Abstract
The calponin family proteins are expressed in both muscle and non-muscle cells and involved in the regulation of cytoskeletal dynamics and cell contractility. In the nematode Caenorhabditis elegans, UNC-87 and CLIK-1 are calponin-related proteins with 42 % identical amino acid sequences containing seven calponin-like motifs. Genetic studies demonstrated that UNC-87 and CLIK-1 have partially redundant function in regulating actin cytoskeletal organization in striated and non-striated muscle cells. However, biochemical studies showed that UNC-87 and CLIK-1 are different in their ability to bundle actin filaments. In this study, I extended comparison between UNC-87 and CLIK-1 and found additional differences in vitro and in vivo. Although UNC-87 and CLIK-1 bound to actin filaments similarly, UNC-87, but not CLIK-1, bound to myosin and inhibited actomyosin ATPase in vitro. In striated muscle, UNC-87 and CLIK-1 were segregated into different subregions within sarcomeric actin filaments. CLIK-1 was concentrated near the actin pointed ends, whereas UNC-87 was enriched towards the actin barbed ends. Restricted localization of UNC-87 was not altered in a clik-1-null mutant, suggesting that their segregated localization is not due to competition between the two related proteins. These results suggest that the two calponin-related proteins have both common and distinct roles in regulating actin filaments.
Keywords: Actin, calponin-related proteins, muscle, sarcomere, thin filaments
Introduction
The calponin family proteins, including calponins and calponin-related proteins, are expressed widely in muscle and non-muscle cells and involved in the regulation of cell contractility and cytoskeletal stability (Carmichael, Winder, Walsh, & Kargacin, 1994; Gimona, Djinovic-Carugo, Kranewitter, & Winder, 2002; Hsieh & Jin, 2023; J. Liu, Zhang, Li, & Wang, 2020; R. Liu & Jin, 2016; S. Ono, 2021; Rozenblum & Gimona, 2008; Wu & Jin, 2008). The most extensively characterized member of the calponin family is vertebrate smooth muscle calponin (also known as calponin 1 or CNN1) that directly binds to actin filaments and inhibits actomyosin ATPase (Abe, Takahashi, & Hiwada, 1990; Takahashi, Hiwada, & Kokubu, 1988; Winder & Walsh, 1990). This family of proteins contain variable number of calponin-like (CLIK) motifs acting as actin-binding sequences (Kranewitter, Ylanne, & Gimona, 2001; Takahashi & Nadal-Ginard, 1991). Transgelins (also designated by various names such as SM22, NP25, Drosophila Mp20, and yeast Scp1) contain only one CLIK motif (Ayme-Southgate, Lasko, French, & Pardue, 1989; Prinjha et al., 1994; Ren et al., 1994; Solway et al., 1995; Winder, Jess, & Ayscough, 2003), whereas vertebrate calponins contain three CLIK motifs (Takahashi & Nadal-Ginard, 1991). Comparative studies indicate that the number of CLIK motifs of these proteins correlates with their strength to stabilize actin filaments in cultured cells (Gimona, Kaverina, Resch, Vignal, & Burgstaller, 2003; Lener, Burgstaller, & Gimona, 2004). Interestingly, some invertebrates have proteins with highly expanded repeats of CLIK motifs (up to 23 repeats in mollusks) (S. Ono, 2021), suggesting that the difference in the number of CLIK motifs is one of the mechanisms to adapt the calponin family proteins to different biological functions. However, many species have multiple calponin family isoforms with similar molecular features, and whether these isoforms have different or similar functions remains largely unclear.
The nematode Caenorhabditis elegans has six genes encoding calponin-related proteins with variable molecular features (S. Ono, 2021). Among them, the unc-87 gene encodes a calponin-related protein with seven CLIK motifs (Goetinck & Waterston, 1994a), which bundles actin filaments (Kranewitter et al., 2001), protects actin filaments from severing by actin depolymerizing factor/cofilin (Yamashiro, Gimona, & Ono, 2007), and inhibits actomyosin ATPase (K. Ono, Obinata, Yamashiro, Liu, & Ono, 2015). Mutations in unc-87 cause disorganization of sarcomeric actin filaments in the body wall muscle by affecting the maintenance rather than initial assembly of sarcomeres (Goetinck & Waterston, 1994b). C. elegans also has the clik-1 gene encoding a calponin-related protein with seven CLIK repeats (S. Ono & Ono, 2020; Wang, Park, Liu, & Sternberg, 2018). UNC-87 and CLIK-1 share similar molecular features with 42 % identity in their amino acid sequences containing seven CLIK motifs (S. Ono & Ono, 2020). Both UNC-87 and CLIK-1 are expressed in the body wall muscle and somatic gonad and partially redundant in the regulation of contractility and actin organization in these tissues (K. Ono et al., 2015; S. Ono & Ono, 2020), suggesting that these proteins have overlapping functions in vivo. However, in in vitro experiments, UNC-87 bundles actin filaments (Kranewitter et al., 2001; Yamashiro et al., 2007), whereas CLIK-1 binds to actin filaments without bundling them (S. Ono & Ono, 2020), suggesting that these proteins also have different biological roles. In this study, I extended the comparative studies of UNC-87 and CLIK-1 in vitro and in vivo and found additional similarities and differences between these related proteins. Notably, they have different effects on myosin binding and actomyosin ATPase in vitro and localize to different subregions within sarcomeric actin filaments in vivo. These results suggest that UNC-87 and CLIK-1 have both common and distinct regulatory roles for the actin cytoskeleton.
Results
UNC-87, but not CLIK-1, inhibits actomyosin ATPase
To better understand the biochemical similarities and differences between UNC-87 and CLIK-1, I compared the modes of their interactions with actin under three different in vitro conditions. First, their binding to actin filaments was tested in the presence of various concentrations of potassium chloride (Fig. 1), because many actin-binding proteins bind to actin filaments electrostatically such that their actin binding is weakened in the presence of high salt. For example, tropomyosin can be dissociated from actin filaments in the presence of 0.6 M potassium chloride (S. Ono & Ono, 2002). Actin filaments were incubated with UNC-87 or CLIK-1 in the presence of 0.1 – 0.6 M potassium chloride, and their binding was examined by co-sedimentation assays. The amounts of UNC-87 or CLIK-1 that co-sedimented with F-actin were not significantly different at all tested salt concentrations (Fig. 1). Therefore, UNC-87 and CLIK-1 were similarly insensitive to high-salt conditions in actin binding, suggesting that hydrophobic interactions are involved in their binding.
Figure 1.
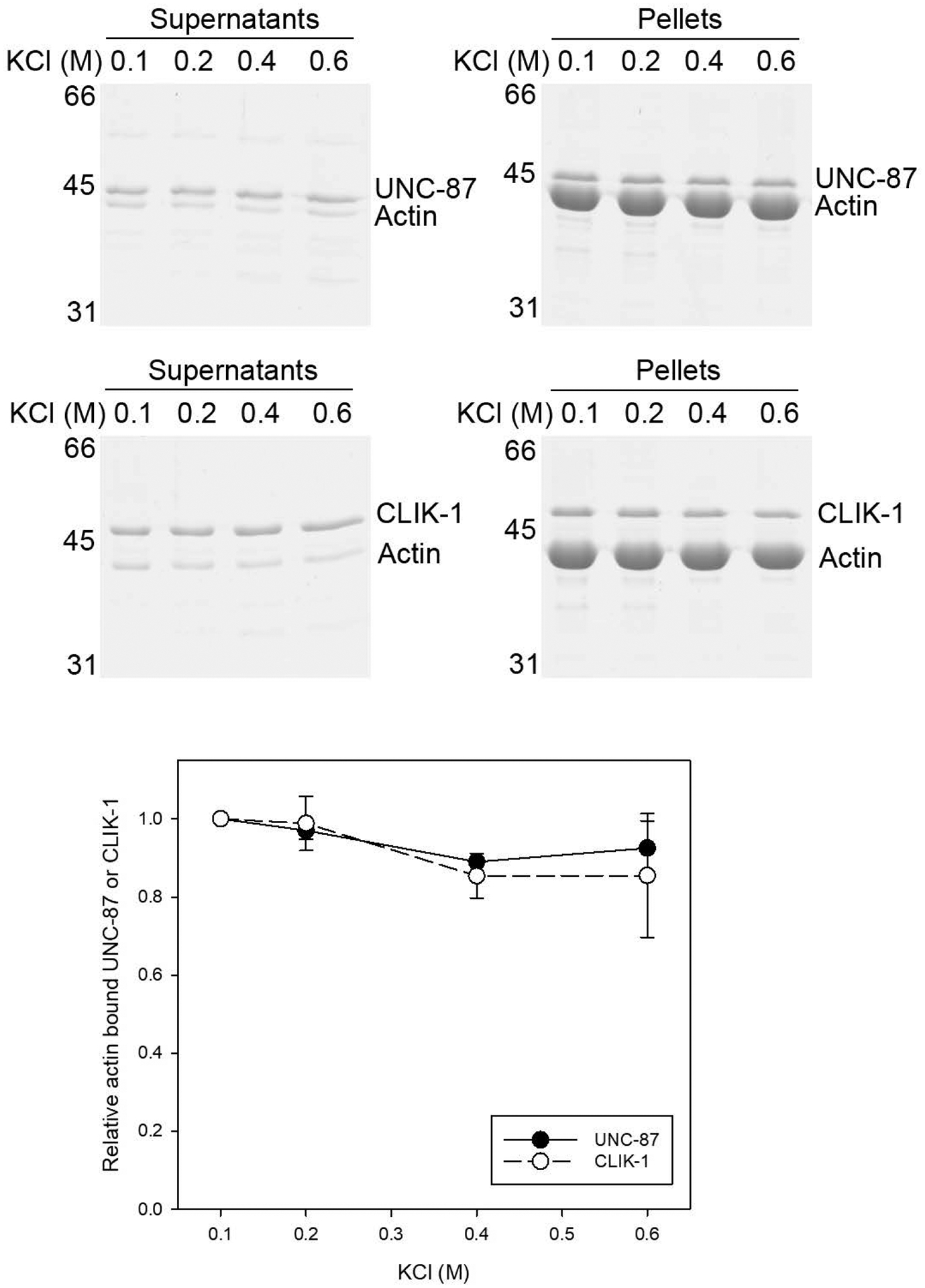
Binding of UNC-87 or CLIK-1 to actin filaments is insensitive to potassium chloride concentrations. UNC-87 or CLIK-1 at 2 μM was incubated with 5 μM F-actin in the presence of 0.1 – 0.6 M KCl for 1 h and ultracentrifuged. Supernatants and pellets were separated and examined by SDS-PAGE. Molecular mass markers (M) in kDa are shown on the left. Positions of UNC-87, CLIK-1, and actin are indicated on the right. Relative amounts of actin-bound UNC-87 (black circles) or CLIK-1 (white circles) were quantified by densitometry and plotted as a function of KCl concentrations. Three independent experiments were performed and plotted as average ± standard deviation.
Second, their actin-filament binding was tested in the presence of various concentrations of inorganic phosphate (Pi) (Fig. 2). Inorganic phosphate binds to ADP-actin subunits within the filaments to generate the ADP-Pi state that is structurally distinct from the ADP state (Carlier & Pantaloni, 1988; Chou & Pollard, 2019; Merino et al., 2018), which can affect actin binding of actin depolymerizing factor/cofilin (Blanchoin & Pollard, 1999; Carlier et al., 1997; Muhlrad, Pavlov, Peyser, & Reisler, 2006; S. Ono & Benian, 1998) and coronin (Cai, Makhov, & Bear, 2007; Gandhi, Achard, Blanchoin, & Goode, 2009; Ge, Durer, Kudryashov, Zhou, & Reisler, 2014). Actin filaments were incubated with UNC-87 or CLIK-1 in the presence of 0 – 25 mM Pi, and their binding was examined by co-sedimentation assays. The amounts of UNC-87 or CLIK-1 that co-sedimented with F-actin were not significantly different at all tested phosphate concentrations (Fig. 2). The results indicate that both UNC-87 and CLIK-1 binds to F-actin similarly in either ADP or ADP-Pi state.
Figure 2.
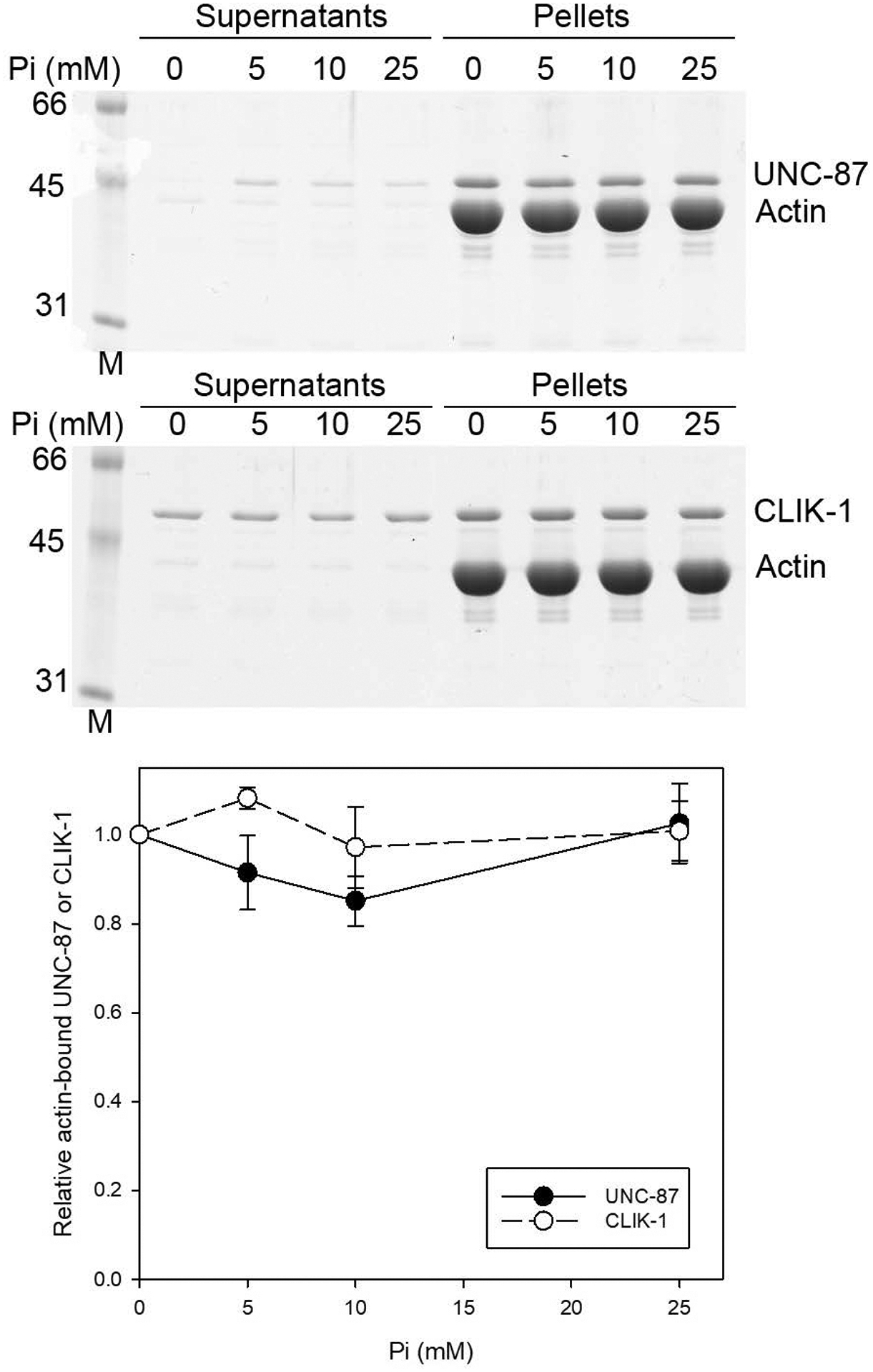
Binding of UNC-87 or CLIK-1 to actin filaments is insensitive to inorganic phosphate concentrations. UNC-87 or CLIK-1 at 2 μM was incubated with 5 μM F-actin in the presence of 0 – 25 mM inorganic phosphate (Pi; in the form of sodium phosphate, pH 7.5) for 1 h and ultracentrifuged. Supernatants and pellets were separated and examined by SDS-PAGE. Molecular mass markers (M) in kDa are shown on the left. Positions of UNC-87, CLIK-1, and actin are indicated on the right. Relative amounts of actin-bound UNC-87 (black circles) or CLIK-1 (white circles) were quantified by densitometry and plotted as a function of Pi concentrations. Three independent experiments were performed and plotted as average ± standard deviation.
Third, competition between UNC-87 and CLIK-1 for F-actin binding was examined at different nucleotide states of F-actin (Fig. 3). UNC-87 and CLIK-1 were mixed at various ratios (total concentrations of UNC-87 plus CLIK-1 were fixed at 5 μM) and tested for their binding to F-actin by co-sedimentation assays at the ADP, ADP-Pi, or adenylyl-imidodiphosphate (AMPPNP; a non-hydrolysable ATP analog) state (Fig. 3A). Then, ratios of CLIK-1 and UNC-87 in the F-actin-bound pellet fractions were quantified [shown as CLIK-1 in pellet (%) in Fig. 3B]. In the ADP state, CLIK-1 was less preferentially bound to F-actin than UNC-87 (Fig. 3B, triangles). However, in the ADP-Pi or AMPPNP state, the preference for UNC-87 in F-actin binding was much less evident, and CLIK-1 was only slightly less favorably bound to F-actin than UNC-87 (Fig. 3B circles and squares). Therefore, when both UNC-87 and CLIK-1 are present, UNC-87 binds to F-actin slightly better than CLIK-1 in the ADP state but not in the ADP-Pi or ATP (as suggested from the AMPPNP actin) state.
Figure 3.
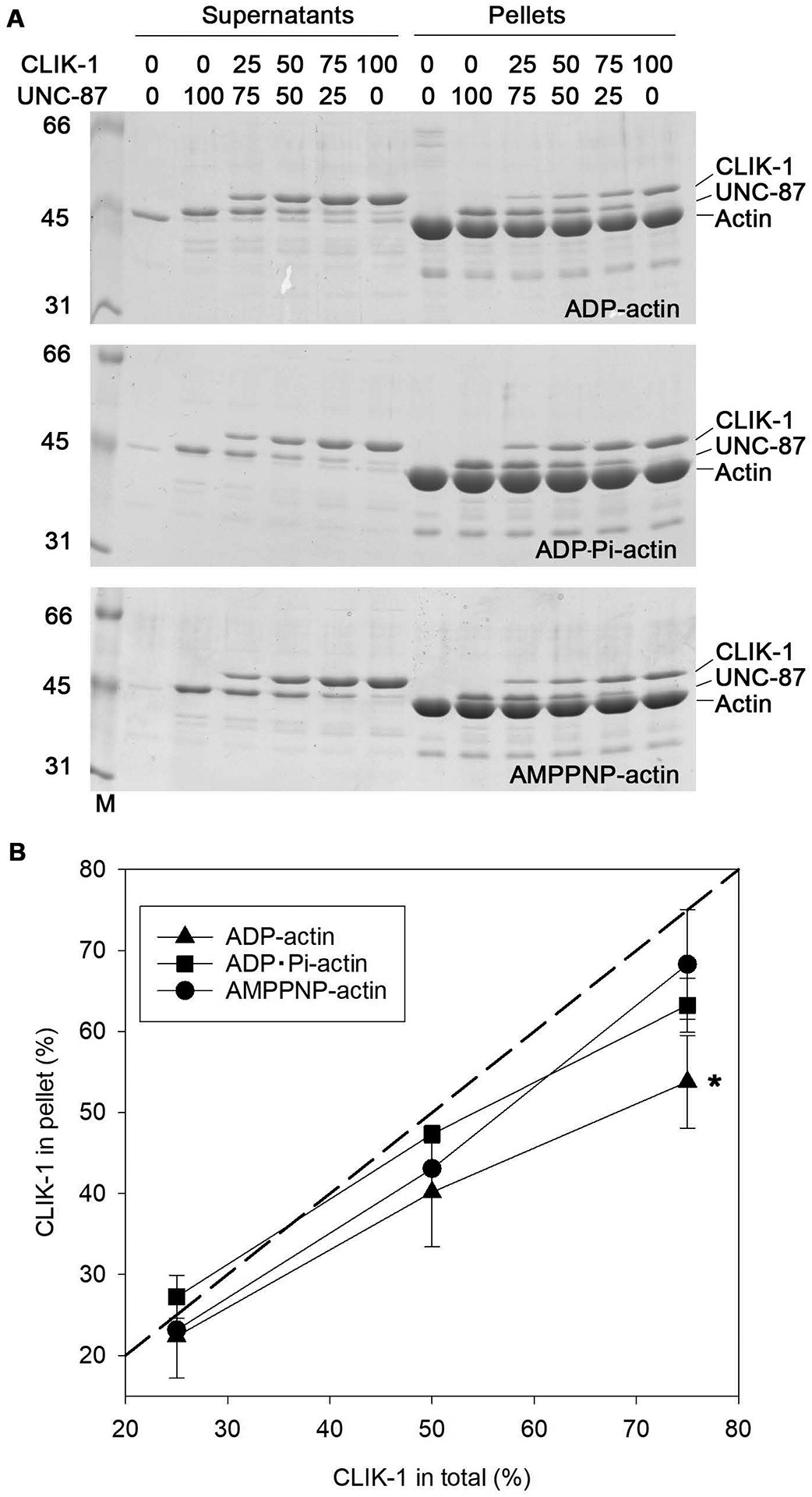
UNC-87 is more preferentially bound to F-actin than CLIK-1 in the ADP, but not ADP-Pi or AMPPNP, state. (A) UNC-87 and CLIK-1 were mixed at total 5 μM (percentages in each mixture are indicated on the top of the gels), incubated with 10 μM F-actin in ADP (top), ADP-Pi (middle), or AMPPNP (bottom) state for 1 h, and ultracentrifuged. Supernatants and pellets were separated and examined by SDS-PAGE. Molecular mass markers (M) in kDa are shown on the left. Positions of UNC-87, CLIK-1, and actin are indicated on the right. (B) Molar ratios of UNC-87 and CLIK-1 in the pellet fractions were quantified by densitometry, and percentages of CLIK-1 in pellets were plotted. A dashed line indicates theoretical percentages of CLIK-1 in pellets (%) when UNC-87 and CLIK-1 binds to F-actin with the same affinity. Three independent experiments were performed and plotted as average ± standard deviation. *, p < 0.05.
Fourth, the effects of UNC-87 and CLIK-1 on myosin were compared (Fig. 4). As previously demonstrated (K. Ono et al., 2015), UNC-87 bound to myosin in the co-sedimentation assays (Fig. 4A). When a constant concentration of UNC-87 was incubated with increasing concentrations of myosin, UNC-87 was shifted from the supernatants to pellets by co-sedimentation with myosin (Fig. 4A and C). However, under the same conditions, CLIK-1 mostly remained in the supernatants even in the presence of myosin (Fig. 4B and C), indicating that CLIK-1 poorly bound to myosin. We next compared the effects of UNC-87 and CLIK-1 on actomyosin ATPase (Fig. 4D). The myosin ATPase was activated in the presence of F-actin (Fig. 4D). At 3 μM, CLIK-1 did not have a significant effect on the actin-activated myosin ATPase activity, whereas UNC-87 partially inhibited the ATPase activity (Fig. 4D), as demonstrated previously (K. Ono et al., 2015). Moreover, an excess amount of CLIK-1 (up to 6 μM) failed to release the inhibitory effect of UNC-87 (Fig. 4D). Therefore, although UNC-87 and CLIK-1 compete for F-actin binding (S. Ono & Ono, 2020), they have distinct effects on actomyosin ATPase because of different affinities for myosin.
Figure 4.
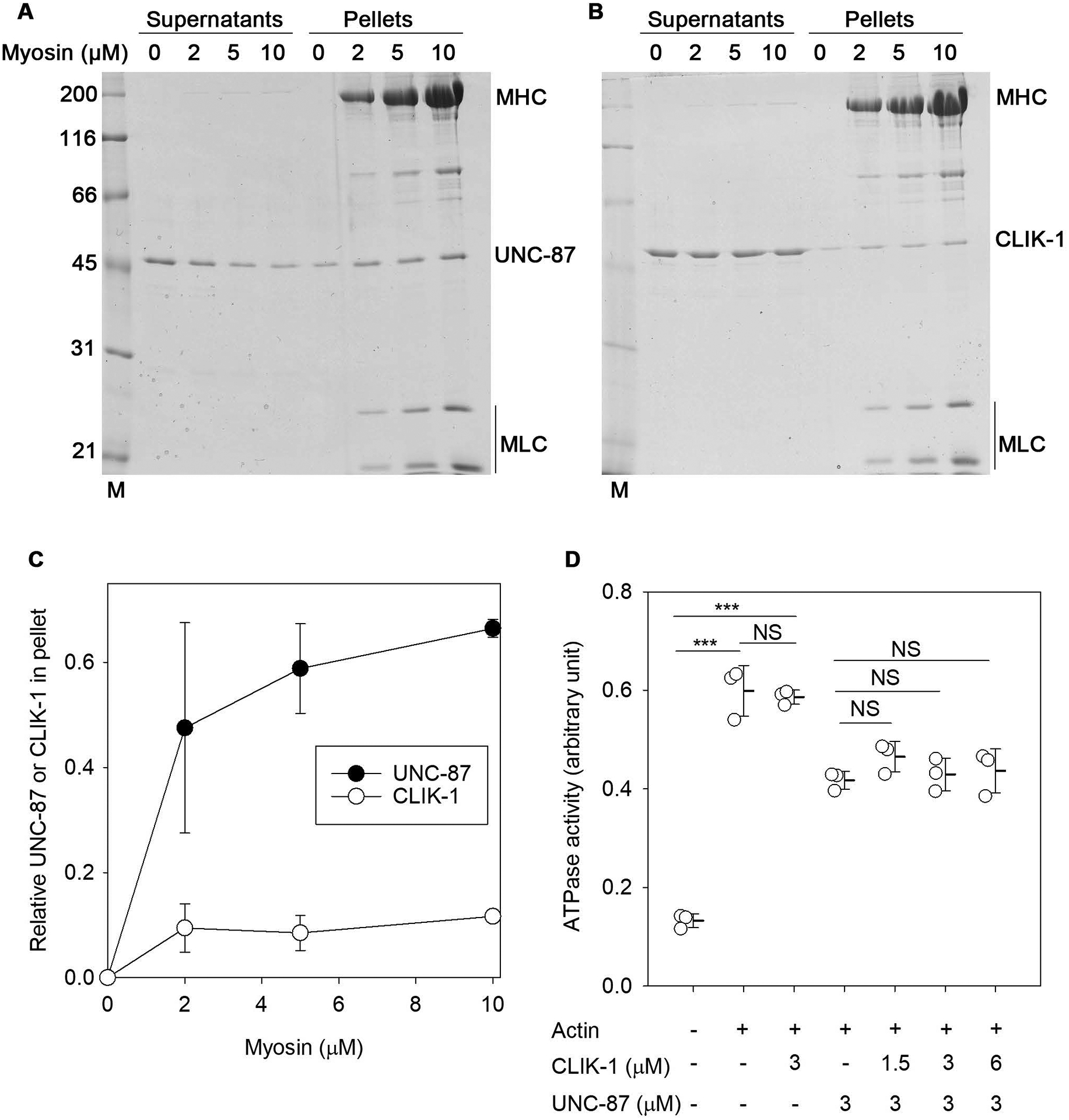
UNC-87, but not CLIK-1, binds to myosin and inhibits actomyosin ATPase. (A, B) Rabbit muscle myosin (0 – 10 μM) was incubated with 2 μM of UNC-87 (A) or CLIK-1 (B) for 1 h and ultracentrifuged. Supernatants and pellets were separated and examined by SDS-PAGE. Molecular mass markers (M) in kDa are shown on the left. Positions of myosin heavy chain (MHC), myosin light chains (MLC), UNC-87, and CLIK-1 are indicated on the right. (C) Relative amounts of myosin-bound UNC-87 (black circles) or CLIK-1 (white circles) in the pellets were quantified by densitometry and plotted as a function of myosin concentrations. Data are average ± standard deviation from three independent experiments. (D) ATPase activity (arbitrary unit) of rabbit muscle myosin (0.4 μM) was determined in the absence or presence of 1.0 μM actin, 1.5 – 6.0 μM UNC-87 and/or 3 μM CLIK-1. Data are average ± standard deviation from three independent experiments. NS, not significant. ***, p < 0.001.
Endogenous tagging of CLIK-1 with GFP does not alter sarcomeres and worm motility
To localize CLIK-1 in vivo, I used a previously reported strain that had been modified to tag endogenous CLIK-1 with green fluorescent protein (GFP) by CRISPR/Cas9-mediated genome editing (S. Ono & Ono, 2020). Effects of GFP tagging on the function of CLIK-1 were tested in sarcomeric F-actin organization and worm motility (Fig. 5, Fig. S1). In wild-type background, CLIK-1::GFP did not alter the sarcomeric pattern of F-actin (Fig. 5A, compare top two rows). However, due to functional redundancy between unc-87 and clik-1, the clik-1 loss-of-function phenotypes appear only in unc-87 mutants (S. Ono & Ono, 2020). Therefore, CLIK-1::GFP was homozygously combined with unc-87(e1216), a loss-of-function allele (Goetinck & Waterston, 1994a), but no detectable enhancement in disorganization of sarcomeric F-actin was observed (Fig. 5A, compare bottom two rows). The localization pattern of CLIK-1::GFP in unc-87 was very similar to that of F-actin in both disorganized sarcomeres and aggregates (Fig. 5A, bottom row). Furthermore, worm motility, as an indication of muscle contractility, was not significantly altered by CLIK-1::GFP in both wild-type and unc-87 backgrounds (Fig. 5B). These results suggest that endogenous tagging of CLIK-1 with GFP does not significantly alter the function of CLIK-1.
Figure 5.
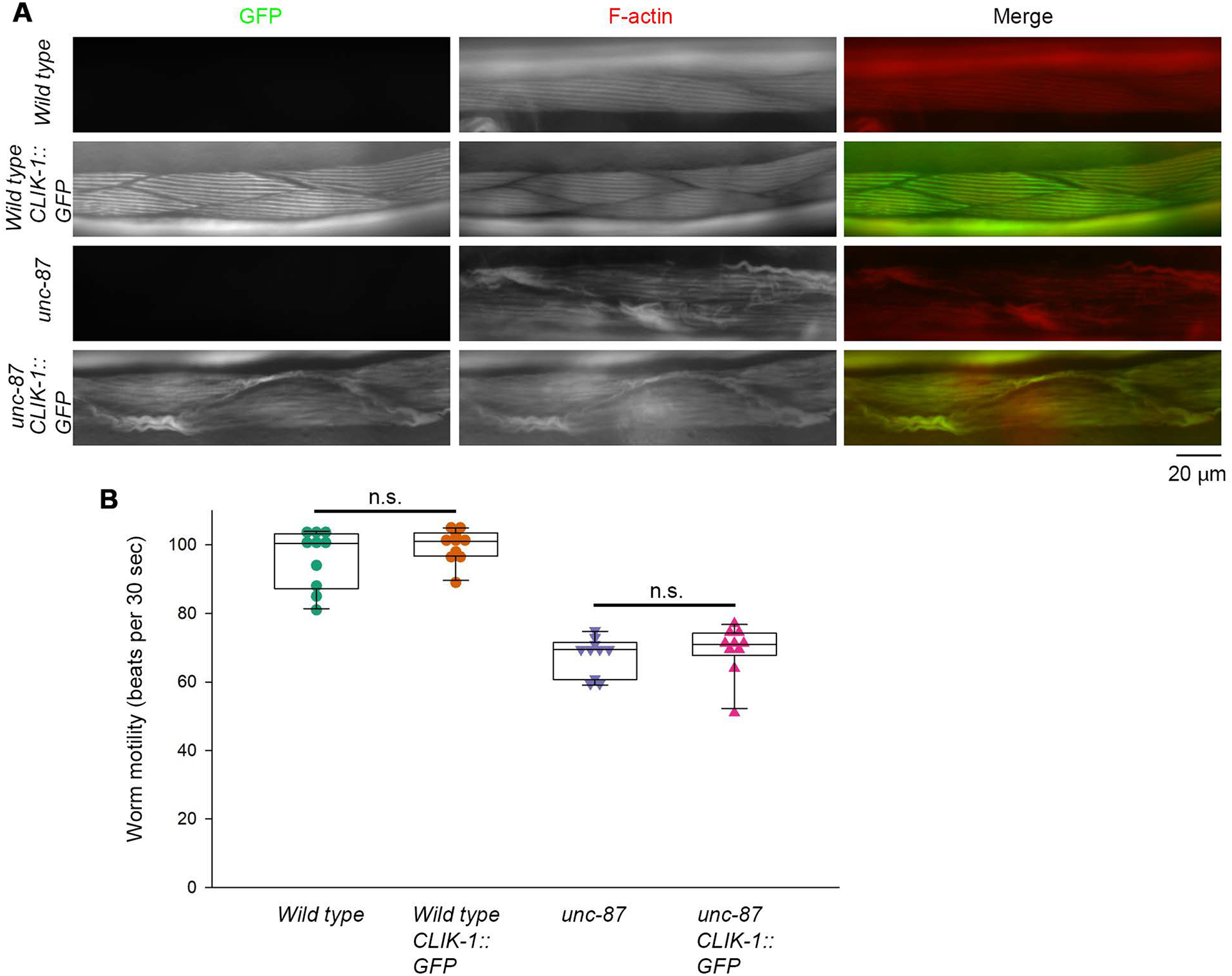
Tagging of endogenous CLIK-1 with GFP does not disturb sarcomere organization and worm motility. (A) Sarcomeric F-actin organization was examined by staining worms with ATTO594-labeled phalloidin. Worms without or with a GFP tag on endogenous CLIK-1 (CLIK-1::GFP) in wild-type or unc-87(e1216) background were tested. Merged images of GFP (green) and F-actin (red) are shown. Bar, 20 μm. (B) Worm motility was quantitatively measured as number of beats per 30 s. n = 10. Boxes represent the range of the 25th and 75th percentiles, with the medians marked by solid horizontal lines, and whiskers indicate the 10th and 90th percentiles. n.s., not significant. A green-magenta version of this figure is presented in Figure S1.
CLIK-1 and UNC-87 are segregated into different subdomains in sarcomeric actin filaments
In our previous studies, we reported that both UNC-87 and CLIK-1 co-localized with actin filaments in the body wall muscle (S. Ono & Ono, 2020; Yamashiro et al., 2007). However, I obtained evidence that UNC-87 and CLIK-1 are segregated into different subdomains in sarcomeric actin filaments in the body wall muscle (Fig. 6, Fig. S2). In immunofluorescence microscopy using anti-UNC-87 antibody, UNC-87 co-localized with actin in sarcomeres (Fig. 6A). In our immunofluorescent staining for actin using anti-actin monoclonal or polyclonal antibody, a portion of sarcomeric actin near the pointed ends are not stained well as reported previously (S. Ono, Lewis, & Ono, 2022). Therefore, closely matched patterns of UNC-87 and actin in immunostaining (Fig. 6A) indicated that UNC-87 was also absent from the edges of sarcomeric actin where the pointed ends of actin filaments were concentrated. In contrast to UNC-87, CLIK-1::GFP was concentrated in sharp lines at the edges of phalloidin-stained sarcomeric actin filaments (Fig. 6B). A similar localization pattern of CLIK-1::GFP was also observed in live worms without fixation, and its concentration to the edges of sarcomeric actin bands was confirmed by comparing with the pattern of mCherry::LifeAct (Fig. 6C), indicating that the localization of CLIK-1::GFP was not an artifact of fixation.
Figure 6.
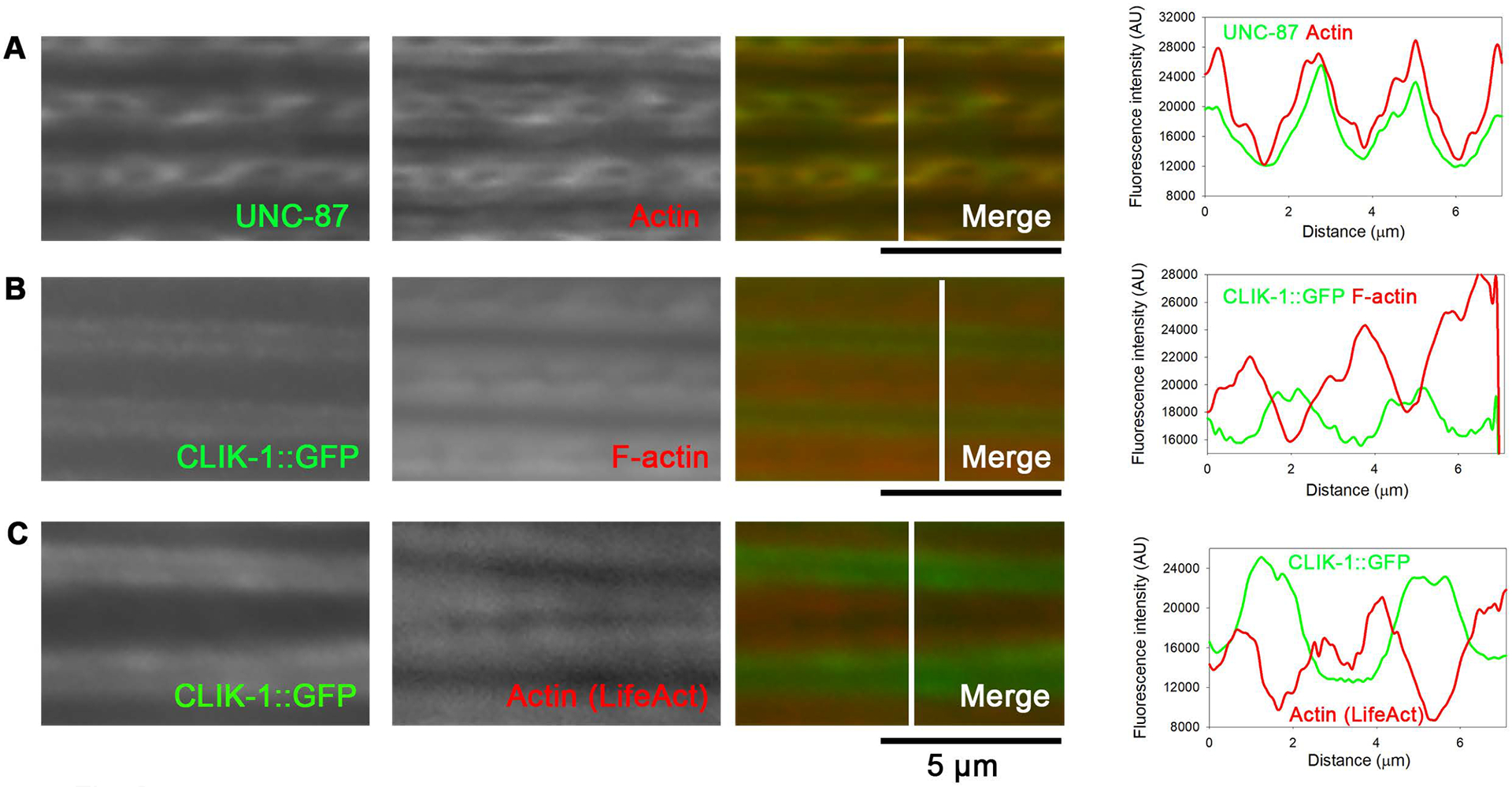
UNC-87 and CLIK-1 are segregated within sarcomeric actin filaments in the body wall muscle. Locations of UNC-87 (A) and CLIK-1 (B, C) were compared by fluorescence microscopy: (A) UNC-87 (antibody-stained) and actin (antibody-stained); (B) CLIK-1::GFP (fixed, no additional staining) and F-actin (phalloidin-stained); (C) CLIK-1::GFP (no fixation, no additional staining) and mCherry::LifeAct (no fixation, no additional staining). Bars, 5 μm. Regions indicated by white lines in the merged images were analyzed for fluorescence intensity (graphs on the right; plotted as a function of the distance from the top). Note that the micrographs are shown with enhanced brightness and contrast for visualization purposes, but the line scans were performed on minimally processed images. A green-magenta version of this figure is presented in Figure S2.
Furthermore, CLIK-1::GFP was not colocalized with ATN-1 α-actinin, which is concentrated at the dense bodies near the barbed ends of actin filaments (Fig. 7A, Fig. S3), suggesting that CLIK-1::GFP localized near the pointed ends of actin filaments. Unfortunately, the linear localization pattern of CLIK-1::GFP was not preserved well under fixation conditions optimized for immunostaining of UNC-87. However, in some instances, although the image quality was not optimal, I obtained images suggesting that UNC-87 and CLIK-1::GFP were segregated into different subdomains within sarcomeric actin filaments (Fig. 7B, Fig. S3). These results demonstrate that the two calponin-related proteins, CLIK-1 and UNC-87, are segregated within sarcomeric actin filaments in C. elegans striated muscle. Since CLIK-1::GFP was localized near the pointed ends of sarcomeric actin filaments (Fig. 6), the location of CLIK-1::GFP was compared with that of UNC-94 tropomodulin that localizes to the pointed ends of sarcomeric actin filaments in the C. elegans body wall muscle (Stevenson et al., 2007; Yamashiro, Cox, Baillie, Hardin, & Ono, 2008) (Fig. 7C, Fig. S3). Comparison of the locations of CLIK-1::GFP and UNC-94 indicated that an overlap between these two proteins was minimum (Fig. 7C), suggesting that CLIK-1 is enriched near the pointed ends of sarcomeric actin filaments but not extended to the pointed ends. Note that UNC-94 appeared as single lines because the pointed ends of the two half sarcomeres are closely located in fixed worms due to contraction. Similarly, tropomodulin appears as single lines in fixed vertebrate muscle cells (Almenar-Queralt, Gregorio, & Fowler, 1999; Gregorio & Fowler, 1995), and separation of two opposing pointed ends can be observed when resting myofibrils are stretched (Fowler, Sussmann, Miller, Flucher, & Daniels, 1993).
Figure 7.
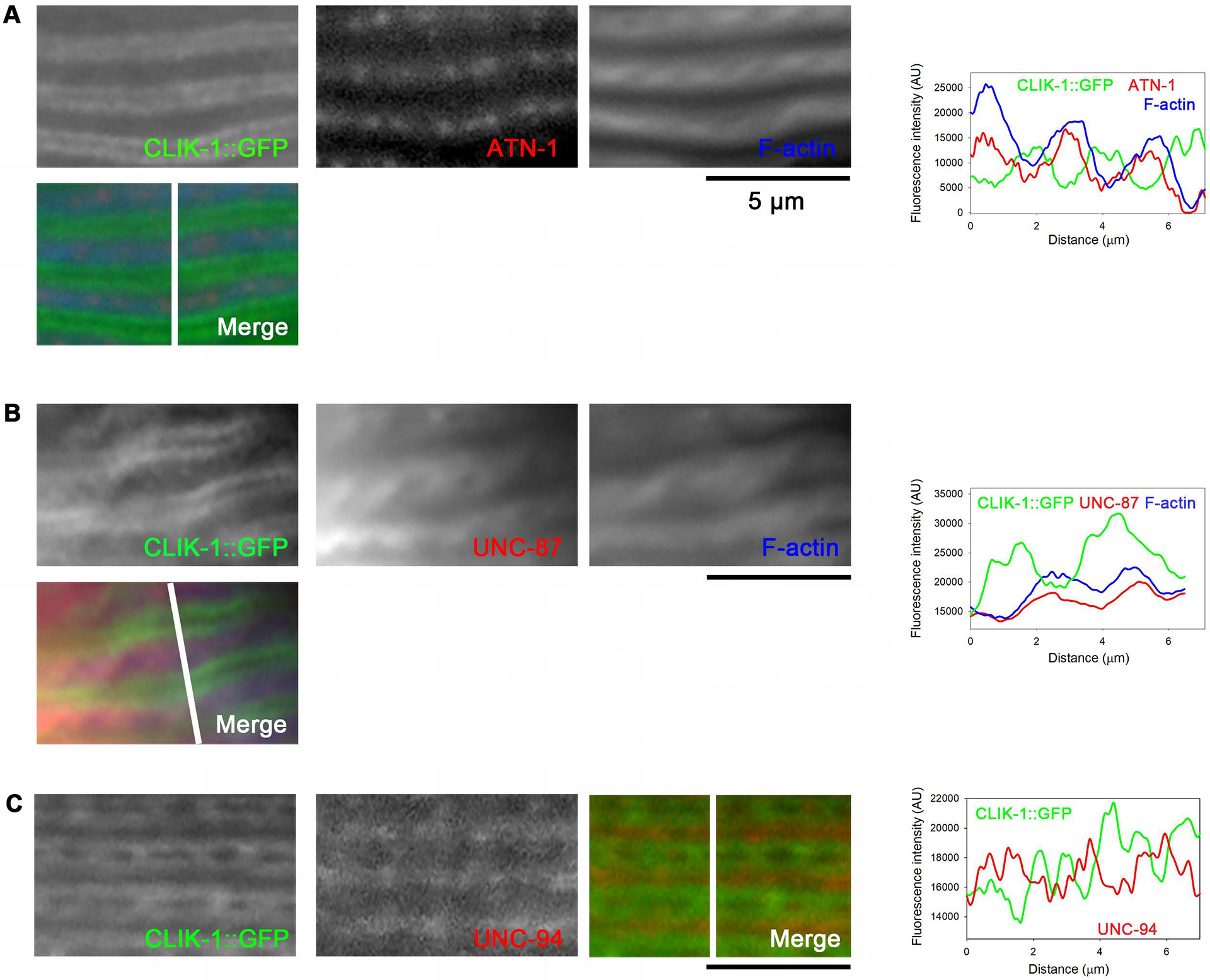
CLIK-1 is enriched near the pointed ends of sarcomeric actin filaments but not extended to the ends. Specific sarcomeric components were visualized by immunofluorescence microscopy: (A) CLIK-1::GFP (fixed, antibody-stained), ATN-1 α-actinin (antibody-stained), and F-actin (phalloidin-stained); (B) CLIK-1::GFP (fixed, antibody-stained), UNC-87 (antibody-stained), and F-actin (phalloidin-stained); (C) CLIK-1::GFP (fixed, antibody-stained) and UNC-94 tropomodulin (antibody-stained). Bars, 5 μm. Regions indicated by white lines in the merged images were analyzed for fluorescence intensity (graphs on the right; plotted as a function of the distance from the top). Note that the micrographs are shown with enhanced brightness and contrast for visualization purposes, but the line scans were performed on minimally processed images. A green-magenta-blue version of this figure is presented in Figure S3.
Limited localization of UNC-87 is independent of CLIK-1
A possible mechanism for the segregated localization of UNC-87 and CLIK-1 is their competitive binding to actin in vivo because UNC-87 and CLIK-1 compete for binding to actin filaments in vitro (S. Ono & Ono, 2020). To test this possibility, I examined localization pattern of UNC-87 in a putative clik-1-null mutant [clik-1(ok2355)], in which a deletion of 1 kb removes a major portion of the clik-1 gene. clik-1(ok2355) homozygotes do not exhibit abnormalities in sarcomeric actin organization in the body wall muscle (S. Ono & Ono, 2020). UNC-87 was still restricted to the proximal portion of the sarcomeric actin filaments in the clik-1 mutant (Fig. 8B, Fig. S4) in a similar manner to the pattern in wild-type (Fig. 8A, Fig. S4). An opposite experiment to test localization of CLIK-1 in unc-87 mutants was not conclusive due to severe disorganization of sarcomeric actin filaments in the mutants (Fig. 5). Nevertheless, these results suggest that segregated localization of UNC-87 and CLIK-1 is not caused by their competitive binding to actin filaments.
Figure 8.
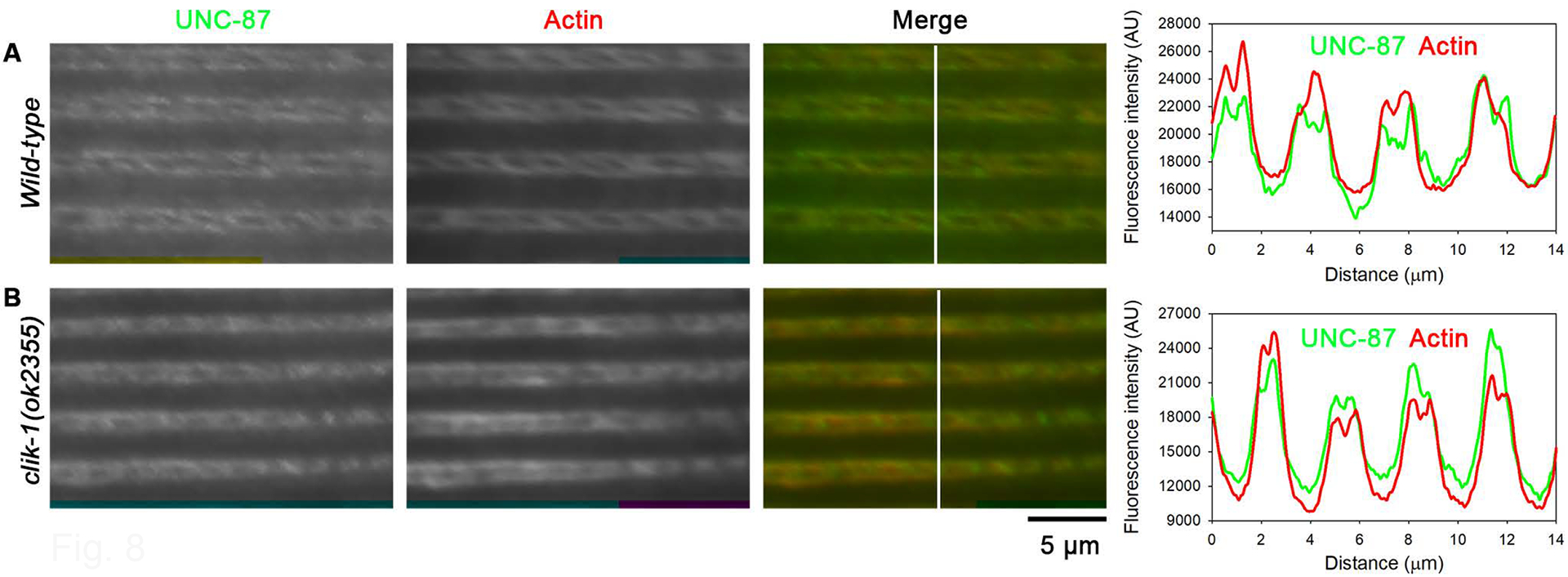
Restricted localization of UNC-87 is not altered in the absence of CLIK-1. Immunofluorescent staining of UNC-87 and actin in wild-type (A) and clik-1(ok2355) (clik-1-null) mutant (B). Bar, 5 μm. Regions indicated by white lines in the merged images were analyzed for fluorescence intensity (graphs on the right; plotted as a function of the distance from the top). A green-magenta version of this figure is presented in Figure S4.
Live imaging of CLIK-1::GFP indicates the contraction states of obliquely striated sarcomeres
Additional evidence for the localization of CLIK-1 near the pointed ends of sarcomeric actin filaments was obtained by live imaging of CLIK-1::GFP at different contraction states of the body wall muscles (Fig. 9). The worms expressing CLIK-1::GFP were embedded in a Pluronic F127 gel that restricts body movement (Krajniak & Lu, 2010). However, in the absence of anesthetics, worms can slightly wiggle within the gel. Since worms move in a sinusoidal bending pattern, we could simply identify the concave side of the body as the contracted state (Fig. 9A) and the convex side as the relaxed and stretched state (Fig. 9F). At a contracted state, single lines of CLIK-1::GFP were detected (Fig. 9A and B, arrows). As the muscle cells were relaxed, many lines of CLIK-1::GFP were split into double lines (Fig. 9C–F, arrows). Previous immunolocalization studies of UNC-94 tropomodulin in C. elegans body wall muscle indicate that UNC-94 tropomodulin at the pointed ends of two opposite thin filaments can coalesce near the M-lines at a contracted state (S. Ono et al., 2022; Stevenson et al., 2007; Yamashiro et al., 2008). Therefore, the transition of CLIK-1::GFP from single to double lines during muscle relaxation is consistent with the location of CLIK-1 at a portion of the sarcomeric thin filaments near the actin pointed ends, as interpreted schematically in Fig. 10. Because of the obliquely striated nature of the sarcomeres in the nematode body wall muscle (Burr & Gans, 1998; Moerman & Fire, 1997; S. Ono, 2014; Rosenbluth, 1965, 1967; Waterston, 1988) (Fig. 10), the separation of the CLIK-1::GFP lines in a relaxed/stretched sarcomere does not appear as wide as the actual distance between the two opposite thin filaments (Fig. 10C).
Figure 9.
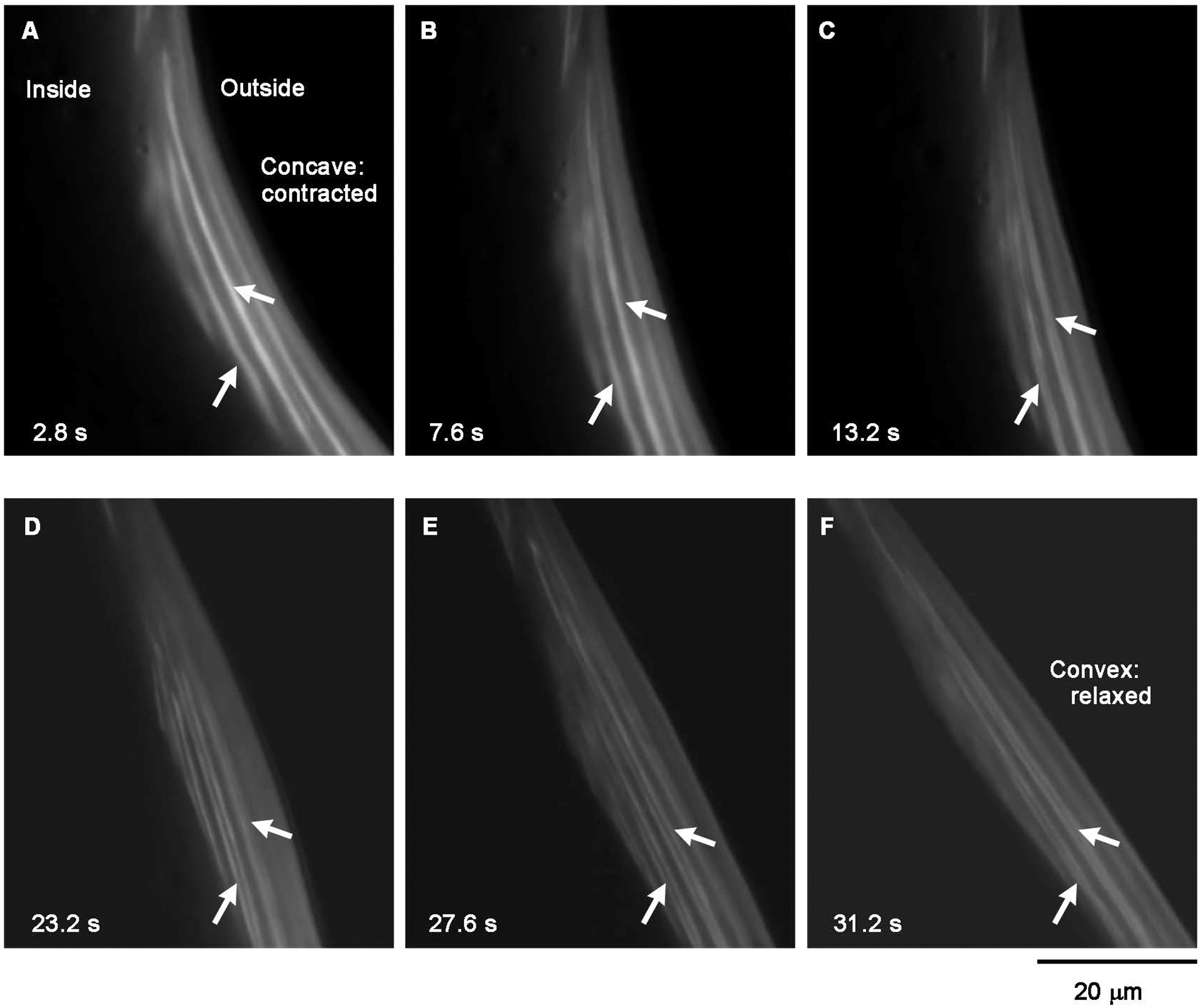
Live dynamics of CLIK-1::GFP are consistent with its localization near the pointed ends of sarcomeric actin filaments. Time-lapse images of CLIK-1::GFP in live worms were recorded, and representative frames at indicated time points are shown. The body wall muscle cells are attached to the body surface, and a selected area on the right side of the body is shown. Therefore, the muscle cells are contracted on the concave side of the body (A) and relaxed/stretched on the convex side of the body (F). Arrows indicate representative CLIK-1::GFP lines in which coalescence and separation were clearly observed. Bar, 20 μm.
Figure 10.
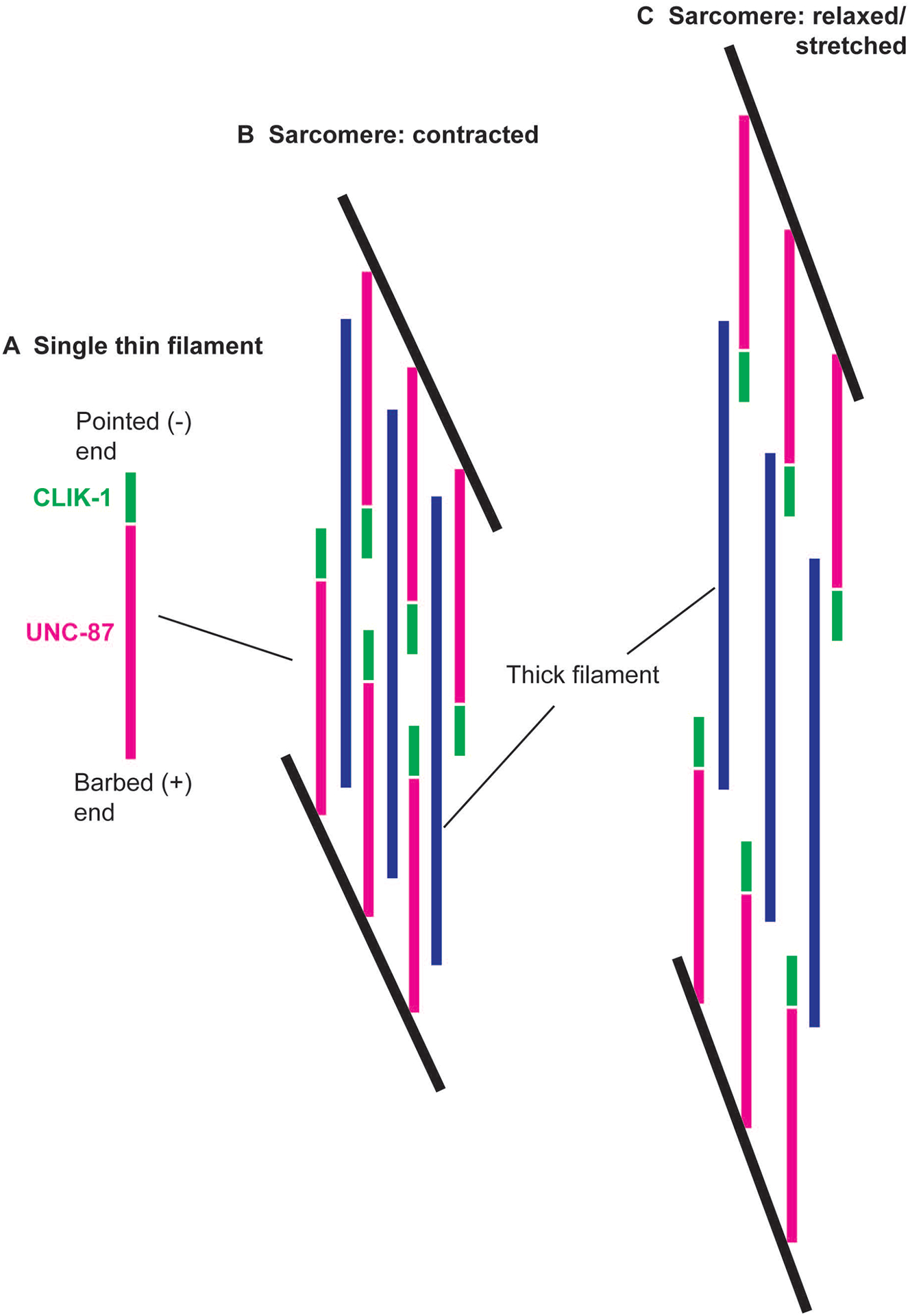
Schematic representation of localization of UNC-87 and CLIK-1 in sarcomeres. (A) In a single thin filament, CLIK-1 (green) is concentrated near the actin pointed (or -) end, whereas UNC-87 (magenta) is enriched towards the actin barbed (or +) end. (B) In an obliquely striated sarcomere of the C. elegans body wall muscle, when the sarcomere is contracted, CLIK-1 in the two oppositely oriented thin filaments is close and appears as a single line (see Fig. 9A). Thick filaments are shown in blue. Black lines indicate the regions where the actin barbed ends are linearly aligned (equivalent to the Z-lines in cross-striated muscles). (C) When the sarcomere is relaxed and stretched, the thin filaments are separated, and CLIK-1 appears as double lines (see Fig. 9F).
Discussion
In addition to the previously described in vitro similarities and differences between CLIK-1 and UNC-87, my current study demonstrates that these proteins are similarly insensitive to high-salt and inorganic phosphate in actin binding but have different effects on actomyosin ATPase. CLIK-1 and UNC-87 share 42 % identical amino acid sequences containing seven CLIK motifs (S. Ono & Ono, 2020). Therefore, the observed similarities in the mode of actin binding were expected. However, the differences in actin bundling activity (S. Ono & Ono, 2020) and effects on actomyosin ATPase (this study) were unexpected. Sequence comparison among CLIK motifs indicates that the fifth, sixth, and seventh CLIK motifs of CLIK-1 contain somewhat diverged sequences (S. Ono & Ono, 2020). Therefore, these CLIK motifs of CLIK-1 may bind to actin with low affinity and not participate in actin-filament bundling and actomyosin regulation. Since how CLIK motifs interact with actin is currently unknown, further comparative biochemical studies of CLIK motifs should help to understand how different calponin-related proteins regulate the actin cytoskeleton.
The difference between CLIK-1 and UNC-87 in the effects on actomyosin ATPase and myosin binding may be functionally related to their segregated localization in sarcomeric actin filaments. CLIK-1 is concentrated near the pointed ends of sarcomeric actin filaments and can reach near the center of sarcomeres at a contracted state (Figs. 6 and 7). Therefore, the location of CLIK-1 may correspond to the bare zone of thick filaments where myosin heads are absent and no actomyosin cross-bridges are formed (Kagawa, Gengyo, McLachlan, Brenner, & Karn, 1989). In contrast, UNC-87 localizes to the region where actin and myosin frequently interact. Although the role of UNC-87 as an inhibitor of actomyosin ATPase is currently unclear, nebulin, a thin-filament protein in vertebrate striated muscles, also inhibits actomyosin ATPase (Root & Wang, 1994, 2001) and is absent from the tips of sarcomeric actin filaments near the pointed ends (Castillo, Nowak, Littlefield, Fowler, & Littlefield, 2009). Thus, UNC-87 may be functionally similar to nebulin in actomyosin regulation.
The segregated localization of CLIK-1 and UNC-87 suggest that sarcomeric thin filaments contain previously unrecognized subdomains (Fig. 10), which might be analogous to a two-segment model that was proposed for skeletal muscle thin filaments (Gokhin & Fowler, 2013). In this model, the sarcomeric architecture of thin filaments is regulated by a nebulin-stabilized proximal segment and a nebulin-free distal segment where actin is more dynamic (Gokhin & Fowler, 2013). Since nebulin is absent in C. elegans, UNC-87 may be functionally similar to nebulin to generate a stable segment of thin filaments. CLIK-1 is less efficient than UNC-87 in inhibiting actin depolymerizing factor/cofilin (S. Ono & Ono, 2020), suggesting that the CLIK-1-localized distal portion may allow dynamic exchange of actin monomers. The nucleotide states of F-actin may promote, but not strongly regulate, sorting of UNC-87 and CLIK-1 (Fig. 3). In striated muscles, a number of actin-regulatory proteins are expressed and influence localization patterns of thin-filament proteins (S. Ono, 2010, 2014). For example, α-actinin is restricted near the actin barbed ends at the Z-discs (Masaki, Endo, & Ebashi, 1967), whereas tropomyosin binds along the sarcomeric thin filaments except at the Z-disc regions (Pepe, 1966). This segregation is most likely due to their competitive binding to actin filaments (Drabikowski, Nonomura, & Maruyama, 1968; Drabikowski & Nowak, 1968). Similarly, in invertebrate muscles, kettin, a connectin/titin-related protein, is associated with sarcomeric actin filaments near the actin barbed ends due to competition with tropomyosin (Lakey et al., 1993; K. Ono, Qin, Johnsen, Baillie, & Ono, 2020; K. Ono, Yu, Mohri, & Ono, 2006; van Straaten et al., 1999). Likewise, there may be an unknown factor(s) that regulates segregation of UNC-87 and CLIK-1.
The nature of a subdomain of sarcomeric thin filaments near the actin pointed ends remains unclear. Drosophila SALS, a WH2-containing protein, transiently localizes near the actin pointed ends in developing sarcomeres (Bai, Hartwig, & Perrimon, 2007). In mouse cardiac muscle, the Fhod3 formin localizes near the actin pointed ends in mature sarcomeres (Fujimoto et al., 2016; Kan-o et al., 2012). Leiomodins are also enriched near, but not at, the actin pointed ends in vertebrate striated muscles (Fowler & Dominguez, 2017). As proposed in the two-segment model, this region may be a segment of sarcomeric thin filaments with relatively rapid actin dynamics (Gokhin & Fowler, 2013). Since sarcomeric actin filaments can elongate at the pointed ends (Littlefield, Almenar-Queralt, & Fowler, 2001; Mardahl-Dumesnil & Fowler, 2001; Tsukada et al., 2010), a portion of the filaments near the pointed ends may contain relatively newly polymerized actin subunits that may be different in the nucleotide states (Chou & Pollard, 2019; Merino et al., 2018) and/or conformation (Huang, Grabarek, & Wang, 2010; Kueh, Brieher, & Mitchison, 2008; Orlova et al., 2004) from actin subunits in the older parts of the filaments, which can recruit a distinct set of actin-binding proteins. To understand the nature of this subdomain/segment and its biological significance, additional investigation to identify proteins that uniquely accumulate at this region should be necessary.
Materials and Methods
Proteins
Actin was purified from rabbit muscle acetone powder (Pel-Freeze Biologicals) as described (Pardee & Spudich, 1982). To prepare ADP-actin filaments, CaATP-G-actin was converted to MgATP-G-actin by incubating with 1 mM glucose and 0.25 units/mL of hexokinase (Worthington Biochemical, catalog no. LS002515) as described by Courtemanche and Pollard (Courtemanche & Pollard, 2013) and polymerized by adding final 0.1 M KCl, 2 mM MgCl2, and 1 mM EGTA overnight at 4 °C. To prepare ADP-Pi-actin filaments, MgATP-G-actin was polymerized by adding final 0.1 M KCl, 2 mM MgCl2, 25 mM inorganic phosphate (potassium phosphate, pH 7.5), and 1 mM EGTA overnight at 4 °C. To prepare AMPPNP-actin filaments, CaATP-G-actin was converted to MgAMPPNP-G-actin by removing nucleotides by Dowex 1X8 resin and incubating with 1 mM AMPPNP (MilliporeSigma, catalog no. 10102547001) for 30 min at room temperature as described by Courtemanche and Pollard (Courtemanche & Pollard, 2013) and polymerized by adding final 0.1 M KCl, 2 mM MgCl2, and 1 mM EGTA overnight at 4 °C. Recombinant UNC-87 (the UNC-87B isoform) (Kranewitter et al., 2001) and CLIK-1 (S. Ono & Ono, 2020) were expressed in Escherichia coli and purified as described previously. Rabbit skeletal muscle myosin (Obinata & Sato, 2012) was a gift from Dr. Takashi Obinata (Chiba University, Chiba, Japan).
F-actin co-sedimentation assays
Filamentous actin (5 or 10 μM) was incubated with proteins of interest in F-buffer (0.1 M KCl, 2 mM MgCl2, 20 mM HEPES-KOH, pH 7.5, 1 mM DTT) with variable KCl or Pi concentrations at room temperature. The samples were ultracentrifuged at 42,000 rpm (200,000 × g) for 20 min using a Beckman 42.2Ti rotor. Supernatants and pellets were separated, adjusted to the same volumes, and examined by SDS–PAGE (12% acrylamide gel) with molecular weight markers (Nacalai USA, catalog no. 29458–24). The gels were stained with Coomassie Brilliant Blue R-250 (National Diagnostics) and scanned by an Epson Perfection V700 scanner at 300 dpi. Band intensity was quantified with ImageJ.
Myosin co-sedimentation assays
Myosin (0 – 10 μM) was incubated with a fixed concentration (2 μM) of UNC-87 or CLIK-1 in a buffer containing 30 mM KCl, 1 mM MgCl2, 1 mM DTT, and 20 mM HEPES-KOH, pH 7.5, for 1 h at room temperature. The reactions were centrifuged at 247,000 × g for 20 min using a Beckman TLA-100 rotor. The supernatants and pellets were fractionated, adjusted to the same volumes, and analyzed by SDS–PAGE. Quantitative analysis was done in the same manner as described for F-actin co-sedimentation assays except that the sedimented portions were calculated from depletion of UNC-87 or CLIK-1 from the supernatants. Myosin-independent sedimentation of UNC-87 or CLIK-1 was determined in the absence of myosin and subtracted from the data.
Myosin ATPase assays
Myosin ATPase activity was determined as described previously (Obinata & Sato, 2012) except that inorganic phosphate was quantitatively determined using BIOMOL® Green (ENZO Life Sciences). Experiments were performed in a modified buffer containing 50 mM KCl, 1 mM MgCl2, and 20 mM imidazole-HCl, pH 7.5. Myosin was used at 0.4 μM. Actin (1.0 μM), CLIK-1 and/or UNC-87 were added in some reactions.
C. elegans strains and culture
The worms were cultured following standard methods (Stiernagle, 2006). Wild-type N2 and CB1216 unc-87(e1216) were obtained from the Caenorhabditis Genetics Center (Minneapolis, MN). ON352 clik-1(kt1 [clik-1::gfp]) and ON225 clik-1(ok2355) were described previously (S. Ono & Ono, 2020). KAG190 Ex[Pmyo-3::mCherry::LifeAct; Pmyo-2::mCherry] (Brouilly et al., 2015) was obtained from Kathrin Gieseler (University of Lyon, Villeurbanne, France) and crossed with ON352 to generate ON383 clik-1(kt1 [clik-1::gfp]); Ex[Pmyo-3::mCherry::LifeAct; Pmyo-2::mCherry]. ON390 unc-87(e1216); clik-1(kt1 [clik-1::gfp]) was generated by crossing CB1216 and ON352 and isolating homozygotes.
Motility assay
Worm motility (beat frequency) in liquid was quantified as described previously (Epstein & Thomson, 1974; S. Ono, Baillie, & Benian, 1999). Briefly, adult worms were placed in M9 buffer. Then, one beat was counted when a worm swung its head to either right or left. The total number of beats in 30 s was recorded.
Fluorescence microscopy
Staining of whole worms with ATTO594–phalloidin (MilliporeSigma) was performed as described previously (S. Ono, 2001, 2022). Immunofluorescent staining of UNC-87, UNC-94, and actin (Fig. 6A, Fig. 7C, Fig. 8) was performed with the method of Nonet et al. (Nonet et al., 1997). Other immunofluorescent staining experiments (Fig. 7A, B) were performed with the freeze-crack method as described previously (K. Ono et al., 2020). Briefly, adult worms were anesthetized by 0.1 % tricaine and 0.01 % levamisole and cut in half using a pair of 28-gauge needles on polylysine-coated glass slides. Glass coverslips were overlaid on cut worms, frozen by dry ice, and then removed while they were frozen using a razor blade. They were immediately fixed by 4% paraformaldehyde in 1× cytoskeleton buffer (10 mm MES-KOH, pH 6.1, 138 mM KCl, 3 mM MgCl2, 2 mM EGTA) containing 0.32 M sucrose for 30 min at room temperature, permeabilized by PBS containing 0.5 % Triton X-100 and 30 mM glycine (PBS-TG) for 10 min, and stained with primary antibodies and ATTO647N-phalloidin (MilliporeSigma) in PBS with 1 % bovine serum albumin for 1 h at room temperature. They were washed three times with PBS (5 min each), treated with fluorescently labeled secondary antibodies in PBS with 1 % bovine serum albumin for 1 h, washed with PBS and mounted with ProLong Gold Antifade Mounting Medium (Thermo Fisher, Waltham, MA, USA). Primary antibodies used were mouse anti-actin monoclonal (C4, MP Biomedicals), guinea pig anti-UNC-87 (Yamashiro et al., 2007), mouse anti-GFP monoclonal (GF200, Nacalai USA), rabbit anti-UNC-94 (Yamashiro et al., 2008), and mouse anti-ATN-1 monoclonal (MH40) (Francis & Waterston, 1985). Secondary antibodies used were Alexa 488-labeled goat anti-mouse IgG (Life Technologies), Alexa 488-labeled goat anti-guinea pig IgG (Life Technologies), Alexa 555-labeled goat anti-guinea pig IgG (Life Technologies), Cy3-labeled goat anti-mouse IgG (Jackson ImmnoResearch), and Cy3-labeled goat anti-rabbit IgG (Jackson ImmnoResearch).
Live worms were either anesthetized in M9 buffer containing 0.1% tricaine and 0.01% tetramisole for 30 min on 2% agarose pads (Fig. 6C) or embedded in 30 % Pluronic F127 (BioVision Inc.) in M9 buffer without anesthetics (Fig. 9).
Samples were observed by epifluorescence using a Nikon Eclipse TE2000 inverted microscope (Nikon Instruments, Tokyo, Japan) with a CFI Plan Fluor ELWD 40× (dry; NA 0.60) or CFI Plan Apo Lambda 100x (oil, NA 1.45) objective. Images were captured by a Hamamatsu ORCA Flash 4.0 LT sCMOS camera (Hamamatsu Photonics, Shizuoka, Japan) and processed by NIS-Elements (Nikon Instruments) and Adobe Photoshop CS3. Line scans of fluorescent images were performed using ImageJ. In some datasets, baselines of fluorescence intensity were adjusted such that multiple fluorophores could be compared within a similar range.
Statistics
Data were tested by one-way analysis of variance (ANOVA) using SigmaPlot 14.5. (Systat Software).
Supplementary Material
Acknowledgements
Some C. elegans strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This work was supported by a grant from the National Institutes of Health (R01 AR048615) to S. O.
References
- Abe M, Takahashi K, & Hiwada K (1990). Effect of calponin on actin-activated myosin ATPase activity. J. Biochem, 108(5), 835–838. [DOI] [PubMed] [Google Scholar]
- Almenar-Queralt A, Gregorio CC, & Fowler VM (1999). Tropomodulin assembles early in myofibrillogenesis in chick skeletal muscle: evidence that thin filaments rearrange to form striated myofibrils. J. Cell Sci, 112 ( Pt 8), 1111–1123. [DOI] [PubMed] [Google Scholar]
- Ayme-Southgate A, Lasko P, French C, & Pardue ML (1989). Characterization of the gene for mp20: a Drosophila muscle protein that is not found in asynchronous oscillatory flight muscle. J. Cell Biol, 108(2), 521–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Hartwig JH, & Perrimon N (2007). SALS, a WH2-domain-containing protein, promotes sarcomeric actin filament elongation from pointed ends during Drosophila muscle growth. Dev. Cell, 13(6), 828–842. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, & Pollard TD (1999). Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J. Biol. Chem, 274(22), 15538–15546. [DOI] [PubMed] [Google Scholar]
- Brouilly N, Lecroisey C, Martin E, Pierson L, Mariol MC, Qadota H, … Gieseler K (2015). Ultra-structural time-course study in the C. elegans model for Duchenne muscular dystrophy highlights a crucial role for sarcomere-anchoring structures and sarcolemma integrity in the earliest steps of the muscle degeneration process. Hum. Mol. Genet, 24(22), 6428–6445. [DOI] [PubMed] [Google Scholar]
- Burr AH, & Gans C (1998). Mechanical significance of obliquely striated architecture in nematode muscle. Biol. Bull, 194(1), 1–6. [DOI] [PubMed] [Google Scholar]
- Cai L, Makhov AM, & Bear JE (2007). F-actin binding is essential for coronin 1B function in vivo. J. Cell Sci, 120(Pt 10), 1779–1790. [DOI] [PubMed] [Google Scholar]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, … Pantaloni D (1997). Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol, 136(6), 1307–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, & Pantaloni D (1988). Binding of phosphate to F-ADP-actin and role of F-ADP-Pi-actin in ATP-actin polymerization. J. Biol. Chem, 263(2), 817–825. [PubMed] [Google Scholar]
- Carmichael JD, Winder SJ, Walsh MP, & Kargacin GJ (1994). Calponin and smooth muscle regulation. Can. J. Physiol. Pharmacol, 72(11), 1415–1419. doi: 10.1139/y94-204 [DOI] [PubMed] [Google Scholar]
- Castillo A, Nowak R, Littlefield KP, Fowler VM, & Littlefield RS (2009). A nebulin ruler does not dictate thin filament lengths. Biophys. J, 96(5), 1856–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou SZ, & Pollard TD (2019). Mechanism of actin polymerization revealed by cryo-EM structures of actin filaments with three different bound nucleotides. Proc. Natl. Acad. Sci. U S A, 116(10), 4265–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtemanche N, & Pollard TD (2013). Interaction of profilin with the barbed end of actin filaments. Biochemistry, 52(37), 6456–6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabikowski W, Nonomura Y, & Maruyama K (1968). Effect of tropomyosin on the interaction between F-actin and the 6S component of alpha-actinin. J. Biochem, 63(6), 761–765. [DOI] [PubMed] [Google Scholar]
- Drabikowski W, & Nowak E (1968). The interaction of alpha-actinin with F-actin and its abolition by tropomyosin. Eur. J. Biochem, 5(2), 209–214. [DOI] [PubMed] [Google Scholar]
- Epstein HF, & Thomson JN (1974). Temperature-sensitive mutation affecting myofilament assembly in Caenorhabditis elegans. Nature, 250(467), 579–580. [DOI] [PubMed] [Google Scholar]
- Fowler VM, & Dominguez R (2017). Tropomodulins and leiomodins: actin pointed end caps and nucleators in muscles. Biophys. J, 112(9), 1742–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler VM, Sussmann MA, Miller PG, Flucher BE, & Daniels MP (1993). Tropomodulin is associated with the free (pointed) ends of the thin filaments in rat skeletal muscle. J. Cell Biol, 120(2), 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis GR, & Waterston RH (1985). Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J. Cell Biol, 101(4), 1532–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto N, Kan OM, Ushijima T, Kage Y, Tominaga R, Sumimoto H, & Takeya R (2016). Transgenic expression of the formin protein Fhod3 selectively in the embryonic heart: role of actin-binding activity of Fhod3 and its sarcomeric localization during myofibrillogenesis. PLoS One, 11(2), e0148472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi M, Achard V, Blanchoin L, & Goode BL (2009). Coronin switches roles in actin disassembly depending on the nucleotide state of actin. Mol Cell, 34(3), 364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge P, Durer ZA, Kudryashov D, Zhou ZH, & Reisler E (2014). Cryo-EM reveals different coronin binding modes for ADP- and ADP-BeFx actin filaments. Nat. Struct. Mol. Biol, 21(12), 1075–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimona M, Djinovic-Carugo K, Kranewitter WJ, & Winder SJ (2002). Functional plasticity of CH domains. FEBS Lett, 513(1), 98–106. [DOI] [PubMed] [Google Scholar]
- Gimona M, Kaverina I, Resch GP, Vignal E, & Burgstaller G (2003). Calponin repeats regulate actin filament stability and formation of podosomes in smooth muscle cells. Mol. Biol. Cell, 14(6), 2482–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetinck S, & Waterston RH (1994a). The Caenorhabditis elegans muscle-affecting gene unc-87 encodes a novel thin filament-associated protein. J. Cell Biol, 127(1), 79–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetinck S, & Waterston RH (1994b). The Caenorhabditis elegans UNC-87 protein is essential for maintenance, but not assembly, of bodywall muscle. J. Cell Biol, 127(1), 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhin DS, & Fowler VM (2013). A two-segment model for thin filament architecture in skeletal muscle. Nat. Rev. Mol. Cell Biol, 14(2), 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio CC, & Fowler VM (1995). Mechanisms of thin filament assembly in embryonic chick cardiac myocytes: tropomodulin requires tropomyosin for assembly. J. Cell Biol, 129(3), 683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TB, & Jin JP (2023). Evolution and function of calponin and transgelin. Front. Cell Dev. Biol, 11, 1206147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Grabarek Z, & Wang CL (2010). Differential effects of caldesmon on the intermediate conformational states of polymerizing actin. J. Biol. Chem, 285(1), 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa H, Gengyo K, McLachlan AD, Brenner S, & Karn J (1989). Paramyosin gene (unc-15) of Caenorhabditis elegans. Molecular cloning, nucleotide sequence and models for thick filament structure. J. Mol. Biol, 207(2), 311–333. [DOI] [PubMed] [Google Scholar]
- Kan-o M, Takeya R, Taniguchi K, Tanoue Y, Tominaga R, & Sumimoto H (2012). Expression and subcellular localization of mammalian formin Fhod3 in the embryonic and adult heart. PLoS One, 7(4), e34765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajniak J, & Lu H (2010). Long-term high-resolution imaging and culture of C. elegans in chip-gel hybrid microfluidic device for developmental studies. Lab. Chip, 10(14), 1862- [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranewitter WJ, Ylanne J, & Gimona M (2001). UNC-87 is an actin-bundling protein. J. Biol. Chem, 276(9), 6306–6312. [DOI] [PubMed] [Google Scholar]
- Kueh HY, Brieher WM, & Mitchison TJ (2008). Dynamic stabilization of actin filaments. Proc. Natl. Acad. Sci. U S A, 105(43), 16531–16536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakey A, Labeit S, Gautel M, Ferguson C, Barlow DP, Leonard K, & Bullard B (1993). Kettin, a large modular protein in the Z-disc of insect muscles. EMBO J, 12(7), 2863–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lener T, Burgstaller G, & Gimona M (2004). The role of calponin in the gene profile of metastatic cells: inhibition of metastatic cell motility by multiple calponin repeats. FEBS Lett, 556(1–3), 221–226. [DOI] [PubMed] [Google Scholar]
- Littlefield R, Almenar-Queralt A, & Fowler VM (2001). Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat. Cell Biol, 3, 544–551. [DOI] [PubMed] [Google Scholar]
- Liu J, Zhang Y, Li Q, & Wang Y (2020). Transgelins: cytoskeletal associated proteins implicated in the metastasis of colorectal cancer. Front. Cell Dev. Biol, 8, 573859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, & Jin JP (2016). Calponin isoforms CNN1, CNN2 and CNN3: Regulators for actin cytoskeleton functions in smooth muscle and non-muscle cells. Gene, 585(1), 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardahl-Dumesnil M, & Fowler VM (2001). Thin filaments elongate from their pointed ends during myofibril assembly in Drosophila indirect flight muscle. J. Cell Biol, 155(6), 1043–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki T, Endo M, & Ebashi S (1967). Localization of 6S component of a alpha-actinin at Z-band. J. Biochem, 62(5), 630–632. [DOI] [PubMed] [Google Scholar]
- Merino F, Pospich S, Funk J, Wagner T, Kullmer F, Arndt HD, … Raunser S (2018). Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM. Nat. Struct. Mol. Biol, 25(6), 528–537. [DOI] [PubMed] [Google Scholar]
- Moerman DG, & Fire A (1997). Muscle: structure, function, and development. In Riddle DL, Blumenthal T, Meyer BJ, & Priess JR (Eds.), C. elegans II (pp. 417–470). Plainview, NY: Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- Muhlrad A, Pavlov D, Peyser YM, & Reisler E (2006). Inorganic phosphate regulates the binding of cofilin to actin filaments. FEBS J, 273(7), 1488–1496. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Staunton JE, Kilgard MP, Fergestad T, Hartwieg E, Horvitz HR, … Meyer BJ (1997). Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J. Neurosci, 17(21), 8061–8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obinata T, & Sato N (2012). Comparative studies on troponin, a Ca2+-dependent regulator of muscle contraction, in striated and smooth muscles of protochordates. Methods, 56(1), 3–10. [DOI] [PubMed] [Google Scholar]
- Ono K, Obinata T, Yamashiro S, Liu Z, & Ono S (2015). UNC-87 isoforms, Caenorhabditis elegans calponin-related proteins, interact with both actin and myosin and regulate actomyosin contractility. Mol. Biol. Cell, 26(9), 1687–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Qin Z, Johnsen RC, Baillie DL, & Ono S (2020). Kettin, the large actin-binding protein with multiple immunoglobulin domains, is essential for sarcomeric actin assembly and larval development in Caenorhabditis elegans. FEBS J, 287(4), 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Yu R, Mohri K, & Ono S (2006). Caenorhabditis elegans kettin, a large immunoglobulin-like repeat protein, binds to filamentous actin and provides mechanical stability to the contractile apparatuses in body wall muscle. Mol. Biol. Cell, 17(6), 2722–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S (2001). The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J. Cell Biol, 152(6), 1313–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S (2010). Dynamic regulation of sarcomeric actin filaments in striated muscle. Cytoskeleton (Hoboken), 67(11), 677–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S (2014). Regulation of structure and function of sarcomeric actin filaments in striated muscle of the nematode Caenorhabditis elegans. Anat. Rec. (Hoboken), 297(9), 1548–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S (2021). Diversification of the calponin family proteins by gene amplification and repeat expansion of calponin-like motifs. Cytoskeleton (Hoboken), 78(5), 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S (2022). Imaging of actin cytoskeleton in the nematode Caenorhabditis elegans. Methods Mol. Biol, 2364, 149–158. [DOI] [PubMed] [Google Scholar]
- Ono S, Baillie DL, & Benian GM (1999). UNC-60B, an ADF/cofilin family protein, is required for proper assembly of actin into myofibrils in Caenorhabditis elegans body wall muscle. J. Cell Biol, 145(3), 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, & Benian GM (1998). Two Caenorhabditis elegans actin depolymerizing factor/cofilin proteins, encoded by the unc-60 gene, differentially regulate actin filament dynamics. J. Biol. Chem, 273(6), 3778–3783. [DOI] [PubMed] [Google Scholar]
- Ono S, Lewis M, & Ono K (2022). Mutual dependence between tropomodulin and tropomyosin in the regulation of sarcomeric actin assembly in Caenorhabditis elegans striated muscle. Eur. J. Cell Biol, 101(2), 151215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, & Ono K (2002). Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J. Cell Biol, 156(6), 1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, & Ono K (2020). Two Caenorhabditis elegans calponin-related proteins have overlapping functions that maintain cytoskeletal integrity and are essential for reproduction. J. Biol. Chem, 295(34), 12014–12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova A, Shvetsov A, Galkin VE, Kudryashov DS, Rubenstein PA, Egelman EH, & Reisler E (2004). Actin-destabilizing factors disrupt filaments by means of a time reversal of polymerization. Proc. Natl. Acad. Sci. U S A, 101(51), 17664–17668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee JD, & Spudich JA (1982). Purification of muscle actin. Methods Enzymol, 85, 164–181. [DOI] [PubMed] [Google Scholar]
- Pepe FA (1966). Some aspects of the structural organization of the myofibril as revealed by antibody--staining methods. J. Cell Biol, 28(3), 505–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinjha RK, Shapland CE, Hsuan JJ, Totty NF, Mason IJ, & Lawson D (1994). Cloning and sequencing of cDNAs encoding the actin cross-linking protein transgelin defines a new family of actin-associated proteins. Cell Motil. Cytoskeleton, 28(3), 243–255. [DOI] [PubMed] [Google Scholar]
- Ren WZ, Ng GY, Wang RX, Wu PH, O’Dowd BF, Osmond DH, … Liew CC (1994). The identification of NP25: a novel protein that is differentially expressed by neuronal subpopulations. Brain Res. Mol. Brain Res, 22(1–4), 173–185. [DOI] [PubMed] [Google Scholar]
- Root DD, & Wang K (1994). Calmodulin-sensitive interaction of human nebulin fragments with actin and myosin. Biochemistry, 33(42), 12581–12591. [DOI] [PubMed] [Google Scholar]
- Root DD, & Wang K (2001). High-affinity actin-binding nebulin fragments influence the actoS1 complex. Biochemistry, 40(5), 1171–1186. [DOI] [PubMed] [Google Scholar]
- Rosenbluth J (1965). Ultrastructural organization of obliquely striated muscle fibers in Ascaris lumbricoides. J. Cell Biol, 25(3), 495–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluth J (1967). Obliquely striated muscle. 3. Contraction mechanism of Ascaris body muscle. J. Cell Biol, 34(1), 15–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblum GT, & Gimona M (2008). Calponins: adaptable modular regulators of the actin cytoskeleton. Int. J. Biochem. Cell Biol, 40(10), 1990–1995. doi: 10.1016/j.biocel.2007.07.010 [DOI] [PubMed] [Google Scholar]
- Solway J, Seltzer J, Samaha FF, Kim S, Alger LE, Niu Q, … Parmacek MS (1995). Structure and expression of a smooth muscle cell-specific gene, SM22 alpha. J. Biol. Chem, 270(22), 13460–13469. [DOI] [PubMed] [Google Scholar]
- Stevenson TO, Mercer KB, Cox EA, Szewczyk NJ, Conley CA, Hardin JD, & Benian GM (2007). unc-94 encodes a tropomodulin in Caenorhabditis elegans. J. Mol. Biol, 374(4), 936–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T (2006). Maintenance of C. elegans. WormBook, 1–11. doi: 10.1895/wormbook.1.101.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Hiwada K, & Kokubu T (1988). Vascular smooth muscle calponin. A novel troponin T-like protein. Hypertension, 11(6 Pt 2), 620–626. [DOI] [PubMed] [Google Scholar]
- Takahashi K, & Nadal-Ginard B (1991). Molecular cloning and sequence analysis of smooth muscle calponin. J. Biol. Chem, 266(20), 13284–13288. [PubMed] [Google Scholar]
- Tsukada T, Pappas CT, Moroz N, Antin PB, Kostyukova AS, & Gregorio CC (2010). Leiomodin-2 is an antagonist of tropomodulin-1 at the pointed end of the thin filaments in cardiac muscle. J. Cell Sci, 123(Pt 18), 3136–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Straaten M, Goulding D, Kolmerer B, Labeit S, Clayton J, Leonard K, & Bullard B (1999). Association of kettin with actin in the Z-disc of insect flight muscle. J. Mol. Biol, 285(4), 1549–1562. [DOI] [PubMed] [Google Scholar]
- Wang H, Park H, Liu J, & Sternberg PW (2018). An efficient genome editing strategy to generate putative null mutants in Caenorhabditis elegans using CRISPR/Cas9. G3 (Bethesda), 8(11), 3607–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH (1988). Muscle. In Wood WB (Ed.), The Nematode C. elegans (pp. 281–335): Cold Spring Harbor Laboratory. [Google Scholar]
- Winder SJ, Jess T, & Ayscough KR (2003). SCP1 encodes an actin-bundling protein in yeast. Biochem. J, 375(Pt 2), 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder SJ, & Walsh MP (1990). Smooth muscle calponin. Inhibition of actomyosin MgATPase and regulation by phosphorylation. J. Biol. Chem, 265(17), 10148–10155. [PubMed] [Google Scholar]
- Wu KC, & Jin JP (2008). Calponin in non-muscle cells. Cell Biochem. Biophys, 52(3), 139–148. [DOI] [PubMed] [Google Scholar]
- Yamashiro S, Cox EA, Baillie DL, Hardin JD, & Ono S (2008). Sarcomeric actin organization is synergistically promoted by tropomodulin, ADF/cofilin, AIP1 and profilin in C. elegans. J. Cell Sci, 121(Pt 23), 3867–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S, Gimona M, & Ono S (2007). UNC-87, a calponin-related protein in C. elegans, antagonizes ADF/cofilin-mediated actin filament dynamics. J. Cell Sci, 120(Pt 17), 3022–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


