Abstract
The vaccinia virus WR A5L open reading frame (corresponding to open reading frame A4L in vaccinia virus Copenhagen) encodes an immunodominant late protein found in the core of the vaccinia virion. To investigate the role of this protein in vaccinia virus replication, we have constructed a recombinant virus, vA5Li, in which the endogenous gene has been deleted and an inducible copy of the A5 gene dependent on isopropyl-β-d-thiogalactopyranoside (IPTG) for expression has been inserted into the genome. In the absence of inducer, the yield of infectious virus was dramatically reduced. However, DNA synthesis and processing, viral protein expression (except for A5), and early stages in virion formation were indistinguishable from the analogous steps in a normal infection. Electron microscopy revealed that the major vaccinia virus structural form present in cells infected with vA5Li in the absence of inducer was immature virions. Viral particles were purified from vA5Li-infected cells in the presence and absence of inducer. Both particles contained viral DNA and the full complement of viral proteins, except for A5, which was missing from particles prepared in the absence of inducer. The particles prepared in the presence of IPTG were more infectious than those prepared in its absence. The A5 protein appears to be required for the immature virion to form the brick-shaped intracellular mature virion.
Vaccinia virus, the archetypal member of the poxvirus family, is a large DNA virus which encodes approximately 185 proteins (30). The cascade of temporally related gene expression, the replication of the genome, and the assembly and maturation of the infectious virion all occur in the cytoplasm of the infected cell. The earliest observed specific structures generated via infection were regions of granular electron-dense material, or viral factories, enriched in vaccinia virus DNA, which are thought to be the site of viral DNA replication and transcription (4, 18, 21, 30). The initial step in virion morphogenesis is the accumulation of crescent-shaped membranes at the cytoplasmic sites of viral replication (7). These membranes encircle the granular electron-dense material to form spherical immature virion (IV) particles. The spherical IV particles develop an internal core surrounding the DNA and mature into brick-shaped intracellular mature virion (IMV) particles. This maturation coincides with the cleavage of several core proteins (24, 25, 32, 44, 48–50). Three basic approaches have been used to delineate the molecular details of vaccinia virus morphogenesis: the characterization of temperature-sensitive mutants with defects in assembly, the use of drugs which inhibit the process and the characterization of mutant viruses resistant to such drugs, and the construction of inducer-dependent conditional mutants in which the expression of a specified gene can be controlled. By using these approaches, proteins directly or indirectly involved in the early stages of morphogenesis (36, 37, 46, 47, 51, 54, 55), in the transition from IV to IMV particles (5, 13, 22, 35, 57), and in the process leading to the formation of extracellular enveloped virus (3, 9, 12, 34, 38, 39, 42, 53) have been identified.
A vaccinia virus protein, designated p39, was shown to be expressed late in infection, to elicit a strong persistent antigenic response in humans, and to correspond to the product of the A4L gene in vaccinia virus Copenhagen or A5L in vaccinia virus WR (8, 27). The protein can be detected in the virosomes, the regions of DNA replication, and becomes encapsidated in the virion core during maturation as shown by immunolabeling and electron microscopy (6). Vaccinia virions contain a nuclease activity (15, 26, 28, 40, 41) which was shown to copurify with the A5 protein over chromatographic columns (29a). The colocalization of A5 with replicated DNA in the virosomes and its copurification with nuclease activity suggested a possible role for A5 during DNA processing or packaging. To determine the role A5 plays in the replication of vaccinia virus, we have taken an in vivo genetic approach by constructing and characterizing a conditional lethal mutant of vaccinia virus with an inducible A5L gene.
MATERIALS AND METHODS
Cells and viruses.
BSC-1 cells (ATCC CCL6) were grown in Eagle’s minimal essential medium (Gibco or Cellgro) supplemented with 10% fetal bovine serum (HyClone). Wild-type and recombinant vaccinia viruses were derived from the WR strain (ATCC Vr119). To prepare stocks of vA5Li, monolayers of BSC-1 cells were infected with 1 PFU/cell in the presence of 50 μM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested 72 h postinfection, and virus stocks were prepared by three freeze-thaw cycles. Partially purified virus, prepared by centrifugation of infected cytoplasm through a 36% sucrose cushion (11), was routinely used to infect cells for experiments.
Plasmid construction.
A copy of the A5L open reading frame was generated by PCR with vaccinia virus WR DNA and the oligonucleotide primers 5′-GGGGGGCCCATGGACTTCTTTAACAAGTTCTCACAG (the NcoI site is underlined and the initiation codon is shown in boldface) and 5′-GGGGGGCTCGAGAGCGTGATTTTAATATCC (the XhoI site is underlined). The PCR product was digested with NcoI and XhoI and inserted into the plasmid pTM-1 (31), generating pTM/A5. The NcoI-XhoI fragment encoding for the A5L open reading frame from pTM/A5 was excised and used to replace the NcoI-XhoI fragment containing the copy of β-galactosidase present in pT7lacZ (1), generating pVOTE/A5. The nucleotide sequence of the open reading frame corresponding to A5L in pVOTE/A5 was determined by using a ABI PRISM Dye Terminator Cycle Sequencing kit (Perkin-Elmer) and shown to be identical to the nucleotide sequence of the A5 open reading frame in the vaccinia virus WR genome.
The plasmid used to delete the endogenous copy of A5L was constructed in three steps. First, an 834-bp fragment preceding the gene was generated by PCR using vaccinia virus WR DNA and the oligonucleotide primers 5′-GGGGGGGCATGCTAAGATTGGATATTAAAATCACGC and 5′-GGGGGGAAGCTTCTGGATTAGGCTATAGGTGTC containing SphI and HindIII restriction sites (underlined), respectively. The PCR product was ligated into an intermediate vector by using the TA cloning kit (Invitrogen), excised with SphI and HindIII, and inserted into pGEM-3, generating pA5delR. Second, the neomycin resistance gene (neo) was excised from pGEM-NEO (20) by using SalI and BamHI and inserted into pA5delR, generating pA5delR/neo. Finally, an 1,188-bp fragment derived from the region following the A5 gene was generated by PCR using oligonucleotide 5′-GGGGGGGAGCTCGAGCGTCGACTACAG AGAACG and 5′-GGGGGGGGATCCAAGGCTTTAAAATTGAATTGC containing SacI and BamHI sites (underlined), respectively. The PCR product was ligated into an intermediate vector by using the TA cloning kit (Invitrogen), excised with SacI and BamHI, and inserted into pA5delR/neo, generating pA5del/neo.
Construction of recombinant viruses.
The vA5Li recombinant virus was constructed in two steps. First, the intermediate virus containing two copies of the A5L gene was generated by homologous recombination between vT7lacOI (1) and pVOTE/A5. Approximately 106 BSC-1 cells were infected with vT7lacOI at a multiplicity of infection (MOI) of 0.05 PFU/cell for 2 h at 37°C, rinsed with Opti-Mem (Life Technologies), and transfected with 5 μg of pVOTE/A5 by using Lipofectamine (Life Technologies) as suggested by the manufacturer. After 4 h, the volume was increased fivefold with complete medium, and after 48 h, the cells were harvested and the virus was released by three freeze-thaw cycles. The vA5Lint was selected by five rounds of plaque purification in BSC-1 cells in the presence of mycophenolic acid, xanthine, and hypoxanthine (11) under an overlay of 0.5% methylcellulose (M-0387; Sigma) in complete medium. Small virus stocks were prepared, and the recombinational changes were confirmed by PCR and Southern blotting. The structure of vA5Lint was also confirmed by infecting BSC-1 cells at a low MOI. Upon infection, the double-crossover vA5Lint produced large multinucleated cells typical of a hemagglutinin gene deletion phenotype.
The recombinant virus vA5Li, containing an inducible copy of A5L, was generated by homologous recombination between vA5Lint and pA5del/neo. The procedure was the same as that described above except that selective pressure was maintained by incubation in the presence of G418 (1 mg/ml; Life Technologies). Plaques were identified by staining with neutral red, and the genomic structure was confirmed by PCR and Southern blot analysis.
One-step virus growth.
BSC-1 cells were infected with 1 PFU of virus/cell for 1 h at 37°C. The incubations were continued with IPTG where applicable, and cells were harvested at various times. Virus was released by three freeze-thaw cycles and stored at −80°C. Virus titers were determined by plaque assays (11) under 0.5% methylcellulose in complete medium in the presence of 25 μM IPTG.
Western blot analysis.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Novex), transferred to Protran (Schleicher & Schuell), and blocked with 5% nonfat dry milk in phosphate-buffered saline. The filters were probed with antibodies specific to vaccinia virus proteins (a gift of B. Moss) and visualized by subsequent incubations with an anti-rabbit immunoglobulin G conjugated with alkaline phosphatase and Western Blue reagent (Promega).
SDS-PAGE analysis of [35S]methionine-labeled polypeptides.
BSC-1 cells were infected at 5 PFU/cell in the presence or absence of IPTG. At the indicated times after infection, the cells were incubated with methionine-free medium for 15 min, labeled for 30 min with 100 μCi of [35S]methionine/ml, harvested in 1% SDS–50 mM Tris-HCl (pH 8.0), and incubated with PAGE loading buffer prior to SDS-PAGE. After electrophoresis, the gels were dried and visualized by exposure on Bio-Max (Kodak) film.
Analysis of viral DNA.
Monolayers were infected with 1 PFU of vA5Lint or vA5Li/ml in the presence or absence of 50 μM IPTG. After 24 h, the cells were harvested and resuspended in cell suspension buffer (10 mM Tris HCl [pH 7.2], 20 mM NaCl, 50 mM EDTA) at 107 cells/ml. An equal volume of 2% CleanCut agarose (Bio-Rad), preincubated at 50°C, was added, and the suspension was formed into 100-μl plugs. After solidification at 4°C, the plugs were treated with proteinase K as previously described (29). The equilibrated agarose plugs were treated by electrophoresis on a Bio-Rad clamped homogeneous electric field DRII apparatus for 22 h at 6 V/cm with a switching time of 70 s. Agarose gels were stained with ethidium bromide.
Purification of virus.
Monolayers of BSC-1 cells were infected at 1 PFU/cell with vA5Lint or vA5Li in the presence of 50 μM IPTG where indicated. The cells were incubated at 37°C for 72 h and harvested, and viral particles were purified as described for vaccinia virus (10). The infecting stock for most experiments was the pellet fraction obtained by centrifugation through a 36% sucrose cushion. For electron microscopy, material banded in two sequential 24 to 40% sucrose gradients was used.
Electron microscopy.
BSC-1 cells were infected with vA5Lint or vA5Li at an MOI of 3 PFU/cell. After 24 h, the cells were fixed in 2% glutaraldehyde in 0.1 M Na cacodylate (pH 7.4) buffer. Purified viral particles were incubated with an equal volume of 4% glutaraldehyde in 0.2 M Na cacodylate (pH 7.4) buffer and collected by centrifugation at 14,000 × g in a microcentrifuge. Samples were embedded in Embed-812 (Electron Microscopy Sciences, Fort Washington, Pa.) as previously described (54). Ultrathin sections of infected cells and virions were viewed with a Phillips CM100 electron microscope.
RESULTS
Construction of a recombinant vaccinia virus with an inducible A5L gene.
Attempts to eliminate the vaccinia virus WR A5L open reading frame (corresponding to A4L in vaccinia virus Copenhagen) by the insertion of color markers or antibiotic resistance genes were unsuccessful (data not shown), implying that A5 is essential for viral replication in BSC-1 cells. The inducible bacterial system for the Escherichia coli lac repressor has been adapted for the regulation of genes in vaccinia virus (14, 38, 56). Recently, an improved version of the system in which the expression of both T7pol and a target gene behind a T7 promoter are controlled by lacO has been described (52, 54). Constitutive expression of the lacI repressor leads to stringent inhibition of both the T7pol and target genes. Infections in the presence of inducer IPTG result in the derepression of T7 RNA polymerase, which transcribes the target gene.
The recombinant virus designed to regulate the expression of the A5L gene was constructed in two steps, starting with the parent virus (vT7lacOI), which contains the lacI and T7 RNA polymerase genes. Initially, a second copy of the A5L gene was inserted by homologous recombination between vT7lacOI and pVOTE/A5, a plasmid with a copy of A5L under the control of the T7 promoter containing lacO sequences, the E. coli gpt gene (for mycophenolic acid selection), and flanking sequences derived from A56R, into the nonessential A56R gene (hemagglutinin locus). The resulting virus, vA5Lint, contains two copies of the A5L gene, the endogenous copy and an inducible copy under the control of the T7 promoter. The structure of the viral genome was confirmed by Southern blot analysis and PCR (data not shown). Also, the vA5Lint was shown by Western blot analysis to express higher levels of A5 than vT7lacOI or vaccinia virus WR when infections were performed in the presence of 50 μM IPTG (data not shown). In the second step, the endogenous copy of A5L in vA5Lint was deleted by insertion of the drug resistance gene neo by homologous recombination between vA5Lint and pA5del/neo, a plasmid containing the neo gene (for selection) flanked by sequences proximal to the A5L open reading frame. Recombinant viruses were selected by plaque isolation in BSC-1 cells treated with G418 and, after multiple rounds, individual plaques were selected and expanded by infecting BSC-1 cells. The genomes of candidate viruses were then analyzed by Southern blot analysis and PCR on isolated DNA (data not shown). A drug-resistant virus, containing only the inducible copy of vaccinia virus WR A5L, and with the genomic structure outlined in Fig. 1, was isolated and designated vA5Li.
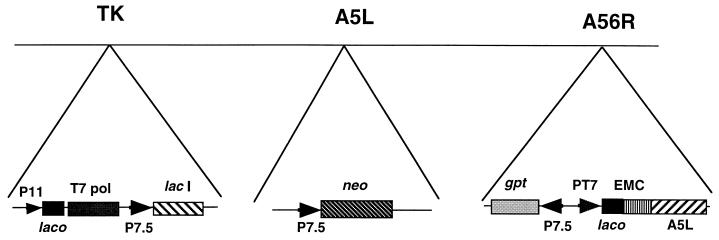
FIG. 1.
Inducible expression of the A5L gene. Portions of the genome for vA5Li are illustrated. DNA insertions have been made in the vaccinia virus thymidine kinase (TK), the A5L, and the hemagglutinin (A56R) genes. Abbreviations: P11 and P7.5, vaccinia virus promoters; PT7 and T7 pol, a bacteriophage T7 promoter and the bacterial T7 RNA polymerase gene, respectively; EMC, a cDNA copy of the untranslated leader of encephalomyocarditis virus which provides cap-independent translation; lacI and lacO, the E. coli lac repressor gene and the lac operator, respectively; neo and gpt, antibiotic selection genes.
Replication of mutant viruses.
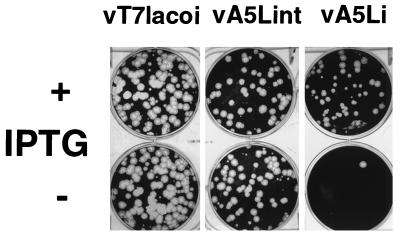
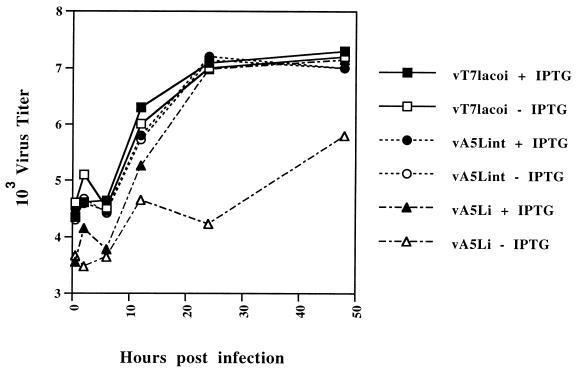
Virus replication was measured by plaque assay with one-step growth conditions in the presence or absence of IPTG. Each virus was used to infect monolayers of BSC-1 cells at an MOI of 1 in the presence or absence of 50 μM IPTG, harvested after 24 h, and analyzed by plaque assay on BSC-1 cells (Fig. 2). The yield of virus was independent of inducer for vT7lacOI and vA5Lint, whereas the yield of vA5Li was directly dependent on the presence of IPTG. The rates of growth for each virus were also compared (Fig. 3), and all viruses except vA5Li that were incubated in the absence of IPTG grew at the same rate. Growth profiles identical to those observed for vA5Lint and vT7lacOI were observed for vaccinia virus WR infections (data not shown). Primary infection of BSC-1 cells with vA5Li in the presence of IPTG produced plaques which were indistinguishable in size and morphology from the vaccinia virus wild type and vA5Lint, while infections of BSC-1 cells with vA5Li in the absence of IPTG produced tiny pinpoint plaques. The yield of infectious virus 24 h after infection from cells infected with vA5Li was reproducibly about 100-fold lower than that from infections with vA5Li performed in the presence of 50 μM IPTG. A limited infection from the input vA5Li, especially at 48 h, may result from the A5 protein present in the input virus stocks grown in the presence of IPTG or from a low level of expression by vA5Li, even in the absence of IPTG.
FIG. 2.
Inducer dependence of virus production. Monolayers of BSC-1 cells were infected with viruses vT7lacOI, vA5Lint, and vA5Li at an MOI of 1 PFU per cell in the presence (+) or absence (−) of 50 μM IPTG. After 24 h, the monolayers were harvested, the virus was released by three cycles of freezing-thawing, and BSC-1 monolayers were infected in the presence of 25 μM IPTG, incubated for 4 days at 37°C, stained with crystal violet, and photographed.
FIG. 3.
Inducer-dependent formation of infectious virus. BSC-1 monolayers were infected with the viruses vT7lacOI, vA5Lint, or vA5Li at an MOI of 1 PFU per cell in the presence (+) or absence (−) of 50 μM IPTG. At various time points up to 48 h, the cells were harvested and virus titers were determined by plaque assay on BSC-1 cells.
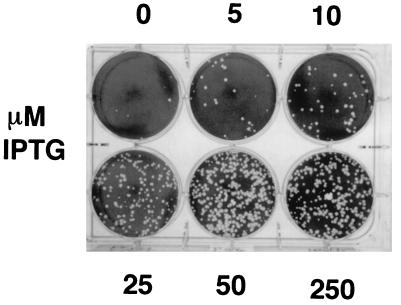
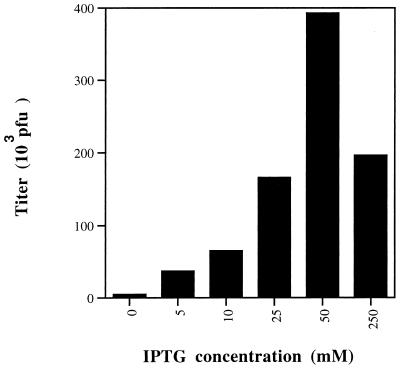
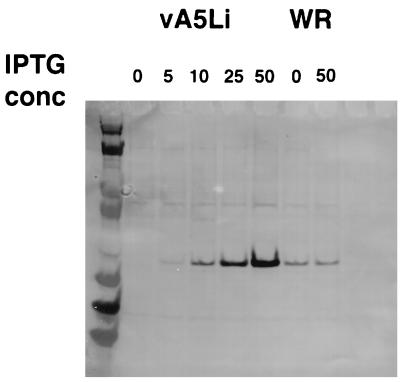
This T7 RNA polymerase inducible system was designed to scale the level of expression of the target gene to the concentration of IPTG. This property allows one to express the target gene at the optimum level, as overexpression of the target gene is sometimes deleterious to viral growth (5). The relationship between inducer concentration and vA5Li virus yield was determined by plaque assay of infected cells as illustrated in Fig. 4. In each infection, BSC-1 cells were infected at an MOI of 1 PFU/cell, harvested after 24 h, and analyzed by plaque assay on BSC-1 cells. The yield of virus was directly proportional to IPTG concentration up to 50 μM. The absolute number of plaques at 250 μM IPTG was approximately half that observed at 50 μM IPTG (Fig. 5). The levels of expression of A5L in cells infected with vA5Li at different IPTG concentrations were also compared by Western blot analysis (Fig. 6). The accumulation of A5 protein from cells infected with vA5Li was directly proportional to the concentration of IPTG. As expected, the production of A5 protein in cells infected by vaccinia virus WR was insensitive to IPTG concentration. The amount of A5 protein synthesized in vA5Li-infected cells at approximately 10 μM IPTG was equivalent to the amount synthesized during an infection with WR. Upon overexposure of the Western blots, a low level of A5 (less than 10% of a WR infection) was noted in the absence of IPTG.
FIG. 4.
Inducer concentration and virus production. Monolayers of BSC-1 cells were infected with virus vA5Li at an MOI of 1 PFU per cell in the presence of 0, 5, 10, 25, 50, or 250 μM IPTG. After 24 h, the monolayers were harvested, the virus was released by three cycles of freezing-thawing, and BSC-1 monolayers were infected in the presence of 25 μM IPTG, incubated for 2 days at 37°C, stained with crystal violet, and photographed.
FIG. 5.
Inducer dependence of virus yield. Monolayers of BSC-1 cells were infected with virus vA5Li at an MOI of 1 PFU per cell in the presence of 0, 5, 10, 25, 50, or 250 μM IPTG. After 24 h, the monolayers were harvested, the virus was released by three cycles of freezing-thawing, and the titers were determined by plaque assay on BSC-1 monolayers infected in the presence of 25 μM IPTG.
FIG. 6.
Synthesis of A5L depends on IPTG concentration. BSC-1 cells were infected with either vA5Li at an MOI of 1 PFU/cell in the presence of 0, 5, 10, 25, or 50 μM IPTG or vaccinia virus WR at an MOI of 1 PFU/cell in the presence of 0 or 50 μM IPTG. After 24 h, the cells were harvested by scraping, concentrated by low-speed centrifugation, and lysed in RIP lysis buffer (100 mM Tris HCl [pH 8.0], 100 mM NaCl, 0.5% Nonidet P-40). Samples were analyzed by Western blotting by using a rabbit antibody against A5L supplied by Bernard Moss followed by incubation with an anti-rabbit antibody–alkaline phosphatase conjugate (Promega) and visualized by using Western Blue Stabilized substrate for alkaline phosphatase (Promega).
The highest level of vA5Li virus production occurred at an IPTG concentration of 50 μM. At this inducer concentration, there appeared to be approximately 10 times the A5 normally found in a vaccinia virus WR infection. Since the protein coding sequence for A5 was identical to the sequence of WR, the optimal virus production at the elevated level of A5 reflects a less efficient utilization of the protein, either directly or by some posttranslational process in the cells infected with the recombinant virus. Similar elevated levels of inducer gene expression for optimum virus production has been observed previously by using the VOTE expression system (5, 19, 54). At the inducer concentration of 250 μM IPTG, even though a higher level of A5 production was observed (data not shown), a decrease in vA5Li production was noted (Fig. 5). A decrease in virus yield at high inducer concentrations has been previously observed (5, 19) and attributed to a toxic effect resulting from the overexpression of the target protein.
Synthesis of viral proteins.
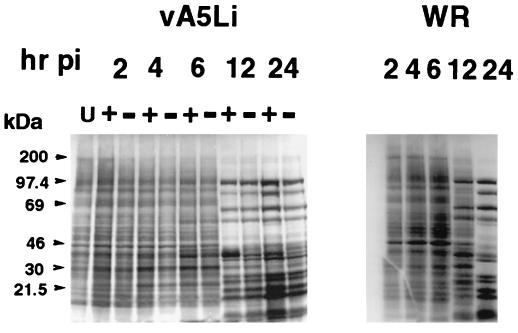
When BSC-1 cells were infected with vA5Li in the absence of A5 production, the yield of infectious virus was greatly reduced. The influence of A5 on the general pattern and timing of viral protein synthesis was investigated by SDS-PAGE of infected cells pulse-labeled with [35S]methionine (Fig. 7). Vaccinia virus early proteins are difficult to resolve from the background of host cell synthesis. At late times after infection, virus-specific bands are clearly discernible since host protein synthesis is inhibited. The pattern of viral late protein expression was nearly identical in cells infected with vA5Li, irrespective of inducer concentration. The major difference was an additional band with an approximate molecular mass of 39 kDa (most obvious at the 12-h time point), which corresponds to the molecular mass of A5, in the presence of inducer. A few other minor changes (an additional band between the 69- and 46-kDa markers in the presence of inducer and an additional protein band between the 30- and 46-kDa markers in the absence of inducer) were also observed. Therefore, the defect in replication of vA5Li was not due to a general perturbation in protein synthesis.
FIG. 7.
Protein synthesis in cells infected with vaccinia virus WR or vA5Li. BSC-1 cells were infected with vaccinia virus WR or vA5Li at an MOI of 5 PFU per cell in the presence (+) or absence (−) of 50 μM IPTG. At the indicated times after infection, the cells were labeled for 30 min with [35S]methionine. Cell lysates were analyzed by SDS-PAGE, and protein products were visualized by exposure on Kodak Bio-Max film. U, uninfected.
Synthesis and processing of viral DNA.
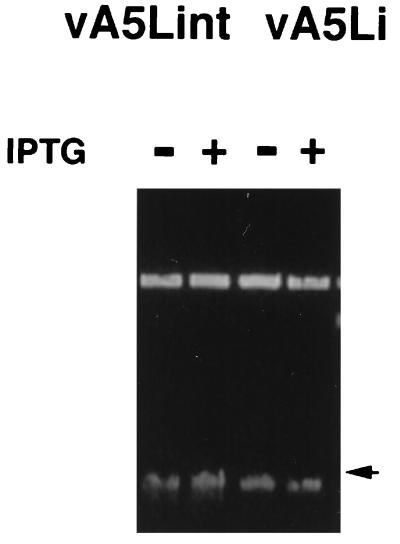
Localization of A5 during the course of an infection demonstrated that the protein colocalizes with the viral DNA, initially in the sites of viral DNA replication and later becoming packaged in the mature infectious virion (6). The synthesis and resolution of viral DNA from vA5Li infections were measured by pulsed-field analysis of DNA isolated from infected cells. Monolayers of BSC-1 cells were infected with vA5Lint or vA5Li and incubated in the presence or absence of 50 μM IPTG, and the DNA was analyzed 24 h postinfection (Fig. 8). The amount of DNA and distribution of full-length monomer genomes were the same in cells infected with either virus irrespective of the presence or absence of inducer. The band migrating at approximately 200 kbp was shown by Southern blotting to be vaccinia virus DNA (data not shown). Also, telomere resolution occurred in the presence or absence of inducer as determined by the appearance of the 1.3-kbp resolved telomere fragment after BstEII digestion (29) and Southern blot analysis (data not shown). Therefore, the absence of A5 has no effect on the synthesis and processing of vaccinia virus DNA.
FIG. 8.
Synthesis and processing of viral DNA. BSC-1 cells were infected with vA5Lint or vA5Li at an MOI of 1 PFU/cell in the presence (+) or absence (−) of 50 μM IPTG. Total DNA was purified 48 h after infection and resolved by pulsed-field gel electrophoresis. The arrow corresponds to 200 kbp as determined by a multimeric lambda DNA marker.
Morphogenesis of vA5Li.
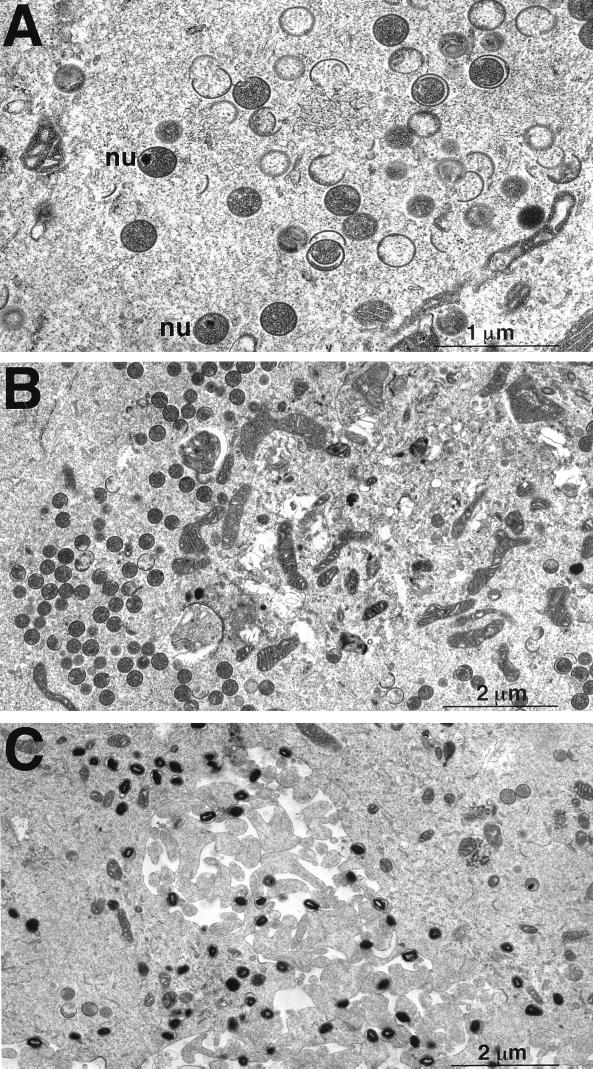
The A5 protein was not expressed in BSC-1 cells infected with vA5Li in the absence of IPTG. The lack of A5 did not influence the general details of protein expression or DNA replication. A potential explanation for the defect in virus replication is that the lack of A5 affects viral assembly or morphogenesis. The constituents of infected cells were investigated by electron microscopy. Electron microscopic images of cells infected with vA5Li in the presence of IPTG (Fig. 9C) contained representatives of all stages of viral morphogenesis, with a majority of the observed structures being brick-shaped IMV particles. In cells infected with vA5Li in the absence of IPTG, representative structures of each stage of morphogenesis were also observed. However, the most common structures detected were the spherical IV particles (Fig. 9A and B). Some IV particles also contained electron-dense nucleoids thought to be packaged DNA (13, 16). In addition, we noted an increased frequency of aberrant IV structures (Fig. 9A) in which crescents and nascent IVs appeared to be enclosed by a second IV membrane. However, these abnormal IVs represented a minority of the IVs generated (Fig. 9B). Therefore, the electron microscopy data suggests that repression of A5 synthesis leads to an accumulation of IVs blocked in the normal progression to IMV.
FIG. 9.
Morphogenesis of mutant viruses. BSC-1 cells were infected with vA5Li in the absence (A and B) or presence (C) of 50 μM IPTG at an MOI of 3 PFU per cell. After 24 h, the cells were fixed in glutaraldehyde and embedded in Epon, and ultrathin sections were prepared for electron microscopy. nu, nucleoid.
Infectivity, composition, and morphology of purified particles.
The accumulation of IV particles in cells infected with vA5Li provides us with an opportunity to purify viral particles predominantly at one stage of viral morphogenesis. Cells infected with vA5Li in the presence (vA5Li+) or absence (vA5Li−) of IPTG were harvested after 72 h, and viral particles were purified by sucrose density gradient centrifugation. A cloudy band was observed in similar positions for vA5Li+ and vA5Li−. The infectivity of the particles, as measured by plaque assay on BSC-1 cells, was compared by using matched protein concentrations of WR, vA5Li+, and vA5Li− virion preparations. The infectivity of vA5Li−, per its optical density at 280 nm (OD280) was 7% of that of vA5Li+. The infectivity of the vA5Li+ particles was 82%, essentially identical to the infectivity of particles derived from WR.
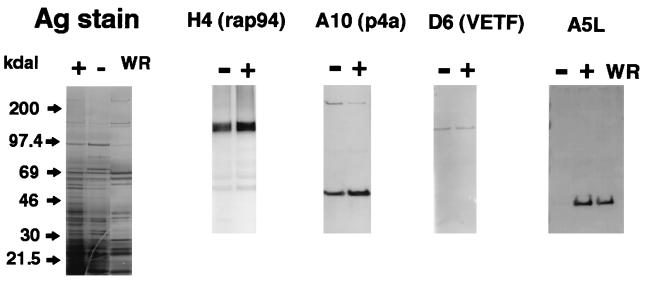
The composition of the purified particles was determined by SDS-PAGE and silver staining (Fig. 10). The protein patterns of vA5Li+ and vA5Li− were almost identical, with the observed differences consisting of a few additional high-molecular-mass bands and an absent band of approximately 39 kDa in vA5Li−. Also, a more concentrated 97-kDa band appears in vA5Li− and may correspond to uncleaved p4a. The lower band probably corresponds to A5, as Western blot analysis of vA5Li− particles confirmed that A5 was greatly reduced in vA5Li− compared to vA5Li+ or WR virion particles (Fig. 10).
FIG. 10.
Protein content and Western blot analysis of purified virus particles. Particles purified by sucrose gradient centrifugation from BSC-1 cells infected with vA5Li in the presence (+) or absence (−) of 50 μM IPTG or infected with wild-type vaccinia virus in the absence of IPTG (WR) were adjusted to similar protein concentrations and analyzed by SDS-PAGE and silver staining (Integrated Separation Systems kit). Analogous protein samples were separated by SDS-PAGE, and the proteins were transferred to Protran (Schleicher & Schuell) and blocked with 5% nonfat dry milk in phosphate-buffered saline. The filters were probed with rabbit antibodies directed against specific vaccinia virus proteins (gift of B. Moss) and visualized by subsequent incubations with an anti-rabbit immunoglobulin G conjugated with alkaline phosphatase and Western Blue reagent (Promega).
The prevalence of specific viral proteins in vA5Li− and vA5Li+ particles was determined by Western blot analysis using antibodies directed against specific viral proteins. Equivalent amounts of protein, as determined by their OD280, were separated by PAGE, transferred to membranes, and treated with antibodies directed against the RNA polymerase-associated protein H4L, a subunit of the viral early transcription factor D6R, and core protein p4a, or A10L (Fig. 10). The only observed difference between vA5Li+ and vA5Li−, other than A5L, was the higher percentage of uncleaved precursor of A10L (p4a) in the vA5Li− sample, indicative of incomplete maturation. The relative distribution of the two forms of A10L was determined by image analysis, and the higher-molecular-mass band was shown to correspond to 12% and 2% of the A10L in the vA5Li− and vA5Li+ particles, respectively. The process of purification appears to enrich for mature virions, as the percentage of mature particles in purified preparations of vA5Li− (14 to 21%) was higher than the percentage of mature particles observed in infected cells in the absence of IPTG (less than 5%). In addition to the H4L and D6R proteins illustrated in Fig. 10, no differences in protein concentrations were noted for the A17L gene, a membrane protein, A8R, a subunit of the viral early transcription factor, and I7L, an enzyme (data not shown).
The particles were tested to determine if they contained viral DNA by applying matched protein concentrations of WR, vA5Li+, and vA5Li− to a Nytran membrane, hybridizing the viral DNA with fluorescein-labeled viral DNA, and visualizing the hybridized product with an anti-fluorescein horseradish peroxidase antibody and luminol (ECL kit). All three samples hybridized equally well to probe (data not shown), demonstrating that all three types of particles contained the same amount of viral DNA. Therefore, the loss of A5 expression does not interfere with viral DNA packaging.
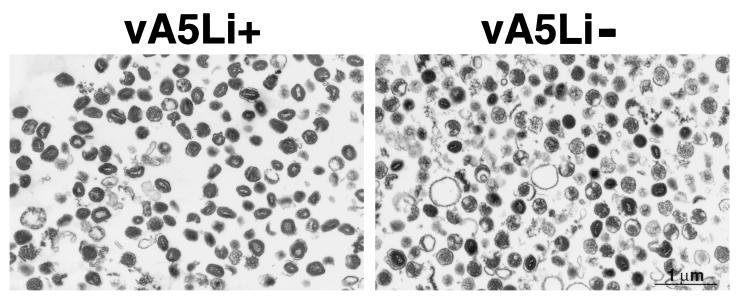
The purified particles were sedimented, and thin sections were examined by electron microscopy. The vA5Li+ particles were primarily brick shaped with clearly visible dumbbell-shaped cores, similar in appearance to WR. The vA5Li− preparation contained some wild-type particles but was composed mainly of irregular roundish particles lacking a clear dumbbell-shaped core structure (Fig. 11). The irregular shapes of the vA5Li− particles, in contrast to the spherical shapes observed in the infected cells, presumably arose during purification or preparation for microscopy.
FIG. 11.
Electron microscopy of purified virus particles. Particles purified by sucrose gradient centrifugation from BSC-1 cells infected with vA5Li in the presence (vA5Li+) or absence (vA5Li−) of 50 μM IPTG were collected by high-speed centrifugation, fixed in glutaraldehyde, and embedded in Epon. Ultrathin sections were examined by electron microscopy.
Although the protein composition of the vA5Li+ and vA5Li− viral particles appears to be almost identical in the distribution and amounts of most viral proteins, the functional state of proteins in the viral particles was unknown. An infectious virion can be activated in vitro to transcribe viral genes (23, 33). The ability of the vA5Li+ and vA5Li− particles to incorporate [α-32P]UTP was measured as a function of protein concentration (5) by using a series of protein concentrations, and vA5Li− particles were shown to elicit about 75% of the activity determined for vA5Li+ particles. Also, virion extracts were prepared (17) from vA5Li+ and vA5Li− particles and assayed for virion nuclease activity (28). Both types of particles were shown to possess equivalent levels of nuclease activity (data not shown).
DISCUSSION
We used a genetic approach to discern the role of A5 in a vaccinia virus infection by the construction and characterization of vA5Li, a vaccinia virus recombinant with an inducible A5 gene. The correct genomic structure of the recombinant was confirmed by Southern blotting and PCR. The expression of A5 in vA5Li was demonstrated to be dependent on and directly proportional to the concentration of IPTG inducer, with maximum yields of virus realized at concentrations of 50 μM IPTG. The residual low yield of infectious virus in the absence of inducer may result from the inefficient production of virus at the low levels of basal A5 expression in the absence of IPTG or reflect an inherent low infectivity of virions lacking A5. Also, some virus may be produced by using the residual A5 carried into the infected cell by the virus grown in the presence of IPTG.
In an attempt to determine the stage of viral replication affected by the absence of A5, metabolic steps involved in virion replication were investigated by analysis of viral protein expression as well as DNA replication and resolution from infections of vA5Li in the presence or absence of inducer. The overall pattern of protein expression was the same for infections performed in the presence or absence of IPTG. The viral DNA was replicated and completely resolved in the absence of inducer. These results suggest that the role of A5 in viral replication is not manifested in viral transcription or replication. However, since A5 is a virion component and stocks of vA5Li are grown in the presence of inducer, we cannot rule out a role for A5 in early gene expression.
Electron-microscopic analysis of viral morphogenesis in cells infected with vA5Li in the presence of IPTG demonstrated the presence of all of the usual stages of morphogenesis, including the mature, brick-shaped infectious virions. Cells infected with vA5Li in the absence of inducer also contained structures representative of all stages of vaccinia morphogenesis. However, only a minority of the structures observed were the mature infectious brick-shaped particles; most of the structures were spherical IVs. Nucleoids were observed in some of the immature particles, implying that DNA packaging is unaffected by the lack of A5. Interestingly, we also observed what appeared to be an accumulation of aberrant immature forms in which crescent structures of various degrees of development were enclosed within a normally sized IV. In some cases, it appeared as if a virion had acquired two complete membranes. Although the number of aberrant forms may well be underrepresented due to the plane of sectioning, it appeared that the majority of the IVs were normal. The accumulation of these abnormal structures may be indicative of a direct role of the A5 protein in the appropriate formation and development of the crescent membranes and IVs but may also be the result of a delay in or decreased efficiency of this stage of morphogenesis due to the role of A5 protein in another process. Further study is required to address this point.
The particles arising from vA5Li infections were purified 72 h postinfection by banding in sucrose gradients. The protein and DNA components of the vA5Li+ and vA5Li− particles were nearly identical. The only obvious differences detected in the protein pattern were the amount of A5L and the form of p4a present. Specific protein constituents assayed by Western blotting demonstrated that the structural proteins A17L and I7L, the two subunits of the early transcription factor, A8R and D6R, and the RNA polymerase component H4L were found in equal amounts in vA5Li+ and vA5Li− preparations. The vA5Li− particles contained more of the unprocessed form of A10L (p4a), implying that inhibition of A5L gene expression delays or partially blocks proteolytic processing of core protein precursors.
The infectivity of the vA5Li− particles was consistently lower than for vA5Li+, as measured by PFU on BSC-1 cells. The in vitro transcriptional activity of vA5Li− particles was consistently about 75% of the level obtained with a protein equivalent of vA5Li+. Since the particles contain a full complement of viral transcriptional components, the difference in infectivity may be a reflection of the relative integrity of the particles. Partially purified preparations of vA5Li− left in sucrose were observed to become viscous and, in contrast to vA5Li+, did not give a discrete band upon centrifugation in sucrose gradients. The fragile nature of the vA5Li− particles may lead to their more frequent rupture, explaining the irregular shape observed by electron microscopy and the lower level of nonspecific transcriptional activity. The data does not unambiguously demonstrate whether the lowered infectivity of vA5Li− results from an impaired ability to infect the cell or from a more fragile viral particle which is less likely to survive the purification process intact. The level of infectivity for each preparation of vA5Li− was proportional to the percentage of mature brick-shaped particles. The percentage of brick-shaped mature viral particles in different preparations of vA5Li− ranged from approximately 14 to 21% and was directly proportional to the amount of A5 detected by Western blotting. Preparations of vA5Li+ consistently contained about 80% brick-shaped mature viral particles.
The progression from IV to IMV particle uses site-dependent protein-protein, lipid-protein, and nucleic acid-protein interactions to generate a sequence of specific structures. The core proteins form the structural internal lattice of the infectious virion, and the loss of any of these proteins may lead to aberrant intermediate structures which are unable to progress to the subsequent stage on the pathway to infectious IMV. Conditional lethal viruses have been useful in identifying several viral core proteins important for vaccinia virus morphogenesis. Conditional repression of functional A5L (this study), F18R (p11) (57), the I7L gene product (13, 22), or membrane-bound core protein L1R (35) led to the accumulation of IV particles.
Other proteins which have not yet been identified as structural core proteins affect the transition from IV to IMV. The conditional repression of the two components of the viral early transcription factor, A8R and D6R, leads to the accumulation of IV particles (19, 20). These proteins are present in the viral core and are relatively abundant but do not have a previously documented structural role. Also, the conditional repression of the A32L protein resulted in the accumulation of IV particles (5) which did not contain DNA. The location or presence of A32L, which is not an abundant protein, is unknown.
Previous studies of the p39 protein, or A5L, are consistent with its apparent role in virion morphogenesis. Biochemical studies indicated that the protein was present in the viral factories and the central region of IV particles (6). During the transition from IV to IMV, the location of A5 changes and it becomes localized between two membranes proximal to the core, possibly forming the spikes found on the outside of the viral core (6). This location is consistent with a role for A5 in maturation at a late step in IMV formation.
The requirement for A5 in poxvirus replication is underscored by the retention of its homologous open reading frame in the widely diverged poxviruses molluscum contagiosum (43) and modified virus ankara (MVA) (2). Compared to the nucleotide sequence for vaccinia virus Copenhagen, the analogue to the vaccinia virus WR A5L open reading frame is missing 27 nucleotides in MVA, leading to an internal deletion of nine amino acids (2). Infections of MVA in non- and semipermissive cell lines produce a nonpropagating virus blocked in morphogenesis at the accumulation of IV particles (45). The role for A5 in vaccinia virus replication described in this article suggests that the altered protein in MVA may contribute to the phenotype of restricted host range in morphogenesis.
The viral gene A5L is essential for virus growth and is synthesized late in the viral life cycle. The association of A5L with the site of viral DNA replication and processing and its copurification with the viral nicking-closing enzyme led to our initial interest in the role of this protein in vaccinia virus infection. Using the viral construct vA5Li, which contains an inducible copy of A5L, we have clearly demonstrated that A5L does not encode a protein with nuclease activity. The protein is required for the progression of IV to infectious IMV particles. This data is consistent with the time course of synthesis and location of the protein during infection.
ACKNOWLEDGMENTS
We thank Jerry Weir, Miles Carroll, and Xiaolei Hu for helpful discussions and Bernie Moss and Norman Cooper for reagents.
REFERENCES
- 1.Alexander W A, Moss B, Fuerst T R. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J Virol. 1992;66:2934–2942. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoine G, Scheiflinger F, Dorner F, Falkner F G. The complete genomic sequence of the modified vaccinia ankara strain: comparison with other orthopoxviruses. Virology. 1998;244:365–396. doi: 10.1006/viro.1998.9123. [DOI] [PubMed] [Google Scholar]
- 3.Blasco R, Moss B. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-dalton outer envelope protein. J Virol. 1991;65:5910–5920. doi: 10.1128/jvi.65.11.5910-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cairns J. The initiation of vaccinia infection. Virology. 1960;11:603–623. doi: 10.1016/0042-6822(60)90103-3. [DOI] [PubMed] [Google Scholar]
- 5.Cassetti M C, Merchlinsky M, Wolffe E J, Weisberg A S, Moss B. DNA packaging mutant: repression of the vaccinia virus A32 gene results in noninfectious, DNA-deficient, spherical, enveloped particles. J Virol. 1998;72:5769–5780. doi: 10.1128/jvi.72.7.5769-5780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cudmore S, Blasco R, Vincentelli R, Esteban M, Sodeik B, Griffiths G, Locker J K. A vaccinia virus core protein, p39, is membrane associated. J Virol. 1996;70:6909–6921. doi: 10.1128/jvi.70.10.6909-6921.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dales S, Siminovitch L. The development of vaccinia virus in Earle’s L strain cells as examined by electron microscopy. J Biophys Biochem Cytol. 1961;10:475–498. doi: 10.1083/jcb.10.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demkowicz W E, Maa J-S, Esteban M. Identification and characterization of vaccinia genes encoding proteins that are highly antigenic in animals and are immunodominant in vaccinated humans. J Virol. 1992;66:386–398. doi: 10.1128/jvi.66.1.386-398.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duncan S A, Smith G L. Identification and characterization of an extracellular envelope glycoprotein affecting vaccinia virus egress. J Virol. 1992;66:1610–1621. doi: 10.1128/jvi.66.3.1610-1621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Earl P L, Cooper N, Moss B. Preparation of cell cultures and vaccinia virus stocks. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1991. pp. 16.16.1–16.16.7. [Google Scholar]
- 11.Earl P L, Moss B. Generation of recombinant vaccinia viruses. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1991. pp. 16.17.1–16.17.16. [Google Scholar]
- 12.Engelstad M, Smith G L. The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology. 1993;194:627–637. doi: 10.1006/viro.1993.1302. [DOI] [PubMed] [Google Scholar]
- 13.Ericsson M, Cudmore S, Shuman S, Condit R C, Griffiths G, Locker J K. Characterization of ts16, a temperature-sensitive mutant of vaccinia virus. J Virol. 1995;69:7072–7086. doi: 10.1128/jvi.69.11.7072-7086.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuerst T R, Fernandez M P, Moss B. Transfer of the inducible lac repressor/operator system from Escherichia coli to a vaccinia virus expression vector. Proc Natl Acad Sci USA. 1989;86:2549–2553. doi: 10.1073/pnas.86.8.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 16.Grimley P M, Rosenblum E N, Mims S J, Moss B. Interruption by rifampin of an early stage in vaccinia virus morphogenesis: accumulation of membranes which are precursors of virus envelopes. J Virol. 1970;6:519–533. doi: 10.1128/jvi.6.4.519-533.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gross C H, Shuman S. Vaccinia virions lacking the RNA helicase nucleoside triphosphate phosphohydrolase II are defective in early transcription. J Virol. 1996;70:8549–8557. doi: 10.1128/jvi.70.12.8549-8557.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harford C G, Hamlin A, Riders E. Electron microscopic autoradiography of DNA synthesis in cells infected with vaccinia virus. Exp Cell Res. 1966;42:50–57. doi: 10.1016/0014-4827(66)90318-1. [DOI] [PubMed] [Google Scholar]
- 19.Hu X, Carroll L J, Wolffe E J, Moss B. De novo synthesis of the early transcription factor 70-kilodalton subunit is required for morphogenesis of vaccinia virions. J Virol. 1996;70:7669–7677. doi: 10.1128/jvi.70.11.7669-7677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu X, Wolffe E J, Weisberg A S, Carroll L J, Moss B. Repression of the A8L gene, encoding the early transcription factor 82-kilodalton subunit, inhibits morphogenesis of vaccinia virions. J Virol. 1998;72:104–112. doi: 10.1128/jvi.72.1.104-112.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joklik W K, Becker Y. The replication and coating of vaccinia DNA. J Mol Biol. 1964;10:452–474. doi: 10.1016/s0022-2836(64)80066-8. [DOI] [PubMed] [Google Scholar]
- 22.Kane E M, Shuman S. Vaccinia virus morphogenesis is blocked by a temperature-sensitive mutation in the I7 gene that encodes a virion component. J Virol. 1993;67:2689–2698. doi: 10.1128/jvi.67.5.2689-2698.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kates J R, McAuslan B R. Poxvirus DNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 1967;58:134–141. doi: 10.1073/pnas.58.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katz E, Moss B. Formation of a vaccinia virus structural polypeptide from a high molecular weight precursor: inhibition by rifampicin. Proc Natl Acad Sci USA. 1970;66:677–684. doi: 10.1073/pnas.66.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katz E, Moss B. Vaccinia virus structural polypeptide derived from a high-molecular-weight precursor: formation and integration into virus particles. J Virol. 1970;6:717–726. doi: 10.1128/jvi.6.6.717-726.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lakritz N, Foglesong P D, Reddy M, Baum S, Hurwitz J, Bauer W R. A vaccinia DNase preparation which cross-links superhelical DNA. J Virol. 1985;53:935–943. doi: 10.1128/jvi.53.3.935-943.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maa J-S, Esteban M. Structural and functional studies of a 39,000-Mr immunodominant protein of vaccinia virus. J Virol. 1987;61:3910–3919. doi: 10.1128/jvi.61.12.3910-3919.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merchlinsky M, Garon C F, Moss B. Molecular cloning and sequence of the concatemer junction from vaccinia virus replicative DNA. Viral nuclease cleavage sites in cruciform structures. J Mol Biol. 1988;199:399–413. doi: 10.1016/0022-2836(88)90613-4. [DOI] [PubMed] [Google Scholar]
- 29.Merchlinsky M, Moss B. Resolution of vaccinia virus DNA concatemer junctions requires late gene expression. J Virol. 1989;63:1595–1603. doi: 10.1128/jvi.63.4.1595-1603.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Merchlinsky, M. Unpublished results.
- 30.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2637–2672. [Google Scholar]
- 31.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. New mammalian expression vectors. Nature (London) 1990;348:91. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 32.Moss B, Rosenblum E N. Protein cleavage and poxvirus morphogenesis: tryptic peptide analysis of core precursors accumulated by blocking assembly with rifampicin. J Mol Biol. 1973;81:267–269. doi: 10.1016/0022-2836(73)90195-2. [DOI] [PubMed] [Google Scholar]
- 33.Munyon W E, Paoletti E, Grace J T., Jr RNA polymerase activity in purified infectious vaccinia virus. Proc Natl Acad Sci USA. 1967;58:2280–2288. doi: 10.1073/pnas.58.6.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkinson J E, Smith G L. Vaccinia virus gene A36R encodes a Mr 43-50 K protein on the surface of extracellular enveloped virus. Virology. 1994;204:376–390. doi: 10.1006/viro.1994.1542. [DOI] [PubMed] [Google Scholar]
- 35.Ravanello M P, Hruby D E. Conditional lethal expression of the vaccinia virus L1R myristylated protein reveals a role in virion assembly. J Virol. 1994;68:6401–6410. doi: 10.1128/jvi.68.10.6401-6410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez D, Esteban M, Rodriguez J R. Vaccinia virus A17L gene product is essential for an early step in virion morphogenesis. J Virol. 1995;69:4640–4648. doi: 10.1128/jvi.69.8.4640-4648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez D, Risco C, Rodriguez J R, Esteban M. Inducible expression of the vaccinia virus A17L gene provides a synchronized system to monitor sorting of viral proteins during morphogenesis. J Virol. 1996;70:7641–7653. doi: 10.1128/jvi.70.11.7641-7653.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez J F, Smith G L. IPTG-dependent vaccinia virus: identification of a virus protein enabling virion envelopment by Golgi membrane and egress. Nucleic Acids Res. 1990;18:5347–5351. doi: 10.1093/nar/18.18.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roper R L, Payne L G, Moss B. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J Virol. 1996;70:3753–3762. doi: 10.1128/jvi.70.6.3753-3762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosemond-Hornbeak H, Moss B. Single-strand deoxyribonucleic acid-specific nuclease from vaccinia virus. Endonucleolytic and exonucleolytic activities. J Biol Chem. 1974;249:3292–3296. [PubMed] [Google Scholar]
- 41.Rosemond-Hornbeak H, Paoletti E, Moss B. Single-strand deoxyribonucleic acid-specific nuclease from vaccinia virus. Purification and characterization. J Biol Chem. 1974;249:3287–3291. [PubMed] [Google Scholar]
- 42.Schmutz C, Payne L G, Gubser J, Wittek R. A mutation in the gene encoding the vaccinia virus 37,000-Mr protein confers resistance to an inhibitor of virus envelopment and release. J Virol. 1991;65:3435–3442. doi: 10.1128/jvi.65.7.3435-3442.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Senkevich T G, Bugert J J, Sisler J R, Koonin E V, Darai G, Moss B. Genome sequence of a human tumorigenic poxvirus: prediction of specific host response-evasion genes. Science. 1996;273:813–816. doi: 10.1126/science.273.5276.813. [DOI] [PubMed] [Google Scholar]
- 44.Silver M, Dales S. Biogenesis of vaccinia: interrelationship between posttranslational cleavage, virus assembly, and maturation. Virology. 1982;117:341–356. doi: 10.1016/0042-6822(82)90474-3. [DOI] [PubMed] [Google Scholar]
- 45.Sutter G, Moss B. Nonreplicating vaccinia vector efficiently expresses recombinant genes. Proc Natl Acad Sci USA. 1992;89:10847–10851. doi: 10.1073/pnas.89.22.10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tartaglia J, Paoletti E. Physical mapping and DNA sequence analysis of the rifampin resistance locus in vaccinia virus. Virology. 1985;147:394–404. doi: 10.1016/0042-6822(85)90141-2. [DOI] [PubMed] [Google Scholar]
- 47.Traktman P, Caligiuri A, Jesty S A, Liu K, Sankar K. Temperature-sensitive mutants with lesions in the vaccinia virus F10 kinase undergo arrest at the earliest stage of virion morphogenesis. J Virol. 1995;69:6581–6587. doi: 10.1128/jvi.69.10.6581-6587.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.VanSlyke J K, Franke C A, Hruby D E. Proteolytic maturation of vaccinia virus core proteins: identification of a conserved motif at the N termini of the 4b and 25K virion proteins. J Gen Virol. 1991;72:411–416. doi: 10.1099/0022-1317-72-2-411. [DOI] [PubMed] [Google Scholar]
- 49.VanSlyke J K, Lee P, Wilson E M, Hruby D E. Isolation and analysis of vaccinia virus previrions. Virus Genes. 1993;7:311–324. doi: 10.1007/BF01703388. [DOI] [PubMed] [Google Scholar]
- 50.VanSlyke J K, Whitehead S S, Wilson E M, Hruby D E. The multistep proteolytic maturation pathway utilized by vaccinia virus p4a protein: a degenerate conserved cleavage motif within core proteins. Virology. 1991;183:467–478. doi: 10.1016/0042-6822(91)90976-i. [DOI] [PubMed] [Google Scholar]
- 51.Wang S, Shuman S. Vaccinia virus morphogenesis is blocked by temperature-sensitive mutations in the F10 gene, which encodes protein kinase 2. J Virol. 1995;69:6376–6388. doi: 10.1128/jvi.69.10.6376-6388.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ward G A, Stover C K, Moss B, Fuerst T R. Stringent chemical and thermal regulation of recombinant gene expression by vaccinia virus vectors in mammalian cells. Proc Natl Acad Sci USA. 1995;92:6773–6777. doi: 10.1073/pnas.92.15.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolffe E J, Isaacs S N, Moss B. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J Virol. 1993;67:4732–4741. doi: 10.1128/jvi.67.8.4732-4741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolffe E J, Moore D M, Peters P J, Moss B. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J Virol. 1996;70:2797–2808. doi: 10.1128/jvi.70.5.2797-2808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, Moss B. Immature viral envelope formation is interrupted at the same stage by lac operator-mediated repression of the vaccinia virus D13L gene and by the drug rifampicin. Virology. 1992;187:643–653. doi: 10.1016/0042-6822(92)90467-4. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, Moss B. Inducer-dependent conditional-lethal mutant animal viruses. Proc Natl Acad Sci USA. 1991;88:1511–1515. doi: 10.1073/pnas.88.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, Moss B. Vaccinia virus morphogenesis is interrupted when expression of the gene encoding an 11-kilodalton phosphorylated protein is prevented by the Escherichia coli lac repressor. J Virol. 1991;65:6101–6110. doi: 10.1128/jvi.65.11.6101-6110.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]