Abstract
The progress in lung cancer treatment is closely interlinked with the progress in diagnostic methods. There are four steps before commencing lung cancer treatment: estimation of the patient's performance status, assessment of disease stage (tumour, node, metastasis), recognition of histological subtype, and detection of biomarkers. The resection rate in lung cancer is <30% and >70% of patients need systemic therapy, which is individually adjusted. Accurate histological diagnosis is very important and it is the basis of further molecular diagnosis. In many cases only small biopsy samples are available and the rules for their assessment are defined in this review. The use of immunochemistry with at least thyroid transcription factor 1 (TTF1) and p40 is decisive in distinction between lung adenocarcinoma and squamous cell carcinoma. Molecular diagnosis and detection of known driver mutations is necessary for introducing targeted therapy and use of multiplex gene panel assays using next-generation sequencing is recommended. Immunotherapy with checkpoint inhibitors is the second promising method of systemic therapy with best results in tumours with high programmed death-ligand 1 (PD-L1) expression on cancer cells. Finally, the determination of a full tumour pattern will be possible using artificial intelligence in the near future.
Shareable abstract
More than 70% of patients with lung cancer need systemic therapy. Accurate histological diagnosis and molecular diagnosis, with detection of known driver mutations and PD-L1 expression on cancer cells, is necessary for introducing personalised therapy. https://bit.ly/3xfXy1X
Introduction
Lung cancer is a global health challenge, with its incidence and mortality rates consistently ranking among the highest for cancer. As one of the most prevalent cancers globally, lung cancer exerts a significant toll on public health systems and communities. According to recent statistics from the World Health Organization (WHO), ∼2.21 million new lung cancer cases were diagnosed in 2020 with an estimated 1.8 million deaths in the same year, ranking lung cancer as the leading cause of cancer-related deaths [1]. These figures emphasise the urgent need for comprehensive strategies to address the burden of this disease. Despite advancements in treatment modalities, lung cancer continues to pose challenges in terms of prognosis and survival outcomes. The 5-year survival rate for lung cancer remains low, largely due to late-stage diagnoses and limited treatment options [1]. Early detection strategies, coupled with access to timely and effective diagnostics and treatments pave the way in improving survival rates and clinical outcomes.
As researchers and clinicians strive to enhance diagnostic precision and treatment efficacy, the spotlight has shifted towards unravelling the complex details of diagnostic pathology and molecular biomarkers. Understanding precise staging, pathology and molecular profile is essential for improving patient outcomes.
The molecular characterisation of lung cancer has changed the classification and treatment of these tumours and it has become an essential component of pathology diagnosis and oncologic therapy decisions [2]. The success of targeted anti-cancer therapies and new immunotherapy approaches has created a new paradigm of personalised therapy, and has also led to accelerated development of new drugs for lung cancer treatment. The choice of treatment depends on factors such as the type of lung cancer, its stage, the patient's overall health and performance status.
The TNM (tumour, node, metastasis) staging system, which classifies tumours based on the extent of the primary tumour (T), the involvement of regional lymph nodes (N) and the presence of distant metastasis (M), developed by the American Joint Committee on Cancer and the Union for International Cancer Control is the cornerstone in the management of lung cancer [3]. The progress in imaging techniques guarantees precise determination of clinical status. Computed tomography (CT), positron emission tomography-CT and ultrasonography offer valuable information regarding the optimal route to tissue.
Traditionally, lung cancer diagnosis relied heavily on histopathological examination, classifying tumours into broad categories such as nonsmall cell lung cancer (NSCLC) and small cell lung cancer (SCLC). However, the emergence of molecular diagnostics has triggered a paradigm shift, demanding a deeper understanding of the genetic and molecular underpinnings of lung cancer. One notable development is the identification of driver mutations that fuel the growth of cancer cells. Epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK) and ROS proto-oncogene 1 (ROS1) gene alterations are among the well-studied driver mutations in lung cancer [4]. Targeting these mutations with specific therapies has shown remarkable success, emphasising the importance of accurate pathological diagnosis in selecting appropriate treatment options.
This review aims to provide a succinct overview of the diagnostic pathology and molecular biomarkers applied in lung cancer and explore how they contribute to reshaping the landscape of lung cancer diagnosis and treatment.
Methods of histopathological diagnosis
Lung cancer diagnosis is based on histopathological examination. “Tissue is the issue” is a strong theme in thoracic oncology. Lung tumour diagnosis and classification is based first on morphology, immunohistochemistry (IHC) and on molecular techniques. There are two main histological subtypes of lung cancer differing in morphology, biology and aggressiveness: SCLC and NSCLC. The treatment of lung cancer until the 2000s was based on these two main histological types. The most frequent type is NSCLC (85% of all cases) with domination of adenocarcinoma (60% of NSCLC) [5]. The introduction of targeted therapies and immunotherapy imposed the differentiation of NSCLC subtypes to adenocarcinoma (ADC) and squamous cell carcinoma (SCC). Pathologists can differentiate these subtypes using IHC markers.
The first histological classification for lung cancer was published in 1967 in the first WHO edition for lung cancer. Subsequently, histological classification guidelines were updated and progressed leading to the 2021 WHO classification of lung tumours, which emphasises a broad spectrum of molecular tests, focusing on evaluation of small biopsies and recognition of histological features with prognostic significance [6].
Surgical specimens are valuable diagnostic materials for pathologists enabling histological subclassification and molecular profiling; however, they are available in only 25–30% of lung cancer cases [1]. The majority of pathology samples are categorised as small biopsies as ∼70% of lung cancer patients present in an advanced stage unsuitable for surgery. A small biopsy (i.e. tissue biopsy) is significantly smaller than surgical one and it is obtained during bronchoscopy from bronchial mucosa, core biopsy or less frequently, by cryobiopsy. Cytological samples of lung tumours are obtained through a variety of methods, one of them being fine-needle aspiration biopsy, for example, endobronchial ultrasound-guided transbronchial needle aspiration (EBUS/TBNA), endoscopic ultrasound fine-needle aspiration, transthoracic needle aspiration. These samples have a better quality for processing and diagnosis than the exfoliative specimens (e.g. bronchial washings, bronchial brushings, bronchoalveolar lavage fluid or pleural fluid). It should be noted that due to its accuracy, EBUS/TBNA and transbronchial needle biopsy (TBNB) has become a method of evaluation for enlarged lymph nodes for staging and also for histological diagnosis of peripheral lung tumours that cannot be reached with the conventional bronchoscope. Current diagnostic procedures require multiple biopsy samples for precise diagnosis and determination of predictive factors. Thus, the cell blocks from cytological specimens are recommended as additional material replacing cell smears.
IHC is widely incorporated in precise malignant tumour diagnosis. In the case of lung cancer, a limited panel of IHC markers is recommended. Cellular morphology allows the recognition of lung cancer type, mainly SCLC and SCC (figure 1). Lung ADC could pose a diagnostic challenge as the lung is often a site of metastases from other organs. Thus, in the case of morphological features of ADC being present, it is vital to identify whether the lung is the primary site of malignancy. Therefore, clinical information should be given to the pathologist to ensure an appropriate diagnosis is reached.
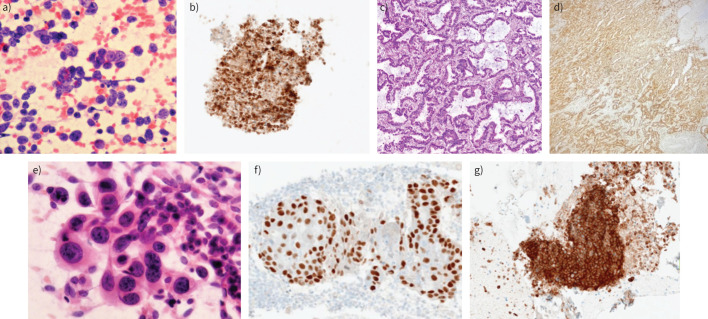
FIGURE 1.
Small cell lung cancer in a cytological smear: a) haematoxylin and eosin staining, b) positive immunohistochemistry (IHC) reaction with CD56, cell block. Adenocarcinoma with acinar pattern, tissue biopsy: c) haematoxylin and eosin staining, d) positive IHC reaction with napsin A. Squamous cell carcinoma, with typical features in a cytological smear: e) haematoxylin and eosin staining, f) positive IHC reaction with p40. g) Programmed death-ligand 1 (PD-L1) expression on nonsmall cell lung cancer cells in cellblock. Tumour proportion score >50, antibody: Ventana SP-263 (author's material).
SCLC is one of the neuroendocrine carcinomas [5]. Neuroendocrine differentiation markers (e.g. CD56, chromogranin, synaptophysin) are useful in diagnosis of SCLC and other neuroendocrine cancers. An assessment of Ki67 proliferation index is helpful in the differential diagnosis of carcinoid and SCLC [5, 6].
In the case of NSCLC, the recognition of SCC and ADC is of vital importance before further molecular diagnosis is performed. Both NSCLC subtypes are not uniform and they may exhibit low differentiation as well as features of anaplasia. In general, the use of the thyroid transcription factor 1 (TTF1) marker for confirmation of lung ADC and p40 for SCC is recommended (table 1). Positive reaction with CK7 and napsin A additionally supports the primary pulmonary ADC diagnosis. The negative reaction with TTF1 and p40 markers suggests “NSCLC not otherwise specified (NOS)”. This term should be used only in small biopsies and with caution.
TABLE 1.
Terminology for nonsmall cell lung cancer (NSCLC) histological diagnosis using immunohistochemistry on resection specimens versus small/ cytological biopsy
| Immunohistochemistry marker | Biopsy sample | ||
|---|---|---|---|
| TTF1 | p40 | Resection specimen | Small biopsy, cytology |
| + | −/+ | ADC | NSCLC, favour ADC |
| − | + (diffuse) | SCC | NSCLC, favour SCC |
| − | − | LCC | NOS |
| No stains available | No stains available | LCC with no additional stains | NOS (no stains available) |
TTF1: thyroid transcription factor 1; ADC: adenocarcinoma; SCC: squamous cell carcinoma; LCC: large cell carcinoma; NOS: not otherwise specified; +: positive; −: negative.
There is a broad panel of IHC markers used for the differential diagnosis of metastatic tumours to the lung. In turn, markers for epithelial versus nonepithelial differentiation and/or markers of haematopoietic neoplasms should be used. In the case of ADC, different types of cytokeratin and specific organ markers are IHC markers of choice [7].
The precise recognition of the histological type of primary lung tumour is the most appropriate direction towards systemic therapy. The relationship of NSCLC subtypes, especially ADC, with molecular alterations and response to treatment has been described in many studies [8].
Pathology reports are the tangible product of the pathologist's work. To be meaningful they must provide clear, consistent information and all elements necessary for decision making. A lung pathology report should include:
a pathological diagnosis with the results of IHC stains,
comments about the differential diagnosis,
information regarding sample adequacy for further molecular testing and information that material was submitted for evaluation of predictive markers.
Diagnosis of molecular alterations
Basis of targeted therapy
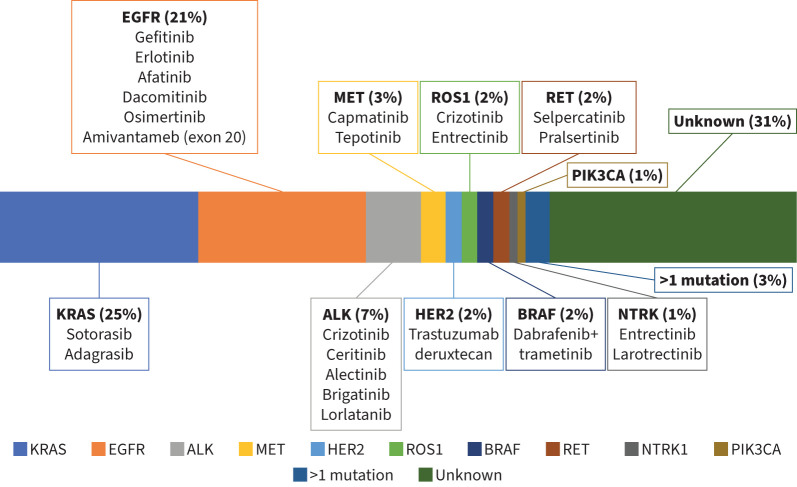
Advances in understanding of the molecular biology of lung cancer and the hallmarks of tumour progression and metastasis have led to the development of novel targeted agents that have significantly altered the prognosis of lung cancer patients [9]. The anti-tumour efficacy of targeted therapies depends on the presence of specific molecular alterations that drive tumour progression [9]. The widespread use of such agents has resulted in significant clinical benefits while avoiding the toxicity of traditional cytotoxic chemotherapy [10]. Targeted therapies include tyrosine kinase inhibitors, monoclonal antibodies, and targeted chemotherapies delivered to tumour cells using liposomes, nanoparticles or antibody–drug conjugates. The frequency of common driver molecular alterations and their targeted therapies in lung ADC are summarised in figure 2.
FIGURE 2.
Frequency of common driven molecular alterations and their targeted therapies in lung adenocarcinoma. KRAS: Kirsten rat sarcoma viral oncogene homolog; EGFR: epidermal growth factor receptor; ALK: anaplastic lymphoma kinase; MET: MET proto-oncogene receptor tyrosine kinase; HER2: human epidermal growth factor receptor 2; ROS1: ROS proto-oncogene 1; BRAF: B-Raf proto-oncogene; RET: rearranged during transfection; NTRK: neurotrophic tropomyosin kinase receptor; PIK3CA: phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α. Adapted from [2].
Molecular diagnosis
Techniques for molecular testing for driver mutations include single-gene assays such as PCR, fluorescence in situ hybridisation (FISH) and IHC, as well as multiplex gene panel assays using next-generation sequencing (NGS), based on either DNA or RNA sequencing. Different diagnostic techniques offer varying sensitivity and specificity depending on the specific molecular alteration (table 2).
TABLE 2.
Techniques for molecular testing and their clinical implications
| Technique | What does it do? | Sample types used | What is the clinical significance? |
|---|---|---|---|
| IHC | Detection of protein expression | Tissue | Therapeutic decision |
| PCR | Detection of gene mutations | Tissue/blood | Therapeutic decision/mechanism of resistance |
| NGS | Detection of gene mutations Amplification Skipping mutations |
Tissue/blood | Therapeutic decision/mechanism of resistance |
| FISH | Gene rearrangements Amplification Skipping mutations |
Tissue | Therapeutic decision/mechanism of resistance |
IHC: immunohistochemistry; NGS: next-generation sequencing; FISH: fluorescence in situ hybridisation.
For single-gene assays, PCR-based testing can identify specific gene mutations with high sensitivity and specificity, for example, EGFR exon 19 deletions or L858R mutations [11], while FISH is generally used for the detection of gene fusion and amplification [12]. ALK rearrangements represent a notable exception, as IHC is an equivalent alternative for treatment decisions and predicting response to ALK inhibitors [13]. Other targetable gene fusions, such as neurotrophic tropomyosin kinase receptors 1–3 (NTRK1–3) and ROS1, require FISH or NGS confirmation when initially detected using IHC [4]. IHC staining is also used to identify programmed death-ligand 1 (PD-L1) expression, which is associated with higher response rates of immune checkpoint inhibitors (ICIs) [13].
NGS allows the simultaneous study of multiple genes/variants in a cost-effective and time-efficient manner for individualising patient care [14]. There are several approaches for a custom NGS panel, with hybridisation and amplicon sequencing being the most common. Although hybridisation capture has several advantages over amplicon sequencing, such as the need for lower DNA input, larger gene content (typically more than 50) and a comprehensive gene profile of all variants, the complexity, and the laborious workflow restrict its use to investigational and clinical trial uses only. In contrast, amplicon sequencing, although typically containing no more than 50 genes, has a simple and faster workflow, affordable cost, and is ideal for analysing single nucleotide variants and insertions/deletions [15]. To reduce turnaround time, which is crucial for treatment decisions in lung cancer, single-gene or smaller multiplex assays may be performed in parallel with larger gene panels [16].
Recommendations: from EGFR to HER2
In clinical practice guidelines from the European Society for Medical Oncology (ESMO) and National Comprehensive Cancer Network (NCCN), molecular testing for alterations with an approved targeted therapy is mandatory for patients with non-squamous NSCLC. In both guidelines, a specific recommendation is made for EGFR and ALK based on large randomised clinical trials, and for ROS1, NTRK, BRAF, MET, KRAS, RET and HER2 based on smaller randomised clinical trials [4, 17]. Seven genes are considered mandatory according to the ESMO scale for clinical actionability of molecular targets: EGFR (common mutations, Del 19 and L858R; acquired T790M; uncommon EGFR mutations G719X in exon 18, L861Q in exon 21, S768 in exon 20, and exon 20 insertions), ALK fusions (mutations only as a mechanism of resistance), MET (mutations ex14 skipping; focal amplifications as acquire resistance mechanism on EGFR-tyrosine kinase inhibitors in EGFR-mutated patients), BRAF mutations, ROS1 fusions, NTRK fusions, and RET fusions, while KRAS mutations and HER2 mutations are strongly recommended [18, 19]. Molecular testing is not recommended for patients with squamous NSCLC, except in unusual cases such as younger age (<50 years) or never/light smokers [4]. Reflex molecular testing rather than bespoke can preserve tissue and speed up results. Although it has appealing benefits, it has not been universally adopted, mostly due to reimbursement difficulties. The recommended diagnostic methods for targetable molecular alterations in non-squamous NSCLC are summarised in table 3.
TABLE 3.
Diagnostic methods for detecting molecular alterations in non-squamous nonsmall cell lung cancer
| IHC | PCR | FISH | NGS | HC-NGS | |
|---|---|---|---|---|---|
| PD-L1 | + | − | − | − | − |
| EGFR sensitising | + | + | + | ||
| EGFRex20ins | −/+ | − | + | + | |
| ALK | + | − | + | + | + |
| ROS1 | −/+# | − | + | + | + |
| BRAF | − | + | − | + | + |
| MET | −/+¶ | +/− | + | + | + |
| HER2 | −/+§ | + | + | + | + |
| RET | +/− | + | + | + | |
| NRTK | − | − | + | + | + |
IHC: immunohistochemistry; FISH: fluorescence in situ hybridisation; NGS: next-generation sequencing; HC-NGS: hybrid capture-based NGS; PD-L1: programmed death-ligand 1; EGFR: epidermal growth factor receptor; ALK: anaplastic lymphoma kinase; ROS1: ROS proto-oncogene 1; BRAF: B-Raf proto-oncogene; MET: MET proto-oncogene receptor tyrosine kinase; HER2: human epidermal growth factor receptor 2; RET: rearranged during transfection; NTRK: neurotrophic tropomyosin kinase receptors. +: indicates clinical utility. #: positive IHC for ROS1 needs further confirmation with FISH or NGS; ¶: IHC for MET overexpression can be used as a predictive marker for response to therapy (experimental use only); §: IHC for HER2 protein is not recommended for lung cancer patients.
Single-gene PCR-based assays can be used to guide decision making for the use of tyrosine kinase inhibitors targeting point mutations in several genes, including EGFR, KRAS and BRAF [20–22]. However, over 40% of patients harbouring EGFRex20ins may go undiagnosed by any PCR, and additional NGS-based testing should be considered to complete gene coverage [23]. IHC for overexpression of HER2 protein is not recommended for lung cancer patients. PCR or NGS for HER2 mutation detection can be predictive for response to targeted therapies [24]. Regarding gene fusions, RNA-based NGS or DNA-based NGS designed for such fusions are the preferred methods. IHC may be used in certain circumstances for screening or validation purposes, as mentioned previously [4, 25]. RNA-based NGS is similarly preferred for detecting targetable skipping mutations, most notably in exon 14 of MET [26]. Given the equivalence or superiority of NGS in the detection of most genetic alterations, the ability to cover all targetable mutations in addition to other genetic alterations with potential clinical significance, and the consumption of less tissue in comparison with conducting multiple single-gene assays, the use of NGS is preferred [4, 17, 18, 27]. NGS or PCR of cell-free DNA (liquid biopsy) can be considered if the initial evaluation of tissue is uninformative or unavailable. Positive results can be used for treatment decisions, but negative results must be confirmed by tissue analysis [4, 17].
ICIs and their role in the treatment of lung cancer: focusing on their diagnosis, biomarkers and immune-related side-effects
Since nivolumab's initial approval for treating advanced NSCLC in 2015, the use of ICIs in lung cancer treatment has significantly expanded [28]. Table 4 illustrates the ICIs currently endorsed by the European Medicines Agency for treating both SCLC and NSCLC. Initially employed as a second-line therapy for advanced NSCLC, these inhibitors are now widely used in first-line systemic treatment for NSCLC, extensive SCLC and recently, in (neo)adjuvant therapy for earlier stages of both resectable and non-resectable lung cancer. Consequently, it is foreseeable that the majority of lung cancer patients will receive at least one ICI during their illness in the future.
TABLE 4.
Immune checkpoint inhibitors currently endorsed by the European Medicines Agency (EMA) for treating lung cancer, information presented is based on the approval documents of the respective immune checkpoint inhibitors, publicly available on the EMA website; as with nivolumab, atezolizumab and pembrolizumab can also be given as second-line treatment after chemotherapy
| Small cell lung cancer | Nonsmall cell lung cancer | ||
|---|---|---|---|
| Neoadjuvant treatment | Adjuvant treatment | Metastatic disease | |
| Atezolizumab in extensive disease Durvalumab in extensive disease |
Nivolumab combined with chemotherapy in resectable disease prior to surgery Pembrolizumab combined with chemotherapy in resectable disease in a perioperative approach# |
Durvalumab after chemoradiotherapy in non-resectable disease Pembrolizumab in resectable disease after surgery Atezolizumab in resectable disease after surgery Nivolumab in resectable disease after surgery, in a perioperative approach# |
Nivolumab as second-line after chemotherapy or together with ipilimumab combined with chemotherapy as first line Durvalumab in combination with tremelimumab as first-line treatment combined with chemotherapy Pembrolizumab as first line treatment in PD-L1>50% or combined with chemotherapy Cemiplimab as first-line treatment in PD-L1>50% or combined with chemotherapy Atezolizumab as first line treatment in PD-L1>50% or combined with chemotherapy |
PD-L1: programmed death-ligand 1. #: EMA approval for perioperative use of pembrolizumab and nivolumab is expected in 2024.
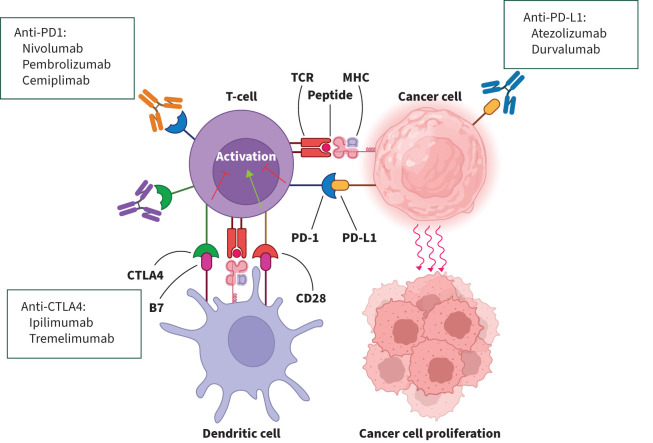
ICIs operate differently from conventional cancer treatments, causing a shift in treatment paradigms. Cancer cells exploit the body's immune control pathways, using certain proteins to escape the typical T-cell driven immune response that would otherwise control them. ICIs work by blocking these specific protein interactions, such as PD-L1 and cytotoxic T-lymphocyte antigen 4 (CTLA4), found on T-cells crucial for immune regulation, thus reversing immune evasion. This heightens the body's immune response against cancer cells, inducing T-cell triggered tumour cell apoptosis [29]. Figure 3 visually illustrates how T-cells engage with tumour cells and antigen presenting cells, demonstrating how ICIs modify these interactions to bolster the immune-driven anti-tumour response.
FIGURE 3.
The interaction between T-cells, dendritic cells and cancer cells is shown, with the role of the pathways programmed death-ligand 1 (PD-L1)–programmed death-1 (PD1) and cytotoxic T-lymphocyte antigen 4 (CTLA4)–B7/CD28 binding as negative regulators of T-cell immune function. Pharmacological agents that alter these interactions (see boxes) enhance the T-cell driven anti-tumour response. Reproduced and modified from [29] with permission.
Assessing PD-L1 protein expression in biopsy samples is crucial in identifying patients who might benefit from some kinds of ICIs. Higher PD-L1 expression in cancer cells correlates theoretically with greater benefit from anti-programmed death 1(PD1)/anti-PD-L1 treatment, as confirmed by different research studies. However, PD-L1 low or negative tumours can also respond to these inhibitors [30]. Nevertheless, PD-L1 expression remains the primary biomarker in lung cancer treatment, influencing drug approval and reimbursement, typically measured via IHC as the tumour proportion score [31]. Recently the usefulness of the combined positive score has been investigated, but not validated. Other biomarkers like tumour mutational burden (TMB) and microsatellite instability have been developed to complement PD-L1 expression in predicting treatment efficacy. Currently these markers require standardisation.
According to the ESMO guidelines, ICIs are recommended as first-line treatments for stage IV NSCLCs without oncogenic drivers and extensive stage SCLCs, either alone or in combination with platinum-based chemotherapy [4]. They are also recommended as consolidation therapy after chemoradiotherapy for stage III lung cancer [32]. Additionally, their use as (neo)adjuvant treatment for earlier stages of lung cancer is expanding, supported by promising trial outcomes [33, 34].
ICIs are administered intravenously every 3–6 weeks. The unique mechanism of action of ICIs can lead to immune-related adverse events (irAEs), by stimulating the immune system and disrupting self-regulation. These events can affect various organs, with combinations of CTLA4 and PD-L1 inhibitors causing more irAEs. Commonly, mild side-effects include skin rashes and autoimmune thyroiditis, necessitating thyroid hormone replacement. More severe irAEs include colitis, hepatitis, nephritis, pneumonitis and myocarditis, which might require treatment interruption and systemic corticosteroids. In severe or corticosteroid-resistant cases, biologic disease-modifying drugs may be considered [35].
Artificial intelligence and future directions for lung cancer pathology and molecular biomarkers
Recent developments in artificial intelligence (AI) have opened new horizons in the fight against lung cancer. With the ability to analyse large datasets and swiftly identify various patterns, AI has made significant advances in lung cancer screening, diagnosis, staging and treatment. Although there are errors reported in some studies, superior detection results to humans in early diagnosis and screening, especially for small lesions, are reported [36–38]. Significant breakthroughs in lung cancer are expected in the future by combining AI applications such as machine learning, deep learning, convolutional neural networks (CNNs), radiomics, pathomics and multi-omics [39–41].
Machine learning is a branch of AI that allows computers to learn from data without being explicitly programmed for specific tasks. In the context of lung cancer pathology and molecular biomarkers, machine learning can help in identifying patterns from vast amounts of pathology slides and biological samples and combine them with medical data, such as patient health records, to predict disease progression, clinical outcomes and optimise treatment plans [42].
Deep learning is a more advanced subset of machine learning that uses structures called neural networks, inspired by the human brain, to process data in complex ways. It is particularly good at handling unstructured data like images and text. For lung cancer pathology and biomarkers, deep learning can analyse pathology or NGS images and combine them with radiology images (e.g. CT scans) to inform treatment plans. An important application is the differentiation between benign and malignant lesions with high accuracy [42].
CNNs can also be applied in lung cancer pathology and molecular biomarkers testing. A CNN is a type of deep learning algorithm specifically designed to process pixel data from images. In lung cancer, CNNs can be trained to recognise patterns associated with the disease in pathology slides and imaging, improving early detection and helping doctors understand disease progression better [42].
AI algorithms, especially deep learning models, have shown remarkable efficiency in analysing diagnostic images and in the future, AI-based radiology imaging could ideally be combined with pathology imaging [43]. AI has been shown to be highly accurate in the identification of colon, prostate, lung, breast and kidney tumours from histopathology slides [44, 45]. AI models have been developed that can classify ADC and SCC of the lung and differentiate lung cancer from healthy tissue [46, 47]. A retrospective study used a VGG-16 neural network to extract deep features from CT images of 157 NSCLC patients and classify them with fully connected layers as either ADC or squamous cell lung cancer. They also independently evaluated the extracted features using three machine learning classification models. The results showed that all models were able to classify tumour histology, and the neural network achieved the highest performance with an area under the curve of 0.751 [48]. Combining deep learning applied to pretreatment CTs with TNM staging is expected to improve prognosis and risk stratification in lung cancer patients [49].
Better survival outcomes are expected with personalisation of lung cancer treatment. AI is expected to make a significant contribution to this improvement with the specialised data it offers for lung cancer treatment and management. AI applications can predict drug resistance in patients undergoing targeted therapy and immunotherapy. TMB is an important determinant of the efficacy of ICIs [50]. Repeated tumour sampling and invasive procedures limit the clinical utility of TMB. Therefore, a noninvasive approach to TMB calculation needs to be developed. In a study by He et al. [51], high TMB was distinguished from low TMB by combining deep learning technology and CT images. These may influence decisions regarding the use of ICIs in patients with advanced NSCLC [51].
Despite these advances, AI in lung cancer faces challenges. Since patient data is sensitive, privacy and security are important concerns. Furthermore, the risk of bias in AI algorithms due to unrepresentative training data poses a challenge to the provision of equitable healthcare. It is recommended that the entire process of the AI sector should be regulated and reviewed to include ethical rules [52, 53].
As a result, the emergence of AI in lung cancer treatment has been the centrepiece of a change process to improve the sensitivity of screening methods, the accuracy of diagnoses, and the effectiveness of treatment planning. In the future, the collection and integration of clinical, radiological and pathological data for the diagnosis and treatment of lung cancer will provide more accurate and comprehensive results. Further advancement of algorithms such as deep learning, machine learning and CNNs will play an essential role in early diagnosis of lung cancer, staging of the disease, and prediction of response to treatment. Genetic and molecular data integration will lead to better treatment plans and fewer side-effects. AI-based tools need to be tested and validated in clinical settings to prove reliability and effectiveness. In addition, studies need to be carried out to resolve legal and ethical handicaps. These developments will usher in a groundbreaking era in oncology, where innovative technology seamlessly integrates with medical expertise, offering unique opportunities for better patient outcomes and personalised care strategies.
Conclusion
Lung cancer diagnosis, treatment and management are undergoing significant transformations with advances in diagnostic pathology, molecular biomarkers, targeted therapies, immunotherapy, and the emerging field of AI. As we navigate the intricacies of the molecular basis of lung cancer, the importance of early detection, accurate pathological diagnosis and identification of genetic alterations is becoming increasingly apparent. It offers a glimmer of hope for improving patient outcomes. The strategic application of ICIs and the integration of new diagnostic techniques such as NGS and multiplex gene panel testing underline a shift towards personalised medicine, where treatments are tailored to the unique genetic profile of each patient's tumour. By embracing these innovations, fostering interdisciplinary collaboration and prioritising patient-centred care, we stand on the brink of a new frontier in oncology. With these advancements, we can significantly reduce the negative impact of lung cancer on lives and ultimately work towards a cure.
Footnotes
Conflict of interest: The authors have nothing to disclose.
References
- 1.Leiter A, Veluswamy RR, Wisnivesky JP. The global burden of lung cancer:mcurrent status and future trends. Nat Rev Clin Oncol 2023; 20: 624–639. doi: 10.1038/s41571-023-00798-3 [DOI] [PubMed] [Google Scholar]
- 2.Araghi M, Mannani R, Heidarnejad Maleki A,et al. Recent advances in non-small cell lung cancer targeted therapy; an update review. Cancer Cell Int 2023; 23: 162. doi: 10.1186/s12935-023-02990-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Detterbeck FC, Boffa DJ, Kim AW, et al. The eighth edition lung cancer stage classification. Chest 2017; 151: 193–203. doi: 10.1016/j.chest.2016.10.010 [DOI] [PubMed] [Google Scholar]
- 4.Hendriks LE, Kerr KM, Menis J,et al. Oncogene-addicted metastatic non-small-cell lung cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol 2023; 34: 339–357. doi: 10.1016/j.annonc.2022.12.009 [DOI] [PubMed] [Google Scholar]
- 5.International Agency for Research on Cancer , World Health Organization . WHO classification of tumours online. https://tumourclassification.iarc.who.int/chaptercontent/35/2
- 6.Nicholson AG, Tsao MS, Beasley MB, et al. The 2021 WHO classification of lung tumors: impact of advances since 2015. J Thorac Oncol 2022; 17: 362–387. doi: 10.1016/j.jtho.2021.11.003 [DOI] [PubMed] [Google Scholar]
- 7.Selves J, Long-Mira E, Mathieu MC,et al. Immunohistochemistry for diagnosis of metastatic carcinomas of unknown primary site. Cancers (Basel) 2018; 10: 108. doi: 10.3390/cancers10040108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inamura K. Clinicopathological characteristics and mutations driving development of early lung adenocarcinoma: tumor initiation and progression. Int J Mol Sci 2018; 19: 1259. doi: 10.3390/ijms19041259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018; 553: 446–454. doi: 10.1038/nature25183 [DOI] [PubMed] [Google Scholar]
- 10.Lee YT, Tan YJ, Oon CE. Molecular targeted therapy: treating cancer with specificity. Eur J Pharmacol 2018; 834: 188–196. doi: 10.1016/j.ejphar.2018.07.034 [DOI] [PubMed] [Google Scholar]
- 11.Malapelle U, Sirera R, Jantus-Lewintre E,et al. Profile of the Roche cobas® EGFR mutation test v2 for non-small cell lung cancer. Expert Rev Mol Diagn 2017; 17: 209–215. doi: 10.1080/14737159.2017.1288568 [DOI] [PubMed] [Google Scholar]
- 12.Pecciarini L, Brunetto E, Grassini G,et al. Gene fusion detection in NSCLC routine clinical practice: targeted-NGS or FISH? Cells 2023; 12: 1135. doi: 10.3390/cells12081135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inamura K. Update on immunohistochemistry for the diagnosis of lung cancer. Cancers (Basel) 2018; 10: 72. doi: 10.3390/cancers10030072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drilon A, Wang L, Arcila ME, et al. Broad, hybrid capture-based next-generation sequencing identifies actionable genomic alterations in lung adenocarcinomas otherwise negative for such alterations by other genomic testing approaches. Clin Cancer Res 2015; 21: 3631–3639. doi: 10.1158/1078-0432.CCR-14-2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruno R, Fontanini G. Next generation sequencing for gene fusion analysis in lung cancer: a literature review. Diagnostics (Basel) 2020; 10: 521. doi: 10.3390/diagnostics10080521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sholl LM. Molecular diagnostics in non-small cell lung carcinoma. Semin Respir Crit Care Med 2020; 41: 386–399. doi: 10.1055/s-0039-3399564 [DOI] [PubMed] [Google Scholar]
- 17.NCCN Guidelines . Non-Small Cell Lung Cancer, Version 1.2024. Plymouth Meeting, National Comprehensive Cancer Network, 2024. https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1450 [Google Scholar]
- 18.Bar J, Peled N, Schokrpur S,et al. Uncommon EGFR mutations: international case series on efficacy of osimertinib in real-life practice in First-LiNe Setting (UNICORN). J Thorac Oncol 2023; 18: 169–180. doi: 10.1016/j.jtho.2022.10.004 [DOI] [PubMed] [Google Scholar]
- 19.Mosele F, Remon J, Mateo J,et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol 2020; 31: 1491–1505. doi: 10.1016/j.annonc.2020.07.014 [DOI] [PubMed] [Google Scholar]
- 20.Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med 2018; 378: 113–125. doi: 10.1056/NEJMoa1713137 [DOI] [PubMed] [Google Scholar]
- 21.Hong DS, Fakih MG, Strickler JH,et al. KRAS G12C Inhibition with Sotorasib in Advanced Solid Tumors. N Engl J Med 2020; 383: 1207–1217. doi: 10.1056/NEJMoa1917239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Planchard D, Smit EF, Groen HJM,et al. Dabrafenib plus trametinib in patients with previously untreated BRAFV600E-mutant metastatic non- small-cell lung cancer: an open-label, phase 2 trial. Lancet Oncol 2017; 18: 1307–1316. doi: 10.1016/S1470-2045(17)30679-4 [DOI] [PubMed] [Google Scholar]
- 23.Ou SI, Hong JL, Christopoulos P,et al. Distribution and detectability of EGFR exon 20 insertion variants in NSCLC. J Thorac Oncol 2023; 18: 744–754. doi: 10.1016/j.jtho.2023.01.086 [DOI] [PubMed] [Google Scholar]
- 24.Vathiotis IA, Charpidou A, Gavrielatou N, et al. HER2 aberrations in non-small cell lung cancer: from pathophysiology to targeted therapy. Pharmaceuticals (Basel) 2021; 14: 1300. doi: 10.3390/ph14121300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerr KM, Bibeau F, Thunnissen E,et al. The evolving landscape of biomarker testing for non-small cell lung cancer in Europe. Lung Cancer 2021; 154: 161–175. doi: 10.1016/j.lungcan.2021.02.026 [DOI] [PubMed] [Google Scholar]
- 26.Fujino T, Suda K, Mitsudomi T. Lung Cancer with METexon 14 skipping mutation: genetic feature, current treatments, and future challenges. Lung Cancer (Auckl) 2021; 12: 35–50. doi: 10.2147/LCTT.S269307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harada G, Yang SR, Cocco E, et al. Rare molecular subtypes of lung cancer. Nat Rev Clin Oncol 2023; 20: 229–249. doi: 10.1038/s41571-023-00733-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.European Medicines Agency . EMA/CHMP/606649/2015 – Summary of opinion (post authorisation) on Opdivo (nivolumab). Date last accessed: December 2023. Date last updated 24September 2015. www.ema.europa.eu/en/documents/smop/chmp-post-authorisation-summary-positive-opinion-opdivo_en.pdf-0
- 29.Shiravand Y, Khodadadi F, Kashani S,et al. Immune checkpoint inhibitors in cancer therapy. Curr Oncol 2022; 29: 3044–3060. doi: 10.3390/curroncol29050247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X, Guo C, Tou F,et al. Association of PD-L1 expression status with the efficacy of PD-1/PD-L1 inhibitors and overall survival in solid tumours: a systematic review and meta-analysis. Int J Cancer 2020; 147: 116–127. doi: 10.1002/ijc.32744 [DOI] [PubMed] [Google Scholar]
- 31.Vranic S, Gatalica Z. PD-L1 testing by immunohistochemistry in immuno-oncology. Biomol Biomed 2023; 23: 15–25. doi: 10.17305/bjbms.2022.7953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Antonia S, Villegas A, Daniel D,et al. Durvalumab after chemoradiotherapy in stage III non-small-cell lung cancer. N Engl J Med 2017; 377: 1919–1929. doi: 10.1056/NEJMoa1709937 [DOI] [PubMed] [Google Scholar]
- 33.Forde PM, Spicer J, Lu S,et al. Neoadjuvant nivolumab plus chemotherapy in resectable lung cancer. N Engl J Med 2022; 386: 1973–1985. doi: 10.1056/NEJMoa2202170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Brien M, Paz-Ares L, Marreaud S,et al. Pembrolizumab versus placebo as adjuvant therapy for completely resected stage IB-IIIA non-small-cell lung cancer (PEARLS/KEYNOTE-091): an interim analysis of a randomised triple-blind, phase 3 trial. Lancet Oncol 2022; 23: 1274–1286. doi: 10.1016/S1470-2045(22)00518-6 [DOI] [PubMed] [Google Scholar]
- 35.Haanen J, Obeid M, Spain L,et al. Management of toxicities from immunotherapy: ESMO clinical practice guideline for diagnosis, treatment and follow-up. Ann Oncol 2022; 33: 1217–1238. doi: 10.1016/j.annonc.2022.10.001 [DOI] [PubMed] [Google Scholar]
- 36.Gandhi Z, Gurram P, Amgai B, et al. Artificial intelligence and lung cancer: impact on improving patient outcomes. Cancers (Basel) 2023; 15: 5236. doi: 10.3390/cancers15215236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cellina M, Cacioppa LM, Cè M, et al. Artificial intelligence in lung cancer screening: the future is now. Cancers (Basel) 2023; 15: 4344. doi: 10.3390/cancers15174344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nam JG, Hwang EJ, Kim J, et al. AI improves nodule detection on chest radiographs in a health screening population: a randomized controlled trial. Radiology 2023; 307: e221894. doi: 10.1148/radiol.221894 [DOI] [PubMed] [Google Scholar]
- 39.Zhao Z, Yin W, Peng X, et al. A machine-learning approach to developing a predictive signature based on transcriptome profiling of ground-glass opacities for accurate classification and exploring the immune microenvironment of early-stage LUAD. Front Immunol 2022; 13: 872387. doi: 10.3389/fimmu.2022.872387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Chen P, Su M, et al. Integrative modeling of multiomics data for predicting tumor mutation burden in patients with lung cancer. Biomed Res Int 2022; 2022: 2698190. doi: 10.1155/2022/2698190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang S, Cao X, Wang Y, et al. Metabolomics analysis and diagnosis of lung cancer: insights from diverse sample types. Int J Med Sci 2024; 21: 234–252. doi: 10.7150/ijms.85704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taye MM. Understanding of machine learning with deep learning: architectures, workflow, applications and future directions. Computers 2023; 12: 91. Doi: 10.3390/computers12050091 [DOI] [Google Scholar]
- 43.Thong LT, Chou HS, Chew HSJ, et al. Diagnostic test accuracy of artificial intelligence-based imaging for lung cancer screening: a systematic review and meta-analysis. Lung Cancer 2023; 176: 4–13. doi: 10.1016/j.lungcan.2022.12.002 [DOI] [PubMed] [Google Scholar]
- 44.Liu M, Wu J, Wang N, et al. The value of artificial intelligence in the diagnosis of lung cancer: a systematic review and meta-analysis. PLoS ONE 2023; 18: e0273445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta S, Gupta MK, Shabaz M, et al. Deep learning techniques for cancer classification using microarray gene expression data. Front Physiol 2022; 13: 952709. doi: 10.3389/fphys.2022.952709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bębas E, Borowska M, Derlatka M, et al. Machine-learning-based classification of the histological subtype of non-small-cell lung cancer using MRI texture analysis. Biomed Signal Process Control 2021; 66: 102446. doi: 10.1016/j.bspc.2021.102446 [DOI] [Google Scholar]
- 47.Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 2018; 24: 1559–1567. doi: 10.1038/s41591-018-0177-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torres FS, Akbar S, Raman S, et al. End-to-end non–small-cell lung cancer prognostication using deep learning applied to pretreatment computed tomography, JCO Clin Cancer Inform 2021; 5: 1141–1150. doi: 10.1200/CCI.21.00096 [DOI] [PubMed] [Google Scholar]
- 49.Chaunzwa TL, Christiani DC, Lanuti Met al. Using deep-learning radiomics to predict lung cancer histology. J Clin Oncol 2018; 36: Suppl., 8545. doi: 10.1200/JCO.2018.36.15_suppl.8545 [DOI] [Google Scholar]
- 50.Rizvi H, Sanchez-Vega F, La K, et al. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J Clin Oncol 2018; 36: 633– 641. doi: 10.1200/JCO.2017.75.3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He B, Dong D, She Y, et al. Predicting response to immunotherapy in advanced non-small-cell lung cancer using tumor mutational burden radiomic biomarker. J Immunother Cancer 2020; 8: e000550. doi: 10.1136/jitc-2020-000550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murdoch B. Privacy and artificial intelligence: challenges for protecting health information in a new era. BMC Med Ethics 2021; 22: 122. doi: 10.1186/s12910-021-00687-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J, Zhang ZM. Ethics and governance of trustworthy medical artificial intelligence. BMC Med Inform Decis Mak 2023; 23: 7. doi: 10.1186/s12911-023-02103-9 [DOI] [PMC free article] [PubMed] [Google Scholar]