Abstract
Cerebral small vessel disease (cSVD) is a leading cause of stroke and dementia with no specific mechanism-based treatment. We used Mendelian randomization to combine a unique cerebrospinal fluid (CSF) and plasma pQTL resource with the latest European-ancestry GWAS of MRI-markers of cSVD (white matter hyperintensities, perivascular spaces). We describe a new biological fingerprint of 49 protein-cSVD associations, predominantly in the CSF. We implemented a multipronged follow-up, across fluids, platforms, and ancestries (Europeans and East-Asian), including testing associations of direct plasma protein measurements with MRI-cSVD. We highlight 16 proteins robustly associated in both CSF and plasma, with 24/4 proteins identified in CSF/plasma only. cSVD-proteins were enriched in extracellular matrix and immune response pathways, and in genes enriched in microglia and specific microglial states (integration with single-nucleus RNA sequencing). Immune-related proteins were associated with MRI-cSVD already at age twenty. Half of cSVD-proteins were associated with stroke, dementia, or both, and seven cSVD-proteins are targets for known drugs (used for other indications in directions compatible with beneficial therapeutic effects. This first cSVD proteogenomic signature opens new avenues for biomarker and therapeutic developments.
Introduction
Characterized by changes in the structure and function of small brain vessels, cerebral small vessel disease (cSVD) is a leading cause of ischemic and hemorrhagic stroke, cognitive decline and dementia. cSVD is extremely common with increasing age and most often covert, namely detectable on brain imaging in the absence of clinical symptoms. Covert cSVD portends a considerably increased risk of stroke and dementia, thus represents a major target to prevent these disabling conditions and promote healthier brain aging1. The most common and heritable MRI-markers of cSVD (MRI-cSVD) are white matter hyperintensities of presumed vascular origin (WMH) and perivascular spaces (PVS)2.
Hypertension is the strongest known risk factor for cSVD, representing a major target for prevention1. However, vascular risk factors explain only a small fraction of MRI-cSVD variability in older age3, and drugs specifically targeting pathological processes underlying cSVD are lacking. Genomics can provide a strong foundation for mechanistic studies and drug target discovery4. Recent genetic studies have identified > 70 genetic risk loci associated with cSVD5,6; however, causal genes and underlying molecular pathways remain poorly understood.
As disease occurrence reflects the complex interplay of factors beyond DNA sequence, there is growing interest in identifying circulating biomarkers, such as proteins, capturing these downstream factors, to enhance our understanding of the underlying biology, accelerate omics-driven drug discovery, and potentially generate circulating biomarkers for clinical use7. While large-scale proteomic investigations have recently been conducted for stroke and dementia, with promising findings,7–13 studies on proteomics of cSVD have been conducted on limited sets of proteins, in small studies of European ancestry (N < 5,000), and in plasma only14–18. We hypothesize that, while plasma may enable easy-access biomarker measurements, CSF, the fluid circulating in perivascular spaces, could reveal a more accurate biological fingerprint of cSVD.
Here we used two-sample Mendelian randomization (2SMR), leveraging large proteomic and genomic resources, to investigate the relation of circulating protein levels in CSF and plasma with WMH and PVS burden and to explore its causal relation and directionality. We further used a multipronged approach for the follow-up of identified associations in independent samples, across fluids, proteomics platforms, ancestries and the lifespan, using both 2SMR and individual-level data. We also explored the ability of proteogenomics to predict extensive cSVD and tested the relation of cSVD-associated proteins with risk of stroke and Alzheimer’s disease (AD). Using single-cell sequencing resources we deciphered cell-types and pathways involved. Finally, we combined our results with pharmacological databases for proteomics-driven drug discovery.
Results
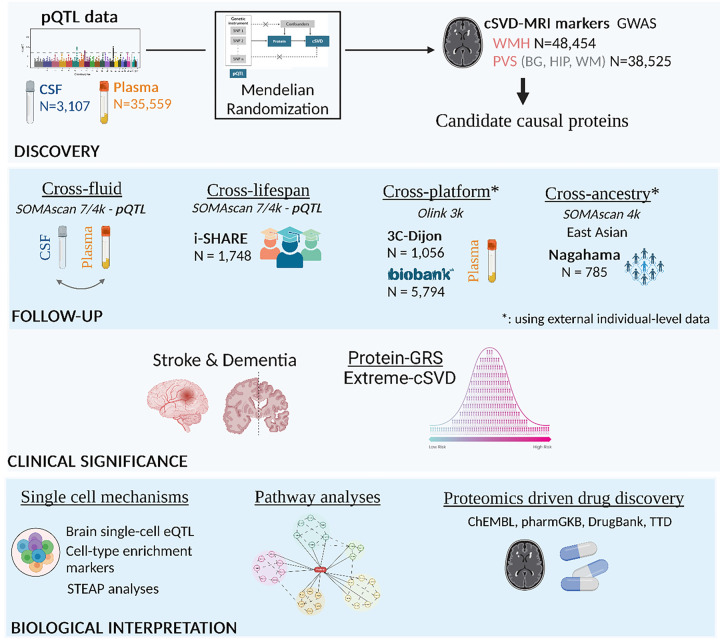
The study design is summarized in Fig. 1.
Figure 1. Summary of the analysis plan.
pQTL: protein quantitative trait loci, CSF: Cerebrospinal fluid, WMH: White matter hyperintensities, PVS: Perivascular Spaces burden, BG: basal ganglia, HIP: hippocampus, WM: white matter. # Cross-platform follow-up analyses have been conducted using a meta-analysis of 3C and UK Biobank
Discovery of protein-cSVD associations
We used 2SMR to test associations of circulating CSF and plasma protein levels with MRI-cSVD. We leveraged summary statistics of large protein quantitative trait loci (pQTL) resources in European-ancestry participants from CSF12 (N = 3,107; aptamer-based Somascan 7k assay) and plasma19 (N = 35,559; Somascan 5K), as well as the largest published GWAS of WMH volume (N = 48,454, mean age 66.0 years)20 and PVS burden (N up to 40,095, mean age 66.3 years)5. PVS were studied in three sublocations, white matter (WM), basal ganglia (BG) and hippocampus (HIP), for which risk factors, including genetic, were shown to differ5. Cis- and trans-genetic instruments could be derived for 1,121 CSF and 1,731 plasma proteins. (Methods)
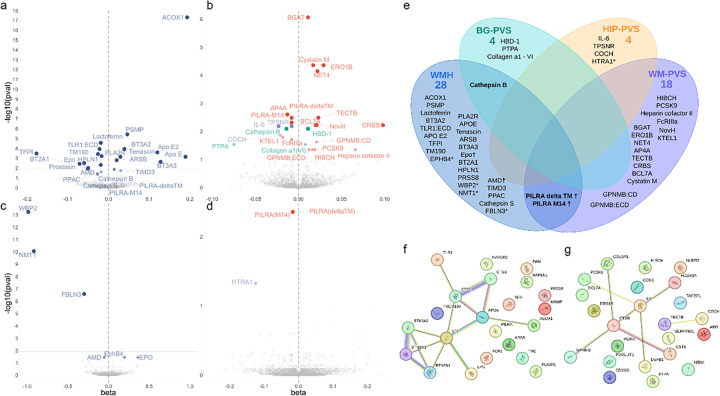
Focusing our primary analyses on cis-pQTLs (Supplementary Table 1), we identified 46 of 1,121 CSF proteins associated with at least one MRI-cSVD (pFDR<0.05): 24 with WMH, and 25 with PVS (18 WM-PVS, 4 BG-PVS, and 3 HIP-PVS, Fig. 2A–B, Supplementary Table 2–3). In parallel we identified 9 of 1,731 plasma proteins associated with MRI-cSVD (pFDR<0.05): 6 with WMH and 3 with PVS (2 WM-PVS, 1 HIP-PVS). Of these, 4 were also significantly associated with MRI-cSVD in CSF (AMD, EPO [WMH], PILRA-M14 and PILRA-deltaTM [WM-PVS], Fig. 2C–D, Supplementary Tables 4–5). For pQTL with multiple instruments (42 proteins), associations were robust to sensitivity analyses (MR-Egger, weighted median and MR-PRESSO); for single-instrument pQTL (14 proteins), there was no evidence of colocalization for two proteins, ACOX1 and WBP2 with PP4 < 0.7 & PP3 > 0.7, which were removed from subsequent analyses (Supplementary Tables 3 and 5). None of the single variant pQTL were non-synonymous variants, which could have resulted in structural changes at the aptamer protein binding site and thus biased its measurement (Supplementary Table 6). Bidirectional MR ruled out reverse causation, except for an association of genetically determined larger WM-PVS burden with higher PCSK9 CSF levels (pFDR=0.011, Supplementary Table 3). In total, 49 proteins were associated robustly with MRI-cSVD in CSF (41), plasma (4), or both (4), including three associated with both WMH and PVS: CTSB (Cathepsin B), a lysosomal protease involved in extracellular matrix (ECM) degradation, and two soluble isoforms deltaTM and M14 of PILRA (Paired Immunoglobin Like Type 2 Receptor Alpha), a microglial immunoreceptor.
Figure 2. Discovery protein-cSVD associations in CSF and plasma using cis-pQTL mendelian randomization.
A. Volcano plots of proteins associated with white matter hyperintensities (WMH) using cis-pQTL MR in CSF. B. Volcano plots of proteins associated with perivascular spaces burden (PVS) using cis-pQTL MR in CSF. C. Volcano plots of proteins associated with WMH using cis-pQTL MR in plasma. D. Volcano plots of proteins associated with PVS using cis-pQTL MR in plasma. Each dot represents the MR results for proteins. Each dot represents the MR results for proteins. FDR-corrected p-values are represented in this graph. Represented proteins are significantly associated with MRI-marker at pFDR (Benjamini-Hochberg false discovery rate threshold) < 0.05. The dotted line in each volcano plot represents the corrected threshold after additionally correcting for the number of phenotypes tested (p<0.0125). E. Venn diagram of identified causal proteins associated with MRI-cSVD. * proteins identified in plasma; † proteins associated in both plasma and CSF; other proteins are associated in CSF. F. String plot of proteins associated with WMH. G. String plot of proteins associated with PVS (WM, BG and HIP). Network nodes represent proteins: colored nodes query proteins and first shell of interactors. Edges represent protein-protein associations. Cyan and pink edges are known interactions, cyan: from curated databases, and pink: experimentally determined. Green and blue edges correspond to predicted interactions. Green: gene neighborhood, and blue: gene co-occurrence. Purple corresponds to protein homology, yellow to text mining and black to co-expression.
In secondary analyses including both cis- and trans-pQTLs, we found 340 proteins associated with at least one MRI-cSVD in CSF or plasma (pFDR<0.05), of which 176 were driven by two trans-hotspots at APOE (147 proteins) and chr16q24 (29 proteins). Although most protein-cSVD associations revealed novel pathways not previously identified, some relate to previous cSVD GWAS findings. Two cis-pQTL were associated with WMH volume at genome-wide significance, for FBLN3 (encoded by EFEMP1) at chr2p16 and NMT1 (NMT1) at chr17q21. Additionally, HTRA1, of which lower genetically determined plasma levels were associated with extensive HIP-PVS, is encoded by a gene harboring both rare mutations causing monogenic cSVD21 and common variants associated with small vessel stroke and suggestively WMH20,22. From secondary analyses, eight trans-pQTL for 29 proteins at the chr16q24 hotspot were associated at genome-wide significance with WMH. The APOE hotspot included four proteins encoded by genes in genome-wide or gene-wide significant risk loci for WMH20 and extreme-cSVD23 (APOE, MRPL38, SULT1B1, and MSRA; Supplementary Tables 6–9, Extended Data Fig. 1).
To assess the independence of observed associations, we used LD-score regression (LDSC)24 to quantify the genetic correlation between protein levels. Only one genetic correlation was significant after multiple testing correction (EPHB4 with PILRA-M14 in plasma at p < 5×10− 5, Methods, Extended Data Fig. 2). Several protein-protein interactions were identified using the STRING database (Fig. 2F).
Follow-up of significant protein-cSVD associations
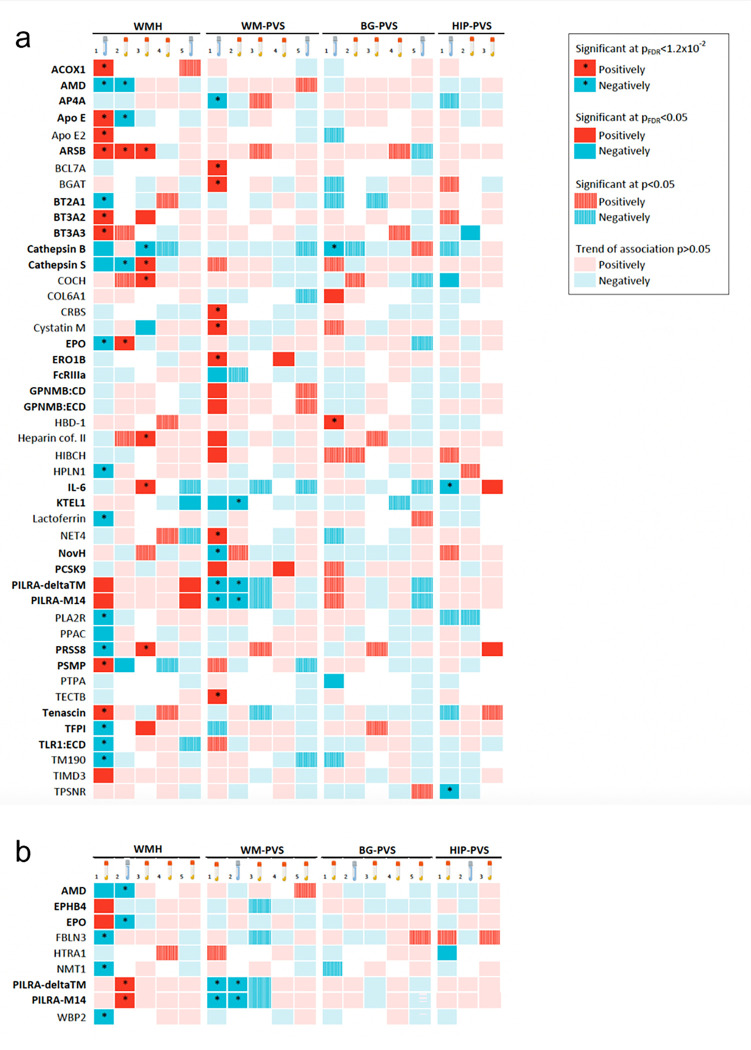
We used a multi-pronged approach to follow-up protein-cSVD associations based on cis-pQTL with significant MR results and colocalization evidence, across fluids, platforms, and ancestries (Figs. 1 and 3).
Figure 3. Summary of proteomics follow-up (discovery, cross-fluid, cross-platform, cross-ancestry and lifespan).
A. Heatmap of proteomic findings using CSF discovery analysis. B. Heatmap of proteomic findings using plasma discovery analysis. 1. Discovery Mendelian randomization using cis-pQTL from CSF (A) and plasma (B). 2. Cross-fluid follow-up Mendelian randomization using cis-pQTL from plasma (A) and CSF (B). 3. Cross-platform follow-up using plasma individual-level data measured with Olink in independent samples (3C, UKB). 4. Cross-ancestry follow-up using plasma individual-level data measured with Somascan in an independent sample (Nagahama). 5. Lifespan follow-up Mendelian randomization using cis-pQTL from CSF (A) and plasma (B). Dark squares correspond to significant results after FDR correction (pFDR<0.05). * corresponds to significant associations after correction for the 4 phenotypes tested (pFDR<0.0125). Hatched squares correspond to p<0.05 results. Red squares correspond to a positive association and blue to negative association. Proteins missing for one of the follow-up analyses are represented with a white square. # results of the analysis of 3C only. Proteins in bold are those showing at least one nominally significant association (p<0.05) in follow-up analyses, with the same MRI-cSVD marker as in the discovery.
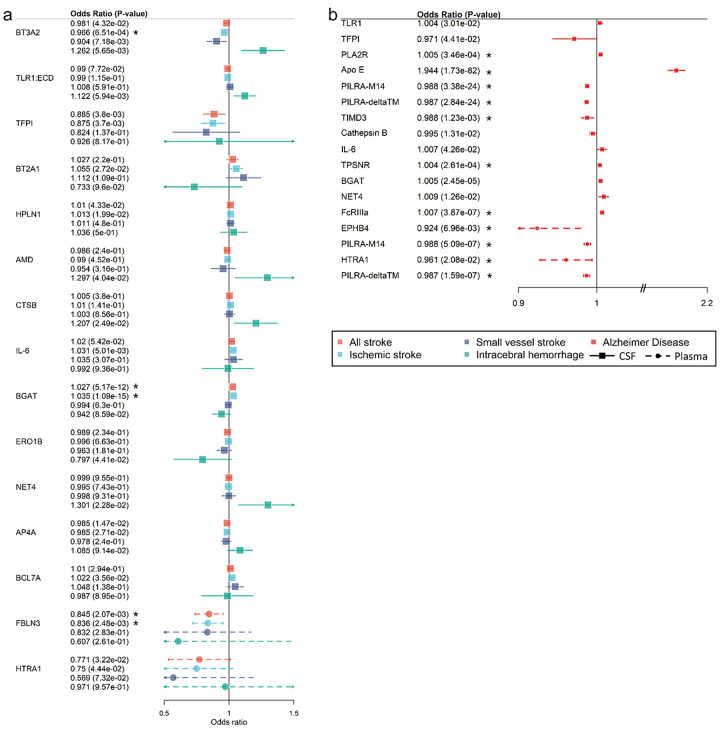
First, using 2SMR, we tested whether cSVD-proteins associations observed in CSF showed some indication of association in plasma, and vice-versa, with a less stringent multiple testing correction than in the discovery analysis, considering significant associations in the original fluid only. Thirty-seven cSVD-associated CSF proteins had plasma pQTL available. Nine of these (24%) were associated with the same MRI-cSVD phenotype in plasma at pFDR<0.05 (APOE, ARSB, EPO, AMD, CTSS, PSMP with WMH, PILRA-M14, PILRA-deltaTM, KTEL1 with WM-PVS, Methods, Fig. 3A and 6, Supplementary Table 10). Six cSVD-associated plasma proteins had CSF cis-pQTL available. Four of these (67%) were associated with the same MRI-cSVD phenotype in CSF at pFDR<0.05 (AMD, EPO with WMH and PILRA-M14, PILRA-deltaTM with WM-PVS, Fig. 3B and 6, Supplementary Table 11). Directions of association were mostly concordant except for EPO, APOE and PSMP, which showed opposite direction of association in CSF and plasma, in line with previous observations12 and highlighting the importance of studying multiple tissues to capture the complexity of underlying biology.
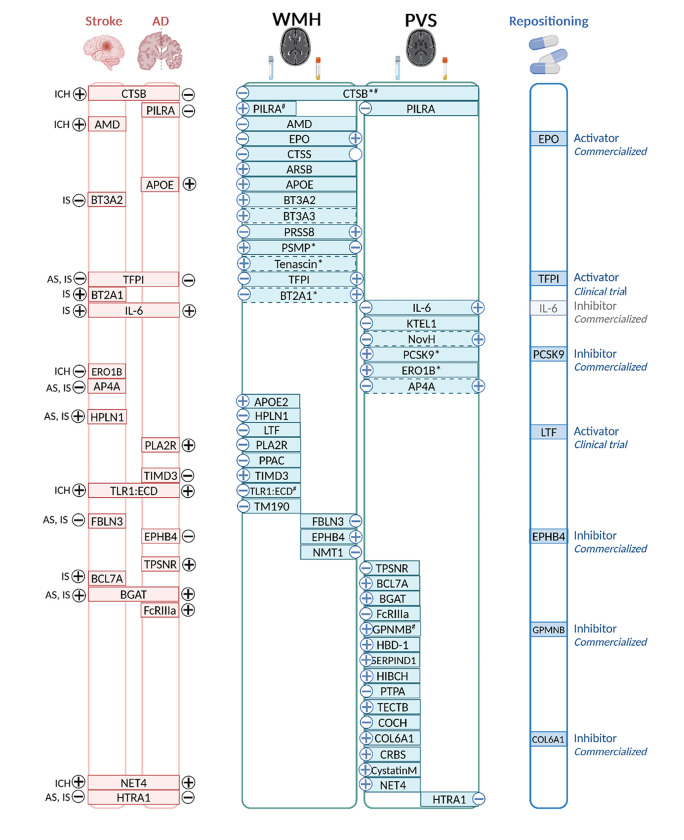
Figure 6. Integrated summary of our findings.
Proteins associations with WMH, PVS, or both are represented in the middle. For each MRI-marker, the left side corresponds to CSF findings and the right side to plasma findings. * proteins with cross-ancestry association. # proteins with lifespan association. Associations with stroke Alzheimer’s disease (AD), or both are represented on the left of the figure. Subtypes of stroke are as follows: AS: Any stroke, IS: Ischemic stroke, SVS: Small vessel stroke, ICH: Intracerebral hemorrhage. − and + signs correspond to the direction of association referring to higher level of the protein. Drug repositioning is represented on the right of the figure.
Second, a cross-platform follow-up was performed by testing the association with MRI-cSVD of plasma protein levels measured on the Olink Explore-3072 platform in two independent population-based studies, 3C-Dijon (N = 1,056; mean age 72.5 years) and UK Biobank (N = 5,494; mean age 63.5 years, Supplementary Table 12). Twenty-nine of the 49 cSVD-associated proteins (59%) were available; 26 were used after quality control and their plasma level was tested against WMH volume and PVS burden using linear regression followed by inverse variance weighted meta-analysis (N = 6,550). Of these, 7 proteins (27%, all identified in CSF 2SMR), showed association with the same MRI-cSVD marker at pFDR<0.05 (ARSB, PRSS8, CTSS, CTSB, TFPI and BT3A2 with WMH, IL-6 with HIP-PVS, Figs. 3 and 6, Supplementary Tables 13–14). Directionality of association with MRI-cSVD was inconsistent between CSF pQTL and plasma protein levels for PRSS8, TFPI, IL-6, and CTSS. Inter-platform correlations for these proteins between Somascan and Olink were moderate to good in plasma and CSF respectively (Supplementary Table 1525); however correlations were not available between plasma and CSF.
Third, we conducted a cross-ancestry exploratory follow-up, testing associations of MRI-cSVD with plasma protein levels measured on the Somascan 4K platform in the Japanese population-based Nagahama study (N = 785; mean age 68 years). Thirty-eight of the 49 cSVD-associated proteins (77%) were available and their plasma level was tested against WMH volume and extensive PVS burden. Two proteins (both identified in CSF 2SMR in Europeans) were associated at pFDR<0.05 with the same MRI-marker (WM-PVS), with consistent directionality (ERO1B and PCSK9); given the small sample size we also considered nominally significant associations, observed for four additional proteins, with WMH (BT2A1, CTSB, TNC, PSMP, Figs. 3 and 6, Supplementary Table 16).
Fourth, we took an exploratory lifespan approach by testing the relation of cSVD-associated proteins with MRI-cSVD in young adults (i-Share study, N = 1,748; mean age 22.1 years). Here we used 2SMR with the same cis-pQTL as for discovery analyses. Consistent with findings in older adults, higher genetically determined CSF levels of PILRA-M14, PILRA-deltaTM were associated with larger WMH volume at pFDR<0.05. In addition, higher genetically determined CSF protein levels of GPNMB:CD and GPNMB:ECD (cellular and extracellular domain of a transmembrane glycoprotein upregulated upon tissue damage and inflammation) and TLR1:ECD (extracellular domain of toll-like receptor 1, which plays a fundamental role in activation of innate immunity) were associated with BG-PVS and WMH volume respectively at p < 0.05, in a direction consistent with older adults (Fig. 3, Supplementary Table 17).
Overall, of 49 cSVD-associated proteins (Supplementary Table 18, Fig. 6), (i) 16 CSF proteins showed associations with the same MRI-cSVD marker in plasma in at least one analysis (pQTL or direct protein measurement) at pFDR<0.05, with consistent directionality in 63%; (ii) 24 CSF proteins were not associated with the same MRI-marker in plasma (p ≥ 0.05) and may be considered as CSF-specific; (iii) 4 proteins were identified in plasma pQTL analyses only, with non-significant follow-up in association with direct plasma protein measurements; (iv) 5 proteins had no follow-up available apart from the lifespan exploration; (iii) and 6 proteins had evidence for lifespan effects at p < 0.05 (2 at pFDR<0.05).
Predictive performance of protein genetic risk scores (GRS)
We assessed the ability of selected cis-based protein-GRS to predict a composite extreme-cSVD phenotype (extensive WMH volume ± lacunes vs. minimal WMH volume without lacunes) in the 3C-Dijon cohort, benchmarking it against a previously validated WMH-GRS20 (Methods). Using the WMH-GRS only, we achieved an AUC of 0.568 (95% Bootstrap CI 0.501–0.634). Adding any of the four selected protein GRS slightly improved the AUC, while adding them all achieved a maximum improvement of + 0.04 (AUC = 0.608; 95% CI [0.544–0.672], Extended Data Fig. 4 Supplementary Table 19).
Clinical significance
We explored the relation of the 49 cSVD-associated proteins with stroke (any, ischemic, and small vessel stroke; intracerebral hemorrhage) and AD (Methods). We leveraged the aforementioned CSF and plasma pQTL, as well as European-ancestry summary statistics of GWAS for stroke and its subtypes (N ≤ 73,652 cases) and AD (N = 71,880 cases). Twenty-four proteins (49%) showed associations with at least one clinical outcome at p < 0.05 (Figs. 4 and 6). At pFDR<0.05, eight CSF proteins (APOE, PILRA-M14, PILRA-deltaTM, FcRIIIa, BGAT, PLA2R, TIMD3 and TPSNR) and four plasma proteins (EphB4, HTRA1, PILRA-M14, PILRA-deltaTM) were significantly associated with AD, while one CSF protein (BGAT, measuring histo-blood group ABO system glycosyltransferase activity) and one plasma protein (FBLN3) were associated with any stroke and ischemic stroke (Supplementary Tables 20–21). Nineteen of 49 proteins were available for partial follow-up in plasma using 2SMR in East-Asian participants in relation with ischemic and small vessel stroke, leveraging plasma pQTL from Biobank Japan (N = 2,886) and an East-Asian stroke GWAS meta-analysis (N ≤ 17,493). Overall, despite substantially smaller sample size for exposure and outcome in East-Asians, correlation of effect sizes was moderate to high (Extended Data Fig. 5). Higher plasma levels of NovH (encoded by CCN3), an ECM associated protein involved in cardiovascular development, were associated with increased risk of small vessel stroke at pFDR<0.05 (Supplementary Table 22).
Figure 4. Clinical significance of protein-cSVD findings in CSF and plasma.
A. Forest plot of protein-cSVD associations with stroke and its subtypes (ischemic stroke, small vessel stroke and intracerebral hemorrhage). B. Forest plot of protein-cSVD association with Alzheimer’s disease. All proteins associated with MRI-cSVD identified in the discovery analysis in CSF and plasma were used for this analysis. Full lines represent proteins measured in CSF. Dotted lines represent proteins measured in plasma. Proteins significant at least at p<0.05 for at least one of the outcomes tested are represented (for stroke, associations with all (sub)types are represented when one or more was significant). * Results significant after multiple testing correction (pFDR<0.05)
Biological interpretation
Using FUMA pathway enrichment analyses, cSVD-associated proteins overall were significantly enriched in proteins involved in proteoglycan binding and extracellular matrix (organization and collagen containing: CTSS, EFEMP1, HAPLN1, CTSB, HTRA1, NTN4, COL6A1, TNC, COCH, APOE, pFDR<0.05, Supplementary Table 23A). Among CSF proteins associated with cSVD, proteins involved in regulation of immune response signaling and activation of immune response were overrepresented (BT2A1, BT3A2, BT3A3, CTSB, CTSS, LTF, TLR1, HAVCR2, pFDR<0.05, Supplementary Table 23B).
To explore enrichment of observed protein-cSVD associations in particular cell-types we first conducted single-cell enrichment analyses using STEAP, leveraging multiple publicly available single-cell sequencing resources (Methods, Supplementary Table 24). Genes encoding several cSVD-associated proteins showed significant enrichment in microglia for several CSF proteins (BT2A1, BT3A2, BT3A3, CTSS, HIBCH) and in immune cells for plasma protein (EPO, Supplementary Table 25, Extended data Fig. 6). Next, we used unique resources of single nucleus RNA sequencing (snRNAseq) derived from up to 443 post-mortem brain samples (dorsolateral prefrontal cortex) from the ROSMAP older population-based cohort26–29. In silico sorting of human cortical tissue samples was used to derive vascular brain cells27,28. From these snRNAseq resources we could derive cell-type specific brain eQTLs for 19 and 10 genes encoding cSVD-associated proteins, in non-vascular and vascular cells respectively (Methods). Using MR, we found lower genetically determined expression levels of TLR1 in oligodendrocytes (pFDR=2.24×10− 4) and CTSS in smooth muscle cells (pFDR=2.3×10− 3) to be associated with larger WMH volume, both consistent with directionality of associations in CSF (Supplementary Table 26–27). Higher genetically determined expression of ABO (encoding BGAT) in pericytes was protective for extensive WM-PVS (pFDR=2.3×10− 3, opposite direction compared to CSF). All three associations showed evidence for colocalization (PP.H4 > 0.7). Genes encoding cSVD-associated proteins showed distinct cerebrovascular cell-specific gene expression patterns (e.g. with EFEMP1 expression dominating in a new subtype of perivascular fibroblasts) and we observed a non-significant trend towards an overall enrichment in pericytes (Extended data Fig. 7). We also tested enrichment of our genes of interest in different microglial states (Methods, Extended data Fig. 7), given the aforementioned results observed with STEAP, and observed significant enrichment in a microglial state type previously found to be itself enriched in processes such as ribosome biogenesis, amyloid fibril formation, and positive regulation of T-cell mediated immunity29.
Proteomics-driven drug discovery
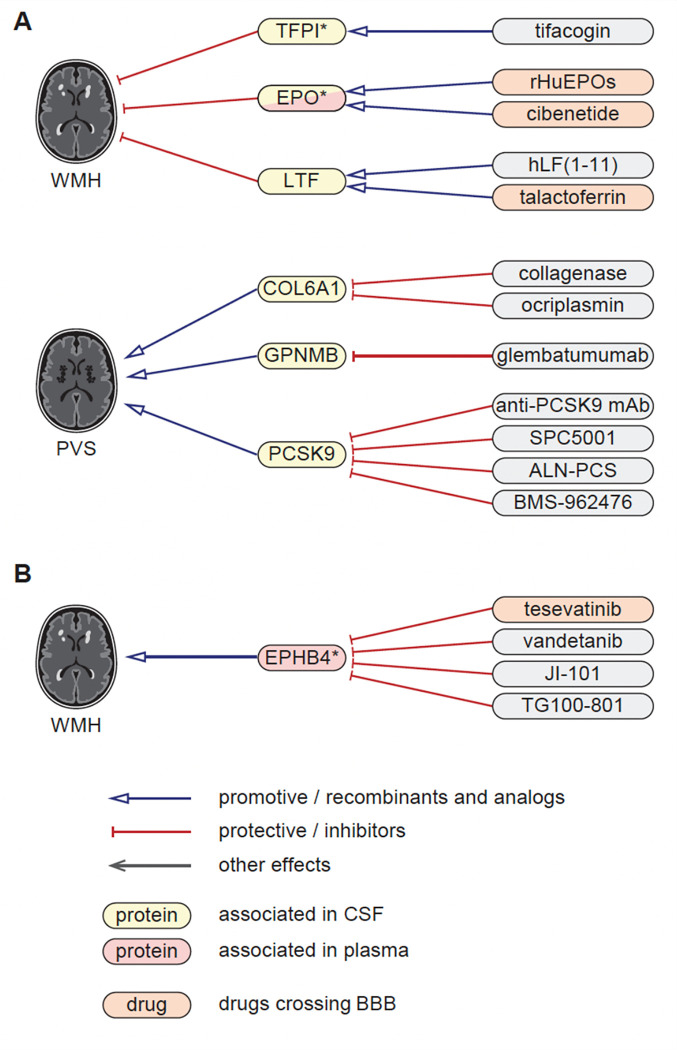
We used MR estimates from the 49 CSF and plasma proteins with MRI-cSVD to support drug discovery. Using public drug databases (Methods), we curated drugs (commercialized for other indications or under investigation in clinical trials) targeting these proteins in a direction compatible with beneficial therapeutic effects against cSVD based on MR estimates. We identified such drugs for EPO, LTF, TFPI, and EPHB4 for WMH; COL6A1, GPNMB, PCSK9 for PVS, most of which were associated with MRI-cSVD in the CSF only, except EPHB4 (plasma), EPO and TFPI (CSF and plasma, Fig. 5, Supplementary Table 28). Some of these proteins have predicted or experimentally proven interactions with each other (Fig. 2E–F), suggesting that identified drugs may impact related pathways. Of note, drugs targeting EPO and LTF as agonists and EPHB4 as inhibitors cross the blood-brain barrier (Supplementary Table 28).
Figure 5. Proteomics-driven drug discovery.
A. Drug-discovery analysis conducted using CSF protein-cSVD Mendelian randomization estimates for WMH and PVS findings. B. Drug-discovery analysis conducted using plasma protein-cSVD Mendelian randomization estimates for WMH. Proteins in yellow correspond to proteins associated with the MRI-cSVD marker in CSF and in red in plasma, in discovery analyses. * proteins with significant associations in at least one of the follow-up modalities (at p<0.05). Red arrows correspond to a protective effect of a protein on MRI-cSVD (reducing cSVD burden) or an inhibitor effect of a drug on the cSVD-associated protein; blue arrows correspond to a of deleterious effects of a protein on MRI-cSVD (promoting cSVD burden) or an analog effect of a drug on the cSVD-associated protein. Drugs in orange cross the blood brain barrier. CSF: cerebrospinal fluid, BBB: blood brain barrier.
Results of protein-cSVD associations along with clinical significance, pathway or cell-type enrichment and drug target identification are summarized in Supplementary Table 18 and Fig. 6.
Discussion
By combining a unique CSF and plasma pQTL resource with the latest GWAS of MRI-cSVD in a Mendelian randomization framework, we describe a new biological fingerprint of cSVD comprising 49 protein-cSVD associations with a putative causal relation, predominantly in the CSF. To assess robustness and specificity of our findings we implemented a multipronged follow-up approach, across fluids, proteomic platforms, and ancestries, which included testing associations of direct plasma protein measurements with MRI-cSVD. We highlight 16 proteins robustly associated in both CSF and plasma, of which 12 are in the same direction, while 24 and four proteins were identified in CSF or plasma only, with no evidence for association in the other fluid. Strikingly, several cSVD-associated proteins already showed associations with WMH and PVS burden at age 20 with consistent directionality. The fact that half of cSVD-associated proteins show at least nominally significant associations with stroke, AD, or both highlights their clinical relevance. Pathway and cell-type enrichment analyses suggest an important role of extracellular matrix and immune response pathways, with single-cell RNA-sequencing analyses pointing predominantly to microglia, but also oligodendrocytes, vascular smooth muscle cells and pericytes. Finally, besides revealing potential novel biomarkers and drug targets to be investigated, our findings also provide genetic support for repositioning of seven drugs for cSVD.
Previous explorations of cSVD proteomics were mainly conducted on focused protein panels30,31, mostly in plasma14–17,32 (except a recent study on 16 CSF proteins)33 and in relatively small cohorts (usually N < 1,000)34. Here we analyzed over 2,500 plasma and CSF proteins in relation with WMH and PVS burden in over 40,000 participants. In recent years, CSF biomarkers have emerged as pivotal for unraveling the intricate mechanisms underlying neurodegenerative and neuroinflammatory diseases, given their proximity to the central nervous system35–37. Our findings suggest that this also holds true for cSVD. Indeed, CSF-based MR analyses revealed five times more protein-cSVD associations than plasma-based MR, despite ten times smaller sample size to derive pQTL. Among proteins with pQTL available in both plasma and CSF resources, 67% of cSVD-associated plasma proteins also showed associations with the same MRI-cSVD markers in CSF, whereas only 24% of cSVD-associated CSF proteins showed associations in plasma. Even when accounting for follow-up with direct protein measurements, only 43% of cSVD-associated CSF proteins were associated with MRI-cSVD in plasma, suggesting that some protein-cSVD associations are specific to CSF, as described for other neurological disorders12,13.
Some proteins associated with MRI-cSVD were particularly robust, with consistent directionalities of their association across fluids and platforms, using both pQTL-based and direct measurements, especially, PILRA-deltaTM, PILRA-M14, ARSB and CTSB.
PILRA (paired immunoglobin like type 2 receptor alpha) is a microglial immunoreceptor involved in β-amyloid uptake and herpes simplex virus 1 infection38. Somascan measures soluble PILRA isoforms lacking the transmembrane domain39 (PILRA-deltaTM and PILRA-M14) while Olink detects the full protein. Higher genetically determined CSF levels of PILRA-M14 and PILRA-deltaTM were associated with larger WMH volume across the lifespan, notably already in young adults in their twenties. In contrast, higher genetically determined CSF and plasma levels of PILRA isoforms were associated with smaller WM-PVS burden and lower risk of AD (p < 10− 23 for high CSF levels). Higher plasma levels of PILRA (Olink direct measurements) were also protective for WM-PVS. This could potentially indirectly point to a protective effect of PILRA on cSVD caused by cerebral amyloid angiopathy (CAA), as WM-PVS was recently proposed as a novel CAA biomarker40, and CAA is associated with a strongly increased risk of AD41. Interestingly, previous experimental work has supported PILRA as the likely causal gene at the chr7q21 AD risk locus,42 suggesting that a common missense variant in this gene (rs1859788, r2 = 0.3 with PILRA pQTL) may protect against AD via reduced inhibitory signaling in microglia and reduced microglial infection during HSV-1 recurrence. The opposite effect we observed on WMH is intriguing, requiring further explorations, such as an examination of differential associations with WMH spatial patterns.
ARSB (arylsulfatase B) plays an important role in ECM degradation, regulation of neurite outgrowth and neuronal adaptability in the central nervous system43, where it is expressed predominantly in the microglia44,45. ARSB deficiency causes a lysosomal storage disorder (mucopolysacharidosisc)46. Here higher ARSB levels in CSF and plasma were associated with greater WMH volume based on both Somascan pQTL and direct Olink protein measurements, making ARSB a compelling candidate to explore as a circulating cSVD biomarker. CTSB (cathepsin B) is a cerebrovascular matrisome-associated protein identified in brain microvessels31. This lysosomal cysteine protease is involved in proteolysis of ECM components and enhanced vessel wall permeability47, as well as in proteolysis of amyloid precursor protein, implicated in AD48. Genetically determined higher CTSB levels in CSF were associated with smaller WMH and BG-PVS burden, replicating in plasma, across platforms (pQTL and direct measurements) and ancestries, and with lower AD risk at nominal significance. Similar associations were observed between higher genetically determined CSF and plasma levels of CTSS (cathepsin S, another cysteine protease) and smaller WMH volume, but higher direct plasma CTSS measurements were associated with larger WMH volume. A potential explanation for such discrepancies could be that pQTL and direct measurements capture different isoforms (Olink assays have been developed for the canonical CTSS isoform 1). Noteworthy, rare mutations in CTSA (encoding cathepsin A, a serine protease like HTRA1) cause a rare monogenic autosomal recessive cSVD known as CARASAL49. Our findings thus expand the involvement of cathepsins to complex cSVD, and to cysteine in addition to serine proteases.
We also show for the first time an association of lower plasma levels of HTRA1 (High-Temperature Requirement A serine peptidase 1), another cerebrovascular matrisome protein, with extensive HIP-PVS, consistent with loss-of-function mechanisms underlying monogenic cSVD caused by rare mutations in HTRA1 (CARASIL, autosomal dominant HTRA1 mutations)50. Rare and common variants at HTRA1 have been associated with larger WMH volume and increased stroke risk in the general population51,52, with recent findings suggesting loss-of-function mechanisms through both reduced HTRA1 expression and lower serine protease enzyme activity. The association of lower genetically determined plasma levels of HTRA1 with extensive HIP-PVS provides additional evidence for an impact of HTRA1 loss-of-function on brain health. Interestingly, lower genetically determined HTRA1 plasma protein levels were also associated with higher risk of stroke (any, ischemic) and AD at p < 0.05.
Overall, our proteogenomic analyses lend support to a prominent role of the cerebrovascular matrisome (extracellular matrix and associated proteins) in both monogenic and multifactorial cSVD, corroborating and expanding findings from large genomic studies5,6 and preclinical work on monogenic cSVD models31. In parallel, our findings also reveal prominent associations of immune response pathways with MRI-cSVD. Intriguingly, associations with proteins involved in immunity and inflammation (with PILRA, TLR1, GPNMB, all three expressed predominantly in microglia) were already detectable in young adults in their twenties. We also found expression of genes encoding CSF cSVD-associated proteins to be significantly enriched in microglial cells, the brain’s primary resident immune cells. The interplay between cSVD and inflammation has gained recent interest, with emerging evidence from focused biomarker studies and experimental models, suggesting that activation of immune cells and in particular microglial cells could play an important role53–57. Co-registration of MRI images with (immune-)histopathological data has shown that WMH volume was associated with higher microglial activation, supporting that the latter could be involved in cSVD etiology58. Our results lend further support to this, suggesting that this could be one of the earliest processes involved, as demonstrated for AD59. Given growing evidence that changes in microglial transcriptional profiles play a crucial role in brain aging and AD and that blood proteins can mediate neurotoxic microglial functions60, the proteogenomic signature we describe might contribute to revealing biological underpinnings of the intricate relation between cSVD and AD29,61,62.
Some cSVD-associated proteins are encoded by genes in cSVD GWAS loci, strengthening evidence for their involvement in disease pathogenesis. At chr17q21, lower plasma levels of NMT1 (N-Myristoyltransferase1), a protein involved in vascular instability and endothelial cell damage63–65, were associated with larger WMH volume, aligning with prior associations of lower arterial NMT1 expression with larger WMH burden66. At chr2p16, lower plasma levels of FBLN3 (Fibulin-3, encoded by EFEMP1), a glycoprotein essential for maintaining ECM and vessel integrity and involved in cell proliferation and migration, were associated with larger WMH volume23,67. Furthermore, beyond genetic risk scores derived from cSVD GWAS, genetic risk scores for cSVD-associated proteins may have added predictive value for identifying those with extensive cSVD burden, highlighting the potential of multiomics approaches for enhancing risk prediction and stratification.
This work further unveiled new prospects for therapeutic repositioning and development, with the identification of seven drugs (targeting EPO, LTF, TFPI, EphB4, COL6A1, GPNMB, and PCSK9) with cSVD MR results compatible with potential beneficial therapeutic effects, warranting further investigation. Of these, agonists for EPO and LTF and inhibitors of EphB4, which are either approved or studied in phase II clinical trials for other indications (Supplementary Table 28) present evidence of successfully crossing the blood brain barrier (BBB), although it is unclear whether this is required to treat cSVD. EPO is a neuroprotective protein safeguarding the BBB against VEGF-induced permeability68, acting through the Keap1/Nrf2 pathway in ischemia reperfusion injury69. LTF has anti-inflammatory and neuroprotective properties and can upregulate EPO69 and downregulate IL-670,71, both associated with MRI-cSVD in our study. EPO and LTF were reported to show strong protein-protein interaction with collaborative anti-inflammatory properties69 and modified, optimized versions of both these proteins have been tested experimentally as neuroprotective agents in ischemic stroke and intracerebral hemorrhage, and, for some, patented (WO2006120030A1)72–74. Erythropoietin-producing hepatocellular receptor B4 (EphB4), a tyrosine kinase receptor expressed in vascular endothelial cells, plays a crucial role in vascular development and adult vascular biology, influencing blood vessel permeability, inflammation, and angiogenesis through interaction with the Notch pathway75. Drugs inhibiting PCSK9, COL6A1, or GPNMB and enhancing TFPI may hold promise for cSVD as well (Supplementary Table 28). PCSK9 is a convertase strongly linked to lipid homeostasis but also involved in neuronal apoptosis, neurogenesis, and brain inflammation76. Elevated PCSK9 levels have been associated with ischemic stroke (plasma) and AD (CSF)76. A protective effect of PCSK9 inhibitors on ischemic stroke has been demonstrated77. More recently, PCSK9 was shown to regulate amyloid beta clearance from the brain and peripheral PCSK9 inhibition reduced Aβ pathology in prefrontal cortex and hippocampus in mice78. Here, the robust association of high PCSK9 levels with larger WM-PVS burden, both in Europeans (CSF, Somascan pQTL) and East-Asians (plasma Somascan direct measurements), could suggest an association with the CAA subtype of cSVD40, characterized by Aβ deposition in the brain vasculature. The bi-directional MR result suggesting not only a putative causal association of higher PCSK9 levels with WM-PVS, but also an association of larger genetically determined WM-PVS burden with higher CSF PSCK9 levels is intriguing. Extensive WM-PVS burden is believed to reflect underlying glymphatic dysfunction, involved in impaired clearance of amyloid beta, but also other substances from the brain79.
Strengths of our study include the large-scale proteogenomics approach in plasma and CSF, using a Mendelian randomization framework that provides evidence for potential causality. The multipronged follow-up strategy across fluids and platforms strongly enhances the robustness of our findings. Although limited by smaller sample size, the extension across the lifespan and to East-Asian ancestry groups is unique and provides crucial insights on early life mechanisms underlying cSVD, while enabling transportability of findings to East-Asian populations where cSVD is particularly prevalent80. We acknowledge limitations. pQTL were derived from a population enriched in neurologically impaired individuals (especially AD patients), however we previously showed that pQTL are only marginally influenced by disease status12; moreover, follow-up samples were not enriched in AD patients. Although we have used the largest available commercial panel, discovery was limited to proteins quantified by Somascan, for which valid pQTL instruments could be derived, representing less than 10% of known proteins (without accounting for isoforms). We had no available sample for following up associations in the CSF, given the scarcity of CSF proteomics resources, and the fact that lumbar puncture is typically not done in the context of cSVD. Non-significant follow-up of associations discovered using Somascan pQTL with Olink direct plasma protein measurements may reflect spurious findings but also lack of power or modest correlation across platforms due to distinct technology. Inconsistent directionality of some significant associations between pQTL analyses and direct measurements or between both platforms requires further exploration but could reflect that distinct isoforms are being captured. Overall, these complexities highlight the importance of multiple follow-up and validation steps when interpreting association results from high-throughput proteomics assays.
Conclusion
Our work provides an extensive, first in vivo biological fingerprint of cSVD derived from large-scale proteogenomics studies in CSF and blood. The results highlight important biological processes underlying cSVD at the molecular and cellular levels, pointing to shared pathways between cSVD and AD of potential therapeutic relevance and early life mechanisms involving immunity and inflammation. This proteogenomic signature paves the way for deriving circulating biomarkers and exploring drug development and repositioning opportunities.
Methods
Discovery of protein-cSVD associations
We applied two-sample Mendelian randomization (MR) analyses to explore the relation of genetically predicted cerebrospinal fluid (CSF) and plasma protein levels with MRI-markers of cerebral small vessel disease (MRI-cSVD).
Deriving genetic instruments for circulating protein levels (instrumental variables for the exposure) using protein quantitative trait loci (pQTL)
pQTL were generated from genome-wide association studies (GWAS) of circulating protein levels. CSF pQTL summary statistics were obtained from 7,028 proteins measured on the aptamer-based Somascan 7k platform in 3,107 research participants of European ancestry. Of these, 1,076 participants were cognitively normal, 1,001 had clinically determined late-onset Alzheimer’s disease (AD), 118 had early-onset AD, 281 non-AD dementia, and 631 Parkinson’s disease.12 Plasma pQTL summary statistics were obtained from 4,907 proteins measured on the Somascan 5k platform in 35,559 cognitively normal individuals of European ancestry19 participating in either the Icelandic cancer project (52%) or deCODE genetics (48%). Cis-pQTL were defined as genetic variants within 1 Mb of the corresponding protein coding gene. Genetic variants were selected based on genome-wide significant associations (p<5×10−8) with protein abundance after clumping using PLINK281 for linkage disequilibrium at r2<0.01, within 1 Mb. Genetic variants included in the MHC region (chr6:26Mb-34Mb) were removed considering the complex LD structure of the region. The strength of the instrumental variables (IV) was measured using the F-statistic (instruments with an F-statistic > 10 were considered strong). Following these steps, we selected up to 1,121 CSF and 1,732 plasma proteins with cis-acting pQTLs; as well as 2,805 CSF and 4,614 plasma proteins with cis- and trans-acting pQTLs for MR analyses.
Genetic associations with MRI-cSVD (outcome)
We used summary statistics from the latest, largest GWAS meta-analyses of white matter hyperintensity (WMH) volume, in 48,454 participants (mean age 66.0 years), and of extensive perivascular space burden (PVS) in white matter (WM), basal ganglia (BG) and hippocampus (HIP), in 38,525 participants (mean age 68.3 years), from the general population, of European ancestry, and free of stroke (described in detail elsewhere5,20). Importantly, the cohorts from which the pQTL were derived were not included in these WMH and PVS GWAS meta-analyses.
Analytical steps for Mendelian randomization analyses
MR analysis was performed using R version 4.1.0, the “TwoSampleMR” package version 0.5.682. We applied two-sample MR analyses to assess the causal association between genetically predicted CSF and plasma protein levels and MRI-cSVD. pQTL obtained after instrument selection for each protein were used as instrumental variables (IVs). We extracted the association estimates between the variants and the exposures or the outcomes and aligned the effect alleles using the TwoSampleMR R package.
For proteins with multiple IVs we computed MR estimates with random-effect Inverse Variance Weighted (IVW) analysis83 that rely on distinct assumptions for validity: (i) Heterogeneity across the MR estimates was assessed for each instrument using Cochran’s Q statistic (p<0.05 was considered significant)83; (ii) Horizontal pleiotropy was assessed using MR-Egger intercept as a measure of directional pleiotropy (p<0.05 was considered significant)84. We further conducted various sensitivity analyses85:
The identification of outlier IVs and their removal from analyses was conducted using MR Pleiotropy residual Sum and Outlier (MR-PRESSO)86 (p<0.05 was considered significant)
Reverse MR was run by reversing the direction of inference, using the MRI-cSVD markers as the exposure and proteins as the outcome, to formally rule out reverse causation.
MR-Egger regression87 and Weighted median that are more robust to the use of pleiotropic instruments were used as sensitivity analyses. When pleiotropy was observed, we retained results when at least 2 of the 3 sensitivity methods (MR-Egger, Weighted median, MR-PRESSO) were concordant with each other and p<0.05.
For proteins with a single IV we computed MR estimates using the Wald ratio. MR analyses were followed by colocalization analyses using coloc88 including variants ±1Mb surrounding the pQTL of interest. Associations were considered significant when the posterior probability H4 (PPH4; shared association with single causal variant) was ≥0.70 and suggestive for PPH4<0.70 when posterior probability H3 (PPH3; shared association with different causal variant)<0.7089. Associations with PPH4<0.70 and PPH3 >0.70 were removed from further analyses.
Discovery MR results were considered significant when passing the FDR Benjamini-Hochberg corrected significance threshold (PFDR<0.05). In sensitivity analyses we additionally corrected for the number of independent phenotypes tested, estimated using correlations between traits in the 3C study applying the Matrix Spectral Decomposition (matSpDlite90) method for WMH volume and each PVS location, (pFDR<1.2×10−2; 0.05/4).
Genetic correlation of identified protein-cSVD
Genetic correlations were performed using LDSC to identify proteins that may have a shared genetic basis leveraging pQTL summary statistics of the 45 proteins identified in CSF and 9 proteins identified in plasma. Only proteins with heritability greater than 20% could be used (NCSF=24, Nplasma=9). (p<5×10−5 was used correcting for the mean of proteins tested and 3 situations: CSF-CSF, CSF-plasma and plasma-plasma; 0.05/18*18*3)
Follow-up of significant protein-cSVD associations
We used a multi-pronged approach to follow-up significant protein-cSVD associations, across fluids, platforms, and ancestries.
Cross-fluid follow-up (pQTL, Somascan, plasma and CSF)
Proteins identified in one fluid were followed up for association with MRI-cSVD in the other fluid. Out of CSF or plasma cSVD-associated proteins, we selected proteins for which genetic instruments were available in both datasets12,19 (N=43). Significant associations were defined by pFDR<0.05. In addition, results of sensitivity analyses at pFDR<1.2×10−2 are displayed, accounting for the 4 phenotypes tested.
Cross-platform follow-up (direct protein measurements, Olink, plasma)
Two large population-based cohort studies were used to follow-up protein-cSVD associations in participants with both MRI-cSVD phenotypes and plasma proteomic measurements, using paired nucleotide-labeled antibody probes (Olink Explore 3072): 3C-Dijon (WMH, BG-PVS, WM-PVS and HIP-PVS) and UK Biobank (WMH, BG-PVS and WM-PVS).
The 3C-Dijon study is a population-based cohort study comprising 4,931 participants aged 65 years and older at inclusion recruited between 1999 and 200191,92. A subset of 1,924 participants aged <80 years took part in an ancillary brain imaging study (1.5T Siemens Magneton scanner). Olink proteomic profiling, based on blood samples obtained at inclusion, was conducted in 1,056 participants selected based on availability of brain MRI and amounts of plasma tubes left (Supplementary Table 12). Protein measurements were conducted on the Olink Explore 3072 panel using Proximity Extension Assay (PEA) technology, following the manufacturer’s protocol93, at McGill Genome Center (Montreal, Canada). This panel measures 2,941 protein analytes and captures 2,923 unique proteins across 8 protein panels (cardiometabolic, cardiometabolic II, inflammation, inflammation II, neurology, neurology II, oncology and oncology II)94. Data pre-processing including plate-based normalization and QC checks were conducted according to standardized Olink protocols. WMH volume was estimated using a multimodal (T1, T2, DP) image processing algorithm92. PVS burden in basal ganglia and white matter was estimated with the previously described machine-learning based SHIVA-PVS algorithm1,2 using T1-weighted images; while PVS burden in hippocampus was estimated using a previously described visual semi-quantitative rating scale95.
The UK Biobank (UKB) study is a British cohort following 502,620 participants recruited between 2006 and 2010. Proteomic profiling was performed on plasma samples collected at baseline from 54,219 UKB participants using Olink Explore 3072 (field ID: 1839), with QC conducted following the protocol implemented by UKB (resource 4658). Of these, 5,494 also underwent a brain MRI (3T Siemens Prisma scanner), with WMH volume measurements (field ID: 25008), and were used for analysis. PVS burden (in basal ganglia and white matter) was estimated with the previously described machine-learning based SHIVA-PVS algorithm96,97 using T1-weighted images from the subset of 5,523 participants with proteomics data (Supplementary Methods).
We conducted multivariable linear and logistic regression of individual proteins with WMH volume and PVS burden adjusted for the delay between age at blood draw and age at the time of MRI, sex, batch effect, total intracranial volume (or mask volume for WMH in 3C-Dijon). WMH volume and PVS burden in basal ganglia and white matter were inverse normal transformed and PVS in hippocampus values were dichotomized, comparing participants in the top quartile of PVS burden distribution to the rest, as previously described5. An inverse variance weighted meta-analysis was performed using metafor R package98 to combine 3C-Dijon and UKB association analyses. The heterogeneity of associations across studies was assessed using the Cochran-Mantel-Haenszel statistical test, only associations with p>1.9×10−3 (0.05/26, correcting for 26 proteins available for follow-up) were considered. Significant associations were defined by pFDR<0.05. In addition, results of sensitivity analyses at pFDR<1.2×10−2 are displayed, accounting for the 4 phenotypes tested.
Correlation analyses between protein levels were conducted in UKB (the largest of the two samples) using the corrplot99 R package. Correlations were defined as significant at the Bonferroni corrected p-value threshold of p<7.7×10−5 (0.05/(26*26)-26).
Cross-ancestry follow-up (direct protein measurements, Somascan, plasma)
Brain imaging and plasma proteomic data from the Nagahama study, a prospective population-based cohort study initiated in 2007 in Nagahama, Japan (N=10,082 at baseline) were used100. Healthy participants (without serious physical impairment and heath issue) aged 30 to 74 years were recruited between 2008 and 2010 from the general population of Nagahama (Japan) and followed-up 5 years after baseline between 2013 and 2015. Plasma proteomic measurements have been conducted on a subset of 2,000 individuals using Somascan 4.0. Of those, 858 had brain MRI measurements. WMH in Nagahama was generated using UBO detector101, a publicly available automated tool which extracts features from T1w and FLAIR input images, such as relative intensity levels, tissue probability, and anatomical location, to classify FLAIR hyperintensities as WMH using k-Nearest Neighbor algorithm. A trained rater reviewed visual quality control report generated by the tool to reject gross failures in tissue probability estimates and WMH classification. PVS burden was estimated using the aforementioned machine-learning based SHIVA-PVS algorithm5,96. QC checks and proteomic measurements transformation (log2) were conducted according to standardized Somascan protocols. After excluding participants for whom the estimation of the MRI-marker was not possible, without proteomics measurements passing QC, with prevalent stroke, missing covariates, or who had withdrawn their consent, a total of 785 participants were available for association analyses. We conducted linear regression for WMH, WM-PVS and BG-PVS as continuous variables inverse normal transformed adjusted for age, sex, batch, total intracranial volume and the first 4 principal components. Significant associations at pFDR<0.05 were reported. Given the exploratory nature of these cross-ancestry analyses on a much small sample size, associations at p<0.05 were also reported.
Follow-up across the lifespan (pQTL, Somascan, plasma and CSF)
We conducted two-sample MR analyses using the aforementioned pQTL in plasma and CSF (instruments) and GWAS for WMH and PVS (outcomes). WMH and PVS GWAS were conducted in the Internet-based Students HeAlth Research Enterprise (i-Share) study, an ongoing prospective population-based cohort study of French-speaking students102. We specifically leveraged the bio-Share ancillary study, a biological platform comprising a collection of blood samples from a subset of the i-Share cohort, and MRi-Share, an ancillary study comprising a brain MRI (3 Tesla Siemens Prisma scanner) and a battery of cognitive tests103–105. Here we used the sub-sample of 1,748 MRi-Share and bio-Share participants aged 18–35 years, recruited in Bordeaux, France, for whom both brain MRI and genome-wide genotype data were available (mean age ± standard deviation (SD): 22.1±2.3 years; 72.2% women)105. MRI protocol, genetic data quality control and imputation procedures are detailed elsewhere5,103–105. For i-Share PVS GWAS summary statistics, we used previously described data5. For i-Share WMH GWAS summary statistics, we performed GWAS using the genome-wide linear mixed model implemented in REGENIE on WMH volume quantified using a recently developed algorithm106 (after excluding 8 participants with multiple sclerosis or radiologically isolated syndrome)107. WMH volume was transformed using an indirect inverse normal transformation (applying inverse normal transformation to residuals from linear regression of WMH adjusted for covariates [age at MRI, sex, total intracranial volume, and the first four principal components of population stratification]). These analyses were restricted to SNPs with an imputation score >0.5 and a MAF>0.01 and were adjusted for age at MRI, sex, intracranial volume and the first four principal components of population stratification.
Following the steps of instrument selection and MR previously described, we performed two-sample MR between each of the 49 proteins associated with MRI-cSVD using the large meta-analyses in older adults and MRI-cSVD measured in young adults. Associations were defined as nominally significant if p<0.05, and significant when pFDR<0.05.
Significant associations at pFDR<0.05 were reported. Given the exploratory nature of these lifespan analyses on a much small sample size, associations at p<0.05 were also reported.
Protein genetic risk scores (protein-GRS)
Quality control of genotypes and summary statistics are detailed in the Supplementary Methods.
Construction of GRS
We constructed GRS for each of the 49 cSVD-associated proteins that passed sensitivity analyses using a standard clumping and thresholding approach100,108. We used PRSice-2 software to clump SNPs with r2<0.1 using the 1000G European subset as a reference panel, and select SNPs from cis-pQTLs reaching genome-wide significance (p<5×10−8)109. A GRS for each protein was derived using the standard weighting method:
where is the count of risk alleles for the -th SNP for the -th individual, and is the effect size for the -th variant in the pQTL summary statistics.
Association analysis with extreme-cSVD
We investigated the ability of protein-GRS to predict extremes of cSVD severity (extreme-cSVD) in the 3C-Dijon cohort92. Briefly, after removing individuals with prevalent stroke, dementia, or brain tumor, we defined a binary phenotype for extreme-cSVD in 1,497 participants with MRI and genome-wide genotype data (N=58 extensive, with WMH burden in the top quartile of the cohort distribution ± presence of lacunes; 253 minimal-cSVD, with WMH burden in the bottom quartile of the cohort distribution and no lacunes or other types of brain infarcts, Supplementary methods).
We performed logistic regression of each of the standardized protein-GRS with extreme-cSVD as the dependent variable, adjusting for the first 5 principal components for population stratification110. We also used a previously derived WMH GRS (weighted sum of independent genome-wide significant risk variants for WMH volume), a strong genetic predictor of WMH volume, for comparison20. The number of SNPs in each GRS is included in Supplementary Table 19. We found five genetically determined CSF and plasma proteins nominally associated with extreme-cSVD, although none remained significant after Bonferroni-correction for the 49 protein-GRS (p<0.001).
Prediction Performance
We assessed the performance of these 5 protein-GRS to predict extreme-cSVD, individually and combined, in models adjusted for the first 5 principal components and WMH-GRS: CSF.Cystatin-M, PLASMA.PILRA-M14, CSF.PPAC, PLASMA.PILRA-deltaTM, and CSF.TLR1-ECD. As PILRA isoforms were extremely correlated (r=0.99), we selected the isoform displaying the strongest association with extreme cSVD (PILRA-M14) for the combined model. Prediction performance was evaluated in 3C-Dijon through internally validated AUC using the optimism bootstrap estimator in the caret R package (2,000 bootstrap replications)111.
Clinical significance
To explore the relation of genetically determined protein levels with clinical complications of cSVD, we used summary statistics of the largest available GWAS (European ancestry subset) for stroke and dementia. Summary statistics for any stroke, ischemic stroke, and small vessel stroke were derived from the GIGASTROKE study (comprising 73,652 patients with any stroke, 62,100 with ischemic stroke, and 6,811 with small vessel stroke112) and the largest publicly available GWAS for intracerebral hemorrhage (ICH, 1,545 patients113). For dementia we used summary statistics of the largest GWAS for Alzheimer’s disease comprising 71,880 AD cases114.
Following the steps of instrument selection and MR described above, we performed two-sample MR to test the relation of each genetically determined levels (in plasma and CSF) of the 49 cSVD associated proteins with stroke (subtypes) and dementia. To capture trends towards clinical significance we considered associations at p<0.05 and reported significant findings after multiple testing correction at pFDR<0.05.
Cross-ancestry
To assess the causal association between serum protein levels of cSVD-associated proteins and stroke, in individuals of East-Asian ancestry, we conducted two-sample MR analyses in BioBank Japan (BBJ, first cohort study115), which recruited around 200,000 participants with at least one of 47 target diseases across 66 hospitals in Japan between 2003 and 2007.. Proteomic profiling was conducted for a total of 2,886 individuals of East-Asian ancestry from two previous studies116,117 with whole genome sequencing datasets, using the Olink Explore 3072 panel following the manufacturer’s protocol. Data pre-processing, including intensity normalization, bridge normalization across batches, and QC, was conducted according to standardized Olink protocols. Rank-based inverse normal transformation was applied to protein level measurements before association tests. pQTL summary statistics of serum protein levels were obtained for 19 available proteins (out of the 49 cSVD-associated proteins from the discovery analysis) by meta-analyzing summary statistics generated in individuals from each study separately using REGENIE v3.2.9107 (adjusted for age, sex, age2,age*sex, age2*sex, batch, and the first 10 genotype principal components) and METAL118 (inverse variance weighted method; fixed effect model). Summary statistics of GWAS for ischemic stroke (N=17,493), large-artery atherosclerotic stroke (N=1,322), cardioembolic stroke (N=747), and small vessel stroke (N=4,876) were obtained in the BBJ first cohort using REGENIE v3.2.9 (adjusted for age, sex, and the first 10 genotype principal components), excluding the samples used for proteomic profiling. Genotyping, quality control, and imputation for BBJ samples used in the stroke GWASs were conducted as previously described119, except that the imputation was performed using a reference panel combining the 1000 Genome Project phase 3 v5 reference panel and 3256 Japanese samples (JEWEL3k) samples120. Individuals without any type of stroke or cerebral aneurysm were used as controls. Instrument selection and MR were conducted following the methods previously described (p-threshold for clumping: 1×10−6, Supplementary methods)
Biological interpretation
Protein-protein interactions
Protein-protein interactions were analyzed using the STRING database with the initial set of 1,121 proteins for CSF and 2,805 for plasma as background.
Pathway enrichment analysis
The GENE2FUNC analysis tool in FUMA (v1.5.4) was employed to conduct gene set enrichment analyses and detect significantly associated GO biological processes121. GENE2FUNC employs a hypergeometric test to assess the over-representation of genes within predefined gene sets, including GO biological processes. The gene IDs used correspond to coding-genes of identified proteins. We tested enrichment of the entire set of genes encoding cSVD-associated proteins identified in CSF and plasma, using the background set of genes encoding proteins tested for MR in each tissue respectively (Supplementary Table 1). Benjamini-Hochberg multiple testing correction was applied to these results (p<0.05).
STEAP enrichment analysis
We performed a cell type enrichment analysis using the Single cell Type Enrichment Analysis for Phenotypes (STEAP) tool (https://github.com/erwinerdem/STEAP/). This tool serves as an extension to CELLECT and integrates stratified LD-score regression (S-LDSC), MAGMA, and H-MAGMA for enrichment analysis. pQTLs summary statistics from the CSF and plasma datasets were preprocessed. Subsequently, expression specificity profiles were computed using single-cell RNA sequencing data from human and mouse databases, including PsychENCODE DER-22, GSE67835, GSE101601, DroNc Human Hippocampus, Allen Brain Atlas MTG and LNG, Mousebrain, Tabula Muris, Descartes Human Cerebrum, and Cerebellum. Cell type enrichment analysis was conducted employing MAGMA, H-MAGMA (which incorporates chromatin interaction profiles from human brain tissues in MAGMA), and S-LDSC. P-values were Bonferroni corrected for the number of independent cell types in each database.
Brain single cell expression quantitative trait loci (eQTL)
Mapping of brain single cell eQTL was described elsewhere26. Briefly, single-nucleus RNA-seq libraries were prepared from dorsolateral prefrontal cortex (dPFC) of 424 participants from the ROSMAP cohort using 10x Genomics Single Cell 3’ kit. Sequencing reads were processed and UMI count matrix was generated using Cell Ranger software (ver.6.0.0, 10x Genomics). Classification of cell types were performed by clustering cells by gene expression using the R package Seurat (ver. 4). “Pseudobulk” gene expression matrix was constructed by aggregating UMI counts of the same cell type of the same donor and normalizing them to the log2 counts per million reads mapped (CPM) values. Genotyping was performed by whole genome sequencing and GATK. Mapping of cis-eQTL was performed using Matrix-eQTL (ver. 2.3) for SNP within 1 Mb from transcription start sites.
Due to the sparsity of vascular cells in brain tissue, a specific dataset from ROSMAP using in silico vasculature enrichment was used for eQTL and expression analysis. Single-nucleus RNA-seq libraries were prepared from brain samples of 409 ROSMAP participants using the 10x Genomics Single Cell 3′ Kit. Read counts were estimated using Cellranger 3.0.1 (10x Genomics) and the UMI count matrix was analysed using the Seurat R package v.3.2.0. Vascular enrichment was conducted in silico using cell sorting from post mortem human samples across seven different brain regions (prefrontal cortex, mid-temporal cortex, angular gyrus, entorhinal cortex, thalamus, hippocampus and mammillary body). Cell-type annotation was performed through clustering, annotating cell-type using a combination of canonical vasculature markers and whole-transcriptome cellular signatures. Detailed methods regarding sRNAseq and in silico vascular enrichment is described elsewhere27,28. Microglia states were defined from 152,459 microglial transcriptomes across 443 individuals (217 AD and 226 controls) identifying 12 transcriptional states. Microglial nuclei were obtained from post-mortem brain samples from the ROSMAP study across 6 brain regions (hippocampus, dPFC, mid-temporal cortex, angular gyrus, entorhinal cortex and thalamus). Using in silico sorting, 174,420 immune cells were collected from snRNA-seq datasets using STAR method forming 12 clusters of microglia. Those clusters were then defined as microglia states based on their molecular signature and function: MG0: hemostatic, MG1: neuronal surveillance, MG2: Inflammatory I, MG3: Ribosome biogenesis, MG4: Lipid Processing, MG5: Phagocytic, MG6: Stress signature, MG7: Glycolytic; MG8: Inflammatory II, MG10: Inflammatory III, MG11: Antiviral, MG12: Cycling. Detailed methods regarding microglial states definition are described elsewhere29.
Proteomics driven drug discovery
Using significant MR results from CSF and plasma, we restricted our analysis to drug-targeting proteins using 4 drug-gene databases (ChEMBL, pharmGKB, DrugBank and TTD). Following this methodology, eight drug-targeting proteins were identified for WMH (EPO, LTF, TFPI, APOE, ARSB, CTSS, CTSB and EPHB4) and seven for PVS (COL6A1, CTSB, GPNMB, PCSK9, FcRIIIA, Heparin co-factor II, IL6). Using public drug databases, we then curated drugs targeting those proteins in a direction compatible with a beneficial therapeutic effect against the corresponding cSVD phenotype based on MR estimates. The desired mode of action (MoA) was defined as the opposite direction of the MR estimate. Once the drugs were identified, we searched the literature for a potential action of the drug.
Acknowledgements
This project is supported by a grant overseen by the French National Research Agency (ANR) as part of the “Investment for the Future Programme” ANR-18-RHUS-0002 and by the Precision and Global Vascular Brain Health Institute (VBHI) funded by the France 2030 IHU3 initiative. The project also received funding from the French National Research Agency (ANR) through the SHIVA project. Computations were performed on the Bordeaux Bioinformatics Center (CBiB) and the CREDIM computer resources, University of Bordeaux. Funding support for additional computer resources has been provided to S.D. by the Fondation Claude Pompidou. The i-Share study has received funding by the French National Agency (Agence Nationale de la Recherche, ANR), via the Investment for the Future program (grant nos. ANR-10-COHO-05 and ANR-18-RHUS-0002) and from the University of Bordeaux Initiative of Exellence (IdEX). The Three City (3C) Study is conducted under a partnership agreement among the Institut National de la Santé et de la Recherche Médicale (INSERM), the University of Bordeaux, and Sanofi-Aventis. The Fondation pour la Recherche Médicale funded the preparation and initiation of the study. The 3C Study is also supported by the Caisse Nationale Maladie des Travailleurs Salariés, Direction Générale de la Santé, Mutuelle Générale de l’Education Nationale (MGEN), Institut de la Longévité, Conseils Régionaux of Aquitaine and Bourgogne, Fondation de France, and Ministry of Research–INSERM Programme “Cohortes et collections de données biologiques.”
We thank all the participants and their families, as well as the many involved institutions and their staff. This work was supported by grants from the National Institutes of Health (R01AG044546 (CC), P01AG003991 (CC, JCM), RF1AG053303 (CC), RF1AG058501 (CC), U01AG058922 (CC), RF1AG074007, R00AG062723, the Chan Zuckerberg Initiative (CZI), the Michael J. Fox Foundation (CC), and the Alzheimer’s Association Zenith Fellows Award (ZEN-22–848604, awarded to CC). GSK provided funding to support the analyses performed in this study. The recruitment and clinical characterization of research participants at Washington University were supported by NIH P30AG066444, P01AG03991, and P01AG026276. This work was supported by access to equipment made possible by the Hope Center for Neurological Disorders, the NeuroGenomics and Informatics Center (NGI: https://neurogenomics.wustl.edu/) and the Departments of Neurology and Psychiatry at Washington University School of Medicine.
This work was supported by grants from the National Institutes of Health (R01AG044546 (CC), P01AG003991 (CC, JCM), RF1AG053303 (CC), RF1AG058501 (CC), U01AG058922 (CC), RF1AG074007 (YJS), R00AG062723 (LI), P30 AG066515 (TWC, MDG), the Chan Zuckerberg Initiative (CZI), the Michael J. Fox Foundation (LI, CC), the Department of Defense (LI- W81XWH2010849), the Alzheimer’s Association Zenith Fellows Award (ZEN-22–848604, awarded to CC), and the Bright Focus Foundation (A2021033S, LI).
The recruitment and clinical characterization of research participants at Washington University were supported by NIH P30AG066444 (JCM), P01AG03991 (JCM), and P01AG026276 (JCM). This work was supported by access to equipment made possible by the Hope Center for Neurological Disorders, the NeuroGenomics and Informatics Center (NGI: https://neurogenomics.wustl.edu/) and the Departments of Neurology and Psychiatry at Washington University School of Medicine.
The Genome Research @ Ace Alzheimer Center Barcelona project (GR@ACE) is supported by Grifols SA, Fundación bancaria ‘La Caixa’, Ace Alzheimer Center Barcelona and CIBERNED. Ace Alzheimer Center Barcelona is one of the participating centers of the Dementia Genetics Spanish Consortium (DEGESCO). The FACEHBI study is supported by funds from Ace Alzheimer Center Barcelona, Grifols, Life Molecular Imaging, Araclon Biotech, Alkahest, Laboratorio de análisis Echevarne and IrsiCaixa. Authors acknowledge the support of the Spanish Ministry of Science and Innovation, Proyectos de Generación de Conocimiento grants PID2021-122473OA-I00, PID2021-123462OB-I00 and PID2019-106625RB-I00. ISCIII, Acción Estratégica en Salud, integrated in the Spanish National R+D+I Plan and financed by ISCIII Subdirección General de Evaluación and the Fondo Europeo de Desarrollo Regional (FEDER “Una manera de hacer Europa”) grants PI13/02434, PI16/01861, PI17/01474, PI19/00335, PI19/01240, PI19/01301, PI22/01403, PI22/00258 and the ISCIII national grant PMP22/00022, funded by the European Union (NextGenerationEU). The support of CIBERNED (ISCIII) under the grants CB06/05/2004 and CB18/05/00010. The support from the ADAPTED and MOPEAD projects, European Union/EFPIA Innovative Medicines Initiative Joint (grant numbers 115975 and 115985, respectively); from PREADAPT project, Joint Program for Neurodegenerative Diseases (JPND) grant N° AC19/00097; from HARPONE project, Agency for Innovation and Entrepreneurship (VLAIO) grant N° PR067/21 and Janssen. DESCARTES project is funded by German Research Foundation (DFG).
Cardiovascular Health Study: This CHS research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, HHSN268201800001C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, 75N92021D00006; and NHLBI grants U01HL080295, R01HL087652, R01HL105756, R01HL103612, R01HL120393, and U01HL130114 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through R01AG023629 from the National Institute on Aging (NIA). A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. This research has been conducted using the UK Biobank Resource under applications no. 94113 and no. 18545.
I.C was supported by the Digital Public Health Graduate Program (DPH), a PhD program supported by the French Investment for the Future Program (grant no. 17-EURE-0019). S.N. was supported by Takeda Science Foundation. Y.O. was supported by JSPS KAKENHI (22H00476), AMED (JP21gm4010006, JP22km0405211, JP22ek0410075, JP22km0405217, JP22ek0109594, JP223fa627002, JP223fa627010, JP233fa627011, JP23zf0127008), JST Moonshot R&D (JPMJMS2021, JPMJMS2024), Takeda Science Foundation, Bioinformatics Initiative of Osaka University Graduate School of Medicine, Institute for Open and Transdisciplinary Research Initiatives and Center for Infectious Disease Education and Research (CiDER), and Center for Advanced Modality and DDS (CAMaD), Osaka University. PMM gratefully acknowledges personal support from the Edmond J Safra Foundation and Lily Safra and receipt of an NIHR Senior Investigator Award. His research is supported by the UK Dementia Research Institute, which is funded primarily by the UKRI Medical Research Council, and by the Imperial College Healthcare Trust NIHR Biomedical Research Centre. L.L. acknowledge Intramural Research Program, National Institutes on Aging.
Support for title page creation and format was provided by AuthorArranger, a tool developed at the National Cancer Institute.
Footnotes
Competing interests
C.C. has received research support from GSK and EISAI and is a member of the advisory board of Circular Genomics and owns stocks in this company. CC is part of the scientific advisory board for ADmit. B.P. serves on the Steering Committee of the Yale Open Data Project funded by Johnson & Johnson. P.M.M. has received an honourarium as Chair of the UKRI Medical Research Council Neuroscience and Mental Health Board until March 2024. He acknowledges consultancy fees from, Biogen, Sudo. Nimbus and GSK. He has received speakers’ honoraria from Sanofi and Redburn, and has received research or educational funds from Biogen, Merck, Bristol Meyers Squibb and Nimbus. J.W declares no commercial COI; various academic research grants and is CI for LACunar Intervention Trials.
The authors declared no potential conflicts of interest with respect to research, authorship, and/or publication of this article. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Code availability
We used publicly available tools from TwoSampleMR (https://mrcieu.github.io/TwoSampleMR/), FUMA (https://fuma.ctglab.nl/), coloc R package (https://chr1swallace.github.io/coloc/), STRING (https://string-db.org/), FUSION (https://github.com/gusevlab/fusion_twas), PLINK2 (https://www.cog-genomics.org/plink/2.0/), OlinkAnalyze R package (https://github.com/Olink-Proteomics/OlinkRPackage), MungeSumstats R package (https://github.com/neurogenomics/MungeSumstats), ggplot2 R package (https://github.com/tidyverse/ggplot2), forestplot R package (https://cran.r-project.org/web/packages/forestplot), Betsholtz database (http://betsholtzlab.org/VascularSingleCells/database.html), metafor R package (https://wviechtb.github.io/metafor/), SHIVA-PVS algorithm (https://github.com/pboutinaud/SHIVA_PVS, T1.PVS/v1), STEAP (https://github.com/erwinerdem/STEAP/). Figures were created using ggplot2 R package (https://ggplot2.tidyverse.org/) and BioRender (https://app.biorender.com).
Contributor Information
Stephanie Debette, University of Bordeaux.
Ilana Caro, University of Bordeaux.
Daniel Western, Department of Psychiatry, Washington University School of Medicine, St. Louis, MO, USA.
Shinichi Namba, Osaka University.
Na Sun, MIT Computer Science and Artificial Intelligence Laboratory; Broad Institute of MIT and Harvard.
Shuji Kawaguchi, Kyoto University Graduate School of Medicine.
Yunye He, Graduate School of Frontier Sciences, The University of Tokyo.
Masashi Fujita, Columbia University Irving Medical Center.
Gennady Roshchupkin, Erasmus Medical Center.
Tim D’Aoust, Bordeaux Population Health, Inserm U1219, University of Bordeaux.
Marie-Gabrielle Duperron, University of Bordeaux, Inserm, Bordeaux Population Health Research Center, UMR 1219.
Murali Sargurupremraj, University of Bordeaux, Inserm, Bordeaux Population Health Research Center, team VINTAGE, UMR 1219, F-33000 Bordeaux, France; Glenn Biggs Institute for Alzheimer’s & Neurodegenerative Diseases Unive.
Masaru Koido, Graduate School of Frontier Sciences, The University of Tokyo.
Marziehsadat Ahmadi, Victor Phillip Dahdaleh Institute of Genomic Medicine, McGill University.
Chengran Yang, Washington University in St. Louis.
Jigyasha Timsina, Department of Psychiatry, Washington University School of Medicine, St. Louis, MO, USA.
Laura Ibanez, Washington University in St. Louis.
Koichi Matsuda, Department of Computational Biology and Medical Sciences, Graduate school of Frontier Sciences, The University of Tokyo.
Yutaka Suzuki, The University of Tokyo.
Yoshiya Oda, Graduate School of Medicine, The University of Tokyo.
Akinori Kanai, The University of Tokyo.
Hans Markus Munter, McGill University.
Dan Auld, Victor Phillip Dahdaleh Institute of Genomic Medicine, McGill University.
Iana Astafeva, Bordeaux Population Health, Inserm U1219, University of Bordeaux; Institute of Neurodegenerative Diseases.
Raquel Puerta, Ace Alzheimer Center Barcelona.
Jerome Rotter, The Lundquist Institute for Biomedical Innovation at Harbor-UCLA Medical Center.
Bruce Psaty, Cardiovascular Health Research Unit.
Joshua Bis, University of Washington.
Will Longstreth, University of Washington.
Thierry Couffinhal, University of Bordeaux, The clinical unit of Exploration, Prevention and Care Center for Atherosclerosis (CEPTA), CHUB, Inserm U1034.
Pablo Garcia-Gonzalez, Ace Alzheimer Center Barcelona.
Vanesa Pytel, Ace Alzheimer Center Barcelona, Universitat Internacional de Catalunya; CIBERNED, Network Center for Biomedical Research in Neurodegenerative Diseases, National Institute of Health Carlos III.
Marta Marquié, ACE Alzheimer Center Barcelona.
Amanda Cano, Ace Alzheimer Center Barcelona.
Mercè Boada, Universitat Internacional de Catalunya.
Marc Joliot, GIN, IMN/UMR5293 UB/CNRS/CEA.
Mark Lathrop, Department of Human Genetics, McGill University, 1205 Dr Penfield Avenue, Montreal, QC, H3A 1B1, Canada.
Quentin Le Grand, University of Bordeaux, Inserm, Bordeaux Population Health Research Center, UMR 1219.
Lenore Launer, National Institute on Aging, National Institutes of Health.
Joanna Wardlaw, University of Edinburgh.
Myriam Heiman, Massachusetts Institute of Technology.
Agustin Ruiz, Glenn Biggs Institute for Alzheimer’s and Neurodegenerative Diseases, University of Texas Health Sciences Center; Ace Alzheimer Center Barcelona, Universitat Internacional de Catalunya;CIBERN.
Paul Matthews, UK Dementia Research Institute Centre at Imperial College London.
Sudha Seshadri, University of Texas Health Science Center.
Myriam Fornage, 1. Institute of Molecular Medicine, McGovern Medical School, The University of Texas Health Science Center 2. Human Genetics Center, Department of Epidemiology, School of Public Health.
Hieab Adams, Department of Human Genetics, Radboud University Medical Center; Latin American Brain Health (BrainLat), Universidad Adolfo Ibáñez.
Aniket Mishra, University of Bordeaux.
David-Alexandre Trégouët, INSERM.
Yukinori Okada, Department of Genome Informatics, Graduate School of Medicine, The Univ. of Tokyo; Department of Statistical Genetics, Osaka Univ. Graduate School of Medicine; Laboratory for Systems Genetic, RIKEN.
Manolis Kellis, MIT.
Philip De Jager, Columbia University Irving Medical Center.
Yoichiro Kamatani, The University of Tokyo.
Fumihiko Matsuda, Kyoto University Graduate School of Medicine.
Carlos Cruchaga, Washington University.
Data availability
We used publicly available data for analyses described in this manuscript, including data from GWAS catalog (https://www.ebi.ac.uk/gwas/, study code: GCST90244151, GCST011947, GCST007320, GCST90104539, GCST90162546), the DECODE project (https://www.decode.com/summarydata/), ChEMBL (https://www.ebi.ac.uk/chembl/), pharmGKB (https://www.pharmgkb.org/), DrugBank (https://go.drugbank.com/), TTD (https://db.idrblab.net/ttd/), CSF pQTL summary statistics available at NIAGADS (full summary statistics available to approved investigators through accession #NG00130), NeuroGenomics and Informatics Center website (https://neurogenomics.wustl.edu/open-science/raw-data/) and ONTIME browser (https://ontime.wustl.edu/).
References
- 1.Wardlaw J. M. et al. ESO Guideline on covert cerebral small vessel disease. Eur. Stroke J. 6, CXI–CLXII (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duering M. et al. Neuroimaging standards for research into small vessel disease-advances since 2013. Lancet Neurol. 22, 602–618 (2023). [DOI] [PubMed] [Google Scholar]
- 3.Wardlaw J. M. et al. Vascular risk factors, large-artery atheroma, and brain white matter hyperintensities. Neurology 82, 1331–1338 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rusina P. V. et al. Genetic support for FDA-approved drugs over the past decade. Nat. Rev. Drug Discov. 22, 864 (2023). [DOI] [PubMed] [Google Scholar]
- 5.Duperron M.-G. et al. Genomics of perivascular space burden unravels early mechanisms of cerebral small vessel disease. Nat. Med. 29, 950–962 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bordes C., Sargurupremraj M., Mishra A. & Debette S. Genetics of common cerebral small vessel disease. Nat. Rev. Neurol. 18, 84–101 (2022). [DOI] [PubMed] [Google Scholar]
- 7.Montaner J. et al. Multilevel omics for the discovery of biomarkers and therapeutic targets for stroke. Nat. Rev. Neurol. 16, 247–264 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Walker K. A. et al. Large-scale plasma proteomic analysis identifies proteins and pathways associated with dementia risk. Nat. Aging 1, 473–489 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker K. A. et al. Proteomics analysis of plasma from middle-aged adults identifies protein markers of dementia risk in later life. Sci. Transl. Med. 15, eadf5681 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L. et al. Systematic Mendelian randomization using the human plasma proteome to discover potential therapeutic targets for stroke. Nat. Commun. 13, 6143 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dammer E. B. et al. Multi-platform proteomic analysis of Alzheimer’s disease cerebrospinal fluid and plasma reveals network biomarkers associated with proteostasis and the matrisome. Alzheimers Res. Ther. 14, 174 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruchaga C. et al. Proteogenomic analysis of human cerebrospinal fluid identifies neurologically relevant regulation and informs causal proteins for Alzheimer’s disease. Res. Sq. rs.3.rs-2814616 (2023) doi: 10.21203/rs.3.rs-2814616/v1. [DOI] [PubMed] [Google Scholar]
- 13.Yang C. et al. Genomic atlas of the proteome from brain, CSF and plasma prioritizes proteins implicated in neurological disorders. Nat. Neurosci. 24, 1302–1312 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuipers S. et al. A cluster of blood-based protein biomarkers reflecting coagulation relates to the burden of cerebral small vessel disease. J. Cereb. Blood Flow Metab. Off. J. Int. Soc. Cereb. Blood Flow Metab. 42, 1282–1293 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan S. et al. Plasma inflammatory biomarkers in cerebral small vessel disease: A review. CNS Neurosci. Ther. 29, 498–515 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fornage M. et al. Biomarkers of Inflammation and MRI-Defined Small Vessel Disease of the Brain: The Cardiovascular Health Study. Stroke 39, 1952–1959 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satizabal C. L., Zhu Y. C., Mazoyer B., Dufouil C. & Tzourio C. Circulating IL-6 and CRP are associated with MRI findings in the elderly: the 3C-Dijon Study. Neurology 78, 720–727 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Jiménez-Balado J. et al. New candidate blood biomarkers potentially associated with white matter hyperintensities progression. Sci. Rep. 11, 14324 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferkingstad E. et al. Large-scale integration of the plasma proteome with genetics and disease. Nat. Genet. 53, 1712–1721 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Sargurupremraj M. et al. Cerebral small vessel disease genomics and its implications across the lifespan. Nat. Commun. 11, 6285 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho B. P. H. et al. Association of Vascular Risk Factors and Genetic Factors With Penetrance of Variants Causing Monogenic Stroke. JAMA Neurol. 79, 1303–1311 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mishra A. et al. Association of variants in HTRA1 and NOTCH3 with MRI-defined extremes of cerebral small vessel disease in older subjects. Brain J. Neurol. 142, 1009–1023 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mishra A. et al. Gene-mapping study of extremes of cerebral small vessel disease reveals TRIM47 as a strong candidate. Brain J. Neurol. 145, 1992–2007 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Consortium ReproGen et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eldjarn G. H. et al. Large-scale plasma proteomics comparisons through genetics and disease associations. Nature 622, 348–358 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujita M. et al. Cell subtype-specific effects of genetic variation in the Alzheimer’s disease brain. Nat. Genet. 56, 605–614 (2024). [DOI] [PubMed] [Google Scholar]
- 27.Garcia F. J. et al. Single-cell dissection of the human brain vasculature. Nature 603, 893–899 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun N. et al. Single-nucleus multiregion transcriptomic analysis of brain vasculature in Alzheimer’s disease. Nat. Neurosci. 26, 970–982 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun N. et al. Human microglial state dynamics in Alzheimer’s disease progression. Cell 186, 4386–4403.e29 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pokhilko A. et al. Global proteomic analysis of extracellular matrix in mouse and human brain highlights relevance to cerebrovascular disease. J. Cereb. Blood Flow Metab. 41, 2423–2438 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joutel A., Haddad I., Ratelade J. & Nelson M. T. Perturbations of the cerebrovascular matrisome: A convergent mechanism in small vessel disease of the brain? J. Cereb. Blood Flow Metab. Off. J. Int. Soc. Cereb. Blood Flow Metab. 36, 143–157 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiménez-Balado J. et al. New candidate blood biomarkers potentially associated with white matter hyperintensities progression. Sci. Rep. 11, 14324 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gertje E. C. et al. Associations Between CSF Markers of Inflammation, White Matter Lesions, and Cognitive Decline in Individuals Without Dementia. Neurology 100, e1812–e1824 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haffner C. Proteostasis in Cerebral Small Vessel Disease. Front. Neurosci. 13, 1142 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaetani L. et al. CSF and Blood Biomarkers in Neuroinflammatory and Neurodegenerative Diseases: Implications for Treatment. Trends Pharmacol. Sci. 41, 1023–1037 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Luebke M., Parulekar M. & Thomas F. P. Fluid biomarkers for the diagnosis of neurodegenerative diseases. Biomark. Neuropsychiatry 8, 100062 (2023). [Google Scholar]
- 37.Robey T. T. & Panegyres P. K. Cerebrospinal fluid biomarkers in neurodegenerative disorders. Future Neurol. 14, FNL6 (2019). [Google Scholar]
- 38.Park Y. H. et al. Association of blood-based transcriptional risk scores with biomarkers for Alzheimer disease. Neurol. Genet. 6, e517 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fournier N. et al. FDF03, a Novel Inhibitory Receptor of the Immunoglobulin Superfamily, Is Expressed by Human Dendritic and Myeloid Cells. J. Immunol. 165, 1197–1209 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Charidimou A. et al. The Boston criteria version 2.0 for cerebral amyloid angiopathy: a multicentre, retrospective, MRI–neuropathology diagnostic accuracy study. Lancet Neurol. 21, 714–725 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greenberg S. M. et al. Cerebral amyloid angiopathy and Alzheimer disease — one peptide, two pathways. Nat. Rev. Neurol. 16, 30–42 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rathore N. et al. Paired Immunoglobulin-like Type 2 Receptor Alpha G78R variant alters ligand binding and confers protection to Alzheimer’s disease. PLOS Genet. 14, e1007427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X. et al. Arylsulfatase B modulates neurite outgrowth via astrocyte chondroitin-4-sulfate: dysregulation by ethanol. Glia 62, 259–271 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanlandewijck M. et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018). [DOI] [PubMed] [Google Scholar]
- 45.He L. et al. Single-cell RNA sequencing of mouse brain and lung vascular and vessel-associated cell types. Sci. Data 5, 180160 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valayannopoulos V., Nicely H., Harmatz P. & Turbeville S. Mucopolysaccharidosis VI. Orphanet J. Rare Dis. 5, 5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hook G. et al. Cathepsin B Gene Knockout Improves Behavioral Deficits and Reduces Pathology in Models of Neurologic Disorders. Pharmacol. Rev. 74, 600–629 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hook G., Kindy M. & Hook V. Cathepsin B Deficiency Improves Memory Deficits and Reduces Amyloid-β in hAβPP Mouse Models Representing the Major Sporadic Alzheimer’s Disease Condition. J. Alzheimers Dis. 93, 33–46 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bugiani M. et al. Cathepsin A–related arteriopathy with strokes and leukoencephalopathy (CARASAL). Neurology 87, 1777–1786 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Mancuso M. et al. Monogenic cerebral small-vessel diseases: diagnosis and therapy. Consensus recommendations of the European Academy of Neurology. Eur. J. Neurol. 27, 909–927 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Malik R. et al. Whole-exome sequencing reveals a role of HTRA1 and EGFL8 in brain white matter hyperintensities. Brain J. Neurol. 144, 2670–2682 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dichgans M. et al. Genetically proxied HTRA1 protease activity and circulating levels independently predict risk of ischemic stroke and coronary artery disease. Res. Sq. rs.3.rs-3523612 (2023) doi: 10.21203/rs.3.rs-3523612/v1. [DOI] [PubMed] [Google Scholar]
- 53.Low A., Mak E., Rowe J. B., Markus H. S. & O’Brien J. T. Inflammation and cerebral small vessel disease: A systematic review. Ageing Res. Rev. 53, 100916 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Low A. et al. In vivo neuroinflammation and cerebral small vessel disease in mild cognitive impairment and Alzheimer’s disease. J. Neurol. Neurosurg. Psychiatry 92, 45–52 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Evans L. E. et al. Cardiovascular comorbidities, inflammation, and cerebral small vessel disease. Cardiovasc. Res. cvab284 (2021) doi: 10.1093/cvr/cvab284. [DOI] [PubMed] [Google Scholar]
- 56.Fu Y. & Yan Y. Emerging Role of Immunity in Cerebral Small Vessel Disease. Front. Immunol. 9, 67 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Y. et al. Epigenetic and integrative cross-omics analyses of cerebral white matter hyperintensities on MRI. Brain 146, 492–506 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Solé-Guardia G. et al. Association between hypertension and neurovascular inflammation in both normal-appearing white matter and white matter hyperintensities. Acta Neuropathol. Commun. 11, 2 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gjoneska E. et al. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature 518, 365–369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mendiola A. S. et al. Defining blood-induced microglia functions in neurodegeneration through multiomic profiling. Nat. Immunol. 24, 1173–1187 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Safaiyan S. et al. White matter aging drives microglial diversity. Neuron 109, 1100–1117.e10 (2021). [DOI] [PubMed] [Google Scholar]
- 62.Leng F. & Edison P. Neuroinflammation and microglial activation in Alzheimer disease: where do we go from here? Nat. Rev. Neurol. 17, 157–172 (2021). [DOI] [PubMed] [Google Scholar]
- 63.Kosciuk T. et al. NMT1 and NMT2 are lysine myristoyltransferases regulating the ARF6 GTPase cycle. Nat. Commun. 11, 1067 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen X., Lu W. & Wu D. Sirtuin 2 (SIRT2): Confusing Roles in the Pathophysiology of Neurological Disorders. Front. Neurosci. 15, 614107 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu B. et al. The role of SIRT2 in vascular-related and heart-related diseases: A review. J. Cell. Mol. Med. 25, 6470–6478 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Persyn E. et al. Genome-wide association study of MRI markers of cerebral small vessel disease in 42,310 participants. Nat. Commun. 11, 2175 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pedrero-Prieto C. M. et al. A comprehensive systematic review of CSF proteins and peptides that define Alzheimer’s disease. Clin. Proteomics 17, 21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martínez-Estrada O. M. et al. Erythropoietin protects the in vitro blood-brain barrier against VEGF-induced permeability. Eur. J. Neurosci. 18, 2538–2544 (2003). [DOI] [PubMed] [Google Scholar]
- 69.Zakharova E. T. et al. Erythropoietin and Nrf2: key factors in the neuroprotection provided by apo-lactoferrin. BioMetals 31, 425–443 (2018). [DOI] [PubMed] [Google Scholar]
- 70.Spagnuolo P. A. & Hoffman-Goetz L. Dietary Lactoferrin Does Not Prevent Dextran Sulfate Sodium Induced Murine Intestinal Lymphocyte Death. Exp. Biol. Med. 233, 1099–1108 (2008). [DOI] [PubMed] [Google Scholar]
- 71.Van De Looij Y. et al. Lactoferrin during lactation protects the immature hypoxic-ischemic rat brain. Ann. Clin. Transl. Neurol. 1, 955–967 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mercier M. International approach to the assessment of chemical risks. Sci. Total Environ. 101, 1–7 (1991). [DOI] [PubMed] [Google Scholar]
- 73.Zhao X. et al. Optimized lactoferrin as a highly promising treatment for intracerebral hemorrhage: Pre-clinical experience. J. Cereb. Blood Flow Metab. Off. J. Int. Soc. Cereb. Blood Flow Metab. 41, 53–66 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kittur F. S., Hung C.-Y., Li P. A., Sane D. C. & Xie J. Asialo-rhuEPO as a Potential Neuroprotectant for Ischemic Stroke Treatment. Pharm. Basel Switz. 16, 610 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou B. et al. Notch signaling pathway: architecture, disease, and therapeutics. Signal Transduct. Target. Ther. 7, 95 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jaafar A. K., Techer R., Chemello K., Lambert G. & Bourane S. PCSK9 and the nervous system: a no-brainer? J. Lipid Res. 64, 100426 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Giugliano R. P. et al. Stroke Prevention With the PCSK9 (Proprotein Convertase Subtilisin-Kexin Type 9) Inhibitor Evolocumab Added to Statin in High-Risk Patients With Stable Atherosclerosis. Stroke 51, 1546–1554 (2020). [DOI] [PubMed] [Google Scholar]
- 78.Mazura A. D. et al. PCSK9 acts as a key regulator of Aβ clearance across the blood-brain barrier. Cell. Mol. Life Sci. CMLS 79, 212 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mestre H., Kostrikov S., Mehta R. I. & Nedergaard M. Perivascular spaces, glymphatic dysfunction, and small vessel disease. Clin. Sci. Lond. Engl. 1979 131, 2257–2274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mok V. et al. Race-ethnicity and cerebral small vessel disease--comparison between Chinese and White populations. Int. J. Stroke Off. J. Int. Stroke Soc. 9 Suppl A100, 36–42 (2014). [DOI] [PubMed] [Google Scholar]
References
- 81.Chang C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hemani G., Tilling K. & Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 13, e1007081 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Burgess S., Butterworth A. & Thompson S. G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Burgess S. & Thompson S. G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 32, 377–389 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Burgess S. et al. Guidelines for performing Mendelian randomization investigations. Wellcome Open Res. 4, 186 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Verbanck M., Chen C.-Y., Neale B. & Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bowden J., Davey Smith G. & Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Giambartolomei C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cronjé H. T. et al. Genetic evidence implicating natriuretic peptide receptor-3 in cardiovascular disease risk: a Mendelian randomization study. BMC Med. 21, 158 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li J. & Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 95, 221–227 (2005). [DOI] [PubMed] [Google Scholar]
- 91.Godin O. et al. White matter lesions as a predictor of depression in the elderly: the 3C-Dijon study. Biol. Psychiatry 63, 663–669 (2008). [DOI] [PubMed] [Google Scholar]
- 92.C Study Group. Vascular factors and risk of dementia: design of the Three-City Study and baseline characteristics of the study population. Neuroepidemiology 22, 316–325 (2003). [DOI] [PubMed] [Google Scholar]
- 93.Lind L. et al. Use of a proximity extension assay proteomics chip to discover new biomarkers for human atherosclerosis. Atherosclerosis 242, 205–210 (2015). [DOI] [PubMed] [Google Scholar]
- 94.Sun B. B. et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature 622, 329–338 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhu Y.-C. et al. Frequency and location of dilated Virchow-Robin spaces in elderly people: a population-based 3D MR imaging study. AJNR Am. J. Neuroradiol. 32, 709–713 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boutinaud P. et al. 3D Segmentation of Perivascular Spaces on T1-Weighted 3 Tesla MR Images With a Convolutional Autoencoder and a U-Shaped Neural Network. Front. Neuroinformatics 15, 641600 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Duperron M.-G. et al. Genomics of perivascular space burden unravels early mechanisms of cerebral small vessel disease. Nat. Med. 29, 950–962 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Viechtbauer W. Conducting Meta-Analyses in R with the metafor Package. J. Stat. Softw. 36, (2010). [Google Scholar]
- 99.Friendly M. Corrgrams: Exploratory Displays for Correlation Matrices. Am. Stat. 56, 316–324 (2002). [Google Scholar]
- 100.Funada S. et al. Longitudinal Analysis of Bidirectional Relationships between Nocturia and Depressive Symptoms: The Nagahama Study. J. Urol. 203, 984–990 (2020). [DOI] [PubMed] [Google Scholar]
- 101.Jiang J. et al. UBO Detector – A cluster-based, fully automated pipeline for extracting white matter hyperintensities. NeuroImage 174, 539–549 (2018). [DOI] [PubMed] [Google Scholar]
- 102.Montagni I., Guichard E. & Kurth T. Association of screen time with self-perceived attention problems and hyperactivity levels in French students: a cross-sectional study. BMJ Open 6, e009089 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tsuchida A. et al. Age-Related Variations in Regional White Matter Volumetry and Microstructure During the Post-adolescence Period: A Cross-Sectional Study of a Cohort of 1,713 University Students. Front. Syst. Neurosci. 15, 692152 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tsuchida A. et al. The MRi-Share database: brain imaging in a cross-sectional cohort of 1870 university students. Brain Struct. Funct. 226, 2057–2085 (2021). [DOI] [PubMed] [Google Scholar]
- 105.Le Grand Q. et al. Genomic Studies Across the Lifespan Point to Early Mechanisms Determining Subcortical Volumes. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 7, 616–628 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tsuchida A. et al. Early detection of white matter hyperintensities using SHIVA-WMH detector. Hum. Brain Mapp. (2023) doi: 10.1002/hbm.26548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mbatchou J. et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 53, 1097–1103 (2021). [DOI] [PubMed] [Google Scholar]
- 108.Choi S. W., Mak T. S.-H. & O’Reilly P. F. Tutorial: a guide to performing polygenic risk score analyses. Nat. Protoc. 15, 2759–2772 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Choi S. W. & O’Reilly P. F. PRSice-2: Polygenic Risk Score software for biobank-scale data. GigaScience 8, giz082 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Price A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- 111.Kuhn M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 28, (2008). [Google Scholar]
- 112.Mishra A. et al. Stroke genetics informs drug discovery and risk prediction across ancestries. Nature 611, 115–123 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Woo D. et al. Meta-analysis of genome-wide association studies identifies 1q22 as a susceptibility locus for intracerebral hemorrhage. Am. J. Hum. Genet. 94, 511–521 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jansen I. E. et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51, 404–413 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nagai A. et al. Overview of the BioBank Japan Project: Study design and profile. J. Epidemiol. 27, S2–S8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Koyama S. et al. Population-specific and trans-ancestry genome-wide analyses identify distinct and shared genetic risk loci for coronary artery disease. Nat. Genet. 52, 1169–1177 (2020). [DOI] [PubMed] [Google Scholar]
- 117.Liu X. et al. Decoding triancestral origins, archaic introgression, and natural selection in the Japanese population by whole-genome sequencing. Sci. Adv. 10, eadi8419 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Willer C. J., Li Y. & Abecasis G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinforma. Oxf. Engl. 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.He Y. et al. East Asian-specific and cross-ancestry genome-wide meta-analyses provide mechanistic insights into peptic ulcer disease. Nat. Genet. 55, 2129–2138 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Terao C. et al. Population-specific reference panel improves imputation quality and enhances locus discovery and fine-mapping. Preprint at 10.21203/rs.3.rs-3194976/v1 (2023). [DOI]
- 121.Watanabe K., Taskesen E., van Bochoven A. & Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We used publicly available data for analyses described in this manuscript, including data from GWAS catalog (https://www.ebi.ac.uk/gwas/, study code: GCST90244151, GCST011947, GCST007320, GCST90104539, GCST90162546), the DECODE project (https://www.decode.com/summarydata/), ChEMBL (https://www.ebi.ac.uk/chembl/), pharmGKB (https://www.pharmgkb.org/), DrugBank (https://go.drugbank.com/), TTD (https://db.idrblab.net/ttd/), CSF pQTL summary statistics available at NIAGADS (full summary statistics available to approved investigators through accession #NG00130), NeuroGenomics and Informatics Center website (https://neurogenomics.wustl.edu/open-science/raw-data/) and ONTIME browser (https://ontime.wustl.edu/).