain.lehmann@umontpellier.fr, Tsuneya Ikezu, tikezu@bu.edu, Aurelie Ledreux, Aurelie.Ledreux@du.edu, Gagan Deep, gdeep@wakehealth.edu, Yael Hirschberg, yael.hirschberg@vito.be
Badhwar AP, Hirschberg Y, Valle‐Tamayo N, et al. Assessment of brain‐derived extracellular vesicle enrichment for blood biomarker analysis in age‐related neurodegenerative diseases: An international overview. Alzheimer's Dement. 2024;20:4411–4422. 10.1002/alz.13823
Charisse N. Winston and Arsalan S. Haqqani contributed equally as co‐authors
DATA AVAILABILITY STATEMENT
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1. GBD 2019 Dementia Forecasting Collaborators . Estimation of the global prevalence of dementia in 2019 and forecasted prevalence in 2050: an analysis for the Global Burden of Disease Study 2019. Lancet Public Health. 2022;7:e105‐e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. GBD 2016 Dementia Collaborators . Global, regional, and national burden of Alzheimer's disease and other dementias, 1990‐2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019;18:88‐106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hustad E, Aasly JO. Clinical and imaging markers of prodromal Parkinson's disease. Front Neurol. 2020;11:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. DeTure MA, Dickson DW. The neuropathological diagnosis of Alzheimer's disease. Mol Neurodegener. 2019;14:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jack CR Jr, Bennett DA, Blennow K, et al. A/T/N: an unbiased descriptive classification scheme for Alzheimer disease biomarkers. Neurology. 2016;87:539‐547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ashton NJ, Hye A, Rajkumar AP, et al. An update on blood‐based biomarkers for non‐Alzheimer neurodegenerative disorders. Nat Rev Neurol. 2020;16(5):265‐284. doi: 10.1038/s41582-020-0348-0 [DOI] [PubMed] [Google Scholar]
- 7. Ashton NJ, Pascoal TA, Karikari TK, et al. Plasma p‐tau231: a new biomarker for incipient Alzheimer's disease pathology. Acta Neuropathol. 2021;141:709‐724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Janelidze S, Teunissen CE, Zetterberg H, et al. Head‐to‐head comparison of 8 plasma amyloid‐β 42/40 assays in Alzheimer disease. JAMA Neurol. 2021;78:1375‐1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thijssen EH, La Joie R, Strom A, et al. Plasma phosphorylated tau 217 and phosphorylated tau 181 as biomarkers in Alzheimer's disease and frontotemporal lobar degeneration: a retrospective diagnostic performance study. Lancet Neurol. 2021;20:739‐752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Janelidze S, Mattsson N, Palmqvist S, et al. Plasma P‐tau181 in Alzheimer's disease: relationship to other biomarkers, differential diagnosis, neuropathology and longitudinal progression to Alzheimer's dementia. Nat Med. 2020;26:379‐386. [DOI] [PubMed] [Google Scholar]
- 11. Karikari TK, Pascoal TA, Ashton NJ, et al. Blood phosphorylated tau 181 as a biomarker for Alzheimer's disease: a diagnostic performance and prediction modelling study using data from four prospective cohorts. Lancet Neurol. 2020;19:422‐433. [DOI] [PubMed] [Google Scholar]
- 12. Sun H‐L, Chen S‐H, Yu Z‐Y, et al. Blood cell‐produced amyloid‐β induces cerebral Alzheimer‐type pathologies and behavioral deficits. Mol Psychiatry. 2021;26:5568‐5577. [DOI] [PubMed] [Google Scholar]
- 13. Zhou B, Xu K, Zheng X, et al. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct Target Ther. 2020;5:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hornung S, Dutta S, Bitan G. CNS‐derived blood exosomes as a promising source of biomarkers: opportunities and challenges. Front Mol Neurosci. 2020;13:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373‐383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gurung S, Perocheau D, Touramanidou L, Baruteau J. The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Commun Signal. 2021;19:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yáñez‐Mó M, Siljander PR‐M, Andreu Z, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Banks WA, Sharma P, Bullock KM, Hansen KM, Ludwig N, Whiteside TL. Transport of extracellular vesicles across the blood‐brain barrier: brain pharmacokinetics and effects of inflammation. Int J Mol Sci. 2020;21:4407. doi: 10.3390/ijms21124407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ding M, Wang C, Lu X, et al. Comparison of commercial exosome isolation kits for circulating exosomal microRNA profiling. Anal Bioanal Chem. 2018;410:3805‐3814. [DOI] [PubMed] [Google Scholar]
- 20. Gomes DE, Witwer KW. L1CAM‐associated extracellular vesicles: a systematic review of nomenclature, sources, separation, and characterization. J Extracell Biol. 2022;1:e35. doi: 10.1002/jex2.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Badhwar A, Haqqani AS. Biomarker potential of brain‐secreted extracellular vesicles in blood in Alzheimer's disease. Alzheimers Dement. 2020;12:e12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fiandaca MS, Kapogiannis D, Mapstone M, et al. Identification of preclinical Alzheimer's disease by a profile of pathogenic proteins in neurally derived blood exosomes: a case‐control study. Alzheimers Dement. 2015;11:600‐607. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kapogiannis D, Mustapic M, Shardell MD, et al. Association of extracellular vesicle biomarkers with Alzheimer disease in the Baltimore Longitudinal Study of Aging. JAMA Neurol. 2019;76:1340‐1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guix FX, Corbett GT, Cha DJ, et al. Detection of aggregation‐competent tau in neuron‐derived extracellular vesicles. Int J Mol Sci. 2018;19:663. doi: 10.3390/ijms19030663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mustapic M, Eitan E. Plasma extracellular vesicles enriched for neuronal origin: a potential window into brain pathologic processes. Front Neurosci. 2017;11:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Winston CN, Goetzl EJ, Akers JC, et al. Prediction of conversion from mild cognitive impairment to dementia with neuronally derived blood exosome protein profile. Alzheimers Dement. 2016;3:63‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jia L, Zhu M, Kong C, et al. Blood neuro‐exosomal synaptic proteins predict Alzheimer's disease at the asymptomatic stage. Alzheimers Dement. 2021;17:49‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fortea J, Zaman SH, Hartley S, Rafii MS, Head E, Carmona‐Iragui M. Alzheimer's disease associated with Down syndrome: a genetic form of dementia. Lancet Neurol. 2021;20:930‐942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. UniProt Consortium T . UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018;46:2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge‐based Human Protein Atlas. Nat Biotechnol. 2010;28:1248‐1250. [DOI] [PubMed] [Google Scholar]
- 31. Darmanis S, Sloan SA, Zhang Y, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci USA. 2015;112:7285‐7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207‐210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Serpente M, Fenoglio C, D'Anca M, et al. MiRNA profiling in plasma neural‐derived small extracellular vesicles from patients with Alzheimer's disease. Cells. 2020;9:1443. doi: 10.3390/cells9061443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sidhom K, Obi PO, Saleem A. A review of exosomal isolation methods: is size exclusion chromatography the best option? Int J Mol Sci. 2020;21:6466. doi: 10.3390/ijms21186466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Théry C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kakarla R, Hur J, Kim YJ, Kim J, Chwae Y‐J. Apoptotic cell‐derived exosomes: messages from dying cells. Exp Mol Med. 2020;52:1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Uges DR. Plasma or serum in therapeutic drug monitoring and clinical toxicology. Pharm Weekbl Sci. 1988;10:185‐188. [DOI] [PubMed] [Google Scholar]
- 38. Witwer KW, Buzás EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2:20360. doi: 10.3402/jev.v2i0.20360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ayers L, Kohler M, Harrison P, et al. Measurement of circulating cell‐derived microparticles by flow cytometry: sources of variability within the assay. Thromb Res. 2011;127:370‐377. [DOI] [PubMed] [Google Scholar]
- 40. Tsamchoe M, Petrillo S, Lazaris A, Metrakos P. Isolation of extracellular vesicles from human plasma samples: the importance of controls. Biotechnol J. 2023;18:e2200575. [DOI] [PubMed] [Google Scholar]
- 41. Karimi N, Dalirfardouei R, Dias T, Lötvall J, Lässer C. Tetraspanins distinguish separate extracellular vesicle subpopulations in human serum and plasma—Contributions of platelet extracellular vesicles in plasma samples. J Extracell Vesicles. 2022;11:e12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha‐granules. Blood. 1999;94:3791‐3799. [PubMed] [Google Scholar]
- 43. Shi M, Kovac A, Korff A, et al. CNS tau efflux via exosomes is likely increased in Parkinson's disease but not in Alzheimer's disease. Alzheimers Dement. 2016;12:1125‐1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shi M, Liu C, Cook TJ, et al. Plasma exosomal α‐synuclein is likely CNS‐derived and increased in Parkinson's disease. Acta Neuropathol. 2014;128:639‐650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Beninson LA, Brown PN, Loughridge AB, et al. Acute stressor exposure modifies plasma exosome‐associated heat shock protein 72 (Hsp72) and microRNA (miR‐142‐5p and miR‐203). PLoS One. 2014;9:e108748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Arakelyan A, Fitzgerald W, Vagida M, Vasilieva E, Margolis L, Grivel J‐C. Addition of thrombin reduces the recovery of extracellular vesicles from blood plasma. J Circ Biomark. 2016;5:1849454416663648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Serrano‐Pertierra E, Oliveira‐Rodríguez M, Rivas M, et al. Characterization of plasma‐derived extracellular vesicles isolated by different methods: a comparison study. Bioengineering. 2019;6:8. doi: 10.3390/bioengineering6010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Veerman RE, Teeuwen L, Czarnewski P, et al. Molecular evaluation of five different isolation methods for extracellular vesicles reveals different clinical applicability and subcellular origin. J Extracell Vesicles. 2021;10:e12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pang B, Zhu Y, Ni J, et al. Quality assessment and comparison of plasma‐derived extracellular vesicles separated by three commercial kits for prostate cancer diagnosis. Int J Nanomedicine. 2020;15:10241‐10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Valle‐Tamayo N, Pérez‐González R, Chiva‐Blanch G, et al. Enrichment of astrocyte‐derived extracellular vesicles from human plasma. J Vis Exp. 2022;(186), e64107. doi: 10.3791/64107 [DOI] [PubMed] [Google Scholar]
- 51. Kiefel H, Bondong S, Hazin J, et al. L1CAM: a major driver for tumor cell invasion and motility. Cell Adh Migr. 2012;6:374‐384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Goetzl EJ, Boxer A, Schwartz JB, et al. Altered lysosomal proteins in neural‐derived plasma exosomes in preclinical Alzheimer disease. Neurology. 2015;85:40‐47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Goetzl EJ, Boxer A, Schwartz JB, et al. Low neural exosomal levels of cellular survival factors in Alzheimer's disease. Ann Clin Transl Neurol. 2015;2:769‐773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Winston CN, Goetzl EJ, Schwartz JB, Elahi FM, Rissman RA. Complement protein levels in plasma astrocyte‐derived exosomes are abnormal in conversion from mild cognitive impairment to Alzheimer's disease dementia. Alzheimers Dement. 2019;11:61‐66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Goetzl EJ, Mustapic M, Kapogiannis D, et al. Cargo proteins of plasma astrocyte‐derived exosomes in Alzheimer's disease. FASEB J. 2016;30:3853‐3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Goetzl EJ, Schwartz JB, Abner EL, Jicha GA, Kapogiannis D. High complement levels in astrocyte‐derived exosomes of Alzheimer disease. Ann Neurol. 2018;83:544‐552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Norman M, Ter‐Ovanesyan D, Trieu W, et al. L1CAM is not associated with extracellular vesicles in human cerebrospinal fluid or plasma. Nat Methods. 2021;18:631‐634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cha DJ, Mengel D, Mustapic M, et al. miR‐212 and miR‐132 are downregulated in neurally derived plasma exosomes of Alzheimer's patients. Front Neurosci. 2019;13:1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chou S‐Y, Chan L, Chung C‐C, Chiu J‐Y, Hsieh Y‐C, Hong C‐T. Altered insulin receptor substrate 1 phosphorylation in blood neuron‐derived extracellular vesicles from patients with Parkinson's disease. Front Cell Dev Biol. 2020;8:564641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dutta S, Hornung S, Kruayatidee A, et al. α‐Synuclein in blood exosomes immunoprecipitated using neuronal and oligodendroglial markers distinguishes Parkinson's disease from multiple system atrophy. Acta Neuropathol. 2021;142:495‐511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Supporting information
Supporting information
Supporting information
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


