Abstract
Context
Multiple common genetic variants have been associated with type 2 diabetes, but the mechanism by which they predispose to diabetes is incompletely understood. One such example is variation in MTNR1B, which implicates melatonin and its receptor in the pathogenesis of type 2 diabetes.
Objective
To characterize the effect of diabetes-associated genetic variation at rs10830963 in the MTNR1B locus on islet function in people without type 2 diabetes.
Design
The association of genetic variation at rs10830963 with glucose, insulin, C-peptide, glucagon, and indices of insulin secretion and action were tested in a cohort of 294 individuals who had previously undergone an oral glucose tolerance test (OGTT). Insulin sensitivity, β-cell responsivity to glucose, and Disposition Indices were measured using the oral minimal model.
Setting
The Clinical Research and Translation Unit at Mayo Clinic, Rochester, MN.
Participants
Two cohorts were utilized for this analysis: 1 cohort was recruited on the basis of prior participation in a population-based study in Olmsted County. The other cohort was recruited on the basis of TCF7L2 genotype at rs7903146 from the Mayo Biobank.
Intervention
Two-hour, 7-sample OGTT.
Main Outcome Measures
Fasting, nadir, and integrated glucagon concentrations.
Results
One or 2 copies of the G-allele at rs10830963 were associated with increased postchallenge glucose and glucagon concentrations compared to subjects with the CC genotype.
Conclusion
The effects of rs10830963 on glucose homeostasis and predisposition to type 2 diabetes are likely to be partially mediated through changes in α-cell function.
Keywords: MTNR1B, TCF7L2, beta-cell function, alpha-cell function, glucagon suppression, insulin secretion
The application of genome-wide association scans to the study of type 2 diabetes (T2DM) has identified multiple common genetic variants regulating pathways not previously associated with the pathogenesis of diabetes [1]. In certain circumstances, these variants can be used as probes to understand their contribution to glucose homeostasis prior to the development of diabetes. For example, the single nucleotide polymorphism (SNP) at rs7903146 in the TCF7L2 locus is associated with impaired α-cell function [2, 3], implicating glucagon abnormalities in the early pathogenesis of prediabetes. Another insight provided by genome-wide association scans is the observation that some variants are associated with increased fasting glucose but not with T2DM risk. Other variants are associated with both increases in fasting glucose and T2DM risk while others have no association with fasting glucose but increase T2DM risk [4]. This implies differential regulation of fasting and postprandial glucose.
Genetic variation in MTNR1B, which encodes the melatonin receptor 2, is associated with T2DM. rs1387153, a SNP approximately 28.3 kb upstream of MTNR1B, is associated with increased fasting glucose and T2DM risk [5]. This SNP is in linkage disequilibrium (r2 = 0.7) with an intronic SNP (rs10830963) shown in other independent cohorts to be associated with fasting glucose, T2DM, and reduced β-cell function [6-8]. This SNP has also been associated with gestational diabetes [9]. Melatonin is an important component of circadian regulation, and its concentrations are highest when insulin secretion is lowest [10]. Circadian disruption and shift work have been associated with increased T2DM risk and impaired insulin secretion [11, 12]. Melatonin prevents these deleterious effects and promotes β-cell survival and function in isolated human islets and rodent models of circadian disruption [13, 14]. Moreover, low nocturnal melatonin levels are associated with increased risk of T2DM in humans [15]. However, it is also important to note that some studies demonstrated that acute administration of melatonin in humans worsens glucose tolerance, an effect exacerbated in people with 1 or more of the diabetes-associated allele at rs10830963. These somewhat conflicting observations have been described as the “melatonin paradox” emphasizing the importance of additional investigations into melatonin's effects on human islet function [16, 17].
The effect of rs10830963 on fasting glucose seems to be greater than its effects on T2DM predisposition [7]. It does not seem to be associated with glucose intolerance and has effects on insulin secretion that are apparent with both oral [6, 8] and intravenous challenges [8]. However, the methodologies used to measure β-cell function in the large cohorts described have been somewhat qualitative and subject to some limitations [18].
Recently, Heianza et al reported an analysis of this SNP in the OmniCarb trial where overweight, middle-aged adults without diabetes were randomized to a sequence of 4 complete study diets that differed in composition by carbohydrate amount and glycemic index. Participants consumed each diet for 5 weeks, with a 2-week washout between each dietary period. They report that the diabetes-associated allele increased fasting glucose and early glucose response to an oral glucose tolerance test (OGTT). However, there were no observable effects on insulin secretion as measured by the insulinogenic index. In addition, overall β-cell function quantified by a disposition index (DI) (insulinogenic index * Matsuda index) was not associated with the SNP [19].
These results are intriguing for several reasons. We have recently shown that impaired fasting glucose is caused in part by impaired suppression of glucagon secretion by glucose. Second, the melatonin receptor 2 receptor is expressed in the α-cell and melatonin augments glucagon secretion in isolated human islets [20], suggesting that genetic variation in MTNR1B could alter glucagon secretion [21]. Finally, the lack of any meaningful change in insulin secretion to explain the observed postchallenge hyperglycemia might also be attributable to α-cell dysfunction.
We therefore used our database [2, 22] to compare subjects homozygous for the diabetes-protective allele (CC at rs10830963) with those having 1 or 2 copies of the diabetes-associated allele (CG/GG at rs10830963) in MTNR1B. We report that there was a small but significant decrease in the dynamic component of β-cell responsivity accompanied by increased fasting and postchallenge glucagon concentrations in people with the CG or GG genotype. Since genotype at rs7903146 was known in these cohorts, we also show that diabetes-associated genetic variation in TCF7L2 seems to have a greater effect on raising peak glucose concentrations as well as impairing the suppression of postchallenge glucagon concentrations.
Methods
Screening
As part of 2 prior published studies [2, 22] approved by the Mayo Clinic Institutional Review Board, potentially eligible subjects who expressed interest in participating were invited to the Clinical Research and Trials Unit for a screening visit. One cohort was identified after participation in a prior population-based cohort study, while the other group was recruited on the basis of TCF7L2 genotype (either CC or TT at rs7903146 but the groups were matched for age, weight, sex, and fasting glucose). After written, informed consent was obtained, subjects underwent a history and physical examination with relevant laboratory testing. This ensured that subjects fulfilled the inclusion criteria for the studies. Body composition was measured at the time of screening using dual-energy X-ray absorptiometry (Lunar, Madison, WI). A 7-sample OGTT was completed on the day of screening or on a separate study day. Subject characteristics are reported in Table 1.
Table 1.
Subject characteristics by genotype at rs10830963
| Genotype at rs10830963 | CC | CG/GG | P-value |
|---|---|---|---|
| n | 148 | 146 | |
| Age (years) | 57 ± 1 | 56 ± 1 | 0.58 |
| M/F | 67/81 | 60/86 | |
| Weight (kg) | 80 ± 1 | 79 ± 1 | 0.61 |
| BMI (kg/M2) | 27.5 ± 0.4 | 27.4 ± 0.4 | 0.86 |
| LBM (kg) | 46 ± 1 | 45 ± 1 | 0.31 |
| Fasting glucose (mmol/L) | 5.52 ± 0.05 | 5.57 ± 0.04 | 0.46 |
| Peak glucose (mmol/L) | 10.5 ± 0.1 | 11.0 ± 0.1 | 0.02 |
| 120-minute glucose (mmol/L) | 8.3 ± 0.2 | 8.5 ± 0.2 | 0.51 |
| Fasting insulin (pmol/L) | 37 ± 2 | 35 ± 2 | 0.46 |
| Peak insulin (pmol/L) | 455 ± 28 | 437 ± 23 | 0.63 |
| Fasting glucagon (ng/L) | 76 ± 2 | 82 ± 2 | 0.02 |
| Nadir glucagon (ng/L) | 54 ± 1 | 58 ± 1 | 0.04 |
Abbreviations: BMI, body mass index; LBM, lean body mass.
Experimental Design—OGTT
The OGTT was performed after an overnight fast. A dorsal hand vein was cannulated at 0900 and placed in a heated Plexiglas box maintained at 55 °C to allow sampling of arterialized venous blood. At 0900 (0 minutes) subjects ingested 75 g of glucose. Blood was collected at 0, 10, 20, 30, 60, 90, and 120 minutes to enable measurement of glucose and hormone concentrations. At the end of the study (1100, 120 minutes), the cannula was removed; participants consumed lunch and left the Clinical Research and Trials Unit.
Analytic Techniques
All blood was immediately placed on ice after collection, centrifuged at 4 °C, separated, and stored at −80 °C until assay. Plasma glucose concentrations were measured using a Yellow Springs glucose analyzer (Yellow Springs, OH). Plasma insulin concentrations were measured using a chemiluminescence assay (Access Assay, Beckman, Chaska, MN). Plasma C-peptide was measured using a 2-site immunenzymatic sandwich assay (Roche Diagnostics, Indianapolis, IN). Plasma glucagon and C-peptide were measured by Radioimmunoassay (Linco Research, St. Louis, MO). Genotyping of the rs10830963 and rs7903146 SNPs were performed using a Taqman™ kit (Applied Biosystems Inc., Foster City, CA).
Calculations and Statistical Analysis
Calculations
Net postprandial insulin action [insulin sensitivity (Si)] and β-Cell responsivity (Ф) were estimated using the oral minimal model and the oral C-peptide minimal model, respectively [23], incorporating age-associated changes in C-peptide kinetics [24]. DI for each subject was subsequently calculated by multiplying Ф by Si.
Statistical Analysis
All continuous data are summarized as means ± SEM. Area under the curve (AUC) and area above basal (AAB) were calculated using the trapezoidal rule. To assess between-group differences, we used a 2-tailed Student's unpaired t-test (parametric) or a Wilcoxon test (nonparametric). To examine the interaction of rs7903146 genotype with rs10830963 genotype, we used 1-way ANOVA and a Tukey's post hoc test to determine between-group differences (parametric data). For nonparametric data, a Kruskal–Wallis test followed by a Dunn's post hoc test was used. Prism 8 (GraphPad Software, San Diego, CA) was utilized for the statistical analysis. A P-value <.05 was considered statistically significant.
Results
Subject Characteristics by Genotype
Genotyping all the study participants demonstrated that 148 subjects had the CC genotype at rs10830963 (Table 1). Twenty subjects had the GG genotype, and the remaining 126 had the CG genotype. The latter 2 genotypes were combined into 1 group as has been done before [19]. Age, weight, body composition, and fasting glucose did not differ between genotype groups.
Glucose, Insulin, C-peptide, and Glucagon Concentrations by Genotype
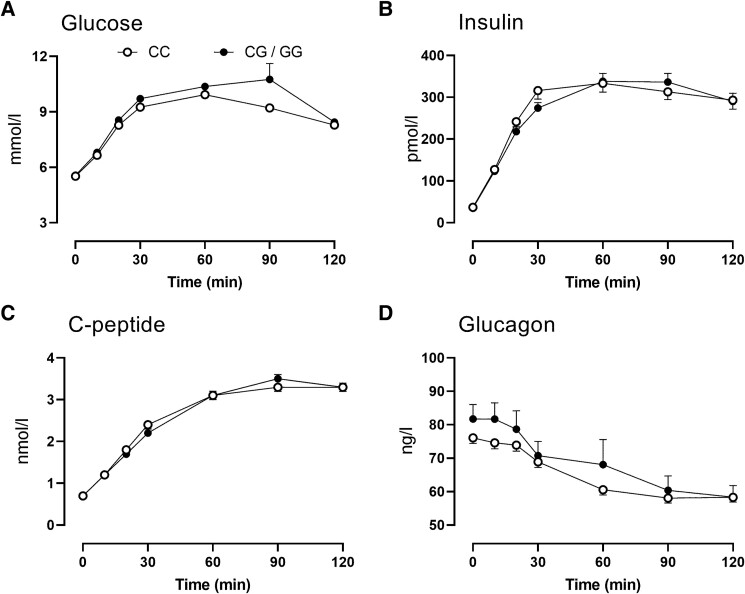
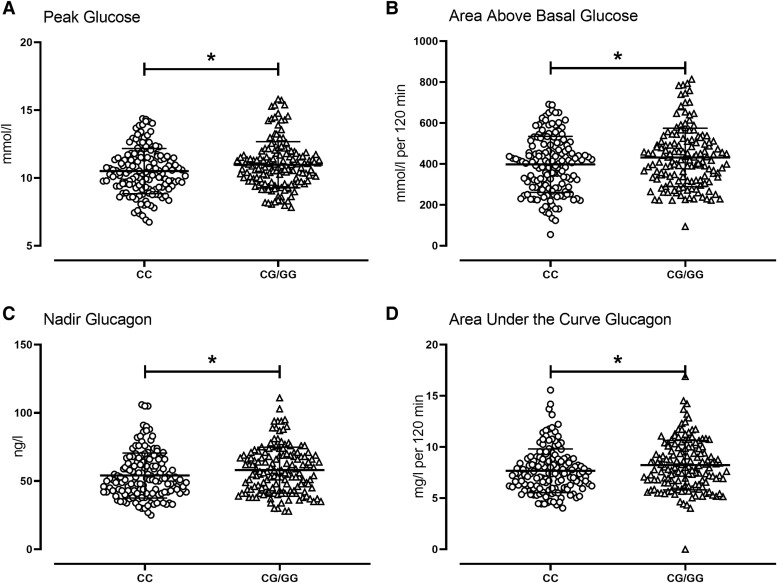
In people with a G-allele, peak and integrated (395 ± 11 vs 468 ± 29 mmol/L per 2 hour, P = .02) glucose concentrations (AAB) in response to the glucose challenge were higher than those with the CC genotype (Fig. 1A). However, at 120 minutes after the glucose challenge there were no between-group differences in glucose concentrations (please also refer to Fig. 2A and 2B).
Figure 1.
Glucose (A), insulin (B), C-peptide (C), and glucagon (D) concentrations during fasting and in response to an oral glucose tolerance test in 294 subjects with the CC (○) or the CG/GG (•) genotype at rs10830963. Values plotted are means ± SEMs.
Figure 2.
Individual values for peak glucose (A), area above basal glucose (B), nadir glucagon (C), and area under the curve glucagon (D) after oral glucose tolerance test in 294 subjects with the CC (○) or the CG/GG (Δ) genotype at rs10830963. The bars and error bars represent means ± SDs. *P < .05 for a Kruskal–Wallis test.
Fasting, peak, and integrated insulin concentrations (AAB) did not differ between genotype groups (Fig. 1B). C-peptide concentrations followed the same pattern and also did not differ between groups (Fig. 1C).
In people with a G-allele, fasting glucagon concentrations were higher compared to people with the CC genotype. Nadir glucagon concentrations were also slightly, but significantly, higher (Fig. 2C). AUC glucagon concentrations over the 120 minutes postglucose challenge were also increased (Fig. 2D) in people with a G-allele (7.7 ± 0.2 vs 8.3 ± 0.2 mg/L per 2 hour, P = .02; Fig. 2D).
Indices of Insulin Secretion and Action by Genotype
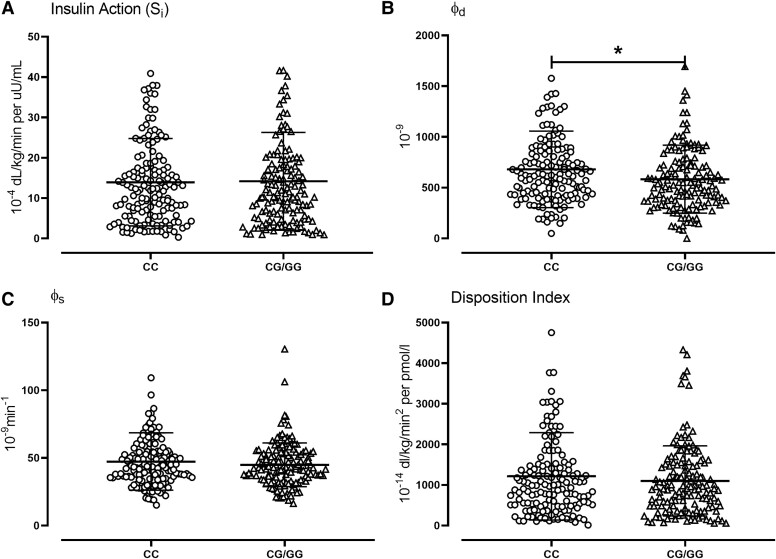
Insulin action (Si) in response to the oral challenge did not differ (14 ± 1 vs 14 ± 1 10−4 dL/kg/min per μU/mL, P = .86) between groups (Fig. 3A). Fasting β-cell responsivity to glucose (ϕb) also did not differ (7.4 ± 0.3 vs 7.3 ± 0.3 10−9 minutes−1, P = .82) between groups (not shown). In contrast, the dynamic component of β-cell responsivity to glucose (ϕd) was decreased in people with a G-allele compared to people with the CC genotype (681 ± 31 vs 583 ± 28 10−9, P = .02; Fig. 3B).
Figure 3.
Individual values for insulin action (Si, A), the dynamic component of β-cell responsivity (ϕd, B), the static component of β-cell responsivity (ϕs, C), and DI (D) after oral glucose tolerance test in 294 subjects with the CC (○) or the CG/GG (Δ) genotype at rs10830963. The bars and error bars represent means ± SDs. *P < .05 for a Kruskal–Wallis test.
Abbreviations: DI, disposition index.
The static component of β-cell responsivity to glucose (ϕs) did not differ between genotype groups (47 ± 2 vs 45 ± 1 10−9 minutes−1, P = .27; Fig. 3C). Total β-cell responsivity to glucose (Φ—55 ± 2 vs 51 ± 1 10−9 minutes−1, P = .15) also did not differ between genotype groups (not shown). The DI also did not differ significantly (1215 ± 89 vs 1099 ± 71 10−14 dL/kg/min2 per pmol/L, P = .31) between genotype groups (Fig. 3D).
Interaction of Hormone and Substrate Concentrations by MTNR1b (rs10830963) and TCF7L2 (rs7903146) Genotype
We examined the effect of the G-allele at rs10830963 on hormone and substrate concentrations in the people with 2 copies of the diabetes-protective allele (CC) and those with 2 copies of the diabetes-associated allele (TT) in the TCF7L2 locus.
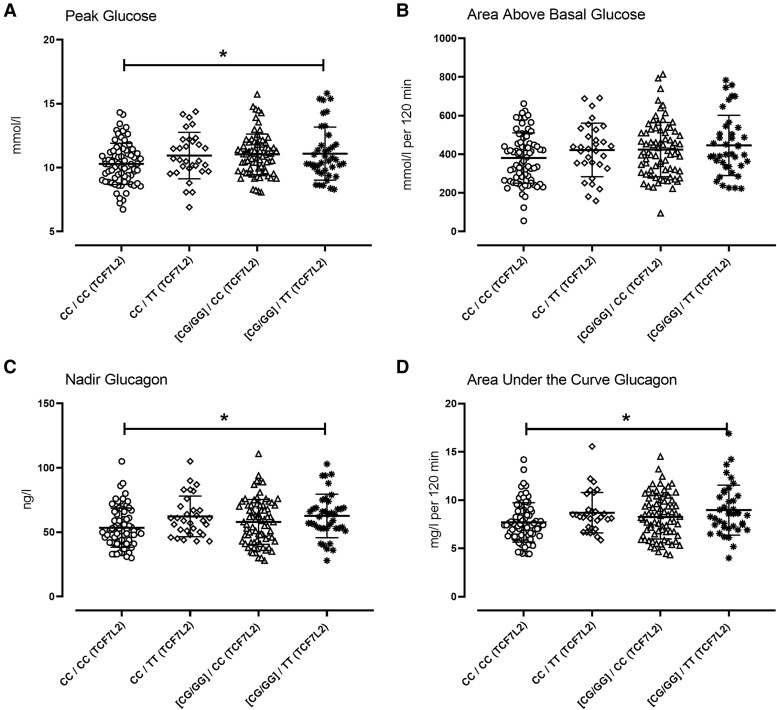
Fasting glucose did not differ between genotype groups. Peak postchallenge glucose concentrations differed (P = .03) among the 4 groups with and without diabetes-associated variation at either SNP (Fig. 4A—10.3 ± 1.6 vs 10.9 ± 1.7 vs 11.1 ± 1.6 vs 11.0 ± 1.9 mmol/L; CC/CC vs CC/TT vs GX/CC vs GX/TT respectively—rs10830963/rs7903146). Integrated (AAB) glucose concentrations did not differ between groups (Fig. 4B).
Figure 4.
Individual values for peak glucose (A), area above basal glucose (B), nadir glucagon (C), and area under the curve glucagon (D), respectively, in people with the CC genotype at rs10830963 and rs7903146 (○, n = 73); the CC genotype at rs10830963 and the TT genotype at rs7903146 (◊, n = 31); the CG/GG genotype at rs10830963 and the CC genotype at rs7903146 (Δ, n = 76); the CG/GG genotype at rs10830963 and the TT genotype at rs7903146 (Δ, n = 40). The bars and error bars represent means ± SDs. *P < .05 for 1-way ANOVA.
Indices of insulin secretion and action did not exhibit any interaction (data not shown). Fasting glucagon also did not differ between genotype groups (data not shown). Nadir glucagon differed significantly between groups (Fig. 4C—54 ± 2 vs 62 ± 3 vs 58 ± 2 vs 60 ± 2 ng/L; P = .03). Integrated glucagon concentrations exhibited a similar relationship (Fig. 4D—7.7 ± 0.2 vs 8.7 ± 0.4 vs 8.2 ± 0.3 vs 8.7 ± 0.3 mg/L per 120 minutes; P = .04).
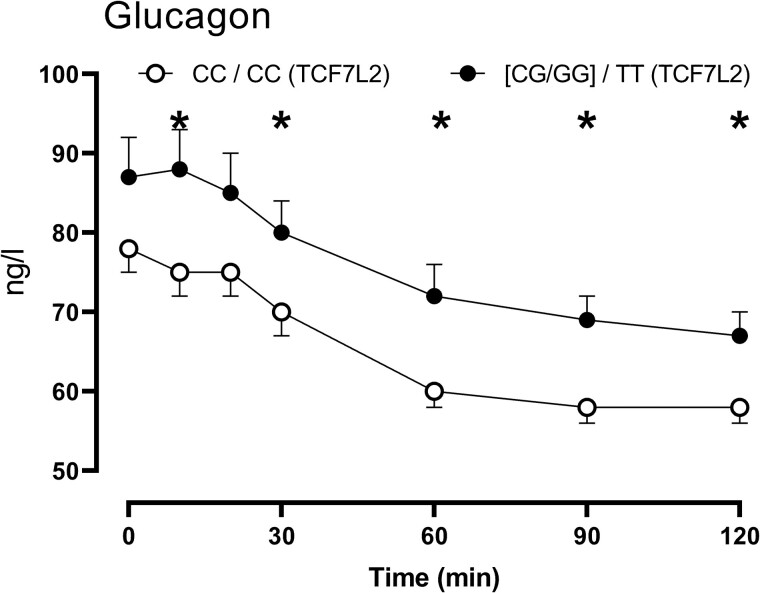
Glucagon Concentrations in People Without and With Diabetes-associated Variation at Both MTNR1b (rs10830963) and TCF7L2 (rs7903146)
A comparison of glucagon concentrations in people with the CC genotype at both rs10830963 and rs790146 with those who had one or two G alleles at rs1083096 and the TT genotype at rs7903146 did not demonstrate changes in fasting glucagon (78 ± 3 vs 86 ± 4 ng/L, P = .10). In keeping with the differences in AUC glucagon (7.7 ± 0.2 vs 8.7 ± 0.3 mg/L per 120 minutes; P = .03) significant differences in glucagon concentrations were observed at various timepoints (Fig. 5). This was of greater magnitude than that attributable to variation at rs10830963 (Fig. 1D) alone.
Figure 5.
Glucagon concentrations over time in people with the CC genotype at rs10830963 and rs7903146 (○, n = 73) compared to those with the CG/GG genotype at rs10830963 and the TT genotype at rs7903146 (•, n = 40). Values plotted are means ± SEMs. *P < .05 for an unpaired, 2-tailed t-test.
Discussion
Postprandial glucose concentrations are dependent on the ability of the endocrine pancreas to secrete insulin in addition to the ability of insulin to suppress endogenous glucose production and to stimulate glucose uptake [23]. The ability of the α-cell to respond to hyperglycemia by suppressing glucagon secretion is also a key factor in the regulation of postprandial glucose [25]. This becomes more important as β-cell function and insulin action decrease [26]. The recent report by Heianza et al examining the effect of diabetes-associated genetic variation on glucose tolerance in the MTNR1B locus did not show an effect on β-cell function. This raised the possibility that effects of this variant on fasting and postprandial glucose concentrations are mediated through effects on α-cell function [19].
Previously, we showed that diabetes-associated variation in TCF7L2 is associated with impaired α-cell function [2]. This illustrated that α-cell dysfunction arises early in the pathogenesis of T2DM. In keeping with this finding, abnormalities in glucagon secretion could predict a decline in glucose tolerance [27]. More recently, impaired fasting glucose has been shown to occur in people with intact β-cell function, and this abnormality can be explained by decreased responsiveness of the α-cell to glucagon [28].
Therefore, we examined the association of variation in rs10830963 with islet function in 2 cohorts studied using a 7-sample OGTT [2, 22]. People with 1 or 2 copies of the diabetes-associated allele at rs10830963 had higher peak and integrated postchallenge glucose concentrations. Fasting and 120-minute glucose concentrations did not differ from those observed in people without diabetes-associated variation in rs10830963. A lack of effect on 120-minute glucose is congruent with reports from several other similarly powered cohorts [8, 19].
On the other hand, we failed to observe an effect on fasting glucose. This might be explained by the fact that a subgroup of our participants was recruited on the basis of rs7903146 genotype—another SNP associated with fasting hyperglycemia. Because of the nature of that experiment, we matched fasting glucose in the cohorts with the CC and TT genotype at rs7903146. As we acknowledged at the time, this may have introduced a conservative error in our estimation of the diabetogenic effects of the T-allele [2]. Nevertheless, we did observe higher fasting glucagon in subjects with diabetes-associated variation at rs10830963.
In addition to higher fasting glucagon, the G-allele at rs10830963 was associated with impaired glucagon suppression in response to glucagon ingestion as implied by the higher nadir and integrated glucagon concentrations observed in this group. The only other accompanying defect in β-cell function that we observed was in the dynamic component of β-cell responsivity to glucose. This is thought to represent the pool of insulin granules that are primed for release in response to rising glucose concentrations [29]. Other investigators have previously suggested an effect of MTNR1B on first-phase insulin secretion in response to an intravenous glucose tolerance test, which would be in keeping with our observations [6, 8, 30].
The regulation of postprandial glucose concentrations is well characterized and depends on insulin secretion and action [23], glucose effectiveness, and glucagon suppression [25]. The rate of gastric emptying also plays a role, although this is less important in the response to an oral liquid glucose challenge [31]. In this analysis, similar time to peak glucose suggests that gastric emptying was not a factor in explaining the differences between genotype groups. Insulin secretion and action also did not differ, strongly suggesting that the differences in glucagon concentrations observed could explain postchallenge glucose. As in other studies, differences in α-cell function can occur independently of β-cell function [2, 32, 33].
Melatonin's action is mediated through high-affinity melatonin receptors [34] expressed in human islets. Persistent (6-12 hours) activation of the melatonin receptor mimicking timing of nightly exposure leads to sensitization and enhancement of adenylate cyclase activity resulting in increased activation of the cAMP, protein kinase A, and cAMP-responsive element binding protein [35]. This phenomenon also occurs in β-cells [13, 36, 37] The ability of melatonin to potentiate the cAMP-dependent signal transduction pathway is important for its actions to regulate circadian clock gene expression [36]. However, this pathway also regulates β-cell function and survival in human islets. Indeed, activation of protein kinase A is essential for glucose-stimulated insulin secretion [38]. Moreover, cAMP-responsive element binding protein expression is essential for maintaining proper β-cell mass and function and preventing β-cell apoptosis [39]. Activation of melatonin signaling in β-cells (with a duration designed to mimic typical nightly exposure) significantly enhances the cAMP-dependent signal transduction pathway and attenuates β-cell oxidative stress and apoptosis [13] restoring glucose-stimulated and incretin-stimulated insulin secretion in islets isolated from subjects with T2DM [13]. In contrast, others noted that genetic overexpression of the melatonin receptor in β-cells attenuates insulin secretion [40]. Unfortunately, much less is known about the effect of melatonin and its receptors on α-cell function.
Since TCF7L2 genotype in both cohorts was known, we examined the effect of diabetes-associated alleles at both loci on glucose and islet function. Unsurprisingly, diabetes-associated variation at rs7903146 was associated with impaired α-cell function. The G-allele at rs10830963 did not interfere with these effects and perhaps may have a small additive effect on postchallenge glucagon concentrations when compared to the cohort. Whether this proves to be the case in larger, independent cohorts remains to be ascertained. If so, it would imply independent mechanisms of the action of TCF7L2 and MTNRB1 on α-cell function.
This study has several limitations, in part arising from its retrospective nature. The potential effect of how 1 cohort was recruited [2] has already been discussed. In addition, the relatively small sample size precludes a better analysis of the interaction, if any, between diabetes-associated variation at rs7903146 and rs10830963. It likely explains why we did not see an effect on β-cell function previously observed in larger studies [6, 8], although it implies that effects on α-cell function are easier to detect than effects on β-cell function. Finally, the glucagon assay used in these studies cross-reacts with other proglucagon-derived peptide fragments [41]. This raises the possibility that genetic variation in MTNR1B alters concentrations of proglucagon peptides other than glucagon. However, this would not explain the hyperglycemia we and others observed in the absence of significant differences in β-cell function. Despite differences in absolute concentrations of fasting and nadir glucagon measured using newer assays, there is good correlation between these assays and the assay used in this experiment [41-44]. Moreover, the association of the T-allele at rs7903146 with higher glucagon concentrations has been observed using other immunoassays in addition to this one [3].
In conclusion, we demonstrate that diabetes-associated genetic variation in MTNR1B can raise glucose concentrations through α-cell dysfunction in addition to having effects on insulin secretion. Future studies in larger cohorts should be better able to characterize an interaction, if any, with diabetes-associated variation at the TCF7L2 locus.
Acknowledgments
The authors wish to acknowledge the excellent editorial assistance of M.M. Davis, Endocrine Research Unit, Mayo Clinic, Rochester, MN.
Contributor Information
Max Vella, Division of Endocrinology, Diabetes & Metabolism, Mayo Clinic College of Medicine, Rochester, MN 55905, USA.
Sneha Mohan, Division of Endocrinology, Diabetes & Metabolism, Mayo Clinic College of Medicine, Rochester, MN 55905, USA.
Hannah Christie, Division of Endocrinology, Diabetes & Metabolism, Mayo Clinic College of Medicine, Rochester, MN 55905, USA.
Kent R Bailey, Division of Biomedical Statistics and Informatics, Mayo Clinic, Rochester, MN 55905, USA.
Claudio Cobelli, Department of Women and Children's Health, University of Padova, 35128 Padova, Italy.
Chiara Dalla Man, Department of Information Engineering, University of Padova, 35128 Padova, Italy.
Aleksey Matveyenko, Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN 55905, USA.
Aoife M Egan, Division of Endocrinology, Diabetes & Metabolism, Mayo Clinic College of Medicine, Rochester, MN 55905, USA.
Adrian Vella, Email: vella.adrian@mayo.edu, Division of Endocrinology, Diabetes & Metabolism, Mayo Clinic College of Medicine, Rochester, MN 55905, USA.
Funding
The authors acknowledge the support of the Center for Clinical and Translational Science, Mayo Clinic (DK TR000135). A.V. is supported by DK78646, DK116231, and DK126206. A.M.E. is supported by DK134767. C.D.M. was supported by MIUR (Italian Minister for Education) under the initiative Departments of Excellence (Law 232/2016).
Author Contributions
M.V. generated the genotyping data and the initial genotype/phenotype analysis, contributed to the discussion, and reviewed/edited the manuscript; S.M. and H.C. researched data and ran the studies, contributed to the discussion, and reviewed/edited manuscript; K.R.B. supervised the statistical analyses; C.C. and C.D.M. supervised the mathematical modeling, contributed to the discussion, and reviewed/edited manuscript; A.M. and A.M.E. contributed to the discussion and reviewed/edited manuscript; A.V. designed the study, oversaw its conduct, researched data, and wrote the first draft of the manuscript. A.V. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Disclosures
A.V. is the recipient of an investigator-initiated grant from Novo Nordisk and has consulted for Crinetics and Rezolute. None of the other authors declare any conflict of interests related to this study.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request. No applicable resources were generated or analyzed during the current study.
References
- 1. Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50(11):1505‐1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shah M, Varghese RT, Miles JM, et al. TCF7L2 genotype and alpha-cell function in humans without diabetes. Diabetes. 2016;65(2):371‐380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adams JD, Egan AM, Laurenti MC, et al. The effect of diabetes-associated variation in TCF7L2 on postprandial glucose metabolism when glucagon and insulin concentrations are matched. Metab Syndr Relat Disord. 2022;20(6):329‐335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smushkin G, Vella A. Genetics of type 2 diabetes. Curr Opin Clin Nutr Metab Care. 2010;13(4):471‐477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bouatia-Naji N, Bonnefond A, Cavalcanti-Proenca C, et al. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet. 2009;41(1):89‐94. [DOI] [PubMed] [Google Scholar]
- 6. Lyssenko V, Nagorny CL, Erdos MR, et al. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet. 2009;41(1):82‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Prokopenko I, Langenberg C, Florez JC, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41(1):77‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sparso T, Bonnefond A, Andersson E, et al. The G-allele of intronic rs10830963 in MTNR1B confers increased risk of impaired fasting glycemia and type 2 diabetes through an impaired glucose-stimulated insulin release: studies involving 19,605 Europeans. Diabetes. 2009;58(6):450‐456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim JY, Cheong HS, Park B-L, et al. Melatonin receptor 1 B polymorphisms associated with the risk of gestational diabetes mellitus. BMC Med Genet. 2011;12(1):82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Korkmaz A, Topal T, Tan D-X, Reiter RJ. Role of melatonin in metabolic regulation. Rev Endocr Metab Disord. 2009;10(4):261‐270. [DOI] [PubMed] [Google Scholar]
- 11. Morris CJ, Purvis TE, Mistretta J, Scheer FA. Effects of the internal circadian system and circadian misalignment on glucose tolerance in chronic shift workers. J Clin Endocrinol Metab. 2016;101(3):1066‐1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sharma A, Laurenti MC, Dalla Man C, et al. Glucose metabolism during rotational shift-work in healthcare workers. Diabetologia. 2017;60(8):1483‐1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Costes S, Boss M, Thomas AP, Matveyenko AV. Activation of melatonin signaling promotes beta-cell survival and function. Mol Endocrinol. 2015;29(5):682‐692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thomas AP, Hoang J, Vongbunyong K, Nguyen A, Rakshit K, Matveyenko AV. Administration of melatonin and metformin prevents deleterious effects of circadian disruption and obesity in male rats. Endocrinology. 2016;157(12):4720‐4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McMullan CJ, Schernhammer ES, Rimm EB, Hu FB, Forman JP. Melatonin secretion and the incidence of type 2 diabetes. JAMA. 2013;309(13):1388‐1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garaulet M, Gomez-Abellan P, Rubio-Sastre P, Madrid JA, Saxena R, Scheer FAJL. Common type 2 diabetes risk variant in MTNR1B worsens the deleterious effect of melatonin on glucose tolerance in humans. Metabolism. 2015;64(12):1650‐1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garaulet M, Qian J, Florez JC, Arendt J, Saxena R, Scheer F. Melatonin effects on glucose metabolism: time to unlock the controversy. Trends Endocrinol Metab. 2020;31(3):192‐204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xiang AH, Watanabe RM, Buchanan TA. HOMA and Matsuda indices of insulin sensitivity: poor correlation with minimal model-based estimates of insulin sensitivity in longitudinal settings. Diabetologia. 2014;57(2):334‐338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heianza Y, Zhou T, Wang X, et al. MTNR1B genotype and effects of carbohydrate quantity and dietary glycaemic index on glycaemic response to an oral glucose load: the OmniCarb trial. Diabetologia. 2024;67(3):506‐515. [DOI] [PubMed] [Google Scholar]
- 20. Ramracheya RD, Muller DS, Squires PE, et al. Function and expression of melatonin receptors on human pancreatic islets. J Pineal Res. 2008;44(3):273‐279. [DOI] [PubMed] [Google Scholar]
- 21. Zibolka J, Bazwinsky-Wutschke I, Muhlbauer E, Peschke E. Distribution and density of melatonin receptors in human main pancreatic islet cell types. J Pineal Res. 2018;65(1):e12480. [DOI] [PubMed] [Google Scholar]
- 22. Sathananthan A, Man CD, Zinsmeister AR, et al. A concerted decline in insulin secretion and action occurs across the spectrum of fasting and postchallenge glucose concentrations. Clin Endocrinol (Oxf). 2012;76(2):212‐219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cobelli C, Dalla Man C, Toffolo G, Basu R, Vella A, Rizza R. The oral minimal model method. Diabetes. 2014;63(4):1203‐1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Van Cauter E, Mestrez F, Sturis J, Polonsky KS. Estimation of insulin secretion rates from C-peptide levels. Comparison of individual and standard kinetic parameters for C-peptide clearance. Diabetes. 1992;41(3):368‐377. [DOI] [PubMed] [Google Scholar]
- 25. Shah P, Basu A, Basu R, Rizza R. Impact of lack of suppression of glucagon on glucose tolerance in humans. Am J Physiol. 1999;277(2):E283‐E290. [DOI] [PubMed] [Google Scholar]
- 26. Adams JD, Egan AM, Laurenti MC, et al. Insulin secretion and action and the response of endogenous glucose production to a lack of glucagon suppression in nondiabetic subjects. Am J Physiol Endocrinol Metab. 2021;321(5):E728‐E736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adams JD, Dalla Man C, Laurenti MC, et al. Fasting glucagon concentrations are associated with longitudinal decline of beta-cell function in non-diabetic humans. Metabolism. 2020;105:154175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kohlenberg JD, Laurenti MC, Egan AM, et al. Differential contribution of alpha and beta cell dysfunction to impaired fasting glucose and impaired glucose tolerance. Diabetologia. 2023;66(1):201‐212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cobelli C, Man CD, Sparacino G, et al. Diabetes: models, signals, and control. IEEE Rev Biomed Eng. 2009;2:54‐96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langenberg C, Pascoe L, Mari A, et al. Common genetic variation in the melatonin receptor 1B gene (MTNR1B) is associated with decreased early-phase insulin response. Diabetologia. 2009;52(8):1537‐1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vella A, Camilleri M. The gastrointestinal tract as an integrator of mechanical and hormonal response to nutrient ingestion. Diabetes. 2017;66(11):2729‐2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Faerch K, Vistisen D, Pacini G, et al. Insulin resistance is accompanied by increased fasting glucagon and delayed glucagon suppression in individuals with normal and impaired glucose regulation. Diabetes. 2016;65(11):3473‐3481. [DOI] [PubMed] [Google Scholar]
- 33. Sharma A, Varghese RT, Shah M, et al. Impaired insulin action is associated with increased glucagon concentrations in non-diabetic humans. J Clin Endocrinol Metab. 2018;103(1):314‐319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gillette MU, McArthur AJ. Circadian actions of melatonin at the suprachiasmatic nucleus. Behav Brain Res. 1996;73(1-2):135‐139. [DOI] [PubMed] [Google Scholar]
- 35. von Gall C, Stehle JH, Weaver DR. Mammalian melatonin receptors: molecular biology and signal transduction. Cell Tissue Res. 2002;309(1):151‐162. [DOI] [PubMed] [Google Scholar]
- 36. Nishiyama K, Hirai K. The melatonin agonist ramelteon induces duration-dependent clock gene expression through cAMP signaling in pancreatic INS-1 beta-cells. PLoS One. 2014;9(7):e102073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kemp DM, Ubeda M, Habener JF. Identification and functional characterization of melatonin Mel 1a receptors in pancreatic beta cells: potential role in incretin-mediated cell function by sensitization of cAMP signaling. Mol Cell Endocrinol. 2002;191(2):157‐166. [DOI] [PubMed] [Google Scholar]
- 38. Kaihara KA, Dickson LM, Jacobson DA, et al. beta-Cell-specific protein kinase A activation enhances the efficiency of glucose control by increasing acute-phase insulin secretion. Diabetes. 2013;62(5):1527‐1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dalle S, Quoyer J, Varin E, Costes S. Roles and regulation of the transcription factor CREB in pancreatic beta -cells. Curr Mol Pharmacol. 2011;4(3):187‐195. [DOI] [PubMed] [Google Scholar]
- 40. Tuomi T, Nagorny CLF, Singh P, et al. Increased melatonin signaling is a risk factor for type 2 diabetes. Cell Metab. 2016;23(6):1067‐1077. [DOI] [PubMed] [Google Scholar]
- 41. Lund A, Bagger JI, Wewer Albrechtsen NJ, et al. Evidence of extrapancreatic glucagon secretion in man. Diabetes. 2016;65(3):585‐597. [DOI] [PubMed] [Google Scholar]
- 42. Bak MJ, Albrechtsen NW, Pedersen J, et al. Specificity and sensitivity of commercially available assays for glucagon and oxyntomodulin measurement in humans. Eur J Endocrinol. 2014;170(4):529‐538. [DOI] [PubMed] [Google Scholar]
- 43. Wewer Albrechtsen NJ, Hartmann B, Veedfald S, et al. Hyperglucagonaemia analysed by glucagon sandwich ELISA: nonspecific interference or truly elevated levels? Diabetologia. 2014;57(9):1919‐1926. [DOI] [PubMed] [Google Scholar]
- 44. Wewer Albrechtsen NJ, Veedfald S, Plamboeck A, et al. Inability of some commercial assays to measure suppression of glucagon secretion. J Diabetes Res. 2016;2016:8352957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request. No applicable resources were generated or analyzed during the current study.