Abstract
Recent serological and molecular surveys of different primate species allowed the characterization of several Kaposi's sarcoma-associated herpesvirus (KSHV) homologues in macaques, African green monkeys, chimpanzees, and gorillas. Identification of these new primate rhadinoviruses revealed the existence of two distinct genogroups, called RV1 and RV2. Using a degenerate consensus primer PCR method for the herpesvirus DNA polymerase gene, the presence of KSHV homologues has been investigated in two semi-free-ranging colonies of eight drill (Mandrillus leucophaeus), five mandrill (Mandrillus sphinx), and two hybrid (Mandrillus leucophaeus-Mandrillus sphinx) monkeys, living in Cameroon and Gabon, Central Africa. This search revealed the existence of not only two distinct KSHV homologues, each one belonging to one of the two rhadinovirus genogroups, but also of two new betaherpesvirus sequences, one being close to cytomegaloviruses and the other being related to human herpesviruses 6 and 7 (HHV-6 and -7). The latter viruses are the first simian HHV-6 and -7 homologues identified to date. These data show that mandrill and drill monkeys are the hosts of at least four novel distinct herpesviruses. Moreover, mandrills, like macaques and African green monkeys, harbor also two distinct gamma-2 herpesviruses, thus strongly suggesting that a second gamma-2 herpesvirus, belonging to the RV2 genogroup, may exist in humans.
Based on biological and molecular criteria, the family Herpesviridae is divided into three subfamilies of Alpha-, Beta-, and Gammaherpesvirinae (13). For humans, eight herpesviruses have been identified to date. Since the discovery of Kaposi's sarcoma-associated herpesvirus (KSHV), also named human herpesvirus 8 (HHV-8) (2), the first human rhadinovirus (Gamma-2 herpesvirinae) (16), the identification of simian rhadinovirus homologues has rapidly become the subject of intense scrutiny. In instances in which particular animal species have been thoroughly analyzed for the presence of herpesviruses, multiple viruses have often been identified. For example, four distinct macaque KSHV homologues have been characterized to date, two for rhesus macaques (Macaca mulatta), namely, RFHVMm (for retroperitoneal fibromatosis herpesvirus of Macaca mulatta) (14) and RRV (5, 17), and two for pig-tailed macaques (Macaca nemestrina), namely, RFHVMn (14) and MneRV2 (for RFHV and rhadinovirus 2 of Macaca nemestrina) (15). Among these viruses, RFHVMm and RFHVMn have been identified in cases of retroperitoneal fibromatosis, a vascular fibroproliferative neoplasm with many morphological and histological similarities to KS (14). Otherwise, RRV have been isolated from simian immunodeficiency (SIV)-infected macaques with a lymphoproliferative disorder reminiscent of human multicentric Castleman's disease (17). Subsequently, two distinct additional KSHV homologues, called ChRV1 and ChRV2 for RV1 and RV2 of Chlorocebus aethiops, respectively, were identified in African green monkeys (C. aethiops) (7). Phylogenetic analyses of these new herpesviral sequences comprizing other known primate gammaherpesviruses suggested the existence of two major and distinct lineages of KSHV-like viruses among Old World primates (1, 7, 15). These two distinct lineages, tentatively named RV1 and RV2 (15), consist of RFHVMm, RFHVMn, ChRV1, and KSHV for RV1 and of RRV, MneRV2, and ChRV2 for RV2. More recently, we reported the identification of three new distinct rhadinoviruses in chimpanzees and gorillas, which are more closely related to KSHV than any other previously identified rhadinovirus (9; V. Lacoste, P. Mauclère, P. Dubreuil, J. Lewis, M.-C. Georges-Courbot, and A. Gessain, submitted for publication). These new viruses, tentatively named PanRHV1a and PanRHV1b for the chimpanzee (Pan troglodytes) rhadino-herpesviruses and GorRHV1 for the gorilla virus, cluster together with KSHV on a distinct branch (9).
We decided to address the possible presence of KSHV homologues in other Old World monkeys, Mandrillus sphinx and Mandrillus leucophaeus. These species originate from Central Africa, an area where both KSHV and KS are highly endemic. Blood specimens from 27 drill and mandrill monkeys were studied. The larger series comprises 11 wild-born animals (8 Mandrillus leucophaeus animals, 2 M. leucophaeus-Mandrillus sphinx animals, and 1 M. sphinx animal) originating from different parts of Cameroon and gathered in a wildlife rescue center in the southwestern province of Cameroon (4). The second group (six M. sphinx animals) originated from a semi-free-ranging colony of mandrills, living in rain forest enclosures, which was established in 1983 at the Centre International de Recherches Médicales de Franceville (CIRMF), Franceville, Gabon (6, 12). The other animals came from three centers in France: the Museum National d'Histoire Naturelle in Paris, La Palmyre Zoo in Les Mathes, and Touroparc Zoo in Romanèche-Thorins (five M. sphinx animals, four M. sphinx animals, and one M. leucophaeus animal, respectively, for which we had only sera). The 6 male mandrills from the CIRMF, belonging to a 102-mandrill colony, of which 16 were caught in the wild and 86 were born in captivity, were all infected by SIV and simian T-cell leukemia virus type 1 (STLV-1). No clinical immunodeficiency syndrome appeared to be associated with such SIV infection, and no specific pathology has ever been associated with STLV infection in these animals (6). Phylogenetic analyses of the mandrillus SIV (SIVmnd) and STLVmnd isolates, together with seroepidemiological and behavioral surveillance of the mandrills within this colony, have suggested that intracolony transmissions of these retroviruses are predominantly the result of male-to-male transmission occurring during bouts of aggression (12). None of the 10 other animals studied was seropositive for any of these two simian retroviruses (Table 1).
TABLE 1.
Epidemiological data and serological STLV-1–SIV and KSHV results
| Country | Name | Genus | Sexa | Estimated age | STLV-1/SIV serologyb | KSHV IFA serologyc | PCR and specific hybridization result ford:
|
Accession no(s) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MndRHV1 | MndRHV2 | MndCMV | MndHVβ | ||||||||
| Cameroon | Mnd201 | Mandrillus leucophaeus | M | Adult (>13 yr) | −/− | − | − | − | − | − | |
| Mnd202 | Mandrillus leucophaeus | F | Adult (>13 yr) | −/− | − | − | − | − | − | ||
| Mnd203 | Mandrillus leucophaeus | M | Approx. 8 yr | −/− | 1/40 | − | − | − | − | ||
| Mnd204 | Mandrillus leucophaeus | F | 4.5–5.5 yr | −/− | − | − | + | + | + | AF282938 | |
| Mnd205 | Mandrillus leucophaeus | F | 2.5–3 yr | −/− | − | − | − | + | − | AF282941 | |
| Mnd206 | Mandrillus leucophaeus | M | 1.75–2 yr | −/− | − | − | + | + | + | ||
| Mnd207 | Mandrillus leucophaeus | M | 0.5–3 yr | −/− | − | − | − | − | − | ||
| Mnd208 | Mandrillus leucophaeus | F | 2.5–3 yr | −/− | − | − | − | − | − | ||
| Mnd301 | Mandrillus sphinx | M | Adult (>13 yr) | −/− | − | − | − | − | + | AF282942 | |
| Mnd401 | Mandrillus leucophaeus- Mandrillus sphinx | F | 5 yr | −/− | − | − | + | − | + | AF282939 | |
| Mnd402 | Mandrillus leucophaeus- Mandrillus sphinx | F | 7 mo | −/− | − | − | + | − | − | AF282940 | |
| Gabon | Mnd9 | Mandrillus sphinx | M | Adult (>17 yr) | +/+ | − | − | − | − | − | |
| Mnd13 | Mandrillus sphinx | M | Adult (>17 yr) | +/+ | − | − | − | − | − | ||
| Mnd15 | Mandrillus sphinx | M | Adult (>16 yr) | +/+ | − | + | + | − | − | AF282937, AF282943 | |
| Mnd18 | Mandrillus sphinx | M | Adult (?) | +/+ | − | − | − | − | − | ||
M, male; F, female.
STLV-1 serology was determined by IFA and Western blotting (10); SIV serology was determined by specific enzyme-linked immunosorbent assay with Western blot confirmation (4).
KSHV serology was determined by IFA at a dilution of 1/40 (3).
Distribution of the different novel herpesviruses isolated from the 15 mandrill and drill monkeys for which DNA was available as determined by specific oligonucleotide probe hybridization of the DFASA and GDTD1B nPCR products. Boldface plus signs correspond to viruses for which herpesvirus DNA polymerase GenBank accession numbers are available.
We first performed a serological analysis of all 27 animals to determine the seroprevalence of KSHV-related viruses. The plasma samples were tested at a 1/40 dilution by an immunofluorescence assay (IFA) which detects unspecified latent and lytic KSHV antigens (Advanced Biotechnology Inc., Columbia, Md.) (3). Serum from only one drill reacted faintly at a 1/40 dilution in this assay (Table 1). No fluorescent reactivity to the KSHV antigen-producing cells (KS-1) was present in the plasma of all the other 8 drill, 16 mandrill, and 2 drill-mandrill hybrid monkeys.
We then attempted to amplify herpesviral sequences from the peripheral blood mononuclear cell DNAs (extracted with a QIAamp DNA Blood Mini kit; Qiagen GmbH, Hilden, Germany) of 15 of the drills and mandrills from Gabon and Cameroon. We used a nested-PCR (nPCR) method with degenerate consensus primers targeted to highly conserved amino acid motifs within the herpesviral DNA polymerase gene according to the method of Rose et al. (14). This assay has been described to be a powerful tool in the search for new animal herpesviruses. We slightly modified the PCR cycling conditions. Briefly, after the DNAs were denaturated at 94°C for 10 min, the reaction mixtures were cycled five times at 94°C for 30 s, 60°C for 1 min, and 72°C for 1 min, followed by 30 cycles at 94°C for 30 s, 46°C for 30 s, and 72°C for 30 s. An extension of 10 min at 72°C was realized on the last cycle (GeneAmp PCR system 9600 thermal cycler; Perkin-Elmer, Branchburg, N.J.). As seen in Fig. 1, DNA samples were initially amplified with the primer pools DFASA and GDTD1B, and an aliquot (2%) of these amplification products was then used as a template in a subsequent nPCR with the VYGA and GDTD1B primer pools. The products of the secondary nPCR amplification were electrophoresed in a 2.5% agarose gel and visualized by irradiation with UV in the presence of ethidium bromide. Amplification products of the predicted size (∼236 bp) were detected and gel purified with the QIAquick gel extraction kit (Qiagen GmbH). The resulting purified fragments were cloned into pCR2.1 vectors using a TA cloning kit from Invitrogen (Carlsbad, Calif.) and sequenced by Eurogentec (Seraing, Belgium) using the BigDye terminator technology. To obtain the nucleotide sequence extending upstream of the VYGA region, a nested set of gene-specific nondegenerate oligonucleotide primers was derived from the complementary sequences of the fragments (Table 2) and used in an nPCR amplification with the DFASA primer pool. The PCR products (DFASA and GDTD1B) from the initial PCR were used as template DNAs in these subsequent amplification reactions. Finally, the upstream nPCR products obtained were subsequently cloned and sequenced. The nucleotide sequences of the DFASA and GDTD1B PCR fragments yielded 476- to 479-bp sequences after exclusion of the 5′ and 3′ primer sequences. Sequencing of seven such PCR products (DFASA and GDTD1B) and searches of the GenBank database by using the BLAST web server revealed the presence of not only two new gamma-2 herpesviruses but also of two new betaherpesvirus sequences in these primates.
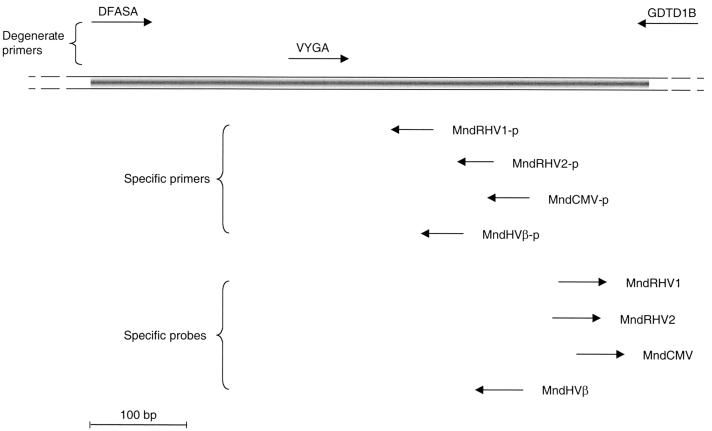
FIG. 1.
Positions and orientations of primers and probes for consensus and virus-specific PCR and for specific Southern hybridization for DNA polymerase. The portion between primers DFASA and GDTD1B of the DNA polymerase gene (open reading frame 9) is shown at the top. Primers above the bar representing the herpesvirus DNA polymerase sequence are the initial herpesvirus degenerate primers used in an nPCR assay to identify novel DNA polymerase sequences. Primers below the bar represent specific primers used in a degenerate (DFASA)-nondegenerate nPCR assay used to amplify the upstream DNA polymerase sequences. Relative positions of the specific oligonucleotide probes used for Southern hybridization on VYGA and GDTD1B nPCR products are shown. The sequences of the oligonucleotides are given in Table 2. A “p” suffix indicates “primer.”
TABLE 2.
Sequences of oligonucleotide primers and probes used for consensus and for gamma-2 and betaherpesvirus-specific PCR for DNA polymerase and Southern hybridization
| Oligonucleotide | Orientationb | 5′→3′ sequencec |
|---|---|---|
| All herpesvirusesa | ||
| DFASA | + | GTG TTC GAC TTY GCN AGY YTN TAY CC |
| VYGA | + | ACG TGC AAC GCG GTG TAY GGN KTN ACN GG |
| GDTD1B | − | CGG CAT GCG ACA AAC ACG GAG TCN GTR TCN CCR TA |
| Specific primers | ||
| MndRHV1-p | − | CCT GTA GCG TGA CCG TCT CG |
| MndRHV2-p | − | CGT AAG ACT TTG ACA TCT CCA GC |
| MndCMV-p | − | CCA GGT TTT CTT CCA CGA AC |
| MndHVβ-p | − | AAC ACC TGA CGC CCC AAA GAT G |
| Specific probes | ||
| MndRHV1 | + | GCG AAC ATG CTG CAG CGA CCC A |
| MndRHV2 | + | AGA CGC GGC TCC GGC GCG CGG TAC G |
| MndCMV | + | TTA ATC GGG AGG ACT ACT CGG |
| MndHVβ | − | TGC TGT GTC AAT GTA ATC GGT G |
Degenerate oligonucleotide primers described in reference 15.
+, sense; −, antisense.
Positions of degeneracy are given. N = A, C, G, and T; Y = C and T; R = A and G; K = G and T.
Regarding the two new gamma-2 herpesvirus sequences, which were tentatively termed MndRHV1 and MndRHV2, comparison of nucleotide and amino acid identities among primate rhadinoviruses indicated that MndRHV1 was most closely related to viruses belonging to the RV1 genogroup, with RFHVMn and RFHVMm being the closest ones (81 and 80% nucleotide identity, respectively) (Table 3). In contrast, MndRHV2 belonged to the RV2 group and was most closely related to ChRV2 and to Macaca fascicularis gammaherpesvirus (84 and 85% nucleotide identity, respectively) (Table 3). Otherwise, our BLAST search for the two other new herpesvirus sequences demonstrated that these sequences were most similar to the DNA polymerases of the Betaherpesvirinae subfamily. One of these, which we termed MndCMV for mandrillus cytomegalovirus, was close to rhesus herpesvirus 5 (RhHV5) (or RhCMV) (18) and human CMV (HCMV), with 79 and 69% nucleotide identity, respectively (Table 3). The latter, tentatively named MndHVβ for mandrillus herpesvirus β, was related to HHV-6 and HHV-7, belonging to the same phylogenetic branch in both the DNA and protein trees (100% bootstrap values) and exhibiting similar levels of nucleotide identity with both of them (58 and 59%, respectively) (Table 3).
TABLE 3.
Nucleotide and amino acid identities between the four novel Mandrillus herpesviruses MndRHV1, MndRHV2, MndCMV, and MndHVβ and the other primate gamma- and betaherpesviruses
| Virusa | % Identityb with:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| MndRHV1
|
MndRHV2
|
MndCMV
|
MndHVβ
|
|||||
| Nucleotide | Amino acid | Nucleotide | Amino acid | Nucleotide | Amino acid | Nucleotide | Amino acid | |
| Gammaherpesviruses | ||||||||
| MndRHV1 | 68 | 69 | 48 | 44 | 43 | 37 | ||
| MndRHV2 | 68 | 69 | 46 | 41 | 43 | 36 | ||
| KSHV | 71 | 82 | 68 | 74 | 46 | 42 | 44 | 37 |
| PanRHV1a | 72 | 81 | 68 | 73 | 45 | 42 | 45 | 37 |
| PanRHV1b | 69 | 78 | 61 | 70 | 46 | 42 | 41 | 35 |
| GorRHV1 | 71 | 80 | 68 | 73 | 48 | 42 | 42 | 35 |
| RFHVMn | 81 | 89 | 66 | 70 | 50 | 43 | 44 | 37 |
| RFHVMm | 80 | 90 | 64 | 68 | 46 | 42 | 44 | 36 |
| ChRV1 | 71 | 81 | 65 | 67 | 48 | 45 | 42 | 36 |
| RRV | 66 | 68 | 84 | 88 | 47 | 41 | 42 | 35 |
| M. fascicularis | 68 | 68 | 85 | 90 | 47 | 41 | 42 | 35 |
| M. nemestrina | 68 | 69 | 84 | 89 | 47 | 42 | 41 | 35 |
| M. mulatta | 66 | 68 | 84 | 88 | 47 | 41 | 42 | 35 |
| MneRV2 | 68 | 68 | 84 | 88 | 48 | 42 | 41 | 37 |
| ChRV2 | 70 | 72 | 84 | 91 | 47 | 41 | 41 | 37 |
| HVA3 | 60 | 64 | 59 | 66 | 45 | 40 | 43 | 35 |
| HVS | 59 | 65 | 61 | 69 | 45 | 39 | 44 | 35 |
| EBV | 63 | 59 | 63 | 59 | 47 | 39 | 43 | 35 |
| Betaherpesviruses | ||||||||
| MndCMV | 47 | 44 | 45 | 40 | 46 | 39 | ||
| MndHVβ | 43 | 37 | 43 | 36 | 46 | 39 | ||
| RhHV5 | 49 | 45 | 45 | 40 | 79 | 81 | 45 | 40 |
| HCMV | 48 | 39 | 47 | 35 | 69 | 70 | 44 | 39 |
| HHV-6 | 46 | 40 | 46 | 41 | 48 | 42 | 58 | 61 |
| HHV-7 | 42 | 41 | 41 | 39 | 45 | 42 | 59 | 61 |
HVA3, ateline herpesvirus 3; HVS, herpesvirus saimiri; EBV, Epstein-Barr virus; RhHV5, rhesus herpesvirus 5.
Numbers refer to values obtained in comparison with the 454-bp fragment, which is available for all these viruses. Boldface indicates sequences of highest identity.
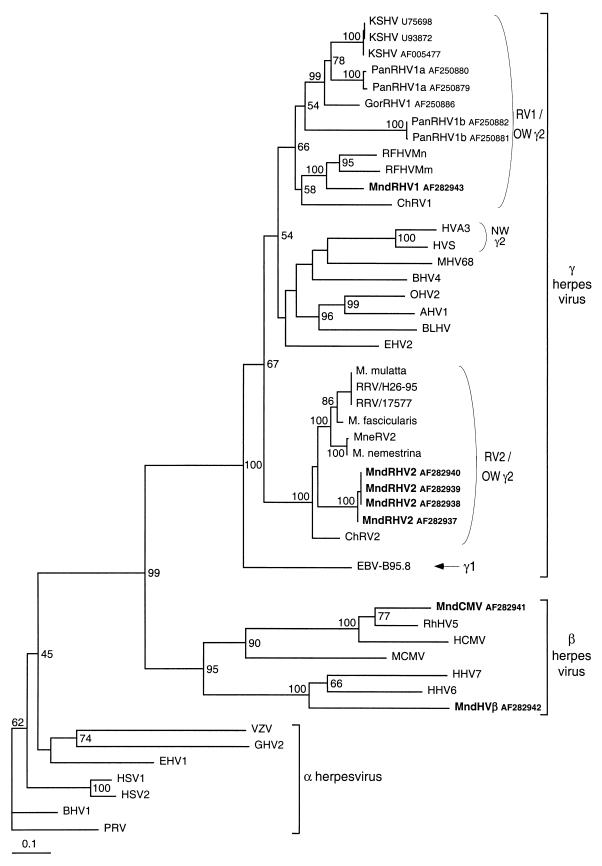
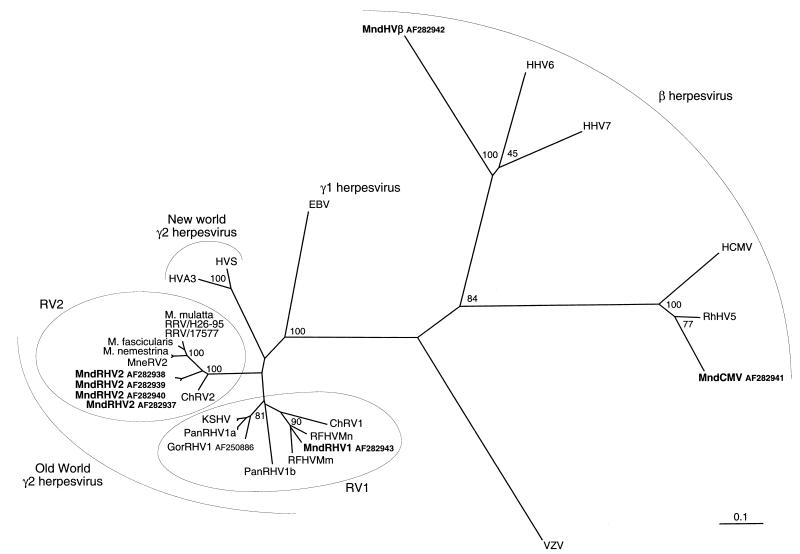
Phylogenetic analyses using different methods (neighbor joining and DNA maximum parsimony) clearly placed these four novel mandrill and drill viruses within the rhadinovirus genus for MndRHV1 and MndRHV2 and within the Betaherpesvirinae subfamily for MndCMV and MndHVβ (Fig. 2 and 3). Regarding the four new Mandrillus herpesviruses reported here, a very similar branching order was obtained and was well supported by high bootstrap values by both phylogenetic methods at the nucleotide level and at the amino acid level. MndRHV1 branches with the human (KSHV), chimpanzee (PanRHV1a and PanRHV1b), gorilla (GorRHV1), macaque (RFHVMm and RFHVMn), and African green monkey (ChRV1) viruses, while MndRHV2 branches separately with the macaque gammaherpesviruses (RRV, MneRV2, and three viruses herein designated simply as M. mulatta, M. fascicularis, and M. nemestrina) and ChRV2. Thus, as for macaque and African green monkeys, mandrill and drill gamma-2 herpesviruses fall, respectively, in the two fairly distinct lineages, RV1 and RV2, among the Old World monkey rhadinoviruses (Fig. 2 and 3). Moreover, sequences derived from each viral species were unique to that species (Fig. 3).
FIG. 2.
Phylogenetic tree resulting from analysis of selected 454-bp fragments (primers QAHNA and GDTD1B (15) of the herpesvirus DNA polymerase gene, which is available for all viruses. The phylogeny was derived by the neighbor-joining method applied to pairwise sequence distances calculated using the Kimura two-parameter method (transition/transversion ratio set to 2). Horizontal branch lengths are drawn to scale, with the bar indicating 0.1 nucleotide replacement per site. Numbers at each node indicate the percentage of bootstrap samples (out of 100) in which the cluster to the right is supported. Brackets on the right indicate previously defined subfamily and genus herpesviral classification. Previously published sequences and their GenBank accession numbers (in parentheses) are as follows: HHV-1 and herpes simplex virus type 1 (HSV1) (X04771); HHV-2 and HSV2 (M16321); HHV-3 and varicella-zoster virus (VZV) (X04370); HHV-4 and Epstein-Barr virus (EBV-B95.8) (V01555); HHV-5 and HCMV (M14709); HHV-6A (X83413); HHV-7 (U43400); HHV-8 and KSHV (U75698, U93872, and AF005477); herpesvirus saimiri (HVS) (M31122); ateline herpesvirus 3 (HVA3) (AF083424); ChRV1 (AJ251573); ChRV2 (AJ251574); RFHVMn (AF005478); RFHVMm (AF005479); RRV and 17577 (AF083501); RRV and H26-95 (AF029302); Macaca gamma virus strains Macaca mulatta (AF159033), Macaca fascicularis (AF159032), and Macaca nemestrina (AF159031), named here M. mulatta, M. fascicularis, and M. nemestrina, respectively; MneRV2 (AF204167); PanRHV1a (AF250879 and AF250880); PanRHV1b (AF250881 and AF250882); GorRHV1 (AF250886); pseudorabies virus (PRV) (L24487); bovine herpesvirus 1 (BHV1) (Z78205), equine herpesvirus 1 (EHV1) (M86664), Marek's disease virus/gallid herpesvirus 2 (GHV2) (L40431); RhCMV (AF0033184); mouse CMV (MCMV) (U68299); EHV2 (U20824); bovine lymphotropic herpesvirus (BLHV) (AF031808); avian herpesvirus 1 (AHV1) (AF005370); ovine herpesvirus 2 (OHV2) (AF031812); murine herpesvirus 68 (MHV68) (U97553); and BHV4 (AF031811). OW, Old World; NW, New World.
FIG. 3.
Neighbor-joining protein distance tree for the 151 amino acid residues encoded by the 454-bp fragment (primers QAHNA and GDTD1B) (15) of DNA polymerase. Sequences were aligned by using CLUSTAL W and analyzed by using the PROTDIST and NEIGHBOR programs in PHYLIP. One hundred replica samplings were subjected to bootstrap analysis (SEQBOOT). The branch lengths are proportional to the evolutionary distances (scale bar) between the taxa. Previously published sequences included and their GenBank accession numbers (in parentheses) are as follows: HHV-6A (X83413); HHV-7 (U43400); HHV-5 and human HCMV (M14709); RhCMV (AF0033184); herpesvirus saimiri (HVS) (M31122); ateline herpesvirus 3 (HVA3) (AF083424); HHV-4 and Epstein-Barr virus (EBV) (V01555); HHV-8 and KSHV (U75698, U93872, and AF005477); ChRV1 (AJ251573); ChRV2 (AJ251574); RFHVMn (AF005478); RFHVMm (AF005479); RRV and 17577 (AF083501); RRV and H26-95 (AF029302); Macaca gamma virus strains Macaca mulatta (AF159033), Macaca fascicularis (AF159032), and Macaca nemestrina (AF159031), named here M. mulatta, M. fascicularis, and M. nemestrina, respectively; MneRV2 (AF204167); PanRHV1a (AF250879 and AF250880); PanRHV1b (AF250881 and AF250882); and GorRHV1 (AF250886).
Although we describe here the first CMV homologue in drills, several CMV-like viruses have already been reported for different species of baboons (11) and macaques (18). In contrast, this study constitutes to our knowledge the first molecular identification of a simian homologue of HHV-6 and -7.
In order to assess the prevalence of these new sequences of herpesviruses in mandrills and drills, specific oligonucleotide probes were designed for Southern hybridizations on VYGA and GDTD1B nPCR products. Hybridization with the MndRHV1-specific probe showed that only one M. sphinx (Mnd15) was infected by this virus, while MndRHV2 probe hybridization revealed that five of our animals (four from Cameroon and one from Gabon) were infected by this novel gamma-2 herpesvirus (Table 1). Interestingly, Mnd15, an STLV-1- and SIV-positive animal, was the only monkey coinfected by the two new representatives of the mandrillus gamma-2 herpesviruses. Multiple infections were also observed for different representatives of the mandrillus gamma-2 and betaherpesviruses. Actually, hybridizations with the MndCMV and MndHVβ probes demonstrated that three and four animals were infected by these viruses, respectively. Among them, several animals were infected by different representatives of these new viruses. For instance, monkeys Mnd204 and Mnd206 were coinfected by three of the four novel herpesviruses described here, MndRHV2, MndCMV, and MndHVβ (Table 1). This situation of multiple herpesviral infection is similar to that observed for humans, who may be infected by different combinations of HHVs. Finally, the DNA sequences obtained for the four MndRHV2 isolates were 99.2% identical (471 of 476 bp) (with an amino acid divergence of 1.3%; 156 of 158 amino acids) between animal Mnd15 and animals Mnd204, Mnd401, and Mnd402. The three MndRHV2 sequences obtained from the last three animals listed above were identical. These data suggest that Cameroonese (Mnd204, Mnd401, and Mnd402) and Gabonese (Mnd15) mandrillus monkeys contained very similar but not identical isolates of MndRHV2. While sequence differences between Macaca mulatta and Macaca nemestrina gamma-2 herpesviruses suggest that cross-species transmission, in a primate center setting, is not common and that macaque rhadinovirus sequences may have thus evolved within their host species, such results are not observed for mandrillus, as MndRHV2 infects either M. leucophaeus or M. sphinx as well as hybrid M. leucophaeus-M. sphinx monkeys. This suggests that cross-species transmission may have occurred perhaps during infancy in these animals, who lived often in close contact in the same enclosure in Cameroon.
Our data clearly show that mandrill and drill monkeys are hosts of not only two novel and distinct herpesviruses belonging to the two known lineages of gamma-2 herpesviruses (as previously reported for macaques and African green monkeys) but also of two new betaherpesviruses, one related to CMVs and the second one related to HHV-6 and -7. We have also shown that members of each of these viral lineages can naturally coinfect the same host animal at least in semi-free-range animal facilities. However, the significant nucleotide and amino acid differences between the members of the gamma-2 herpesvirus lineages suggest that they have separately evolved over a long period of time. Significantly, strain-to-strain sequence variations, observed for the MndRHV2 DNA polymerase coding fragment studied, were found to be much less profound than species-to-species variations (MndRHV1 versus MndRHV2).
The identification of four new herpesviruses in these Central African Old World monkey species indicates that such animals constitute an important reservoir of novel viruses. The close identity of the new mandrill and drill herpesviruses with their human pathogenic counterparts, their presence in peripheral blood mononuclear cells, and the importance in the western part of Central Africa of contacts between such monkeys and humans, especially during hunting, indicate the potential of viral interspecies transmission (8). As an example, it has been recently suggested that contacts between inhabitants and mandrills in these regions might account for recurring episodes of interspecies viral transmission of mandrill STLV-1 to HTLV-1 subtype D (10).
The extensive amount of sequence information available for the primate gamma-2 herpesviruses facilitates studies of phylogenetic relationships. MndRHV1, the mandrillus virus which appears to be more closely related to KSHV, represents the KSHV homologue in this species. This issue will, however, be definitely resolved once more sequence information on this strain becomes available (characterization of the complete DNA polymerase and/or glycoprotein B is ongoing, for example). The identification of MndRHV2, belonging to the RV2 genogroup, confirms that Old World primate rhadinoviruses are broadly grouped in two clusters. The two mandrillus gamma-2 herpesviruses are more distantly related to KSHV, as are the macaque and African green monkey viruses, than the chimpanzee and gorilla viruses (9). With chimpanzee and gorilla species being evolutionary closest to humans, these data support the central hypothesis that evolution of herpesviruses, and in our case gamma-2 herpesviruses, has occurred by cospeciation with their hosts.
Comparative data obtained for all the nonhuman primate species, including those presented here, raise the possibility of the existence of another gamma-2 herpesvirus, belonging to the second rhadinovirus lineage RV2, in humans, for which only KSHV, belonging to the RV1 genogroup, has been identified to date. The ongoing identification of novel RV2 herpesvirus sequences in other nonhuman primate species (V. Lacoste et al., submitted) and the generation of new consensus degenerate primers targeted to the herpesvirus DNA polymerase may be helpful in the detection and identification of this putative human RV2 herpesvirus.
Regarding the names of the new primate herpesviruses described in this paper, we have tentatively and provisionally provided them names such as MndRHV1 for mandrillus rhadinovirus 1. However, among the specialists of the field, a new proposal for primate rhadinovirus nomenclature is being discussed and debated. When new names are approved by the consensus of such specialist groups, we will, of course, modify the names of these new herpesviruses in our papers.
Acknowledgments
Vincent Lacoste is a recipient of a fellowship from the Ligue Nationale Contre le Cancer. This work was partly supported by grants from the Agence Nationale de Recherches sur le SIDA (ANRS), SIDACTION, the Association de Recherches sur le Cancer (ARC), and Action Concertée from the network of the Institut Pasteur.
We acknowledge Peter Jenkins and Liza Gadsby (the Pandrillus Directors) for their great help in obtaining some of the blood samples studied and their continuous interest in this work. We thank also Y. Chaduc for providing the sample from the Touroparc zoo.
REFERENCES
- 1.Bosch M L, Strand K B, Rose T M. Gammaherpesvirus sequence comparisons. J Virol. 1998;72:8458–8459. doi: 10.1128/jvi.72.10.8458-8459.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 3.Chatlynne L G, Lapps W, Handy M, Huang Y Q, Masood R, Hamilton A S, Said J W, Koeffler H P, Kaplan M H, Friedman-Kien A, Gill P S, Whitman J E, Ablashi D V. Detection and titration of human herpesvirus-8-specific antibodies in sera from blood donors, acquired immunodeficiency syndrome patients, and Kaposi's sarcoma patients using a whole virus enzyme-linked immunosorbent assay. Blood. 1998;92:53–58. [PubMed] [Google Scholar]
- 4.Corbet S, Muller-Trutwin M C, Versmisse P, Delarue S, Ayouba A, Lewis J, Brunak S, Martin P, Brun-Vezinet F, Simon F, Barre-Sinoussi F, Mauclere P. env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group N from the same geographic area. J Virol. 2000;74:529–534. doi: 10.1128/jvi.74.1.529-534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desrosiers R C, Sasseville V G, Czajak S C, Zhang X, Mansfield K G, Kaur A, Johnson R P, Lackner A A, Jung J U. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J Virol. 1997;71:9764–9769. doi: 10.1128/jvi.71.12.9764-9769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Georges-Courbot M C, Moisson P, Leroy E, Pingard A M, Nerrienet E, Dubreuil G, Wickings E J, Debels F, Bedjabaga I, Poaty-Mavoungou V, Hahn N T, Georges A J. Occurrence and frequency of transmission of naturally occurring simian retroviral infections (SIV, STLV, and SRV) at the CIRMF Primate Center, Gabon. J Med Primatol. 1996;25:313–326. doi: 10.1111/j.1600-0684.1996.tb00023.x. [DOI] [PubMed] [Google Scholar]
- 7.Greensill J, Sheldon J A, Renwick N M, Beer B E, Norley S, Goudsmit J, Schulz T F. Two distinct gamma-2 herpesviruses in African green monkeys: a second gamma-2 herpesvirus lineage among Old World primates? J Virol. 2000;74:1572–1577. doi: 10.1128/jvi.74.3.1572-1577.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hahn B H, Shaw G M, De Cock K M, Sharp P M. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 9.Lacoste V, Mauclère P, Dubreuil P, Lewis J, Georges-Courbot M-C, Gessain A. KSHV-like herpesviruses in chimps and gorillas. Nature. 2000;407:151–152. doi: 10.1038/35025145. [DOI] [PubMed] [Google Scholar]
- 10.Mahieux R, Chappey C, Georges-Courbot M C, Dubreuil G, Mauclere P, Georges A, Gessain A. Simian T-cell lymphotropic virus type 1 from Mandrillus sphinx as a simian counterpart of human T-cell lymphotropic virus type 1 subtype D. J Virol. 1998;72:10316–10322. doi: 10.1128/jvi.72.12.10316-10322.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michaels M G, Alcendor D J, St. George K, Rinaldo C R, Jr, Ehrlich G D, Becich M J, Hayward G S. Distinguishing baboon cytomegalovirus from human cytomegalovirus: importance for xenotransplantation. J Infect Dis. 1997;176:1476–1483. doi: 10.1086/514144. [DOI] [PubMed] [Google Scholar]
- 12.Nerrienet E, Amouretti X, Muller-Trutwin M C, Poaty-Mavoungou V, Bedjebaga I, Nguyen H T, Dubreuil G, Corbet S, Wickings E J, Barre-Sinoussi F, Georges A J, Georges-Courbot M C. Phylogenetic analysis of SIV and STLV type I in mandrills (Mandrillus sphinx): indications that intracolony transmissions are predominantly the result of male-to-male aggressive contacts. AIDS Res Hum Retroviruses. 1998;14:785–796. doi: 10.1089/aid.1998.14.785. [DOI] [PubMed] [Google Scholar]
- 13.Roizmann B, Desrosiers R C, Fleckenstein B, Lopez C, Minson A C, Studdert M J. The family Herpesviridae: an update. The Herpesvirus Study Group of the International Committee on Taxonomy of Viruses. Arch Virol. 1992;123:425–449. doi: 10.1007/BF01317276. [DOI] [PubMed] [Google Scholar]
- 14.Rose T M, Strand K B, Schultz E R, Schaefer G, Rankin G W, Jr, Thouless M E, Tsai C C, Bosch M L. Identification of two homologs of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J Virol. 1997;71:4138–4144. doi: 10.1128/jvi.71.5.4138-4144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schultz E R, Rankin G W, Jr, Blanc M P, Raden B W, Tsai C C, Rose T M. Characterization of two divergent lineages of macaque rhadinoviruses related to Kaposi's sarcoma-associated herpesvirus. J Virol. 2000;74:4919–4928. doi: 10.1128/jvi.74.10.4919-4928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulz T F. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) J Gen Virol. 1998;79:1573–1591. doi: 10.1099/0022-1317-79-7-1573. [DOI] [PubMed] [Google Scholar]
- 17.Searles R P, Bergquam E P, Axthelm M K, Wong S W. Sequence and genomic analysis of a Rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73:3040–3053. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swanson R, Bergquam E, Wong S W. Characterization of rhesus cytomegalovirus genes associated with anti-viral susceptibility. Virology. 1998;240:338–348. doi: 10.1006/viro.1997.8935. [DOI] [PubMed] [Google Scholar]