Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) is a gammaherpesvirus that has been implicated in the pathogenesis of Kaposi's sarcoma. KSHV encodes K-bZIP (open reading frame K8), a protein that belongs to the basic region-leucine zipper (bZIP) family of transcription factors. Here we show that K-bZIP associates with the cellular transcription factor p53 directly in vitro and in vivo. This interaction requires the bZIP domain of K-bZIP and the carboxy-terminal region (amino acids 300 to 393) of p53. We also show that K-bZIP represses the transcriptional activity of p53 which is required for apoptosis of the host cell. These results imply that K-bZIP blocks p53-mediated host cell death through its interaction with p53.
Kaposi's sarcoma-associated herpesvirus (KSHV; also designated human herpesvirus 8) has been implicated as a major agent in the genesis of Kaposi's sarcoma and several B-cell lymphoproliferative diseases (2, 3, 21). Phylogenetic analysis of the KSHV genome sequence revealed that KSHV belongs to the Gammaherpesvirinae subfamily; thus KSHV shares significant sequence homology with herpesvirus saimiri and Epstein-Barr virus (EBV) (22). The KSHV genome encodes a basic region-leucine zipper (bZIP) protein called K-bZIP (open reading frame K8), which forms a homodimer using its carboxyl-terminal bZIP domain (17). The expression pattern of the K-bZIP gene indicates that it is an early gene (30). The K-bZIP protein typically localizes in the host cell nucleus (11). Tetradecanoyl phorbol acetate (TPA) is reported to induce an authentic lytic program and result in viral DNA replication and lytic gene expression (25).
K-bZIP shows significant homology with EBV Zta (also designated EB1, Zebra, and BZLF1) (5, 6, 10, 15, 17, 18). Zta is known to play a crucial role in the initiation of the EBV lytic cascade, as ectopic expression of Zta in latently infected cells is sufficient to activate the entire EBV replicative cycle (4, 9). Zta is a sequence-specific DNA binding protein that transactivates several EBV early lytic promoters via canonical AP-1 binding sites or Zta-responsive elements (6, 7, 12, 16, 31). In addition to its role in vital transcription and replication, Zta was shown by Zhang et al. (32) to interact directly with the tumor suppressor and cell cycle regulatory protein p53 in vitro as well as in vivo and to repress the ability of p53 to activate to transcription. Flemington and colleagues (1, 26) showed that Zta caused cell cycle arrest through the induction of cyclin-dependent kinase inhibitors p21 and p27. In this study, we show that K-bZIP associated directly with p53 and determine the effects of K-bZIP binding on p53 function.
K-bZIP and p53 interact directly in vitro.
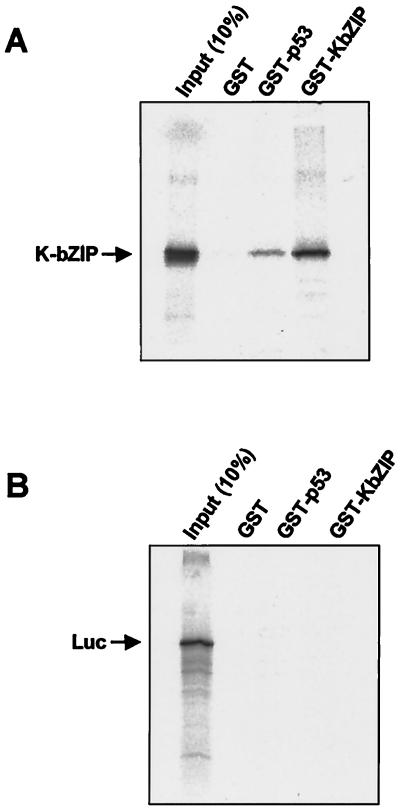
To determine whether the K-bZIP and p53 proteins can interact directly in vitro, we carried out glutathione S-transferase (GST) pull-down assays. First, we cloned the full-length K-bZIP cDNA into various expression vectors. The K-bZIP cDNA was synthesized using reverse transcription-PCR and total RNA from BCBL-1 cells treated with TPA as described previously (25). The PCR-amplified K-bZIP was cleaved with EcoRI and XhoI and inserted into pcDNA3 (Invitrogen, Groningen, The Netherlands), pGEX4T-1 (Amersham Pharmacia Biotech, Uppsala, Sweden), and pME18S (FLAG-tagged SRα promoter plasmid [27]). The cloned plasmids were designated pcDNA3/K-bZIP, pGEX4T-1/K-bZIP, and FLAG-KbZIP, respectively. pGEX4T-1/K-bZIP was cleaved with BamHI and NotI, inserted into pEBG, and designated pEBG/K-bZIP. The splicing sites of K-bZIP cDNA were confirmed by direct DNA sequencing. pcDNA3/K-bZIP was used for the in vitro translation of K-bZIP. The cDNAs encoding various p53 subsequences were generated from full-length p53 cDNA (a gift from B. Vogelstein). PCR-amplified p53 cDNA was cleaved with BamHI and XhoI and inserted into hemagglutinin (HA)-tagged pcDNA3 and designated HA-p53, and p53 cDNA was amplified by another primer set, cleaved with EcoRI and XhoI, and designated GST-p53. The p53 was expressed in bacteria as a GST fusion protein, purified from bacterial cell extracts, and precipitated with in vitro-translated K-bZIP using glutathione-Sepharose beads (Amersham Pharmacia Biotech) as described previously (32). In vitro-translated K-bZIP was retained by the GST-p53 fusion protein (as compared with the GST control) (Fig. 1A). Because the K-bZIP protein forms a homodimer (17), GST-KbZIP served as a positive control to show that in vitro-translated K-bZIP was able to bind tightly to GST-KbZIP. In contrast, no detectable binding was observed with K-bZIP and the in vitro-translated luciferase control (Fig. 1B). These results show that K-bZIP interacts with p53 directly in vitro.
FIG. 1.
K-bZIP interacts with p53 in vitro. Equal amounts of GST, GST-p53, and GST-KbZIP were incubated with 35S-labeled in vitro-translated K-bZIP (A) or luciferase (Luc) (B), and an aliquot [Input (10%)] from each binding reaction was precipitated with glutathione-Sepharose beads. The bead-bound proteins were eluted and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). K-bZIP and Luc (both indicated by an arrow) were visualized by autoradiography. GST-KbZIP was used as a positive control for K-bZIP binding ability.
K-bZIP and p53 proteins interact directly in vivo.
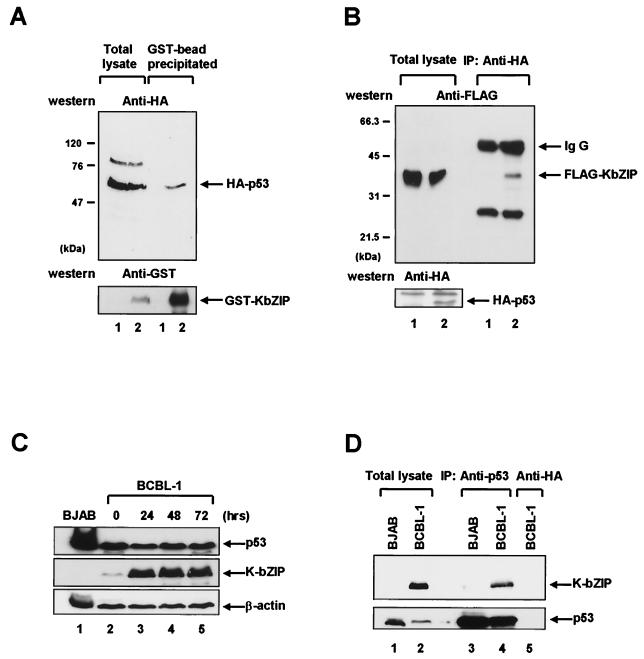
The GST-KbZIP and HA-p53 expression plasmids (pEBG/K-bZIP and pcDNA3/HA-p53, respectively) were cotransfected into 293T cells using the calcium phosphate procedure (8). Forty-eight hours after transfection, the cells were harvested and lysed to yield the cell extract (14). GST-KbZIP was purified from the cell extract using glutathione-Sepharose beads, and GST-KbZIP-bound HA-p53 was detected by immunoblotting using a monoclonal antibody to HA. As shown in Fig. 2A, GST-KbZIP bound to HA-p53 while GST alone did not. In the reverse experiment, Flag-KbZIP (pME18S/KbZIP) and HA-p53 (pcDNA3/HA-p53) expression plasmids were cotransfected into 293T cells and coimmunoprecipitation analyses were performed as described previously (14). Incubation of the transfected 293T cell extracts with a HA-specific antibody resulted in the coimmunoprecipitation of HA-p53 and Flag-KbZIP (Fig. 2B). Flag-KbZIP did not coimmunoprecipitate with HA alone. To confirm this interaction in KSHV-infected cells, we performed the direct coimmunoprecipitation assay for the KSHV-positive BCBL-1 cell line. K-bZIP was expressed during the lytic replication and detected by anti-KbZIP rabbit polyclonal antibody (Fig. 2C). Incubation of the cell extract with a p53 specific antibody, DO-1 (Santa Cruz, Santa Cruz, Calif.), resulted in the coimmunoprecipitation of p53 and K-bZIP (Fig. 2D, lane 4). However, K-bZIP was not detected either in KSHV-negative BJAB cell extract or in the immunoprecipitation with anti-HA antibody (Fig. 2D, lanes 3 and 5). These results demonstrate that K-bZIP interacts with p53 in vivo.
FIG. 2.
K-bZIP interacts with p53 in vivo. (A) 293T cells were transiently transfected with a plasmid encoding HA-p53 (pcDNA3/HA-p53) in combination with either GST (pEBG) (lanes labeled 1) or GST-KbZIP (pEBG/K-bZIP) (lanes labeled 2). Whole-cell lysates were prepared 48 h after transfection and were precipitated with glutathione-Sepharose beads. The bead-bound proteins were separated on a 7% polyacrylamide gel, transferred to a nitrocellulose membrane, and immunoblotted (Western) with anti-HA antibody (upper panel) or anti-GST antibody (lower panel). HA-p53 and GST-K-bZIP are indicated by arrows. (B) 293T cells were transfected with a plasmid encoding FLAG-KbZIP (pME18S/K-bZIP) in combination with either an HA-p53 expression plasmid (pcDNA3/HA-p53) (lanes labeled 2) or a control plasmid (pcDNA3) (lanes labeled 1). Cell lysates were subjected to immunoprecipitation (IP) with anti-HA monoclonal antibody. Immunoprecipitated proteins were separated on a 12% polyacrylamide gel, followed by immunoblotting (Western) with anti-FLAG antibody (upper panel). Immunoglobulin G (IgG) and Flag-K-bZIP are indicated by arrows. The identical membrane was reprobed with anti-HA antibody (lower panel) to confirm the expression of HA-p53 (indicated by an arrow). (C) The level of p53 and K-bZIP during the lytic replication cycle. BCBL-1 cells were treated with TPA as described previously (25). Whole cell lysates were prepared after the indicated number of hours, and Western blots were performed on equal quantities of whole cell extract (50 μg) by using p53 specific monoclonal antibody (upper panel), K-bZIP specific polyclonal antibody (middle panel), and β-actin antibody (lower panel). (D) The direct coimmunoprecipitation assay was performed using the BCBL-1 cell line 48 h after TPA induction. BCBL-1 cells and BJAB cells (107 cells) were harvested, and cell lysates were subjected to IP with p53 specific monoclonal antibody (lanes 3 and 4) and anti-HA antibody (lane 5). K-bZIP was detected using K-bZIP polyclonal antibody (upper panel), and the same membrane was reprobed with p53 antibody (bottom panel).
p53-K-bZIP interaction required the carboxy-terminal region of p53 and the bZIP domain of K-bZIP.
To define the K-bZIP binding domain within p53, in vitro-translated K-bZIP was incubated under the appropriate binding conditions with a series of GST fusion proteins that contained distinct domains of p53 (Fig. 3A and B). Although the transactivation domain (amino acids 1 to 42) of p53 did not appear to interact with K-bZIP, the p53 carboxy-terminal region (amino acids 300 to 393) interacted with K-bZIP. The p53 DNA binding domain (amino acids 100 to 300) also interacted with K-bZIP but binding was weaker than with the p53 carboxy-terminal region (amino acids 300 to 393), indicating that the carboxy-terminal region of p53 is the major binding target of K-bZIP (Fig. 3B). The full-length GST-p53 fusion protein and GST protein were used as positive and negative controls, respectively.
FIG. 3.
Identification of domains involved in K-bZIP–p53 interaction. K-bZIP binds to the carboxy-terminal region of p53. (A) Schematic representation of the domains of human p53. Numbers correspond to the amino acid sequence. Indicated are the positions of the transcriptional activation domain (TAD; amino acids 1 to 42), the DNA binding domain (DBD; amino acids 100 to 300), and the carboxy-terminal region (CT; amino acids 300 to 393). (B) Upper panel, the p53 segments present in each of the GST fusion proteins used in GST pull-down assays with 35S-labeled, in vitro-translated K-bZIP. After subjecting the input (10%) and GST pull-down reaction mixtures to SDS-PAGE, K-bZIP (indicated by arrow) was visualized by autoradiography. Bottom panel, expression of GST-p53 fusion proteins. Purified GST fusion proteins of p53 deletion mutants were electrophoresed on an SDS-13% polyacrylamide gel and visualized by Coomassie brilliant blue staining. p53 associates with the bZIP domain of K-bZIP. (C) Schematic representation of K-bZIP. Indicated are the amino acid positions of the putative transcriptional activation domain (TAD; amino acids 1 to 121), the putative DNA binding domain (DBD; amino acids 122 to 189), and the leucine zipper domain (ZIP; amino acids 190 to 237). (D) Upper panel, an experiment similar to that described above for panel B performed using the GST-KbZIP fusion proteins and 35S-labeled, in vitro-translated p53 (indicated by an arrow). Bottom panel, expression of GST-KbZIP fusion proteins. Purified GST fusion proteins of K-bZIP deletion mutants were electrophoresed on an SDS-13% polyacrylamide gel.
To define the p53 binding domain within K-bZIP, experiments similar to those described above were performed (Fig. 3C and D). The most interesting structural features of K-bZIP are the leucine zipper domain, which is involved in homodimerization, and the adjunct basic region, which presumably functions to bind DNA (17). Three GST-KbZIP fusion proteins were synthesized: GST fused to K-bZIP with the leucine zipper deleted (amino acids 1 to 189), GST fused to the bZIP domain (amino acids 122 to 237), and GST fused to the leucine zipper domain (amino acids 190 to 237). These GST fusion proteins were then used in GST pull-down experiments with 35S-labeled, in vitro-translated p53. The physical interaction between p53 and GST-KbZIP with the leucine zipper domain deleted (amino acids 1 to 189) was so weak that in vitro-translated p53 was barely detected. The interaction between p53 and the GST-bZIP domain (amino acids 122 to 237) fusion protein, however, was similar to that between p53 and the wild-type K-bZIP, and the GST-leucine zipper (amino acids 190 to 237) fusion did not interact with p53. These results indicate that the leucine zipper domain is essential but not sufficient for the interaction of K-bZIP with p53, and the bZIP domain (amino acids 122 to 237) of K-bZIP, which comprises the putative DNA binding domain and leucine zipper domain, is sufficient for its binding to p53.
Expression of K-bZIP in human cells represses the ability of p53 to activate transcription.
The most notable biochemical property of p53 is its DNA sequence-specific transcriptional activation of target genes. In order to determine whether K-bZIP can influence p53 function, a human cell line carrying a mutated p53 gene (C33A) was transiently transfected (calcium phosphate method) with a luciferase reporter plasmid that contained synthetic p53 response elements fused to the luciferase gene (PG13-Luc) with or without HA-p53 (pcDNA3/HA-p53) along with expression plasmid encoding wild-type FLAG-KbZIP (pME18S/K-bZIP). Twenty-four hours after transfection, the cells were rinsed with phosphate-buffered saline, resuspended in cell lysis buffer (Promega, Madison, Wis.), and incubated for 10 min on ice. Insoluble material was removed by centrifugation, and luciferase activity in the cleared supernatant was quantitated in the presence of luciferin (Promega) and ATP using a luminometer (EG&G BERTHOLD, Pforzheim, Germany). Each assay was normalized with total protein concentration in the samples. In the presence of p53, transcription of the luciferase gene was induced up to 50-fold. However, in the presence of K-bZIP, p53-driven transcription of the luciferase gene was reproducibly inhibited by 60% (Fig. 4A). Basal transcription was not inhibited by K-bZIP, indicating that the transcriptional repression by K-bZIP was not the result of general transcriptional repression.
FIG. 4.
K-bZIP represses p53-dependent transcription in C33A cells. (A) A synthetic p53 promoter fused to a luciferase reporter gene (PG13-Luc) was repressed by K-bZIP. C33A cells, which carry a mutated version of p53, were cotransfected with the reporter plasmid PG13-Luc (0.5 μg) in the presence (+) or absence (−) of the p53 expression plasmid pcDNA3/HA-p53 (0.5 μg) together with 2 to 4 μg of the K-bZIP expression plasmid, FLAG-KbZIP (pME18S/K-bZIP). The total amounts of transfected DNA (5 μg) in each dish were kept constant by the addition of a blank plasmid (pME18S). Cell lysates were prepared 24 h after transfection, and luciferase activity was measured in the lysates. Luciferase activity was normalized according to total protein concentration in the sample. Transfections were performed in triplicate, and the standard deviation is shown. (B) Repression of a p53-dependent promoter by a series of K-bZIP mutants. C33A cells were transfected with the PG13-Luc reporter plasmid (0.5 μg) with or without pcDNA3/HA-p53 (0.5 μg), together with expression plasmids encoding wild-type K-bZIP or one of three K-bZIP deletion mutants (4 μg). The total amount of transfected DNA in each dish was kept constant by the addition of a blank plasmid (pME18S). Transfections were performed in triplicate, and the standard deviation is shown. (C) K-bZIP did not influence the unrelated activated transcription. C33A cells were transfected with the Gal4-Luc reporter plasmid (0.5 μg) with or without expression plasmids encoding Gal4-VP16 (0.5 μg), together with K-bZIP expression plasmids. Transfections were performed in triplicate, and the standard deviation is shown. (D) The concentration of p53 and K-bZIP protein-transfected C33A cells described above for panels A and B was assessed by Western blot analysis (proteins are indicated by arrows). Lane 1, 1 μg of PG13-Luc and 9 μg of blank plasmid (pME18S); lane 2, 1 μg of PG13-Luc, 1 μg of HA-p53 (pcDNA3/HA-p53), and 8 μg of blank plasmid; lane 3, 1 μg of PG13-Luc, 8 μg of FLAG-KbZIP (pME18S/K-bZIP), and 1 μg of blank plasmid; lane 4, 1 μg of PG13-Luc, 1 μg of HA-p53, and 8 μg of FLAG-KbZIP; lane 5, 1 μg of PG13-Luc, 1 μg of HA-p53, and 8 μg of FLAG-KbZIP lacking the leucine zipper domain (1 to 189); lane 6, 1 μg of PG13-Luc, 1 μg of HA-p53, and 8 μg of FLAG-KbZIP containing only the bZIP domain (122 to 237). HA-p53 protein was immunoprecipitated with anti-HA monoclonal antibody and assessed by Western blotting with the same antibody. The concentration of the various K-bZIP proteins in the cell lysates was monitored by Western blotting using anti-FLAG monoclonal antibody. Molecular sizes are shown at the left in kilodaltons.
To characterize further the connection between K-bZIP–p53 interaction and inhibition of p53 transcriptional activity, K-bZIP deletion mutants (Fig. 3B) were used in transfection assays similar to those described in Fig. 4A. In the presence of the K-bZIP mutant that lacked the leucine zipper domain (amino acids 1 to 189), transcriptional activation by p53 was inhibited to 20% (Fig. 4B). However, the K-bZIP mutant that contained only the bZIP domain (amino acids 122 to 237) inhibited p53-driven transcriptional activation to the same degree as wild type. To show that K-bZIP did not influence the transcriptional activity of an unrelated transactivator, we used Gal4-VP16 fusion protein with Gal4-Luc. The promoter activated by Gal4-VP16 was not influenced by K-bZIP (Fig. 4C). Expression of p53 and the K-bZIP mutants in the transfected cells was monitored by Western blot assay, which showed that the level of p53 and K-bZIP wild type and mutants did not change significantly (Fig. 4D). These results indicate that the repression of p53-driven transcriptional activity is related to the physical interaction between p53 and K-bZIP.
Experiments described herein reveal that K-bZIP can interact directly with p53 in vitro as well as in vivo. This interaction required the bZIP domain of K-bZIP and the carboxy-terminal region of p53. The expression of K-bZIP in transfected cells repressed the transcriptional activity of p53. In addition, the ability of p53 to interact with the various K-bZIP deletion mutant proteins correlated positively with repression of p53-driven transcription of a reported gene. These findings are consistent with the notion that the repression of p53-driven transcription is caused by the functional association of p53 and K-bZIP.
The tumor suppressor protein p53 is a multifunctional transcriptional regulatory protein that plays an important role in cell cycle arrest and apoptosis (19). Various cellular and viral proteins have been shown to inactivate p53 function via various cellular pathways (19, 29). From our results, we hypothesized mechanisms by which K-bZIP can impede p53 function. Because the carboxy-terminal region of p53 associated with K-bZIP more tightly than did other p53 domains, we hypothesize that the carboxy-terminal region of p53 is the main target of K-bZIP. The p53 carboxy-terminal region contains at least three biological domains, nuclear localization, tetramerization, and both nonspecific DNA binding and recognition of primary DNA damage (28). It is well established that p53 forms tetramers (13) via a tetramerization domain (amino acids 323 to 356) in the carboxy-terminal region. p53 tetramerization appears to be required for efficient transcriptional activation by p53 in vivo and for p53-mediated suppression of the growth of carcinoma cell lines (24). Consistent with our results, we propose a model whereby K-bZIP blocks the tetramerization of p53 and, therefore, inhibits p53 function. However, we cannot exclude the possibility that K-bZIP may hinder the binding of p53 to DNA. This inhibition could occur through the interaction of K-bZIP with the DNA binding domain of p53, even though this interaction is weaker than the interaction of K-bZIP with the tetramerization domain of p53.
p53 is known to be a regulator of cell growth, and the induction of p53 leads to either cell cycle arrest or apoptosis (19). As K-bZIP is expressed during the lytic replication cycle, it does not seem likely that K-bZIP is involved in the host cell proliferation through blocking of p53-mediated cell cycle arrest. Instead, K-bZIP may block host cell death mediated by p53 and help viral replication. It is known that p53 mediates apoptosis using p53-dependent transcription, such as bax and Fas/APO-1 (20, 23). Since K-bZIP inhibits the transcriptional activity of p53, K-bZIP may block host cell death mediated by p53. Further study is required to decipher K-bZIP's exact role in KSHV viral replication.
Acknowledgments
We thank J. Jung for the generous gift of the anti-KbZIP rabbit polyclonal antibody.
This work was supported in part by grants from the National Research Laboratory Program of the Korea Institute of Science and Technology Evaluation and Planning (KISTEP), from the Korea Science and Engineering Foundation (KOSEF) through the Protein Network Research Center at Yonsei University, and from the BK21 Program from the Ministry of Education, Korea.
REFERENCES
- 1.Cayrol C, Flemington E K. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 1996;15:2748–2759. [PMC free article] [PubMed] [Google Scholar]
- 2.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 3.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 4.Countryman J, Miller G. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Countryman J, Jenson H, Seibl R, Wolf H, Miller G. Polymorphic proteins encoded within BZLF1 of defective and standard Epstein-Barr viruses disrupt latency. J Virol. 1987;61:3672–3679. doi: 10.1128/jvi.61.12.3672-3679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farrell P J, Rowe D T, Rooney C M, Kouzarides T. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 1989;8:127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flemington E, Speck S H. Autoregulation of Epstein-Barr virus putative lytic switch gene BZLF1. J Virol. 1990;64:1227–1232. doi: 10.1128/jvi.64.3.1227-1232.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham F L, van der Eb A J. Transformation of rat cells by DNA of human adenovirus 5. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90163-3. [DOI] [PubMed] [Google Scholar]
- 9.Grogan E, Jenson H, Countryman J, Heston L, Gradoville L, Miller G. Transfection of a rearranged viral DNA fragment, WZhet, stably converts latent Epstein-Barr viral infection to productive infection in lymphoid cells. Proc Natl Acad Sci USA. 1987;84:1332–1336. doi: 10.1073/pnas.84.5.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruffat H, Portes-Sentis S, Sergeant A, Manet E. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) encodes a homologue of the Epstein-Barr virus bZip protein EB1. J Gen Virol. 1999;80:557–561. doi: 10.1099/0022-1317-80-3-557. [DOI] [PubMed] [Google Scholar]
- 11.Katano H, Sato T, Kurata T, Mori S, Sata T. Expression and localization of human herpesvirus 8-encoded proteins in primary effusion lymphoma, Kaposi's sarcoma, and multicentric Castleman's disease. Virology. 2000;269:335–344. doi: 10.1006/viro.2000.0196. [DOI] [PubMed] [Google Scholar]
- 12.Kenney S, Holley-Guthrie E, Mar E C, Smith M. The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators. J Virol. 1989;63:3878–3883. doi: 10.1128/jvi.63.9.3878-3883.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraiss S, Quaiser A, Oren M, Montenarh M. Oligomerization of oncoprotein p53. J Virol. 1988;62:4737–4744. doi: 10.1128/jvi.62.12.4737-4744.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee D, Lee B, Kim J, Kim D W, Choe J. CBP binds to human papillomavirus E2 protein and activates E2-dependent transcription. J Biol Chem. 2000;275:7045–7051. doi: 10.1074/jbc.275.10.7045. [DOI] [PubMed] [Google Scholar]
- 15.Lieberman P M, Hardwick J M, Sample J, Hayward G S, Hayward S D. The Zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J Virol. 1990;64:1143–1155. doi: 10.1128/jvi.64.3.1143-1155.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieberman P M, Hardwick J M, Hayward S D. Responsiveness of the Epstein-Barr virus NotI repeat promoter to the Z transactivator is mediated in a cell-type-specific manner by two independent signal regions. J Virol. 1989;63:3040–3050. doi: 10.1128/jvi.63.7.3040-3050.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin S F, Robinson D R, Miller G, Kung H J. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J Virol. 1999;73:1909–1917. doi: 10.1128/jvi.73.3.1909-1917.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manet E, Gruffat H, Trescol-Biemont M C, Moreno N, Chambard P, Giot J F, Sergeant A. Epstein-Barr virus bicistronic mRNAs generated by facultative splicing code for two transcriptional trans-activators. EMBO J. 1989;8:1819–1826. doi: 10.1002/j.1460-2075.1989.tb03576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.May P, May E. Twenty years of p53 research: structural and functional aspects of the p53 protein. Oncogene. 1999;18:7621–7636. doi: 10.1038/sj.onc.1203285. [DOI] [PubMed] [Google Scholar]
- 20.Miyashita T, Krajewski S, Krajewski M, Wang H G, Lin H K, Hoffman B, Lieberman D, Reed J C. Tumor suppressor p53 is a regulator of bcl-2 and bax in gene expression in vitro and in vivo. Oncogene. 1994;9:1799–1805. [PubMed] [Google Scholar]
- 21.Moore P S, Chang Y. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N Engl J Med. 1995;332:1181–1185. doi: 10.1056/NEJM199505043321801. [DOI] [PubMed] [Google Scholar]
- 22.Moore P S, Gao S J, Dominguez G, Cesarman E, Lungu O, Knowles D M, Garber R, Pellett P E, McGeoch D J, Chang Y. Primary characterization of a herpesvirus agent associated with Kaposi's sarcoma. J Virol. 1996;70:549–558. doi: 10.1128/jvi.70.1.549-558.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Owen-Scaub L B, Zhang W, Cusack J C, Angelo L S, Santee S M, Fujiwara T, Roth J A, Deisseroth A B, Zhang W-W, Kruzel E, Radinksy R. Wild-type human p53 and a temperature-sensitive mutant induce Fas/APO-1 expresson. Mol Cell Biol. 1995;15:3032–3040. doi: 10.1128/mcb.15.6.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pietenpol J A, Tokino T, Thiagalingam S, el-Deiry W S, Kinzler K W, Vogelstein B. Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc Natl Acad Sci USA. 1994;91:1998–2002. doi: 10.1073/pnas.91.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renne R, Zhong W, Herndir B, Mcgreath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez A, Armstrong M, Dwyer D, Flemington E. Genetic dissection of cell growth arrest functions mediated by the Epstein-Barr virus lytic gene product, Zta. J Virol. 1999;73:9029–9038. doi: 10.1128/jvi.73.11.9029-9038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shiio Y, Yamamoto T, Yamaguchi N. Negative regulation of Rb expression by the p53 gene product. Proc Natl Acad Sci USA. 1992;89:5206–5210. doi: 10.1073/pnas.89.12.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soussi T, May P. Structural aspects of the p53 protein in relation to gene evolution: a second look. J Mol Biol. 1996;260:623–637. doi: 10.1006/jmbi.1996.0425. [DOI] [PubMed] [Google Scholar]
- 29.Stommel J M, Marchenko N D, Jimenez G S, Moll U M, Hope T J, Wahl G M. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun R, Lin S F, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J Virol. 1999;73:2232–2242. doi: 10.1128/jvi.73.3.2232-2242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Urier G, Buisson M, Chambard P, Sergeant A. The Epstein-Barr virus early protein EB1 activates transcription from different responsive elements including AP-1 binding sites. EMBO J. 1989;8:1447–1453. doi: 10.1002/j.1460-2075.1989.tb03527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q, Gutsch D, Kenney S. Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. Mol Cell Biol. 1994;14:1929–1938. doi: 10.1128/mcb.14.3.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]