Abstract
The ZEBRA protein mediates the switch between the latent and lytic life cycles of Epstein-Barr virus. Z(S186A), a point mutant in ZEBRA’s basic domain in which serine 186 is changed to alanine, is unable to induce expression of lytic cycle mRNAs or proteins from the latent EBV genome even though it retains the ability to activate transcription from reporters bearing known ZEBRA-responsive promoters (A. L. Francis et al., J. Virol. 71:3054–3061, 1997). We now describe three distinct phenotypes of ZEBRA mutants bearing different amino acid substitutions at S186. These phenotypes are based on the capacity of the mutants to activate expression of the BRLF1 and BMRF1 genes, which are targets of ZEBRA’s action, and to synergize with the BRLF1 gene product Rta (R transactivator) in activating expression of downstream genes. One mutant class, represented by Z(S186T), was similar to the wild type, although reduced in the capacity to activate BRLF1 and BMRF1 early lytic cycle genes from the latent virus. A second class, represented by Z(S186C) and Z(S186G), was impaired in transcriptional activation, unable to activate early lytic cycle products from the latent virus, and not rescued by overexpression of Rta. A third class, Z(S186A), although unable by itself to activate BRLF1 or other lytic cycle genes, synergized with Rta. Rta rescued the capacity of Z(S186A) to activate the BMRF1 early lytic cycle gene from the latent virus. All mutant classes bound to DNA in vitro, although their capacity to bind to different ZEBRA response elements varied. Serine 186 of ZEBRA is a critical residue that is required for the distinct activities of induction of BRLF1 expression and for synergy with Rta. Since only Z(S186T) among the mutants behaved similarly to the wild type, activation of BRLF1 likely requires phosphorylation of S186. However, since Z(S186A) could synergize with Rta, synergy with Rta does not appear to be dependent on phosphorylation of S186. S186 likely mediates DNA recognition on the BRLF1 promoter in the context of the latent virus, protein-protein interactions, or both. The Z(S186) mutants define the amino acid side chains required for these functions.
The human gammaherpesvirus Epstein-Barr virus (EBV) establishes a persistent infection in B lymphocytes and remains primarily in a latent state for the lifetime of the host. The basic mechanisms underlying latency of EBV and the switch into viral lytic cycle gene expression can be analyzed in cultured B lymphocytes. Two viral genes, BZLF1, encoding the ZEBRA protein, and BRLF1, encoding the R transactivator (Rta), play critical roles in this switch (10, 11, 22–24). Both genes are activated simultaneously soon after treatment of cultured B cells with agents that induce the lytic cycle (14, 18, 32, 41). Transfer of ZEBRA-expressing plasmids into B-cell lines latently infected with EBV activates BRLF1 and ultimately induces the entire lytic cycle cascade (26, 28, 39). Transfer of Rta expression plasmids also activates EBV lytic gene expression in epithelial cells and in B cells (38, 45). In some permissive B-cell backgrounds, Rta activates ZEBRA expression; lytic viral DNA replication and late gene expression ensue. Thus, ZEBRA and Rta may reciprocally stimulate each other’s expression. The two proteins synergistically activate promoters of several downstream target genes, including BHRF1, encoding the EBV Bcl2 homologue, and BMRF1, encoding a DNA polymerase processivity factor (8, 12, 20, 24, 25, 37).
The ZEBRA protein, a member of the basic-zipper (bZIP) family of transcriptional activators, shows extensive amino acid identity with cellular bZIP proteins such as c-Fos/c-Jun in the basic DNA recognition domain (16, 34). ZEBRA homodimers and c-Fos/c-Jun heterodimers each bind specifically to DNA containing AP-1 heptamer sites (TGAGTCA); however, ZEBRA dimers recognize an array of ZEBRA response elements (ZREs) that are poorly bound by c-Fos/c-Jun (6, 16, 17, 29, 30, 34, 42). Moreover, substitution of ZEBRA’s basic domain with that of c-Fos leads to formation of a chimeric protein that is unable to stimulate EBV lytic cycle gene expression (26). While exploring differences in DNA binding and biologic activity between ZEBRA and c-Fos, we changed serine 186 in the ZEBRA DNA recognition domain to alanine, the amino acid found at the corresponding position of c-Fos/c-Jun. In the crystal structure of c-Fos/c-Jun bound to an AP-1 site, the alanine at this position was shown to make specific contacts with DNA (21). The resultant mutant, Z(S186A), exhibited a remarkable phenotype. While retaining the ability to bind to ZREs and the capacity to activate transcription from chloramphenicol acetyltransferase (CAT) reporters bearing the promoter of a known ZEBRA-responsive gene, BMRF1, Z(S186A) was unable to stimulate expression of three lytic cycle mRNAs, i.e., BRLF1, BMRF1, and BaRF1, from the endogenous virus. The Z(S186A) mutation thus affected a crucial biologic function of ZEBRA, namely, its capacity to disrupt latency by activating lytic gene expression from a latent virus (19).
Several hypotheses were offered to account for this phenotype. The most obvious explanation was that the Z(S186A) mutation altered the DNA binding properties of ZEBRA. Gel mobility shift experiments showed that in vitro binding of Z(S186A) to some DNA sites but not others was altered compared to wild-type ZEBRA. For example, Z(S186A) bound to several ZREs, such as ZIIIB (TTAGCAA) and ZRE-2 (TGAGCAA), approximately as well as wild-type ZEBRA, bound to ZRE-R (TGAGCGA) and ZIIIA (TGAGCCA) less well than ZEBRA, and bound to an octamer AP-1/CREB site (TGACATCA) more efficiently than wild-type ZEBRA. Since the Z(S186A) mutation destroyed a consensus protein kinase C (PKC) phosphorylation site, another hypothesis to account for the phenotype of Z(S186A) was that this mutation eliminated a phosphorylation site that was essential for activation of lytic cycle transcription from the intact latent viral genome. Perhaps this phosphorylation site mediated protein-protein interactions that were essential for activating lytic gene expression from the viral genome but were dispensable for activation of reporter plasmids in transient transfection assays.
The present report provides further delineation of the functional role of amino acid 186 of ZEBRA. We describe the phenotypes of additional Z(S186) mutants that have a threonine (T), glycine (G), cysteine (C), valine (V), aspartate (D), or glutamate (E) at this position. We analyzed the phenotype of this expanded group of mutants by using assays of DNA binding, activation of ZEBRA target promoters fused to reporters in transient transfections, and the capacity to activate lytic cycle mRNAs and proteins. Since mutations in ZEBRA’s DNA binding domain are known to affect synergy with Rta (20), assessment of the phenotypes of these Z(S186) mutants also included studies of transcriptional synergy with Rta on CAT reporters and on genes expressed from the intact viral genome. The diverse phenotype of the mutants emphasize that S186 of ZEBRA is a crucial residue required for transcriptional activation of BRLF1 and for synergy with Rta, the BRLF1 gene product. While mutants such as Z(S186T) preserved both functions, and mutants such as Z(S186G) and Z(S186C) lost both functions, the Z(S186A) mutant demonstrated that there are distinct requirements for activation of BRLF1, the most proximal target of ZEBRA action, and for synergy with Rta on downstream lytic cycle genes such as BMRF1.
MATERIALS AND METHODS
Bacterial expression systems.
Wild-type BZLF1 cDNA and mutant BZLF1 constructs containing substitutions at S186 were cloned into the NcoI and BamHI sites of pET-11d (Stratagene). The mutants were made by PCR, using Taq DNA polymerase and BZLF1 cDNA as a template. The mutations were introduced by two PCRs that produced overlapping DNA fragments. The 5′ fragment was made with one primer, containing an NcoI site, complementary to the 5′ end of BZLF1 cDNA and another primer complementary to BZLF1 cDNA encoding amino acids (aa) 188 to 181 but with a mutation at aa 186. The 3′ fragment was made with one primer, containing a BamHI site, complementary to the 3′ end of BZLF1 cDNA and another primer complementary to aa 184 to 190 with a mutation at aa 186. The two PCR fragments were purified from agarose and used as a template in a second PCR with the primers containing NcoI and BamHI sites. The final PCR product was cloned into pET-11d and transformed into Escherichia coli AG1. Minilysate DNA from individual colonies was screened for the mutation by sequencing across the region encoding aa 186.
E. coli BL21(DE3), featuring inducible T7 RNA polymerase, was transformed with constructs expressing wild-type or mutant ZEBRA. Ampicillin-resistant cultures were grown to an optical density of 0.7 and induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 4 h. Induced cultures were harvested at 4°C, and pellets were resuspended in 1/5 volume of 6 M urea and sonicated on ice for eight rounds of 15 s each. Urea was removed by dialysis against DNA binding buffer, and the bacterial extracts, calibrated by immunoblotting to contain approximately equal amounts of ZEBRA protein, were used in DNA binding assays.
DNA binding assays.
Oligonucleotides containing ZREs and identical top-strand and bottom-strand flanking sequences were used as probes as described elsewhere (19). In other experiments (kindly suggested by S. Kenney), probes contained flanking sequences found in the promoter of the BRLF1 gene. These probes were Rp ZIIIA (5′ GATCTGCCAATGGCTCATAAAAGA) and Rp ZRE-R (5′ GATCAAGCTTATGAGCGATTTTAT) (the ZREs are underlined). A 100-ng aliquot of each top- and bottom-strand oligonucleotide was annealed in TM buffer (0.1 M Tris [pH 7.5], 0.1 M MgCl2, 0.1 M dithiothreitol) at 95°C for 5 min, 65°C for 10 min, 37°C for 10 min, and room temperature for 30 min. The annealed oligonucleotides were labeled by one of two different methods. They were end labeled with 50 μCi of [γ-32P]ATP, using 10 U of T4 polynucleotide kinase (Boehringer Mannheim) for 30 min at 37°C. Alternatively, they were synthetically labeled with 50 μCi of [α-32P]dCTP and 1 mM deoxynucleoside triphosphates, using 2 U of Klenow enzyme (Boehringer Mannheim) for 15 min at 25°C. Unincorporated radioactive nucleotides were removed on a P4 resin column (Bio-Rad). Probes were stored at −20°C until used.
Binding reactions of 10 μl contained protein extracts that were diluted in DNA binding buffer (15 mM HEPES [pH 7.5], 75 mM KCl, 0.2 mM EDTA, 1 mM dithiothreitol), 1 μg of poly(dI-dC), and 3 × 104 cpm of radiolabeled duplex oligonucleotide. The DNA binding mixtures were incubated for 10 min at 25°C and electrophoresed in a 6% acrylamide gel in 0.5× TBE (45 mM Tris, 45 mM boric acid, 1.5 mM EDTA) at 200 V. The gel was dried at 80°C for 2 h and exposed to film overnight. The proportion of bound and free probe was determined by phosphorimagery.
Eukaryotic expression systems.
The expression construct for ZEBRA contained EBV genomic DNA from nucleotides (nt) 102115 to 103181 (a NaeI-to-NcoI subfragment of BamHI-Z) cloned into pHD1013 (15), which utilizes the cytomegalovirus (CMV) immediate-early promoter for constitutive expression of genes in mammalian cells. The preparation of Z(S186A) and Z(S186T) has been described elsewhere (19). The additional ZEBRA constructs containing point mutations at position S186 were constructed by a two-step PCR process identical to that described for expression constructs in E. coli except that EBV genomic DNA was the template and the mutants were constructed as XbaI and BamHI fragments for cloning into pHD1013. The sequence of primers used for additional mutagenesis at this position to create mutants with G, C, or V substitutions are available upon request. All mutants were sequenced in their entirety by automated DNA sequencing. The expression construct for EBV Rta, pRTS15, a kind gift from Diane Hayward, contains the BRLF1 gene linked to the simian virus 40 early promoter and is followed by a polyadenylation signal in the plasmid pRTS2. pRTS Δ HindIII (pRTS) was used as a vector control (38).
Cell culture and transfections.
The ability of ZEBRA and Z(S186) mutants to activate transcription and to synergize with Rta in transient transfection assays using promoter/CAT fusions was determined in the EBV-negative B-cell line BJAB (33). The capacity of wild-type and mutant ZEBRA to activate lytic cycle gene expression from the endogenous latent EBV genome was measured in the EBV-positive Burkitt lymphoma cell line Raji. Cell lines maintained in RPMI 1640 medium containing 8% fetal bovine serum were subcultured to 2 × 105 cells per ml and transfected 48 to 72 h later when the cell counts were approximately 0.7 × 106 to 1.0 × 106/ml. Appropriate amounts of plasmid DNA were mixed with 107 cells in 500 μl of RPMI 1640 plus 5% fetal bovine serum. Cells were electroporated with 0.25 V in a Bio-Rad gene pulse unit. The electroporated cells were subcultured at 106 cells per ml, incubated at 37°C in 5% CO2–air, and harvested after 72 h for CAT assay, or after 24 to 48 h for RNA or immunoreactive protein assays.
Reporters and CAT assays.
Three reporter constructs fused promoters of EBV genes to CAT. These were BMRF1p/CAT, containing EBV nt 79537 to 79871 (41), BRLF1p/CAT (Rp/CAT) containing EBV nt 106123 to 107143 (39), and divergent promoter CAT (Dp/CAT) controlling BHRF1 and BHLF1 (20, 23), containing EBV nt 52568 to 54361. All promoters were cloned into pCAT Basic (Promega). Cells transfected 72 h previously with 10 μg of reporter and 5 to 10 μg of activator DNA were washed in phosphate-buffered saline and resuspended at 8 × 104/μl in reporter lysis buffer (Promega). A 150-μl enzyme reaction mixture, containing 59 μl of cell extract supernatant, 66 μg of acetyl coenzyme A, and 1 μl of [14C]chloramphenicol in 0.5 M Tris (pH 7.8), was incubated for 1 h at 37°C. The reaction was extracted into ethyl acetate and resuspended in 30 μl of ethyl acetate. Each spot on thin-layer chromatography paper (Baker-Flex) contained the equivalent of 4.7 × 106 cells. The thin-layer chromatography plate was developed in 95% chloroform–5% methanol for 1 h. The proportion of [14C]chloramphenicol that was acetylated was determined by liquid scintillation counting. If the reactivity was off scale, the assay was repeated with smaller amounts of cell extract. Each value represents two or more transfections. Fold activation was calculated as percent acetylation of chloramphenicol in the presence of the activator/percent acetylation in the presence of the vector alone.
RNA preparation and Northern blotting.
Total RNA was prepared from 4 × 106 cells 24 to 48 h after transfection, using RNeasy and Qiashredder spin columns (Qiagen) according to the manufacturer’s protocol. Each lane of a gel was loaded with the RNA from 2 × 106 cells. Northern blots were prepared as described previously (26). Blots were probed with a 531-nt restriction fragment of EBV DNA (EBV genomic sequence 80141 to 80672) that detects transcripts from the BMRF1 and BaRF1 genes and with a 720-nt restriction fragment of EBV DNA (EBV genomic sequence 104577 to 105297) that detects transcripts of the BRLF1 gene. To control for RNA loading, the blots were probed with a 370-nt NcoI-to-PstI restriction fragment from the cDNA for the H1 component of human RNase P (3). Probes were labeled by the random-prime method with [α-32P]dCTP and Klenow enzyme (Boehringer Mannheim). The abundance of mRNA was measured by phosphorimagery and standardized to the level of RNase P.
SDS-polyacrylamide gel electrophoresis and immunoblotting.
Protein extracts were prepared from transfected cells that had grown in culture for 24 to 48 h. Cells were harvested by centrifugation, resuspended in sodium dodecyl sulfate (SDS) sample buffer, and sonicated for 15 s. Samples containing 3 × 106 cells were boiled and electrophoresed through a 10% polyacrylamide–SDS gel before proteins were transferred to nitrocellulose. The immunoblots were reacted with a 1:200 dilution of a rabbit antiserum to exon I of ZEBRA (44) or a 1:150 dilution of a rabbit antiserum to the N-terminal 320 aa of Rta (38) in skim milk. To detect the BMRF1 product, also known as diffuse early antigen (EA-D), a 1:1,000 dilution of the R3.1 mouse monoclonal antibody (36) was used, followed by a 1:400 dilution of a rabbit anti-mouse immunoglobulin bridge (Axell; Accurate Chemical and Scientific Corp.). Protein loading was controlled by probing the immunoblot with a 1:100 dilution of affinity-purified antibody to β-actin (Sigma A2066). Immunoreactive bands were detected with 125I-labeled protein A and autoradiographed.
RESULTS
Only wild-type ZEBRA and the Z(S186T) mutant activate the BRLF1 and BMRF1 lytic cycle genes.
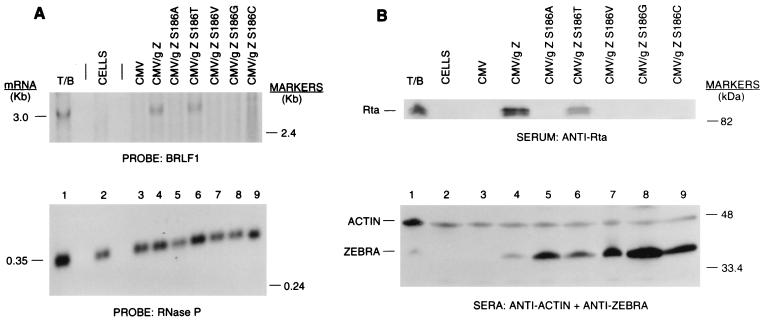
When transfected into EBV-positive Raji cells, wild-type ZEBRA activates expression of the viral BRLF1 gene as well as downstream target genes such as BMRF1, which is known to be a synergistic target of ZEBRA and Rta (26, 28, 37). Of the seven point mutants examined, only Z(S186T) activated expression of the BRLF1 product, Rta, and the BMRF1 product, EA-D (Fig. 1 and 5 and data not shown). In the studies shown in Fig. 1, the mutants were overexpressed, from 2- to 10-fold relative to wild-type ZEBRA, to determine whether this would compensate for their inability to activate Rta. Nonetheless, only wild-type ZEBRA and Z(S186T) induced the appearance of BRLF1 mRNA (Fig. 1A). In the experiment illustrated in Fig. 1B, the amount of Rta induced by transfected wild-type or mutant ZEBRA was measured and corrected for the amount of ZEBRA protein that was expressed and for the level of β-actin present in the cellular extracts. Z(S186T) induced expression of Rta and EA-D less efficiently than wild-type ZEBRA. Wild-type ZEBRA induced a 60-fold increase in Rta protein relative to Z(S186A), which was at background levels, and Z(S186T) induced a 15-fold increase of Rta relative to Z(S186A) (Fig. 1B). The other mutants did not stimulate Rta expression.
FIG. 1.
Capacity of Z(S186) mutants to activate the EBV BRLF1 gene. Raji cells were chemically induced (lane 1), untreated (lane 2), or transfected with various amounts of expression vector (from 1.2 to 14.3 μg) plus pHD1013 to a total of 15 μg of plasmid DNA (lanes 3 to 9) to ensure that the expression level of each of the mutants equaled or exceeded the wild-type ZEBRA expression level. RNA and protein extracts were prepared 24 h after transfection. Each lane of a Northern blot (A) contained RNA from 2 × 106 cells. The Northern blot was probed with BRLF1 and the H1 component of RNase P to control for RNA loading. Each lane of an immunoblot (B) contained an extract of 3 × 106 cells. The immunoblot was probed with monospecific antibodies to Rta, to ZEBRA, and to β-actin to control for protein loading. T/B, tetradecanoylphorbol acetate/n-butyrate.
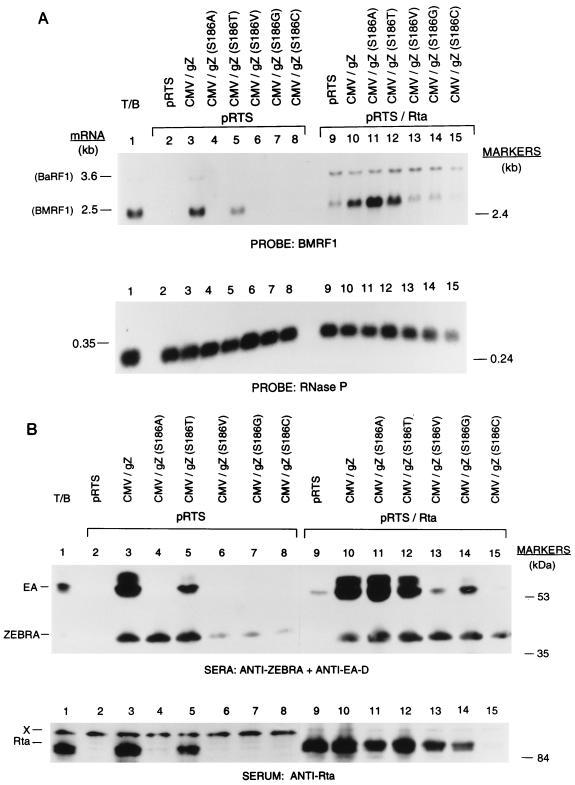
FIG. 5.
Capacity of Rta to rescue BMRF1 expression following cotransfection with various Z(S186) mutants. Raji cells were chemically treated (lane 1) or transfected with 10 μg of pRTS, the expression vector for Rta (lanes 2 and 9), or with wild-type or Z(S186) mutant expression plasmids (lanes 3 to 8 and 10 to 15). In lanes 2 to 10, the cells received an additional 10 μg of empty vector pRTS; in lanes 9 to 15, the cells received pRTS/Rta. For the Northern blot shown in panel A, cells were harvested 46 h after transfection; for the immunoblot in panel B, cells were harvested 48 h after transfection. T/B, tetradecanoylphorbol acetate/n-butyrate.
Comparative DNA binding affinities of ZEBRA and the Z(S186A) mutant on various ZREs.
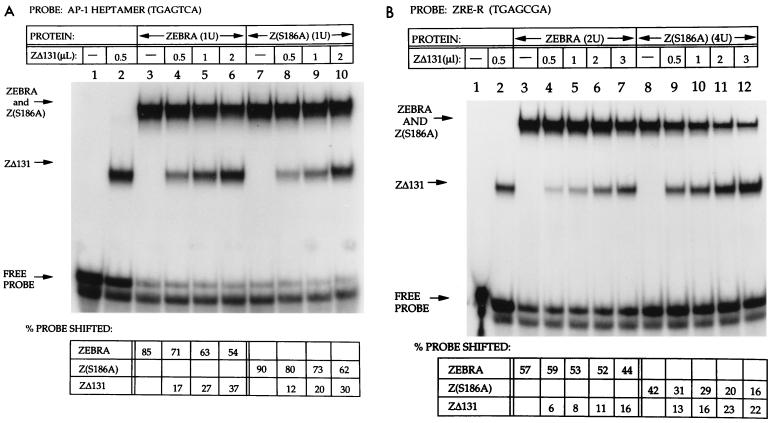
One explanation for the inability of ZEBRA mutants to activate BRLF1 or BMRF1 expression could reside in a diminished capacity to bind ZRE DNA in the promoters of these genes. In previous work we showed, by means of a competition electrophoretic mobility shift assay (EMSA), that wild-type ZEBRA and Z(S186A) bound with similar affinities to ZRE2, one of the ZREs present in the BMRF1 promoter (19). This method involved subjecting constant amounts of ZEBRA and Z(S186A) binding complexes to competition with increasing amounts of the same competitor DNA binding protein, ZΔ131, a truncation mutant lacking the N-terminal 131 aa of ZEBRA but containing a wild-type DNA binding domain. We extended these competition assays to four additional ZEBRA binding sites, including an AP-1 heptamer site found in the BMRF1 promoter, as well as ZRE-R and ZIIIA, the two sites found in the BRLF1 promoter (Fig. 2 and data not shown). When the AP-1 heptamer site was the target, ZΔ131 competed more effectively with ZEBRA than with the Z(S186A) mutant (Fig. 2A). The affinity of wild-type ZEBRA for an AP-1 site was approximately 70% that of the Z(S186A) mutant. However, when a probe containing a ZRE-R site was used in a similar assay, the mutant bound considerably less well than the wild type. On oligonucleotide probes containing AP-1 heptamer and AP-1 octamer sites, the Z(S186A) mutant bound with higher affinity than the wild type. On probes containing ZRE-2 and ZIIIB sites, the wild-type and mutant bound with comparable affinities. On probes containing ZIIIA and ZRE-R, the two sites present in Rp, wild-type ZEBRA bound with higher affinity, 2.4- and 8.5-fold, respectively, than the S186A mutant.
FIG. 2.
Comparison of DNA binding by ZEBRA and Z(S186A) by competition EMSA. The oligonucleotide probe contained an AP-1 heptamer site (A) or the ZRE-R site (B). In panel A, 1 U of ZEBRA and Z(S186A) was determined to be that amount of bacterial extract that contained an approximately equivalent amount of immunoreactive ZEBRA or mutant protein; in panel B, the volume of cell extract required to achieve equivalent immunoreactivity is indicated. The binding of ZEBRA and Z(S186A) was assessed alone or in the presence of increasing amounts of competitor ZΔ131 extract. The proportion of probe that was bound by each protein was quantitated by phosphorimage analysis and is indicated below the gel.
Comparing DNA binding by ZEBRA and an additional group of Z(S186) mutants.
Using EMSA, we compared ZEBRA, Z(S186A), and three additional Z(S186) mutants that were unable to activate BRLF1 or BMRF1 (Fig. 3). Extracts of E. coli in which the mutants were expressed were standardized for the amount of immunoreactive ZEBRA protein (Fig. 3A). The probes, consisting of a panel of radiolabeled oligonucleotides containing ZREs and AP-1-like sequences, were labeled either with polynucleotide kinase or by filling in with Klenow fragment of DNA polymerase, with similar results. To estimate the relative DNA binding ability of the mutants, we tested equal amounts of all mutants at the same time on the same probe and measured the fraction of probe shifted by each mutant by phosphorimagery. No proteins contained in extracts of E. coli BL21(DE3) with or without the expression vector, pET-11d, shifted the mobility of any probe.
FIG. 3.
Comparison of DNA binding activities by ZEBRA and a group of Z(S186) mutant proteins. (A) Bacterial lysates containing ZEBRA or Z(S186) mutant proteins were examined by immunoblotting with rabbit antiserum to ZEBRA. The level of expressed protein was quantitated by phosphorimagery and adjusted so that all DNA binding reactions contained approximately equal amounts of immunoreactive protein. (B) EMSAs of ZEBRA and Z(S186) mutant proteins with duplex oligonucleotide probes harboring one of several known ZEBRA binding sites labeled with polynucleotide kinase. Lane 6 contained extract of bacterial cells transformed with the vector pET-11d; lane 7 contained extract of E. coli BL21(DE3) cells alone. Only the portion of the gel containing probe that was shifted by the protein is shown.
Consistent differences were observed in the capacity of Z(S186) mutants to interact with different sites (Fig. 3B). All mutants were impaired relative to the wild type in the capacity to bind the two sites in the BRLF1 promoter, ZRE-R (TGAGCGA) and ZIIIA (TGAGCCA). The Z(S186V) mutant was particularly deficient in binding to these two sites. Nonetheless, the Z(S186V) mutant bound well to several other sites, including ZIIIB and AP-1 heptamer. All of the mutants bound well to ZIIIB, AP-1, and ZRE-2. As previously reported, the Z(S186A) mutant bound to the octamer site markedly better than the wild type. The Z(S186C) mutant also bound to this site as efficiently as the wild type, but the other mutants bound to the octamer site poorly. In summary, although all of the mutants containing substitutions at S186 retained the capacity to bind DNA, the various substitutions differentially affected their ability to interact with different sites.
When the EMSAs were repeated with probes that contained the ZREs in Rp (ZRE-R and ZIIIA) in the context of naturally occurring flanking sequences, all mutants were reduced in binding relative to wild-type ZEBRA. The G and V mutants were most reduced in the capacity to bind DNA, and the Z(S186A) mutant was affected least (data not shown).
Ability of ZEBRA and Z(S186) mutants to activate transcription of reporter constructs containing viral early lytic cycle promoters.
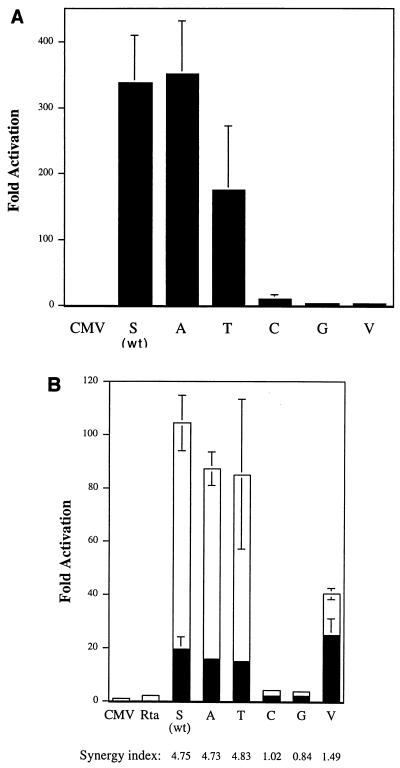
We found that the Z(S186T) mutant, which was reduced in its ability to disrupt latency (Fig. 1 and 5), was also about twofold reduced in activity relative to the wild type in activation of BMRF1p/CAT in transient assays in EBV-negative BJAB cells (Fig. 4A). The activities of the C, G, and V substitution mutants, which were unable to disrupt latency, were only 1 to 3% of those of wild-type and Z(S186A), examined in parallel. Since Rp, the promoter of the BRLF1 gene, is considered one of the most proximal targets of ZEBRA, the panel of Z(S186) substitution mutants was studied for the capacity to activate a CAT reporter bearing Rp (Fig. 4B). The results were generally similar to those for assays using the BMRF1 promoter; the S186A and S186T mutants were slightly less active than the wild type, and the S186C and S186G mutants were markedly reduced in activity. There was, however, one notable exception: Z(S186V), a mutant that was inactive on BMRF1p/CAT (Fig. 4A), was equivalent to wild-type ZEBRA in its ability to activate Rp/CAT (Fig. 4B).
FIG. 4.
Transcriptional activation of promoter/CAT reporters by ZEBRA and a panel of Z(S186) mutants. (A) Activation of BMRF1p/CAT. BJAB cells were transfected with 10 μg of BMRF1 promoter reporter plasmid and 10 μg of wild-type (wt) or mutant BZLF1 expression plasmid. The wild type and mutants are indicated by the amino acid present at position 186. The data represent the mean and standard error of the mean of three separate transfections. Fold activation is percent acetylation in the presence of BZLF1 or mutant expression vector/percent acetylation in the presence of CMV vector alone. (B) Activation of BRLF1p/CAT (Rp/CAT). BJAB cells were cotransfected with 10 μg of Rp/CAT reporter and with a total of 10 μg of activator plasmid. The activator consisted of 5 μg of BZLF1 or mutant expression vector plus 5 μg of vector alone (CMV) (solid bars) or 5 μg of Rta (open bars). Transfection efficiency was corrected by transfection of 1 μg of pGL2 Basic + HMP. Fold activation is relative to the level with reporter together with 10 μg of CMV. Synergy index: percent acetylation by ZEBRA or Z(S186) mutant in the presence of Rta/percent acetylation by ZEBRA or mutant alone plus percent acetylation by Rta alone.
There was a discrepancy between the results from the in vitro DNA binding assays and assays for transcriptional activation. The C, G, and V mutants, which were markedly impaired in their capacity to activate BMRF1 CAT in vivo, were nonetheless able to bind to ZRE2 and AP-1 sites, the two known ZEBRA binding sites in the BMRF1 promoter, efficiently in vitro (Fig. 3B). The Z(S186V) mutant, which activated Rp/CAT as well as wild type in transient reporter assays (Fig. 4B), showed negligible in vitro binding to the two ZEBRA binding sites in Rp, ZIIIA and ZRE-R (Fig. 3B). The S186C mutant, which activated Rp/CAT poorly, bound the two sites in Rp more efficiently than did S186V, which activated Rp/CAT well.
Ability of ZEBRA and Z(S186) mutants to synergize with Rta to activate expression of reporter constructs containing lytic cycle promoters.
ZEBRA synergizes with Rta in activation of certain lytic cycle promoters such as Dp (divergent promoter) that controls expression of BHRF1 and BHLF1 (12). In preliminary experiments we found that wild-type ZEBRA, Z(S186A), and Z(S186T) demonstrated 5- to 20-fold synergy with Rta on the reporter Dp/CAT (not shown). The G, V, and C mutants had markedly diminished ability or were unable to synergize with Rta.
Synergy between ZEBRA and Rta was also clearly evident when Rp/CAT reporter constructs were used (Fig. 4B). Wild-type ZEBRA stimulated Rp/CAT less than 20-fold in EBV-negative BJAB cells and Rta stimulated the promoter less than 5-fold, yet the two activators together stimulated Rp/CAT more than 100-fold. Thus, the level of synergy for wild-type ZEBRA was about 4.8-fold. The Z(S186A) and Z(S186T) mutants could synergize with Rta to the same extent as wild-type ZEBRA. The Z(S186V) mutant manifest increased activity in the presence of Rta. The level of synergy was only 1.5-fold in the experiment illustrated but was 5-fold in additional experiments (not shown). The Z(S186C) and Z(186G) mutants, which manifested low activity on Rp by themselves, were unable to synergize with Rta.
Transcriptional synergy between Z(S186A) and Rta on promoters residing on the viral genome.
The experiments using transient transfection reporter assays showed that while the Z(S186A) mutant was unable to induce expression of Rta, it was nonetheless capable of synergy with Rta on at least two promoters, Dp and Rp (Fig. 4B). Therefore we examined the effect of supplying Rta in trans on the capacity of the Z(S186A) mutant to activate transcription from the viral genome. The Rta protein by itself weakly activated the BMRF1 and BaRF1 genes in Raji cells (not shown). Increasing the input level of Rta expression plasmid to more than 10 μg did not increase the extent of activation of these two lytic cycle genes, as measured by the abundance of stable mRNA. The Z(S186A) mutant by itself activated neither of these two genes. However, if Z(S186A) was cotransfected with Rta, there was massive amplification of expression of the BMRF1 gene, as indicated by the abundance of BMRF1 mRNA and by the level of immunoreactive EA-D protein. In contrast, BaRF1 gene expression did not increase when Z(S186A) was introduced together with Rta. These results indicated that Z(S186A) and Rta acted in synergy on the BMRF1 promoter from the viral genome. The BaRF1 promoter was maximally stimulated by Rta alone; Z(S186A) did not synergize with Rta on this promoter.
Among the panel of Z(S186) mutants, only Z(S186A) and Z(S186T) work in synergy with Rta in activating expression of the BMRF1 gene from the intact viral genome.
A panel of seven Z(S186) mutants was tested for the capacity to activate the BMRF1 gene in Raji cells in the absence or presence of cotransfected Rta (Fig. 5 and data not shown). In the absence of Rta, transfection of Raji cells with constructs expressing wild-type ZEBRA or the Z(S186T) mutant resulted in increased abundance of the 2.5-kb BMRF1 transcript (Fig. 5A, lane 5). Expression of Rta by itself led to weak activation of BMRF1 (lane 9). However, when Rta was coexpressed with the Z(S186A) mutant, the level of BMRF1 mRNA expression was markedly higher than the level induced by Rta alone (lane 11) even though the Z(S186A) mutant by itself failed to activate BMRF1 expression (lane 4). The level of activation of BMRF1 was also increased by coexpression of the Z(S186T) mutant and Rta (lane 12). The other mutants were unable by themselves to activate BMRF1 expression and were not rescued by cotransfection of Rta. Rta by itself also induced the appearance of the BaRF1 mRNA, the abundance of which was unaffected by wild-type ZEBRA or any of the mutants, a finding that emphasizes the primary role played by Rta in control of the BaRF1 ribonucleotide reductase gene.
In the second half of this experiment, immunoblots were probed for the expression of EA-D, the BMRF1 product, for Rta, and for ZEBRA in order to determine if Rta rescued the capacity of any of the mutants to activate the EA-D protein (Fig. 5B). Only wild-type ZEBRA and the Z(S186T) mutant induced expression of EA-D (lane 5). The level of EA-D expression stimulated by the T mutant was reduced, commensurate with its reduced ability to activate BMRF1 mRNA (Fig. 5A). When Rta was supplied in trans, the amount of EA-D that was expressed in the presence of the Z(S186A) mutant was similar to the level expressed by wild-type ZEBRA and Rta, even though Z(S186A) was unable to stimulate EA-D expression in the absence of Rta (compare lanes 11 and 4). The amount of EA-D expressed following cotransfection of Rta and Z(S186T) was also greater than that after cotransfection with Z(S186T) alone (compare lane 12 with lane 5). Cotransfection of Rta with each of the Z(S186C), Z(S186G), and Z(S186V) mutants did not restore their ability to induce EA-D expression.
The experiment illustrated in Fig. 5 demonstrated that the Z(S186A) and Z(S186T) mutants could synergize with Rta in inducing BMRF1 gene expression from the latent virus, while mutants with substitutions of G, C, and V at position 186 were unable to synergize with Rta. In related experiments (not shown), we found that mutants with substitutions of aa 186 with glutamate and aspartate were also unable to synergize with Rta.
DISCUSSION
Serine 186 of ZEBRA is critical for at least two distinct functions of the ZEBRA protein, namely, its ability to activate expression of the most proximal downstream viral target gene, BRLF1, and its capacity to synergize with the BRLF1 product, Rta, in activation of certain early kinetic class genes such as BMRF1.
Classification of the mutants.
Side chain substitutions define three phenotypic classes of mutants with different capacities to activate Rp and to synergize with Rta on the viral genome (Fig. 6). One class, represented by Z(S186T), is similar to wild-type ZEBRA though diminished in the ability to activate Rta expression and downstream lytic cycle genes from the latent EBV genome (Fig. 1 and 5). If Rta is provided in trans, the defect in the capacity of Z(S186T) to activate BMRF1 is overcome. Another class, represented by Z(S186A), is unable to activate BRLF1 expression but appears fully competent to activate BMRF1 if Rta is provided in trans. The third class, represented by the G, V, C, D, and E substitutions at position S186, is unable to activate BRLF1 or BMRF1 from the viral genome, and this defect is not remedied by supplying Rta. Thus, all of the mutants except Z(S186T) are defective in activation of BRLF1; the C, G, V, D and E mutants are additionally defective in synergy with Rta.
FIG. 6.
Summary of the behavior of the panel of Z(S186) mutants. (A) Hypothetical model for behavior of ZEBRA mutants on Rp. ZEBRA mutants that are unable to activate Rp have two possible defects: a low affinity of binding to Rp and an inability to be phosphorylated. (B) Hypothetical model for capacity of ZEBRA mutants to synergize with Rta on the BMRF1 promoter. Synergy requires mutual contact between Rta, ZEBRA, and protein X. ZEBRA mutants that do not synergize with Rta are unable to contact protein X.
If one considers the behavior of the mutants in activating the BMRF1 and BRLF1 promoters linked to CAT in transient transfection assays in EBV-negative cells in the absence and presence of Rta (Fig. 4), a somewhat different classification arises. In transient transfection reporter assays, the mutants Z(S186A) and Z(S186T) behave more or less like wild-type ZEBRA and are fully competent to engage in synergy with Rta. Mutants with C or G substitutions are poor activators, either on Rp/CAT or on BMRF1p/CAT; this low activity is not overcome by providing Rta. The Z(S186V) mutant exemplifies a special case: while it is unable to activate BMRF1p/CAT, it does activate Rp/CAT and can synergize with Rta on this reporter.
Thus, this group of mutants illustrate that the amino acid side chain substitutions at position 186 discriminate between target templates that are present in the chromatinized latent viral genome and the presumably un- or underchromatinized targets present on plasmids in transient transfection assays. Three of the five mutants are concordant in their behavior on the two types of templates: the T substitution mutant is active, and the C and G substitution mutants are inactive or markedly impaired in both assays. Two of the mutants are discordant in their behavior on the two templates: both Z(S186A) and Z(S186V) can activate transcription in reporter assays, but neither by itself induces expression of viral genes in the context of the viral genome.
Abnormalities in binding to DNA.
The most appealing explanation to account for this panoply of phenotypes would be that the mutants display distinct abnormalities in binding to DNA. However, in general there was poor correlation between in vitro DNA binding assays and the capacity of the mutants to activate transcription in cells. All of the mutants were impaired relative to the wild type in the ability to bind to the two sites, ZIIIA and ZRE-R, present in Rp, whether examined in the context of identical flanking sequences or naturally occurring flanking sequences. But this decrease in binding to ZREs in Rp did not account for the behavior of the mutants in the activation of transcription from Rp. For example, the Z(S186A) and Z(S186C) mutants bound ZIIIA and ZRE-R with approximately equal affinities, yet the two mutants were distinctly different in the ability to activate Rp/CAT. Furthermore, the Z(S186V) mutant, which was most impaired in binding to the two ZRE sites in Rp, was nonetheless able to activate Rp/CAT to a higher level than mutants such as Z(S186G) that bound these sites efficiently. As a further example, all mutants bound with similar affinities to ZRE-2 and AP-1 (heptamer), the two sites in the BMRF1 promoter, yet the mutants differed markedly in the ability to activate BMRF1p/CAT.
Several possibilities could account for the discordance between DNA binding and transcriptional activation. One potential defect might lie in cooperative binding to DNA. Positive control mutants in the basic region of MyoD, which bound DNA but lost the capacity to activate transcription, were found to lose the ability to bind DNA in a cooperative fashion (5). It was suggested that such mutants failed to undergo upon binding to DNA conformational changes that allowed them to bind cooperatively. Further in vitro studies using purified ZEBRA mutant proteins would be required to determine whether the S186 mutants display differences in cooperative DNA binding. Alternatively, the mutations may affect the way ZEBRA makes protein-protein interactions that are required for transcriptional activation. Although the two EBV promoters studied in detail, the BMRF1 promoter and Rp, contain ZREs, it is nonetheless possible that they are activated by indirect mechanisms that require protein-protein interactions that are affected by mutations at Z(S186). We have experimentally excluded the explanations that the Z(S186) mutants do not activate transcription because they fail to find their way to the nucleus (not shown) or that the proteins are not expressed (Fig. 1 and 5).
Possible role of phosphorylation of Z(S186).
Our experiments implicate phosphorylation of serine 186 as essential for the ability of ZEBRA to activate expression of BRLF1 from the intact viral genome (Fig. 6A). However, since Z(S186A), which could not be phosphorylated at this position, is competent to synergize with Rta, phosphorylation is not needed for synergy (Fig. 6B). The single mutant that resembles wild-type ZEBRA in its capacity to activate BRLF1 is Z(S186T). A serine-threonine kinase would phosphorylate Z(S186T). Moreover, mutants that substitute acidic residues, glutamate and aspartate, for serine 186 are still unable to activate the lytic cycle, as assessed by the expression of Rta and EA-D (data not shown). Therefore, negative charge at position 186, alone, is not sufficient for activation of BRLF1.
ZEBRA is phosphorylated at multiple sites in vivo (13). Serine at position 186 of ZEBRA fits a consensus for a site that can be phosphorylated by PKC (44), and we have found that bacterially expressed ZEBRA can be phosphorylated in vitro by PKC (18a). Baumann et al. have recently shown that ZEBRA but not Z(S186A) can be phosphorylated in vitro by PKCα (4). Baumann et al. find that tetradecanoyl phorbol acetate treatment of 293 cells causes additional phosphorylation of wild-type ZEBRA but not Z(S186A). Thus, position 186 likely functions as a substrate for PKC-mediated phosphorylation in vivo.
What might be the function of phosphorylation of S186? It is not absolutely required for transcriptional activation, since Z(S186A) can efficiently activate transcription from reporters bearing the BMRF1 and BRLF1 promoters (Fig. 4). However, this requirement for phosphorylation might be specific for certain promoters in some cell backgrounds, since Baumann et al. show that tetradecanoyl phorbol acetate treatment markedly enhances the ability of ZEBRA to activate the BHRF1 promoter in EBV-negative BL41 cells (4). In our view, a likely biologic function of phosphorylation of ZEBRA at position S186 would be to enhance its interaction with a coactivator that is required for the activation of the BRLF1 gene in the context of the viral genome (Fig. 6A).
A candidate coactivator that might interact with ZEBRA is the CREB binding protein known as CBP/p300 (9). CBP/p300 has been found to interact with many different transcriptional activators (27). CBP/p300 not only provides a bridge to the general transcription machinery but also has histone acetylase activity that may modify chromatin to a transcriptionally active state (7, 31). Interactions of transcription factors such as CREB (35), c-Jun (2), and NF-κB p65 (46) with CBP are modulated by phosphorylation and dephosphorylation. Often phosphorylation is required for the association of the transcription factor with CBP. An attractive hypothesis is that phosphorylation of ZEBRA at S186 allows the protein to form a stable interaction with CBP and thus allows ZEBRA to activate gene expression from a chromatinized latent EBV genome. Alternatively, phosphorylation of ZEBRA at S186 may allow interaction with a component of the general transcription apparatus that is needed for activation of BRLF1 from a chromatinized viral genome but not from a transfected plasmid.
ACKNOWLEDGMENTS
This study was supported by grants CA 12055, CA 16038, and CA70036 to G.M. from the NIH.
We are grateful to S. D. Hayward for providing the Rta expression vector pRTS15 and to W. Hammerschmidt and S. Kenney for communicating results before publication.
REFERENCES
- 1.Adamson A L, Kenney S C. Rescue of the Epstein-Barr virus BZLF1 mutant, Z(S186A), early gene activation defect by the BRLF1 gene product. Virology. 1998;251:187–197. doi: 10.1006/viro.1998.9396. [DOI] [PubMed] [Google Scholar]
- 2.Bannister A J, Oehler T, Wilhelm D, Angel P, Kouzarides T. Stimulation of c-Jun activity by CBP: c-Jun residues Ser63/73 are required for CBP-induced stimulation in vivo and CBP binding in vitro. Oncogene. 1995;11:2509–2514. [PubMed] [Google Scholar]
- 3.Bartkiewicz M, Gold H, Altman S. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev. 1989;3:488–499. doi: 10.1101/gad.3.4.488. [DOI] [PubMed] [Google Scholar]
- 4.Baumann M, Mischak H, Dammeier S, Kolch W, Gires O, Pich D, Hammerschmidt W. Activation of the Epstein-Barr virus transcription factor BZLF1 by 12-O-tetradecanoyphorbol-13-acetate-induced phosphorylation. J Virol. 1998;72:8105–8114. doi: 10.1128/jvi.72.10.8105-8114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bengal E, Flores O, Rangarajan P N, Chen A, Weintraub H, Verma I M. Positive control mutations in the MyoD basic region fail to show cooperative DNA binding and transcriptional activation in vitro. Proc Natl Acad Sci USA. 1994;91:6221–6225. doi: 10.1073/pnas.91.13.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang Y-N, Dong D L-Y, Hayward G S, Hayward S D. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J Virol. 1990;64:3358–3369. doi: 10.1128/jvi.64.7.3358-3369.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 8.Chevallier-Greco A, Gruffat H, Manet E, Calender A, Sergeant A. Both Epstein-Barr virus (EBV) encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 1989;5:3243–3249. doi: 10.1002/j.1460-2075.1986.tb04635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 10.Countryman J, Miller G. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Countryman J, Jenson H, Seibl R, Wolf H, Miller G. Polymorphic proteins encoded within BZLF1 of defective and standard Epstein-Barr viruses disrupt latency. J Virol. 1987;61:3672–3679. doi: 10.1128/jvi.61.12.3672-3679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox M A, Leahy J, Hardwick J M. An enhancer within the divergent promoter of Epstein-Barr Virus responds synergistically to the R and Z transactivators. J Virol. 1990;64:313–321. doi: 10.1128/jvi.64.1.313-321.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daibata M, Humphreys R E, Sairenji T. Phosphorylation of the Epstein-Barr virus BZLF1 immediate-early gene product ZEBRA. Virology. 1992;188:916–920. doi: 10.1016/0042-6822(92)90553-2. [DOI] [PubMed] [Google Scholar]
- 14.Daibata M, Speck S H, Mulder C, Sairenji T. Regulation of the BZLF1 promoter of Epstein-Barr virus by second messengers in anti-immunoglobulin-treated B cells. Virology. 1994;198:446–454. doi: 10.1006/viro.1994.1056. [DOI] [PubMed] [Google Scholar]
- 15.Davis M G, Huang E S. Transfer and expression of plasmids containing human cytomegalovirus immediate-early gene 1 promoter-enhancer sequences in eukaryotic and prokaryotic cells. Biotechnol Appl Biochem. 1988;10:6–12. [PubMed] [Google Scholar]
- 16.Farrell P J, Rowe D T, Rooney C M, Kouzarides T. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 1989;8:127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flemington E, Speck S H. Autoregulation of Epstein-Barr virus putative lytic switch gene BZLF1. J Virol. 1990;64:1227–1232. doi: 10.1128/jvi.64.3.1227-1232.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flemington E, Goldfield A E, Speck S H. Efficient transcription of the Epstein-Barr virus immediate-early BZLF1 and BRLF1 genes requires protein synthesis. J Virol. 1991;65:7073–7077. doi: 10.1128/jvi.65.12.7073-7077.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Francis A. Ph.D. thesis. New Haven, Conn: Yale University; 1997. [Google Scholar]
- 19.Francis A L, Gradoville L, Miller G. Alteration of a single serine in the basic domain of the Epstein-Barr virus ZEBRA protein separates its functions of transcriptional activation and disruption of latency. J Virol. 1997;71:3054–3061. doi: 10.1128/jvi.71.4.3054-3061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giot J-F, Mikaelian I, Buisson M, Manet E, Joab I, Nicholas J-C, Sergeant A. Transcriptional interference between the EBV transcription factors EB1 and R: both DNA-binding and activation domains of EB1 are required. Nucleic Acids Res. 1991;19:1251–1258. doi: 10.1093/nar/19.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glover J N, Harrison S C. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature. 1995;373:257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 22.Grogan E J, Jenson J, Countryman J, Heston L, Gradoville L, Miller G. Transfection of a rearranged viral DNA fragment WZhet, stably converts latent Epstein-Barr virus infection to productive infection in lymphoid cells. Proc Natl Acad Sci USA. 1987;84:1332–1336. doi: 10.1073/pnas.84.5.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardwick J M, Liebermann P M, Hayward S D. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J Virol. 1988;62:2274–2284. doi: 10.1128/jvi.62.7.2274-2284.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holley-Guthrie E, Quinlivan E B, Mar E-C, Smith M. The Epstein-Barr virus (EBV) BMRF1 promoter for early antigen (EA-D) is regulated by the EBV transactivators BRLF1 and BZLF1 in a cell-specific manner. J Virol. 1990;64:3753–3759. doi: 10.1128/jvi.64.8.3753-3759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kenney S, Holley-Guthrie E, Mar E-C, Smith M. The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators. J Virol. 1989;63:3878–3883. doi: 10.1128/jvi.63.9.3878-3883.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolman J L, Taylor N, Gradoville L, Countryman J, Miller G. Comparing transcriptional activation and autostimulation by ZEBRA and ZEBRA/c-Fos chimeras. J Virol. 1996;70:1493–1504. doi: 10.1128/jvi.70.3.1493-1504.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 28.Le Roux F, Sergeant A, Corbo L. Epstein-Barr virus (EBV) EB1/Zta protein provided in trans and competent for the activation of productive cycle genes does not activate the BZLF1 gene in the EBV genome. J Gen Virol. 1996;77:501–509. doi: 10.1099/0022-1317-77-3-501. [DOI] [PubMed] [Google Scholar]
- 29.Lehman A M, Ellwood K B, Middleton B E, Carey M. Compensatory energetic relationships between upstream activators and the RNA polymerase II general transcription machinery. J Biol Chem. 1998;273:932–939. doi: 10.1074/jbc.273.2.932. [DOI] [PubMed] [Google Scholar]
- 30.Lieberman P M, Berk A J. In vitro transcriptional activation, dimerization, and DNA binding of the Epstein-Barr virus Zta protein. J Virol. 1990;64:2560–2568. doi: 10.1128/jvi.64.6.2560-2568.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez-Balbas M A, Bannister A J, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mellinghoff I, Daibata M, Humphreys R E, Mulder C, Takada K, Sairenji T. Early events in Epstein-Barr virus genome expression after activation: regulation by second messengers of B cell activation. Virology. 1991;185:922–928. doi: 10.1016/0042-6822(91)90574-u. [DOI] [PubMed] [Google Scholar]
- 33.Menezes J, Leifbold W, Klein G, Clements G. Establishment and characterization of an Epstein-Barr virus (EBV)-negative lymphoblastoid B cell line (BJA-B) from an exceptional, EBV-genome negative African Burkitt’s lymphoma. Biomedicine. 1975;22:276–284. [PubMed] [Google Scholar]
- 34.Packham G, Economou A, Rooney C M, Rowe D T, Farrell P J. Structure and function of the Epstein-Barr virus BZLF1 protein. J Virol. 1990;64:2110–2116. doi: 10.1128/jvi.64.5.2110-2116.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker D, Ferreri K, Nakajima T, LaMorte V J, Evans R, Koerber S C, Hoeger C, Montminy M R. Phosphorylation of CREB at Ser-133 induces complex form with CREB-binding protein via a direct mechanism. Mol Cell Biol. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson G R, Vroman B, Chase B, Sculley T, Hummel M, Kieff E. Identification of polypeptide components of the Epstein-Barr virus early antigen complex with monoclonal antibodies. J Virol. 1983;47:193–201. doi: 10.1128/jvi.47.1.193-201.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quinlivan E B, Holley-Guthrie E A, Norris M, Gutsch D, Bachenheimer S L, Kenney S C. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus early promoter, BMRF1. Nucleic Acids Res. 1993;21:1999–2007. doi: 10.1093/nar/21.8.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ragoczy T, Heston L, Miller G. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J Virol. 1998;72:7978–7984. doi: 10.1128/jvi.72.10.7978-7984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rooney C, Rowe D T, Ragot T, Farrell P J. The spliced BZLF1 gene of Epstein-Barr virus (EBV) transactivates an early EBV promoter and induces the virus productive cycle. J Virol. 1989;663:3109–3116. doi: 10.1128/jvi.63.7.3109-3116.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serio T R, Kolman J L, Miller G. Late gene expression from the Epstein-Barr virus BcLF1 and BFRF3 promoters does not require DNA replication in cis. J Virol. 1997;71:8726–8734. doi: 10.1128/jvi.71.11.8726-8734.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takada K, Ono Y. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J Virol. 1989;63:445–449. doi: 10.1128/jvi.63.1.445-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor N, Flemington E, Kolman J L, Baumann R P, Speck S H, Miller G. ZEBRA and a Fos-GCN4 chimeric protein differ in their DNA-binding specificities for sites in the Epstein-Barr virus BZLF1 promoter. J Virol. 1991;65:4033–4041. doi: 10.1128/jvi.65.8.4033-4041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor N, Countryman J, Rooney C, Katz D, Miller G. Expression of the BZLF1 latency-disrupting gene differs in standard and defective Epstein-Barr viruses. J Virol. 1989;63:1721–1728. doi: 10.1128/jvi.63.4.1721-1728.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woodgett J R, Gould K L, Hunter T. Substrate specificity of protein kinase C. Use of synthetic peptides corresponding to physiological sites as probes for substrate recognition requirements. Eur J Biochem. 1986;161:177–184. doi: 10.1111/j.1432-1033.1986.tb10139.x. [DOI] [PubMed] [Google Scholar]
- 45.Zalani S, Holley-Guthrie E, Kenney S. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc Natl Acad Sci USA. 1996;93:9194–9199. doi: 10.1073/pnas.93.17.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong H, Voll R E, Ghosh S. Phosphorylation of NF-KB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]