Abstract
We have previously shown that Xenopus oocytes require coinjection of both poliovirus RNA and HeLa cell extracts to support a complete cycle of viral replication yielding high levels of infectious viral particles. This novel system provides a tool for identifying host factors and for biochemically dissect individual steps that lead to virus production. Here we demonstrate that Xenopus oocytes are able to support replication of other picornaviruses such as human rhinovirus 14 and mengovirus. Unlike poliovirus, microinjection of mengovirus RNA yields high viral titers (about 107 PFU/oocyte) without the need for coinjection of additional cell extracts. In contrast, formation of infectious rhinovirus particles requires coinjection of human cell extracts. We found that one of these human factors is required for efficient rhinovirus translation. Our findings uncover differences in the host factor requirements among members of the picornavirus family and provide the means to identify the human protein(s) involved in rhinovirus production.
The picornavirus family consists of a large number of animal viruses associated with a plethora of different diseases. Their positive-sense RNA genomes have similar genetic organizations and common structural properties. They are covalently linked to a small viral protein, Vpg, and contain a single open reading frame (10, 17). The 5′ untranslated regions (UTRs) are highly structured and exceptionally long (600 to 1,500 nucleotides), while the 3′ UTRs are relatively short (∼100 nucleotides) and are terminated by a poly(A) tail.
The steps of the life cycle of picornaviruses follow a common scheme. After entry, the genomic RNA functions as mRNA directing the synthesis of a large polypeptide, which is proteolytically processed to yield mature viral proteins. The same RNA molecule is then amplified in a two-step process: first, its complementary negative strand is synthesized, and second, the negative-strand RNA is used as a template to generate new molecules of positive-strand RNA (for a review see reference 16). The synthesis of both negative- and positive-strand RNA is catalyzed by an RNA-dependent RNA polymerase (3Dpol) (7, 9). RNA replication requires viral proteins, cis-acting elements present in the viral genome, host cell membranous structures, and host cell proteins (1, 2, 5, 12, 15). Recently, Paul et al. have shown that 3Dpol, a primer-dependent enzyme, is able to directly uridylate the viral protein Vpg to form VpgpUpU, which in turn serves as a primer for the initiation process (19). VpgpUpU has been proposed to serve as a primer for the initiation of RNA synthesis (reviewed in reference 16). However, the mechanism by which a single viral RNA molecule is selectively amplified into thousands of RNA progeny is not clearly understood.
Picornavirus translation is initiated in a cap-independent manner by an internal ribosome entry mechanism. This process requires an RNA segment of the 5′ UTR (the internal ribosomal entry site) that specifically directs the ribosomes to the viral RNA. A vast amount of information has been accumulated in recent years about this novel mechanism of translation (1, 4, 15). However, the molecular details of how canonical and noncanonical initiation factors recruit the ribosome to the viral RNA remain unclear.
Biochemical analysis of picornavirus replication using in vitro systems is complicated by the fact that the process of viral replication is associated with microsomal membranes. An important contribution was the development by Molla et al. of an in vitro system capable of supporting complete poliovirus replication (18). This cell-free translation and replication system is an extremely powerful tool to dissect each step of the viral life cycle (2, 3, 5, 6). Recently, this in vitro system was employed to study translation and RNA synthesis of human rhinovirus type 14 (HRV 14). However, formation of HRV 14 infectious particles was not observed, presumably due to a deficiency in particle assembly (21).
In our laboratory we have developed an alternative and complementary system to study poliovirus replication using Xenopus oocytes (12). Microinjection of poliovirus RNA into oocytes initiates a complete cycle of viral replication, yielding a high level of infectious particles. At least two cytoplasmic HeLa cell factors are essential for poliovirus replication in oocytes, one which is necessary for translation and one which is necessary for RNA synthesis. This observation provides direct evidence that host factors are required at specific steps during viral replication (1). In recent years, we have successfully used the oocyte system to study several aspects of poliovirus replication. We analyzed the mechanism of how the virus controls the usage of the viral RNA template, which is utilized in translation and RNA synthesis (13), as well as the functional role of host factors during viral replication (11, 14, 20). We report here that the oocyte system can support replication of other members of the picornavirus family, such as mengovirus and rhinovirus type 14.
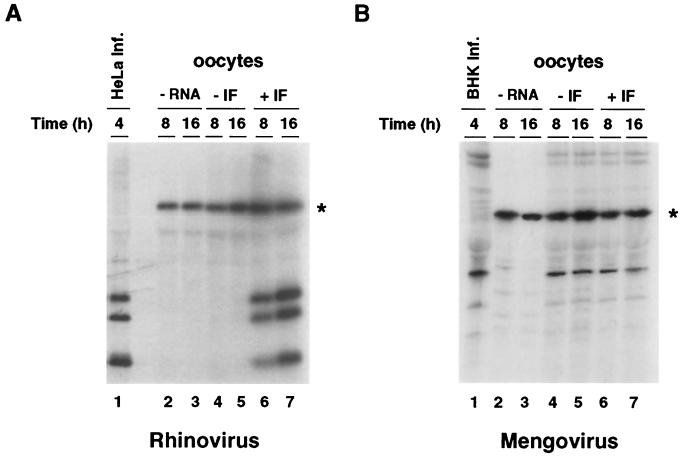
To determine whether the translation machinery of Xenopus oocytes is able to translate picornavirus genomes, we microinjected viral RNA from HRV 14 or mengovirus into oocytes. HRV 14 was obtained from the American Type Culture Collection, and mengovirus was recovered from BHK cells after transfection of an in vitro-transcribed RNA using the cDNA clone pM16 (8). Viruses were purified from 108 infected cells as previously described (12). Viral particles were treated with proteinase K (200 μg/ml), and the RNAs were extracted with phenol-chloroform and precipitated in ethanol. The viral RNAs were microinjected into oocytes either alone or together with HeLa cell extracts. The oocytes were incubated in the presence of [35S]methionine for 8 and 16 h at 20°C. Total cytoplasmic extracts obtained from 30 oocytes were immunoprecipitated with antibodies directed against the respective virus, and the labeled proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Efficient translation of HRV 14 RNA in oocytes required HeLa cell factors (Fig. 1A; compare lanes 4 and 5 with 6 and 7). In contrast, translation of mengovirus RNA in oocytes was readily detectable regardless of the presence or absence of HeLa proteins, and the amounts of viral proteins produced under both conditions were very similar (Fig. 1B, compare lanes 4 and 5 with 6 and 7). It has been previously reported that rhinovirus translation is inefficient in in vitro systems compared with translation of other picornavirus RNAs (15). In oocytes, however, the efficiency of translation observed in the presence of HeLa proteins was similar to that of poliovirus under the same conditions (data not shown).
FIG. 1.
Translation of HRV 14 and mengovirus in Xenopus oocytes. (A) A HeLa cell cytoplasmic factor is required for initiation of HRV 14 translation in Xenopus oocytes. Oocytes were microinjected with buffer (lanes 2 and 3), 10 ng of HRV 14 RNA (lanes 4 and 5), or 10 ng of HRV 14 RNA together with 100 ng of HeLa cell ribosomal salt wash proteins (IF, lanes 6 and 7). The oocytes were incubated in [35S]methionine at 20°C for 8 h (lanes 2, 4, and 6) or 16 h (lanes 3, 5, and 7). Cytoplasmic extracts were immunoprecipitated with antiserum directed against capsid proteins obtained from the American Type Culture Collection and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The asterisk indicates a labeled oocyte protein unspecifically precipitated that was used as an internal control of sample manipulations. Arrows indicate viral capsid proteins. As a marker for viral proteins, HeLa cells were infected with HRV 14, and the [35S]methionine-labeled proteins were immunoprecipitated after 4 h of incubation at 32°C (lane 1). (B) The oocyte translation machinery can translate mengovirus RNA. Oocytes were microinjected and viral proteins were analyzed as described for panel A. As a marker for viral proteins, BHK cells were infected with mengovirus, and the [35S]methionine-labeled proteins were obtained after 4 h of incubation at 37°C (lane 1).
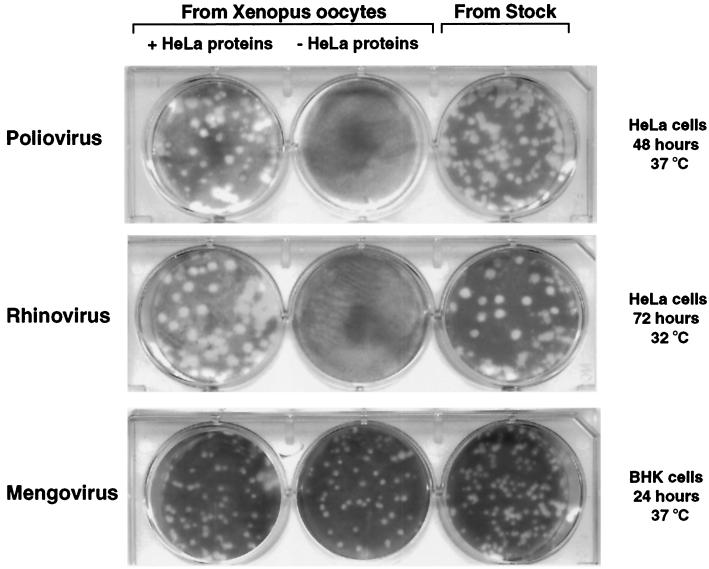
To establish whether HRV 14 and mengovirus can undergo a complete replication cycle in oocytes, we microinjected the viral RNAs with and without HeLa proteins. After 40 h of incubation at 30°C, oocyte cytoplasmic extracts were obtained and sterilized by passage through a 0.22-μm-pore-size filter, and the presence of infectious viral particles was determined by plaque assay. As a control, poliovirus RNA was microinjected under the same conditions. For poliovirus and rhinovirus, plaque assays were performed in HeLa cells and developed after 2 days of incubation at 37°C and 3 days at 32°C, respectively. For mengovirus, the plaque assays were carried out in BHK cells and developed after 1 day of incubation at 37°C. Both HRV 14 and mengovirus replicate in oocytes (Fig. 2). As expected, replication of HRV 14 as well as of poliovirus took place only when HeLa proteins were coinjected. In contrast, mengovirus replication was very efficient with or without the addition of human proteins. These results indicate that oocytes provide the host factors required for translation, efficient RNA synthesis, and particle assembly of mengovirus. In addition, we analyzed the plaque morphology of the viruses generated in oocytes by comparing them to viruses maintained in cell culture (Fig. 2, right panel). In all cases, the plaque phenotypes of viruses produced by oocytes were indistinguishable from those obtained in HeLa and BHK cells (Fig. 2, compare left and right panels).
FIG. 2.
Microinjection of poliovirus, HRV 14, and mengovirus RNAs yields infectious viral particles in Xenopus oocytes. Ten nanograms of viral RNA was injected into oocytes in the presence or absence of 100 ng of HeLa cell cytoplasmic proteins. The presence of infectious viral particles in filter-sterilized oocyte extracts obtained after 40 h of incubation at 30°C was determined by plaque assays. For comparison of plaque phenotype, plaque assays using viruses maintained in cell culture were also included, as indicated on the top (From Stock).
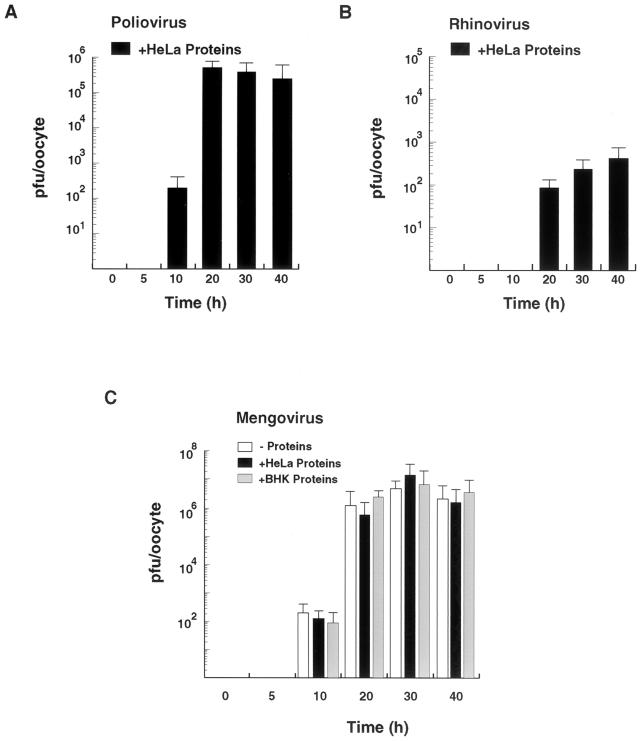
Time course studies of viral replication in oocytes indicate that poliovirus and mengovirus infectious particles are formed more efficiently than those of HRV 14 (Fig. 3). At 10 h after microinjection, titers of 102 to 103 PFU per oocyte were already observable for both poliovirus and mengovirus, while HRV infectious particles were undetectable. For all three viruses, replication in oocytes increased as a function of time, reaching a plateau between 20 and 40 h postinjection. Mengovirus titers at 30 h after microinjection were about 107 PFU per oocyte, which is about 10-fold higher than the highest titers obtained for poliovirus. Addition of HeLa cell proteins did not increase mengovirus titers (Fig. 3C), suggesting that host proteins are not the limiting factor for viral replication in oocytes. Since mengovirus replicates more efficiently in BHK cells than in HeLa cells (data not shown), we analyzed whether BHK cell extracts could improve mengovirus replication in oocytes. Coinjection of BHK cell extracts together with the viral RNA did not improve mengovirus replication in oocytes (Fig. 3C). Thus, mengovirus can efficiently replicate using the host factors provided by the oocytes.
FIG. 3.
Time course studies of picornavirus replication in oocytes. (A) Poliovirus replication in Xenopus oocytes. Ten nanograms of poliovirus RNA was injected into oocytes together with 100 ng of HeLa cell cytoplasmic proteins and incubated at 30°C for 0, 5, 10, 20, 30, and 40 h. The titer of infectious poliovirus particles was determined by plaque assays in HeLa cells. (B) HRV 14 replication in Xenopus oocytes. The experiment was carried out under the conditions used for panel A. (C) Mengovirus replication in Xenopus oocytes. Ten nanograms of mengovirus RNA was microinjected into oocytes alone or together with 100 ng of cytoplasmic proteins from HeLa cells or BHK cells and incubated at 30°C for 0, 5, 10, 20, 30, and 40 h. Infectious-mengovirus titers were determined by plaque assays in BHK cells as for Fig. 2. The standard errors were calculated from three independent microinjections using the same batch of oocytes.
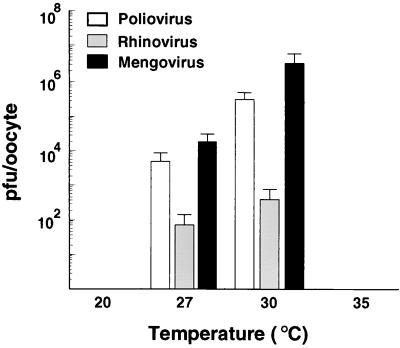
We have previously reported that temperature is a critical parameter for poliovirus assembly in oocytes (12) and that incubation at temperatures below 25°C yields no infectious particles. In addition, it is known that HRV 14 replication in cell culture requires temperatures of about 32 to 34°C, which are lower than those for other picornaviruses. Therefore, we decided to analyze the effect of temperature on picornavirus replication in Xenopus oocytes. We incubated oocytes microinjected with poliovirus, mengovirus, and HRV 14 RNAs at 20, 27, 30, and 35°C. The titers obtained indicate that the optimal temperature for the growth of all three viruses was between 27 and 30°C (Fig. 4). The inability of oocytes to produce viruses at temperatures higher than 30°C was attributed to a deleterious effect of the temperature on the oocytes. We observed a pronounced degradation of rRNA at temperatures above 32°C (data not shown). Thus, the window for optimal viral replication represents a compromise between the low temperature required for oocyte survival and the higher temperature needed for optimal viral replication.
FIG. 4.
Temperature dependency of poliovirus, HRV 14, and mengovirus replication in Xenopus oocytes. The experiment was carried out basically as described in Fig. 2 except that the oocytes were incubated at 20, 27, 30, and 35°C for 30 h. The standard errors were calculated from two independent microinjections.
In summary, we found that microinjection of HRV 14 and mengovirus into oocytes yields newly synthesized infectious particles. Translation of rhinovirus was strictly dependent on the coinjection of mammalian proteins, resembling the requirements of poliovirus in this system. Biochemical characterization suggests that the same factor is required for efficient translation of rhinovirus and poliovirus (data not shown). In contrast, mengovirus replication was very efficient when only the Xenopus oocyte machinery was used. As previously mentioned, poliovirus replication requires two cell host factors, one involved in translation and the other in RNA replication (12). The finding that mengovirus replicates completely independently of additional host factors indicates that enteroviruses and cardioviruses differ not only in the requirements for internal ribosomal entry site function but also in subsequent steps of viral replication.
Finally, the results presented here indicate that the oocyte system can be useful to study in detail the replication of several picornaviruses. Oocytes have the important advantage of representing intact and fully functional cells that at the same time are amenable to biochemical manipulation by microinjection. Currently, we are investigating the possibility of using Xenopus oocytes to study the life cycle of other medically and economically important viruses, especially those that are difficult to grow in cell culture.
Acknowledgments
We are grateful to Ann Palmenberg, who kindly provided antibodies against mengovirus and the plasmid containing the cDNA of this virus. We also thank Shane Crotty, Jens Herold, and Debbie Silvera for useful comments on the manuscript and Amy Corder for graphics.
REFERENCES
- 1.Andino R, Boddeker N, Silvera D, Gamarnik A V. Intracellular determinants of picornavirus replication. Trends Microbiol. 1999;7:76–82. doi: 10.1016/s0966-842x(98)01446-2. [DOI] [PubMed] [Google Scholar]
- 2.Barton D J, Black E P, Flanegan J B. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J Virol. 1995;69:5516–5527. doi: 10.1128/jvi.69.9.5516-5527.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton D J, Flanegan J B. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J Virol. 1997;71:8482–8489. doi: 10.1128/jvi.71.11.8482-8489.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belsham G J, Sonenberg N. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blyn L B, Towner J S, Semler B L, Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuconati A, Molla A, Wimmer E. Brefeldin A inhibits cell-free, de novo synthesis of poliovirus. J Virol. 1998;72:6456–6464. doi: 10.1128/jvi.72.8.6456-6464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dasgupta A, Baron M H, Baltimore D. Poliovirus replicase: a soluble enzyme able to initiate copying of poliovirus RNA. Proc Natl Acad Sci USA. 1979;76:2679–2683. doi: 10.1073/pnas.76.6.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duke G M, Palmenberg A C. Cloning and synthesis of infectious cardiovirus RNAs containing short, discrete poly(C) tracts. J Virol. 1989;63:1822–1826. doi: 10.1128/jvi.63.4.1822-1826.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flanegan J B, Baltimore D. Poliovirus-specific primer-dependent RNA polymerase able to copy poly(A) Proc Natl Acad Sci USA. 1977;74:3677–3680. doi: 10.1073/pnas.74.9.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flanegan J B, Petterson R F, Ambros V, Hewlett N J, Baltimore D. Covalent linkage of a protein to a defined nucleotide sequence at the 5′-terminus of virion and replicative intermediate RNAs of poliovirus. Proc Natl Acad Sci USA. 1977;74:961–965. doi: 10.1073/pnas.74.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamarnik A V, Andino R. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J Virol. 2000;74:2219–2226. doi: 10.1128/jvi.74.5.2219-2226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gamarnik A V, Andino R. Replication of poliovirus in Xenopus oocytes requires two human factors. EMBO J. 1996;15:5988–5998. [PMC free article] [PubMed] [Google Scholar]
- 13.Gamarnik A V, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gamarnik A V, Andino R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson K A, Sarnow P. Viral RNA synthesis. In: Rotbart H, editor. Human enterovirus infections. Washington, D.C.: ASM Press; 1995. pp. 95–112. [Google Scholar]
- 17.Lee Y F, Nomoto A, Detjen B M, Wimmer E. A protein covalently linked to poliovirus genome RNA. Proc Natl Acad Sci USA. 1977;74:59–63. doi: 10.1073/pnas.74.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molla A, Paul A V, Wimmer E. Cell-free, de novo synthesis of poliovirus. Science. 1991;254:1647–1651. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- 19.Paul A V, van Boom B J, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 20.Silvera D, Gamarnik A V, Andino R. The N-terminal K homology domain of the poly(rC)-binding protein is a major determinant for binding to the poliovirus 5′-untranslated region and acts as an inhibitor of viral translation. J Biol Chem. 1999;274:38163–38170. doi: 10.1074/jbc.274.53.38163. [DOI] [PubMed] [Google Scholar]
- 21.Todd S, Towner J S, Semler B L. Translation and replication properties of the human rhinovirus genome in vivo and in vitro. Virology. 1997;229:90–97. doi: 10.1006/viro.1996.8416. [DOI] [PubMed] [Google Scholar]