Abstract
Vaccinia viruses defective in the essential gene coding for the enzyme uracil DNA glycosylase (UDG) do not undergo DNA replication and do not express late genes in wild-type cells. A UDG-deficient vaccinia virus vector carrying the tick-borne encephalitis (TBE) virus prM/E gene, termed vD4-prME, was constructed, and its potential as a vaccine vector was evaluated. High-level expression of the prM/E antigens could be demonstrated in infected complementing cells, and moderate levels were found under noncomplementing conditions. The vD4-prME vector was used to vaccinate mice; animals receiving single vaccination doses as low as 104 PFU were fully protected against challenge with high doses of virulent TBE virus. Single vaccination doses of 103 PFU were sufficient to induce significant neutralizing antibody titers. With the corresponding replicating virus, doses at least 10-fold higher were needed to achieve protection. The data indicate that late gene expression of the vaccine vector is not required for successful vaccination; early vaccinia virus gene expression induces a potent protective immune response. The new vaccinia virus-based defective vectors are therefore promising live vaccines for prophylaxis and cancer immunotherapy.
Vaccinia virus is a useful tool for research and vaccine development, serving as a means to synthesize biologically active proteins and to investigate mechanisms of humoral and cell-mediated immunity. Attenuated vaccinia virus and poxvirus strains are candidates for live vaccines and for cancer therapy (24, 25). Most classical vaccinia virus vaccine strains, however, do not meet modern standards of safety, and experimentation using laboratory strains requires biosafety measures. The selective deletion of virulence factors and the construction of highly attenuated strains have been achieved through genetic engineering (30). A different approach toward the design of safe vaccinia virus-based expression and vaccine vectors, however, is the construction of defective vaccinia viruses in complementing cell lines by deletion of at least one essential gene and further modification of the defective virus by enhancing or decreasing activities of genes modulating the immune response, resulting in vectors suitable for vaccination or gene therapy. We have recently constructed permanent cell lines that fully complement a prototype vaccinia virus defective in an early gene coding for viral uracil DNA glycosylase (UDG) (14). This virus is blocked in its life cycle prior to late gene expression and is therefore further designated as late defective. With the aim of developing late defective vaccinia viruses as vaccine vectors, we now report growth and expression characteristics of such replication-incompetent vaccine vectors. The membrane-bound tick-borne encephalitis (TBE) virus prM/E protein was chosen as a model antigen in the vaccination study because prM/E proteins are good immunogens in many flaviviruses (for reviews, see references 12 and 32). Attempts to develop a live vaccinia virus-based TBE virus vaccine have been reported recently; a vaccinia virus vector expressing the C-prM-E-NS1-NS2A-NS2B-NS3 region was found to efficiently protect mice (6). Protection was also elicited by a replication-defective adenovirus vector expressing the TBE virus glycoprotein NS1 (16). The primary goal of this study was to evaluate the vaccine properties of an engineered late defective vaccinia virus vector with respect to those of a corresponding replicating vector, using TBE virus antigen expression and mouse protection experiments as a model system.
MATERIALS AND METHODS
Virus and cell lines.
The African green monkey kidney cell lines CV-1 (ATCC CCL-70) and Vero (ATCC CCL-81), the rabbit kidney cell line RK13 (ATCC CCL-37), and the Western Reserve (WR) strain of vaccinia virus (ATCC VR119) were obtained from the American Type Culture Collection. The cell line RK-D4R-44.20 is described elsewhere (14).
Construction of plasmids. (i) pTKselP-tSpME.
To delete the internal NcoI site in the gene encoding protein E, a prM/E NcoI gene cassette was created by PCR using cDNA plasmid pBA5 (20) as the template. The 5′ portion encoding prM and the amino-terminal part of protein E was synthesized with the PCR primers pPCR1783 (CTACCATGGC GTGGATGAGC TGGTTGCTGG TC) containing the transcriptional start codon and pPCR524 (CTGGATCCAG TTTTGCGCGC CCTCATGTTT CCAAGGCAGA GCCAG). The region encoding the C-terminal half of protein E was amplified from cDNA with the primers pPCR523 (CTGGCTCTGC CTTGGAAACA TGAGGGCGCG CAAAACTGGAC) and pPCR1784 (CAGTCGACCA TGGACTAGTT AACTACGCCC CCACTCCAAGG GTCA), introducing a translational stop codon. The overlapping primers 523 and 524 destroy the internal NcoI site and introduce a unique BssHII site without affecting the amino acid sequence. Both parts were assembled as NcoI-BssHII fragments in a modified pUC vector and further subcloned as an NcoI cassette in the expression vector pTKgpt-selP (26), resulting in pTKselP-tSpME. The prM/E gene cassette used in the plasmid constructs contains (i) the putative signal sequence of prM starting at position 412 and (ii) the coding regions for the prM protein and glycoprotein E, including its two hydrophobic transmembrane domains ending at position 2439 (numbering according to reference 20).
(ii) pD4-vA.
A 4.4-kb PstI/NotI fragment was excised from plasmid ptk-vA (27) and cloned into PstI/NotI-cleaved pD4Rf (14). The 4.4-kb insert codes for a lacZ and gpt (guanosine-hypoxanthine phosphoribosyltransferase) gene marker cassette flanked by tandem repeats of phage φX sequences to allow for transient dominant selection (9, 27). The resulting plasmid was named pD4-vA.
(iii) pD4-prME.
The 2.2-kb HindIII-NotI fragment, containing the TBE virus prM and E open reading frame (ORF) controlled by a strong early/late promoter (4), was excised from plasmid pTKselP-tSpME and inserted into the corresponding restriction sites of pD4s (13), resulting in pD4-prME.
(iv) pVV-21.
The vaccinia virus thymidine kinase (tk) gene region of the WR strain (bases 83621 to 84761; numbering according to reference 10) was inserted counterclockwise between the PvuII sites of pTZ19R (Pharmacia, Inc.). Bases 84718 to 84738 were deleted by a PCR-based mutagenesis procedure, resulting in pVV-21.
Construction of viruses. (i) vD4-vA.
Twenty micrograms of pD4-vA plasmid DNA was transfected into vaccinia virus WR-infected CV-1 cells and further processed as described previously (8). Plaque assays were performed with the complementing cell line RK-D4R-44.20 under GPT selection according to a transient dominant selection protocol (27). Three isolates, designated vD4-vA#5, vD4-vA#6, and vD4-vA#13, were grown to large scale in the complementing cell line.
(ii) vD4tk-ZG and vD4-prME.
Twenty micrograms of either ptk-ZG plasmid DNA (13) or pD4-prME plasmid DNA was transfected into vD4-vA-infected RK-D4R-44.20 cells. Four rounds of plaque purification were performed in the complementing cell line under GPT selection and blue plaque screening (3).
(iii) varec280.
The virus was constructed according to standard protocols using GPT- and TK-negative selection (8, 19) using plasmid pTKselPtSpME and the vaccinia virus WR strain as recombination partners in CV-1 cells. The viruses used for the vaccinations were grown in large Roux bottles, and cell-bound virus was prepared by trypsinization and centrifugation through a 36% sucrose cushion as described previously (17). No prM/E antigen was detectable in the virion preparations by Western blot analysis.
Western blotting.
Expression of the prM/E protein by the vD4-prME recombinant was assessed by Western blotting. Infections were done at a multiplicity of infection (MOI) of 0.1 for 72 h or at an MOI of 1 for 24 h. A total of 5 × 106 RK-D4R-44 cells or RK13 wild-type (wt) cells were infected with vD4-prME. Infections with the replication-competent recombinant varec280 expressing the prM/E gene were performed in the cell line of interest; 150 ng of glycoprotein E from purified TBE virions served as a positive control. prM/E protein-specific bands were detected with an equal mixture of the anti-glycoprotein E monoclonal antibodies A3, C3, W3, I, N, R, and S (Immuno AG), used in a 1:100 dilution. The second antibody was a 1:2,000 goat-anti-mouse alkaline phosphatase-conjugated immunoglobulin G (Sigma Inc.).
Assay for replicating virus.
Confluent monolayers of RK13 cells grown in 10 Roux bottles (2 × 108 cells) were infected with 2 × 107 PFU (0.1 PFU/cell) of the respective defective viral stock and grown in serum-free medium for 3 to 4 days. Plaques of replicating viruses were readily detectable, if present, after staining with crystal violet.
TBE virus neutralization test and antigen ELISA.
The prechallenge sera of the mice, pooled according to the grouping of the animals, together with a positive control anti-TBE serum, were diluted 1:5, 1:20, and 1:80 with TCM199 medium (Gibco BRL) and incubated with 2 volumes of a standard TBE virus suspension (103 50% tissue culture infective doses/ml in TCM199). After incubation for 2.5 h, the neutralization mixtures (100 μl/well) were layered over confluent Vero cells grown in 96-well plates (8 wells per dilution). The mixture was further diluted 1:10 three times, and the cells were incubated for 7 days under standard conditions to allow virus growth. The neutralization titer, defined as the antiserum dilution that neutralizes TBE virus growth in 50% of the wells, was determined according to reference 18. Since TBE virus does not form plaques, the antigen content in the cell culture supernatant was determined by a TBE virus antigen enzyme-linked immunosorbent assay (ELISA). Micro-ELISA plates coated with guinea pig anti-TBE immunoglobulin G (Immuno AG) were incubated with the virus-containing samples and the controls. After 1 h, the wells were washed three times with phosphate-buffered saline (PBS) and incubated with a 1:1,000 dilution of rabbit anti-TBE serum (Immuno AG) for 1 h; after a second wash (3× PBS), the plates were incubated for 1 h with an anti-rabbit-peroxidase conjugate (Chemical Accurate, Inc.). After a third wash step (5× PBS), the peroxidase substrate was added for 15 min.
Animal protection studies.
Groups of 10 mice (CD1 strain; Charles River, Inc.) were immunized intramuscularly, subcutaneously, and intraperitoneally with a single dose of 102 to 106 PFU of purified virus (see Fig. 5) resuspended in PBS. After 19 days, sera were collected and pooled according to the grouping of the animals. Twenty-one days later, the mice were challenged by an intraperitoneal injection with 100 50% lethal doses (LD50) of virulent TBE virus (strain Neudörfl). Unprotected mice usually began to die 7 days after challenge; survivors were recorded at day 21 after challenge. A dose-response experiment with increasing TBE virus concentrations in nonvaccinated mice was done in parallel to the challenge experiments to confirm the validity of the LD50 titer.
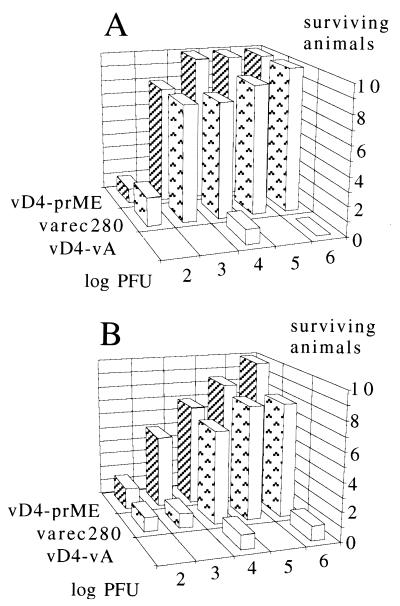
FIG. 5.
Vaccination of mice with defective vaccinia viruses expressing the TBE virus prM/E antigen and protection against a lethal TBE virus challenge. Groups of 10 mice were vaccinated once with the viruses and doses indicated and challenged 21 days later with 100 LD50 of TBE virus. The y axis represents the number of survivors 3 weeks after challenge. Mean values of two consecutive experiments are shown. (A) Intramuscular vaccination; (B) Subcutaneous vaccination.
In vivo replication experiments.
Groups of five mice (CD1 strain) were immunosuppressed (according to reference 30) by intraperitoneal injection of cyclophosphamide (0.2 ml of a 20-mg/ml solution in PBS per animal) at days −2, +1, and +4. Virus varec280 or pD4-prME was injected intramuscularly at day 0 (106 and 108 PFU). At 3, 5, and 7 days postinoculation, total spleen and muscle (injection site) samples were washed and resuspended in PBS, homogenized, and trypsinized. Fetal bovine serum was added, and serial dilutions were titered on CV-1 cells or RK-D4R-44 cells. The marked atrophy of the spleens in the cyclophosphamide-treated mice was an indicator of immunosuppression.
Quantitative vaccinia virus PCR.
Genomic equivalents of vaccinia virus DNA were determined by gene scanning of fluorescence-labelled amplicons relative to an internal standard (11), with the following modifications. Total spleens or muscle were removed, washed once in 4 ml of PBS, and homogenized. A 250-μl sample was mixed with 80 μl of proteinase K mix (11) and 300 copies of pVV-21 internal standard, and water was added to a final volume of 500 μl. Incubation was done at 56°C for 30 min, followed by standard DNA extraction and precipitation of DNA using glycogen as a carrier. The DNA pellet was dissolved in 50 μl of water; 10 μl of the extract was amplified at hot-start conditions in a 50-μl reaction volume containing 2.5 U of Taq DNA polymerase (Boehringer Mannheim), deoxynucleoside triphosphates at 200 μM each, primers opr1-f/B FAM (5-carboxyfluorescein; Applied Biosystems, Inc.)-labelled 5′-ATATTAGATGGTG CCACCGTAG-3′ and opr1-r/B 5′-AAAATAGGAT CATGATGGCG-3′, and 1.5 mM MgCl2–10 mM Tris (pH 8.3)–50 mM KCl. PCR was performed (thermocycler type 9600; Perkin-Elmer) for 33 cycles, each cycle consisting of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s, followed by final incubation at 72°C for 10 min. To determine the PFU/particle ratios, the virus samples (dilutions of 100, 10, 1.0, 0.1, and 0.01 PFU in Tris-buffered saline–1% bovine serum albumin–0.1% Nonidet P-40, [pH 8.0]; original stock sonicated before dilution) were digested with proteinase K and treated as described above. The PCR products of internal standard and wild-type template (116 and 137 bp in length) were quantified after separation on sequencing gels using a 373A DNA sequencer and Genescan software (Applied Biosystems, Inc.).
RESULTS
Construction of the basic defective vaccinia virus expression vector vD4-vA.
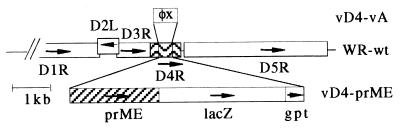
Since the prototype vaccinia virus deficient in the gene coding for the DNA repair enzyme UDG (14) already contained marker genes, it was not suitable as a vector for further gene insertions. We therefore constructed a new defective vaccinia virus vector lacking marker genes, termed vD4-vA, using a transient marker stabilization procedure (27) in the complementing cell line RK-D4R-44 (14). First we generated plasmid pD4-vA, in which the Escherichia coli lacZ and gpt marker genes are located between 450-bp DNA repeats that are flanked by sequences of the D4 gene. Upon recombination, the flanking regions direct the markers transiently into the viral D4 locus, resulting in a marker gene-free and partially deleted D4 locus. Southern hybridizations with a D4R-specific probe confirmed the genomic structure and the purity of the defective virus (data not shown). Due to the deletion of D4-specific sequences and the insertion of one of the 450-bp repeats, the D4 locus of vD4-vA is about 100 bp larger than the corresponding wt gene region. A map of the D4 gene region and the modified D4 locus in viruses vD4-vA and vD4-prME is shown in Fig. 1.
FIG. 1.
Map of the D4R gene regions in wt vaccinia virus (WR-wt) and in the defective viruses vD4-vA and vD4-prME. In vD4-vA, the D4R ORF is partially substituted by a 400-bp DNA fragment of phage φX. In vD4-prME, the substitution comprises the TBE virus prM/E, lacZ, and gpt ORFs. The direction of transcription is indicated by arrows.
The defective virus grows in complementing cells as efficiently as does the wt strain.
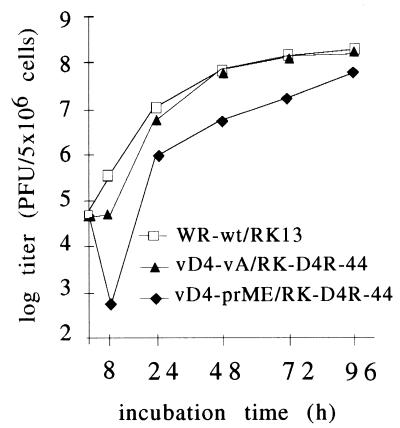
Among the critical parameters for the use of vaccinia virus as a vector are growth and stability of the virus. Therefore, growth of the basic construct vD4-vA in the complementing cell line RK-D4R-44.20 was compared to that of the WR wt virus in RK13 cells. The cells were infected with an MOI of 0.01 and harvested at different time points between 8 and 96 h. The yield of the defective virus vD4-vA, after an incubation period of 96 h in complementing cells, approximated that of the wt virus in RK13 cells (Fig. 2). The growth of the D4R defective virus was not negatively affected, approaching the same titers as obtained with the WR strain. Furthermore, the virus could be purified by standard protocols (17) without substantial loss of infectivity. Thus, we obtained a defective basic virus vector with excellent growth properties in the complementing cells that was physically stable and easy to purify. The virus vD4-prME (see below) grew to lower titers than the parental strain vD4-vA (Fig. 2); since this depression in titer was also observed with the replicating virus varec280 (data not shown), the phenomenon is presumably insert specific. The characteristics of the viruses used in this study are shown in Table 1.
FIG. 2.
Growth of defective and wt viruses. Infected cells were harvested at the indicated time points and titered on the complementing cell line RK-D4R-44.20 (RK44) for the defective viruses vD4-vA and vD4-prME and on wt cells (RK13) for the WR wt virus.
TABLE 1.
Gene inserts and genotypes of viruses
| Virus | Promoter foreign gene cassettea | Insertion site | R/NRb | UDG | TK |
|---|---|---|---|---|---|
| vD4-prME | selP-prM/E, elP1-lacZ, elP1-gpt | D4R locus | NR | − | + |
| varec280 | selP-prM/E, elP1-lacZ, elP1-gpt | tk locus | R | + | − |
| vD4-vA | No foreign gene | NR | − | + | |
| vD4-ZG | elP1-lacZ, elP1-gpt | D4R locus | NR | − | + |
| vD4tk-ZG | elP1-lacZ, elP1-gpt | tk locus | NR | − | − |
| vtk-Z | elP1-lacZ, elP1-gpt | tk locus | R | + | − |
selP, synthetic early/late promoter; elP1, synthetic early promoter.
R, replicating; NR, nonreplicating.
Early gene expression in D4-defective viruses under abortive conditions.
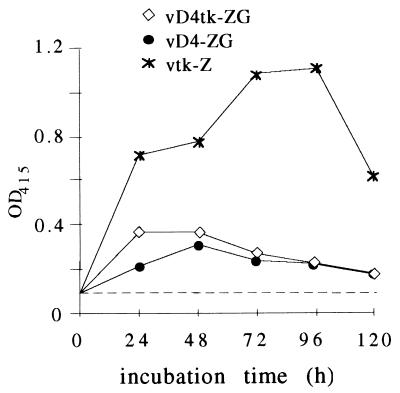
In the UDG-defective viruses, early proteins accumulate to time points exceeding 8 h postinfection (14). Since the duration and extent of expression of a nonreplicating live vector is presumably critical for its vaccine properties, early protein synthesis was examined under noncomplementing conditions. Expression of the E. coli β-galactosidase (β-Gal) gene (22) was used as a reporter system to evaluate the time course of abortive protein synthesis. Two defective viruses expressing β-Gal were used in the study: vD4-ZG, with the lacZ insert in the D4 locus (14); and a newly constructed virus, vD4tk-ZG, with the lacZ insertion in the tk locus of virus vD4-vA (Table 1; Materials and Methods). As shown in Fig. 3, early gene expression in cultures infected with 10 PFU per cell resulted in β-Gal levels that reached a maximum 24 to 48 h postinfection in nonpermissive cells and could still be detected after 120 h. Earlier experiments had shown that from abortively infected nonpermissive cells, no replicating defective virus could be rescued in complementing cells, confirming that the input virus is slowly degraded and finally disappears; the maximal β-Gal levels under nonpermissive conditions were 25 to 30% of the levels induced by the replicating reference virus vtk-Z. The different genomic integration loci of the β-Gal gene in vD4-ZG and vD4tk-ZG had little influence on the expression levels. The relatively high expression under nonpermissive conditions prompted us to use UDG-defective viruses as live vectors for the delivery of antigens.
FIG. 3.
β-Gal production by defective and wt viruses in noncomplementing RK13 cells. The cultures were infected with 10 PFU of the respective virus per cell, and β-Gal was measured at the indicated time points. The viruses contain the same promotor-lacZ gene construct. Average values of three independent experiments are shown. OD415, optical density at 415 nm.
Expression of the TBE virus prM/E protein.
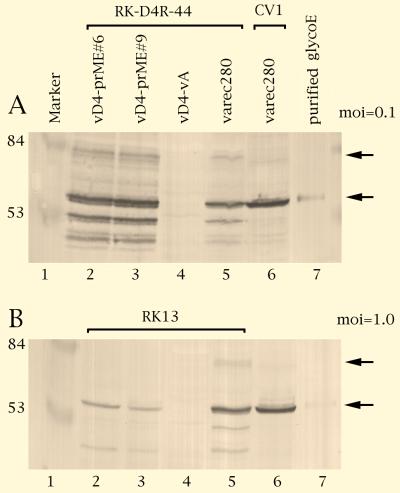
The next step was the construction of a defective and a replicating control virus expressing a vaccine antigen. The membrane-bound TBE virus prM/E protein was chosen as a model antigen because prM/E proteins are good immunogens in many flaviviruses (32). Moreover, the coding region of this protein is free of vaccinia virus early transcription stop signals (34), a prerequisite for efficient early expression (7). The precursor of the prM/E open reading frame is a fusion protein which is processed into the M and E proteins, both of which are sufficient in many flaviviruses to induce a protective immune response. To generate the D4-defective virus expressing the TBE virus proteins, plasmid pD4-prME was inserted into the defective virus vD4-vA, resulting in virus vD4-prME. The replicating reference virus was constructed by recombination of plasmid pTKselPtSpME into the tk locus of the vaccinia virus WR strain, resulting in the TK-negative virus varec280 (Table 1; Materials and Methods). Both viruses contain the same foreign gene cassette in which the TBE virus prM/E ORF is controlled by a strong early/late promoter (4). The viruses were used to infect complementing and wt cells, and expression of the foreign gene was examined. As shown by Western blot analysis, the defective virus vD4-prME induced in the complementing cell line, after an incubation period of 72 h, slightly higher amounts of the membrane-bound foreign protein than did the replicating varec280 virus in RK13 cells (Fig. 4A; compare lanes 2 and 3 with lanes 5 and 6). The prM/E fusion protein precursor (Fig. 4A, lanes 2, 3, 5, and 6; upper arrow) is proteolytically processed by cellular proteases into the mature glycoprotein E (lanes 2, 3, 5, and 6; lower arrow) and further cleavage products. As quantified by scanning of the Western blots (data not shown), in noncomplementing RK13 cells infected with an MOI of 1.0, the expression level of the defective virus reached about 10 to 20% of that of the reference level (Fig. 4B). Expression was not detectable in nonpermissive cells infected with 0.1 PFU/cell, suggesting that expression levels depend on infection dose.
FIG. 4.
Expression of TBE virus prM/E antigens. Cell cultures were infected with 0.1 (A) or 1.0 (B) PFU/cell, and total proteins were harvested 72 h postinfection. Lane 1, size marker (positions are indicated in kilodaltons); lanes 2 to 5, complementing cells (A; RK-D4R-44) or cells nonpermissive for defective virus (B; RK-13) infected with the defective virus clones vD4-prME#6 (lane 2) and vD4-prME#9 (lane 3), negative control defective virus vD4-vA (lane 4), and positive control wt virus varec280 (lane 5); lane 6, CV-1 cells infected with varec280; lane 7, purified TBE virus glycoprotein E. The upper arrow points to the prM/E fusion protein and the lower arrow points to glycoprotein E.
The defective virus induces protective immunity more efficiently than does the replicating control.
The relatively high levels of expression under noncomplementing conditions indicated the potential of the defective viruses as vaccine vectors. The potency test of a conventionally produced inactivated whole-virus TBE virus vaccine was used to evaluate the immunizing properties of the defective virus vaccine (see Materials and Methods). Groups of 10 mice were immunized with a single dose of 102 to 106 PFU by different routes (Fig. 5). Twenty-one days later, the mice were challenged with 100 LD50 of virulent TBE virus. The mice receiving the defective virus vD4-vA alone were not protected. The animals receiving the prM/E antigen-producing viruses, however, were protected against lethal challenge to different degrees depending on dose, route of administration, and type of vaccine virus. Surprisingly, the most efficient protection was achieved by intramuscular injection of the defective virus vD4-prME. A single dose of 104 PFU was sufficient to induce full protection (Fig. 5A). With the replicating virus, full protection was achieved with 106 PFU.
Upon subcutaneous administration of the vaccine virus, full protection was seen with 106 PFU of defective virus. With the replicating virus, full protection was not obtained in the range of titers tested (Fig. 5B). With the intraperitoneal route, full protection was achieved with the defective virus at 105 PFU/animal and with the replicating virus at 106 PFU/animal (data not shown).
Estimation of the PFU/particle ratio of replicating and nonreplicating virus stocks.
An interesting parameter to characterize a virus stock is the ratio of infectious units (PFU) to total viral particles, which is dependent on many factors, including virus strains, host cells, growth conditions, and handling and storage of virus stocks. The viruses used in this study were grown in rabbit kidney cells (varec280 in RK13 cells and vD4-prME in RK13-derived RK-D4R-44 cells) and purified by spinning infected cell lysates through one sucrose cushion (see Material and Methods). The defective viruses were titrated on the complementing cell line RK-D4R-44, whereas the replicating virus (varec280) was titrated on CV-1 cells, because this cell line is among the most sensitive for titrating replicating WR-derived vaccinia viruses. The varec280 and vD4-prME stocks had comparable titers, 1.0 × 1010 and 0.5 × 1010 PFU/ml, respectively. To compare the amounts of total virus in the stocks, 2.5 × 107 PFU of each virus was analyzed by Western blotting with rabbit anti-P11 antibodies (26) to detect the virion phosphoprotein P11. The blot confirmed that about equal amounts of P11 were present in both vD4-prME and varec280 stocks (data not shown). In addition, the genomic equivalents of the virus stocks were determined by a quantitative PCR assay. The virus samples and an internal standard were coprocessed by using fluorescence-labeled primers; the amplicons were separated on sequencing gels and quantified by laser-induced fluorescence (see Materials and Methods). In both vD4-prME and varec280 virus stocks, PFU/genomic equivalent ratios in the range of 1:20 to 1:50 were measured. Assuming that one genomic equivalent corresponds to one particle, the two virus preparations did not differ significantly in PFU/particle ratios.
Virus neutralization titers correlate with protection.
Since neutralizing antibodies play a central role in TBE virus immunity, sera collected at day 19 postvaccination were assayed for neutralizing antibodies (Materials and Methods; Table 2). A neutralizing titer of 1:10 is considered to be protective in humans. No protective neutralizing antibody titers were obtained after vaccination with the defective control virus vD4-vA; with virus vD4-prME, however, neutralizing activity was observed at vaccination doses (intramuscular route) as low as 103 PFU per animal (Table 2), correlating well with the protection experiment (Fig. 5A). Neutralizing antibodies were also obtained with the replicating virus varec280; for unknown reasons, the titers were not as consistent as those obtained with the nonreplicating virus.
TABLE 2.
TBE virus neutralization assay
| Virus | Dose (log PFU) | Neutralization titer |
|---|---|---|
| vD4-vA | 4 | <1:3 |
| 6 | 1:5 | |
| 7 | <1:3 | |
| vD4-prME | 2 | 1:4 |
| 3 | 1:10a | |
| 4 | 1:12 | |
| 5 | 1:14 | |
| 6 | 1:17 | |
| 7 | 1:20 | |
| varec280 | 2 | 1:3 |
| 3 | 1:4 | |
| 4 | 1:40 | |
| 5 | 1:10 | |
| 6 | 1:10 |
Minimal protective titer.
In vitro and in vivo reversion.
The reversion rate to wt virus and the persistence of the inoculated virus in the host are important safety aspects of a defective vaccine vector. Therefore, we performed a sensitive in vitro assay (see Materials and Methods) from which, when the results are negative, it can be concluded that a stock contains less than 1 replicating virus per 2 × 107 defective viruses. In this assay, our vD4-prME stock used for the vaccinations did not contain replicating virus.
Attempts to isolate replicating (revertant) virus were also performed in the vaccinated host. Mice immunosuppressed with cyclophosphamide were infected with the nonreplicating vD4-prME and the control virus varec280, and titration of muscle and spleen samples were performed (see Materials and Methods). With the maximal dose of defective virus used in the vaccination experiment (106 PFU), no replicating virus was detectable in muscle (site of inoculation) or spleen. With a dose of the defective virus 100-fold higher than that used for the vaccinations (108 PFU/animal), replicating virus was found in the muscle (site of inoculation; 102/muscle) in 1 of 15 mice but not in the spleen. Sequencing the D4R locus of this isolate revealed that this virus had a wt D4R gene, indicating that it was a rare contaminant (present in the virus stock below the detection limit of the in vitro reversion assay) rather than a revertant, confirming that reversion in the vaccinated host is an extremely rare event.
Clearance of the vaccine virus in the immunosuppressed mice.
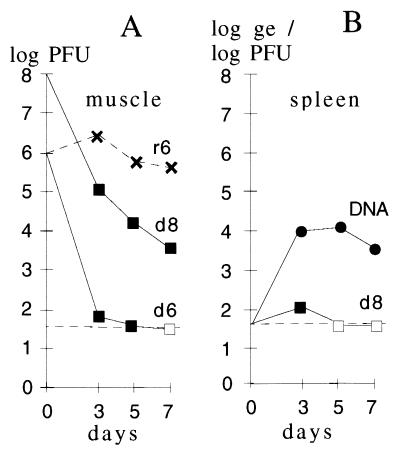
Titering the vD4-prME-inoculated organ samples in the complementing cell line allowed comparison of the clearance kinetics of the defective and the replicating viruses. From an inoculum of 106 PFU per animal, no defective virus could be isolated from muscle (site of inoculation) at day 7, while the varec280 titers remained high (Fig. 6A). In the high-dose experiment (input of 108 PFU/animal), residual 4 × 103 PFU of vD4-prME was found in muscle samples at day 7. Hence, in both experiments, the titers of the defective virus dropped by more than 4 logs within 1 week. In a similar experiment, the immunocompromised mice inoculated with the replicating virus died from day 8 on, but those receiving the defective virus survived (not shown).
FIG. 6.
Rapid clearance of the defective virus in vaccinated animals. (A) Replication of the vaccine preparations in cyclophosphamide-treated mice was assayed by injecting two different doses of the defective virus vD4-prME (106 or 108 PFU/animal [d6 or d8]) and one dose of replicating virus varec280 (106 PFU/animal [r6]). Virus was isolated from muscle preparations (site of inoculation) at the times indicated. Titration of the defective virus was done in the complementing cell line RK-D4R-44; the varec280 virus was titered in RK13 cells. Average titers obtained from three animals are shown. (B) Titer and genomic DNA load in the spleens of vaccinated animals. After injection of defective virus (108 PFU/animal), spleens were isolated at the indicated times and the virus titer was determined (d8). From the same preparations, the amount of vaccinia DNA, given in genomic equivalents (ge), was determined by quantitative PCR. Titers below the detection limit (dashed line) are indicated by open symbols.
Since almost no defective virus could be rescued from spleen samples, even at the high input dose of 108 PFU/animal, the same samples (see above) were screened by quantitative PCR for biologically inactive virus in spleens of the immunosuppressed mice that had received the intramuscular dose of 108 PFU of defective virus (see Materials and Methods). Substantial amounts of vaccinia virus DNA were found in the spleens, suggesting uptake and transport of the virus particles into the spleen by macrophages. Low amounts of defective virus titratable on complementing cells were detectable only on day 3 (Fig. 6B).
DISCUSSION
The concept of “nonreplicating” recombinant poxvirus vectors as vaccines was first described in a study using avipoxvirus recombinants to successfully vaccinate nonpermissive hosts (31). In avipoxvirus-infected monkey and human cell lines, early and late gene expression as well as replication, including viral morphogenesis with the production mainly of immature particles, was observed (28). Nonreplicating poxvirus vectors further include genetically engineered attenuated vaccinia virus strains (30) or classically attenuated viruses, such as the modified vaccinia virus Ankara (MVA) vaccine strain (1, 21, 29). As seen with nonreplicating avipoxviruses, in MVA, expression proceeds through the late phase and virus production is aborted in the vaccinated host.
Nonreplicating late defective vaccine vectors are novel and allow the contribution of early gene expression to the immune response to be evaluated. Late defective viruses, including vD4-prME and corresponding temperature-sensitive mutants at the elevated temperature, do not produce detectable amounts of late gene products and do not replicate their genomic DNA (14, 23). Upon infection of nonpermissive cells, early gene expression in cell culture is measurable over several days. As shown in this study, full protection after a single vaccination with vD4-prME was observed with viral doses as low as 104 PFU. At least a 10-fold higher dose was required with a corresponding replicating virus.
In this connection, the ratio of infectious virus to total viral particles in the virus preparations is of interest. A Western blot analysis of equal amounts of infectious units of the viruses resulted in bands of similar intensity; in addition, quantitative PCR analysis revealed similar PFU/particle ratios. Therefore, the replicating and the nonreplicating viruses do not differ significantly in PFU/particle ratios.
Surprisingly, a single dose of 103 to 104 PFU was sufficient to induce significant neutralizing antibodies. The antibody titers, however, were relatively low and did not increase much depending on the vaccination dose. This may be explained by the experimental design of the protection study. The mice were immunized by a single dose, the serum samples were taken 19 days later, and challenge was at day 21. It is conceivable that boosting with a second dose of virus or with a conventional TBE vaccine would result in much higher neutralizing antibody titers. Since replicating virus was not detectable in the vaccine preparations used for immunizations by in vitro and in vivo assays, early gene expression was sufficient to induce a strong protective immunity in the murine TBE virus challenge model. Therefore, defective viruses expressing exclusively early genes, though not spreading within the host, allow efficient presentation of antigens.
As shown previously, only early vaccinia virus gene expression efficiently induced a cellular immune response (5, 33). In addition, only viruses expressing a tumor-associated antigen under the control of an early promoter were effective candidate cancer vaccines, probably due to the interference of vaccinia virus with the major histocompatibility complex class I presentation late in infection; this effect could be correlated with poor vaccinia virus late promoter activity (abortive expression) in professional antigen-presenting cells, which are necessary for efficient T-cell stimulation (2). Consistent with the results obtained in our study, early gene expression is sufficient for an efficient cellular immune response. The unique difference of our study from previous ones (using early or late promoters to assess the relative contributions to the immune response) is that the immune response is determined in the absence of vaccinia virus late proteins, which also include immunomodulatory molecules. Early antigen expression and lack of immune modulation by late genes seems to be a favorable property of the defective vaccine vector system.
The development of more efficient vaccinia virus vectors is desirable because the dose of vaccinia virus vectors in humans required to elicit an immune response is higher than that required in mice. A critical safety parameter in this context is the rate of reversion of a defective virus to a replicating one, which should be as low as possible. Since vaccinia virus does not splice RNA, reversion of the defective vaccinia virus vector in the vaccinated host by uptake of a cellular UDG gene would require a retrotransposition event or an accidental superinfection with a virus carrying a suitable UDG gene followed by an illegitimate recombination event. A more trivial source of replicating virus is external contamination. In the first generation of defective viruses, vD4-vA and its derivatives, the central part, but not the entire D4R ORF, is deleted. Therefore, the genomes of the complementing cell line and the defective virus share homologous D4R sequences. Since vaccinia virus replicates in the cytoplasm and the potentially complementing UDG cDNA is located in the nucleus, a low reversion rate in the complementing cell of the defective virus was expected. In fact, revertants were observed occasionally; 50% of the viral stocks contain about 1 revertant per 106 defective viruses. The virus stock used in this study contained less than 1 replicating virus in 2 × 107 defective viruses, as judged by an in vitro assay; the more sensitive in vivo assay using immunosuppressed mice, however, revealed a low contamination with replicating virus (approximately 1 replicating in 108 defective viruses). Since the maximal dose injected into the mice was 106 PFU, the virus dilutions used for the vaccinations were practically free of replicating virus. To eliminate the only practical relevant source of reversion, reversion by homologous recombination, the next-generation UDG-defective vector, termed eVAC-1, does not share homologous sequences with the host cell (15). Nevertheless, in contrast to the replicating virus, the defective virus was rapidly cleared in immunosuppressed mice, confirming its enhanced safety.
An important aspect of this study was the excellent growth of the basic defective virus in the complementing cell line and its stability during purification and storage, allowing efficient production of potential vaccines based on this virus. Together with the excellent immunizing potential, UDG-defective viruses are promising nonreplicating vaccine vectors. Furthermore, our current knowledge regarding the structure and function of poxvirus immune evasion genes will serve as a basis for the improvement of future nonreplicating vaccine vectors.
ACKNOWLEDGMENTS
We thank V. Wieser, J. Mayrhofer, S. König, I. Albrecht, and E. Saloukeh for expert technical assistance, A. Mitterer for purified glycoprotein E, and C. P. Gibbs for critically reading the manuscript.
REFERENCES
- 1.Antoine G, Scheiflinger F, Dorner F, Falkner F G. Genomic sequence of the modified vaccinia Ankara (MVA) strain: comparison with other orthopoxviruses. Virology. 1998;244:365–396. doi: 10.1006/viro.1998.9123. [DOI] [PubMed] [Google Scholar]
- 2.Bronte V, Carroll M W, Goletz T J, Wang M, Overwijk W W, Marincola F, Rosenberg S A, Moss B, Restifo N P. Antigen expression by dendritic cells correlates with the therapeutic effectiveness of a model recombinant poxvirus tumor vaccine. Proc Natl Acad Sci USA. 1997;94:3183–3188. doi: 10.1073/pnas.94.7.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakrabarti S, Brechling K, Moss B. Vaccinia virus expression vector: coexpression of beta-galactosidase provides visual screening of recombinant virus plaques. Mol Cell Biol. 1985;5:3403–3409. doi: 10.1128/mcb.5.12.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakrabarti S, Sisler J R, Moss B. Compact, synthetic, vaccinia virus early/late promoter for protein expression. BioTechniques. 1997;23:1094–1097. doi: 10.2144/97236st07. [DOI] [PubMed] [Google Scholar]
- 5.Coupar B E, Andrew M E, Both G W, Boyle D B. Temporal regulation of influenza hemagglutinin expression in vaccinia virus recombinants and effects on the immune response. Eur J Immunol. 1986;16:1479–1487. doi: 10.1002/eji.1830161203. [DOI] [PubMed] [Google Scholar]
- 6.Dmitriev I P, Khromykh A A, Ignatyev G M, Gainullina M N, Ageenko V A, Dryga S A, Vorobyeva M S, Sandakhchiev L S. Immunization with recombinant vaccinia viruses expressing structural and part of the nonstructural region of tick-borne encephalitis virus cDNA protect mice against lethal encephalitis. J Biotechnol. 1996;44:97–103. doi: 10.1016/0168-1656(95)00141-7. [DOI] [PubMed] [Google Scholar]
- 7.Earl P L, Hugin A W, Moss B. Removal of cryptic poxvirus transcription termination signals from the human immunodeficiency virus type 1 envelope gene enhances expression and immunogenicity of a recombinant vaccinia virus. J Virol. 1990;64:2448–2451. doi: 10.1128/jvi.64.5.2448-2451.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falkner F G, Moss B. Escherichia coli gpt gene provides dominant selection for vaccinia virus open reading frame expression vectors. J Virol. 1988;62:1849–1854. doi: 10.1128/jvi.62.6.1849-1854.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falkner F G, Moss B. Transient dominant selection of recombinant vaccinia viruses. J Virol. 1990;64:3108–3111. doi: 10.1128/jvi.64.6.3108-3111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. , 517–563. [DOI] [PubMed] [Google Scholar]
- 11.Hämmerle T, Himmelspach M, Dorner F, Falkner F G. A sensitive PCR assay system for the quantitation of viral genome equivalents: human immunodeficiency virus type 1 (HIV-1) and hepatitis virus (HBV) Arch Virol. 1997;142:1297–1306. doi: 10.1007/s007050050161. [DOI] [PubMed] [Google Scholar]
- 12.Heinz F X, Mandl C W. The molecular biology of tick-borne encephalitis virus. APMIS. 1993;101:735–745. doi: 10.1111/j.1699-0463.1993.tb00174.x. . (Review.) [DOI] [PubMed] [Google Scholar]
- 13.Holzer G. Ph.D. thesis. Vienna, Austria: Universität für Bodenkultur; 1997. [Google Scholar]
- 14.Holzer G W, Falkner F G. Construction of a vaccinia virus deficient in the essential DNA repair enzyme uracil DNA glycosylase by a complementing cell line. J Virol. 1997;71:4997–5002. doi: 10.1128/jvi.71.7.4997-5002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holzer G W, Gritschenberger W, Mayrhofer J A, Wieser V, Dorner F, Falkner F G. Dominant host range selection of vaccinia recombinants by rescue of an essential gene. Virology. 1998;249:160–166. doi: 10.1006/viro.1998.9307. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs S C, Stephenson J R, Wilkinson G W G. Protection elicited by a replication-defective adenovirus vector expressing the tick-borne encephalitis virus non-structural glycoprotein NS1. J Gen Virol. 1994;75:2399–2402. doi: 10.1099/0022-1317-75-9-2399. [DOI] [PubMed] [Google Scholar]
- 17.Joklik W K. The purification of four strains of poxvirus. Virology. 1962;18:9–18. doi: 10.1016/0042-6822(62)90172-1. [DOI] [PubMed] [Google Scholar]
- 18.Kaerber G. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Arch Exp Pathol Pharmakol. 1931;162:480–483. [Google Scholar]
- 19.Mackett M, Smith G L, Moss B. Vaccinia virus: a selectable eukaryotic cloning and expression vector. Proc Natl Acad Sci USA. 1982;79:7415–7419. doi: 10.1073/pnas.79.23.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandl C W, Heinz F X, Kunz C. Sequence of the structural proteins of tick-borne encephalitis virus (Western subtype) and comparative analysis with other flaviviruses. Virology. 1988;166:197–205. doi: 10.1016/0042-6822(88)90161-4. [DOI] [PubMed] [Google Scholar]
- 21.Mayr A, Stickl H, Müller H K, Danner K, Singer H. The smallpox vaccination strain MVA: marker, genetic structure, experience gained with the parenteral vaccination and behavior in organisms with a debilitated defence mechanism. Zentbl Bakteriol Hyg Abt 1 Orig Reihe B. 1978;167:375–390. [PubMed] [Google Scholar]
- 22.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 23.Millns A K, Carpenter M S, DeLange A M. The vaccinia virus-encoded uracil DNA glycosylase has an essential role in viral DNA replication. Virology. 1994;198:504–513. doi: 10.1006/viro.1994.1061. [DOI] [PubMed] [Google Scholar]
- 24.Moss B. Genetically engineered poxviruses for recombinant gene expression, vaccination, and safety. Proc Natl Acad Sci USA. 1996;93:11341–11348. doi: 10.1073/pnas.93.21.11341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paoletti E. Applications of pox virus vectors to vaccination: an update. Proc Natl Acad Sci USA. 1996;93:11349–11353. doi: 10.1073/pnas.93.21.11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfleiderer M, Falkner F G, Dorner F. Requirements for optimal expression of secreted and non-secreted recombinant proteins in vaccinia virus systems. Protein Expression Purif. 1995;6:559–569. doi: 10.1006/prep.1995.1074. [DOI] [PubMed] [Google Scholar]
- 27.Scheiflinger F, Dorner F, Falkner F G. Transient marker stabilisation: a general procedure to construct marker-free recombinant vaccinia virus. Arch Virol. 1998;143:467–474. doi: 10.1007/s007050050303. [DOI] [PubMed] [Google Scholar]
- 28.Somogyi P, Frazier J, Skinner M A. Fowlpox virus host range restriction: gene expression, DNA replication, and morphogenesis in nonpermissive mammalian cells. Virology. 1993;197:439–444. doi: 10.1006/viro.1993.1608. [DOI] [PubMed] [Google Scholar]
- 29.Sutter G, Moss B. Nonreplicating vaccinia vector efficiently expresses recombinant genes. Proc Natl Acad Sci USA. 1992;89:10847–10851. doi: 10.1073/pnas.89.22.10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tartaglia J, Perkus M E, Taylor J, Norton E K, Audonnet J C, Cox W I, Davis S W, van der Hoeven J, Meignier B, Riviere M, Languet B, Paoletti E. NYVAC: a highly attenuated strain of vaccinia virus. Virology. 1992;188:217–232. doi: 10.1016/0042-6822(92)90752-b. [DOI] [PubMed] [Google Scholar]
- 31.Taylor J, Paoletti E. Fowlpox virus as a vector in non-avian species. Vaccine. 1988;6:466–468. doi: 10.1016/0264-410x(88)90091-6. [DOI] [PubMed] [Google Scholar]
- 32.Venugopal K, Gould E A. Towards a new generation of flavivirus vaccines. Vaccine. 1994;12:966–975. doi: 10.1016/0264-410x(94)90329-8. [DOI] [PubMed] [Google Scholar]
- 33.Wachsman M, Aurelian L, Hunter J C, Perkus M E, Paoletti E. Expression of herpes simplex virus glycoprotein D on antigen presenting cells infected with vaccinia recombinants and protective immunity. Biosci Rep. 1988;8:323–334. doi: 10.1007/BF01115223. [DOI] [PubMed] [Google Scholar]
- 34.Yuen L, Moss B. Oligonucleotide sequence signaling transcriptional termination of vaccinia virus early genes. Proc Natl Acad Sci USA. 1987;84:6417–6421. doi: 10.1073/pnas.84.18.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]