Abstract
We report on the construction of a full-length cDNA clone, pBRM33, derived from wild-type rubella virus M33 strain. The RNA transcripts synthesized in vitro from pBRM33 are highly infectious, and the viruses produced retain the phenotypic characteristics of the parental M33 virus in growth rate and plaque size. This cDNA clone was used to study the role of E1 transmembrane and cytoplasmic domains in virus assembly by site-directed mutagenesis. Three different alanine substitutions were introduced in the transmembrane domain of E1. These included substitution of leucine 464, cysteine 466, cysteine 467, and both cysteines 466 and 467 to alanine. In the E1 cytoplasmic domain, cysteine 470 and leucine 471 were altered to alanine. We found that these mutations did not significantly affect viral RNA replication, viral structural protein synthesis and transport, or E2/E1 heterodimer formation. Except for the substitution of cysteine 470, these mutations did, however, lead to a reduction in virus release. Substitution of cysteine 467 in the transmembrane region and of leucine 471 in the cytoplasmic domain dramatically reduced virus yield, resulting in the production of only 1 and 10% of the parental virus yield, respectively, in a parallel infection. These data show that E1 transmembrane and cytoplasmic domains play an important role in late stages of virus assembly, possibly during virus budding, consistent with earlier studies indicating that the E1 cytoplasmic domain may interact with nucleocapsids and that this interaction drives virus budding.
Rubella virus (RV), the etiological agent of German measles, is the only member of the Rubivirus genus in the Togaviridae family (15). This family also includes the Alphavirus genus, containing the Sindbis and Semiliki Forest viruses. RV uses a strategy similar to that of alphaviruses in gene replication and expression (7). The RNA genome of RV is 9,758 nucleotides (nt) in length, exclusive of a cap structure and a poly(A) tract (6, 22). The 5′ two-thirds of the genome encodes nonstructural proteins, and the 3′ one-third encodes structural proteins. The nonstructural proteins are translated from the genomic RNA as a polyprotein precursor, which is cleaved by a virus-encoded protease into two nonstructural proteins, p150 and p90 (3, 29). These two proteins are thought to form a replication complex with host factors and to be involved in RNA replication. The subgenomic RNA corresponding to the 3′ one-third of the genome is synthesized from the genomic RNA and used as a template for translation of structural proteins (4, 5, 18).
RV virions contain an icosahedral nucleocapsid surrounded by a lipid envelope in which two virus-encoded glycoproteins, E1 and E2, are embedded (7, 26). The structural proteins are synthesized as a polyprotein precursor in the order capsid-E2-E1 (17). This is translocated into the endoplasmic reticulum (ER) by two independently functioning signal peptides within the carboxy terminus of capsid and E2 (9, 12). In the ER, the polyprotein precursor is cleaved by a signal peptidase into the three structural proteins, capsid, E2, and E1 (10, 16). Multiple copies of capsid bind to the genomic RNA to form nucleocapsids in the cytoplasm, and the glycoproteins E2 and E1 form a specific heterodimer in the ER shortly after synthesis (1, 7). The E2/E1 heterodimers are transported out of the ER to the Golgi and plasma membranes, where the virus matures by the budding of nucleocapsids through cellular membranes to acquire the viral envelope. In BHK cells, the primary budding site for RV is at the Golgi membrane, although budding at the plasma membrane occurs in Vero cells (7, 11). In this regard, RV is different from alphaviruses, which bud exclusively at the plasma membrane (25). However, like alphaviruses, this budding process appears to be capsid dependent, since formation of virus-like particles by transfection of subgenomic cDNA into BHK cells is strictly dependent on the expression of glycoproteins E2 and E1 and capsid (11, 24). In alphaviruses, genetic studies by construction of chimeric viruses demonstrated that the cytoplasmic domain of glycoprotein E2 interacts with the nucleocapsid in a sequence-specific fashion during virus budding (14). Site-directed mutations with infectious cDNA clones revealed that two amino acids, tyrosine and leucine, at the amino terminus of the E2 cytoplasmic domain play a critical role in the interaction (19, 30). The role of glycoproteins E1 and E2 in RV assembly is not well defined. RV E2 and E1 are anchored in the membrane as a type 1 membrane protein. The putative E2 transmembrane sequence is 39 residues in length, followed by a positively charged sequence, RRACRRR, and a stretch of 20 hydrophobic residues that is reinserted into the ER membrane and serves as the signal sequence for E1. The putative E1 transmembrane domain is 22 residues in length and is followed by a stretch of 13 amino acids that forms the cytoplasmic domain of E1. It is suggested that this 13-amino-acid stretch of E1 may play a role in mediating the interaction of glycoproteins with nucleocapsids in RV assembly (11).
In this study, we describe the construction of a full-length infectious cDNA clone of the M33 strain. RNA transcribed from the cDNA clone produced infectious virus that showed the same characteristics as the parental virus M33 strain in growth and plaque size. We have used this infectious cDNA clone to investigate the role of E1 cytoplasmic and transmembrane domains in viral assembly by introduction of mutations in these regions. Our data showed that mutation of cysteine 467 at the C terminus of the E1 transmembrane domain and leucine 471 at the N terminus of the E1 cytoplasmic domain dramatically reduced virus release.
MATERIALS AND METHODS
Virus and cells.
Vero cells were grown in Eagle’s minimum essential medium (MEM) supplemented with 5% fetal bovine serum, penicillin (100 U/ml), and streptomycin (50 μg/ml). BHK-21 cells were grown in MEM containing 10% fetal bovine serum and 10% tryptose phosphate broth. RV M33 was propagated in Vero cells and used in this study.
Preparation of virion RNA and first-strand cDNA synthesis.
Monolayers of Vero cells grown in 14 150-mm-diameter dishes were infected with RV M33. At day 3 postinfection, the culture medium was harvested and virions were precipitated by using polyethylene glycol (PEG) as described previously (27) and were resuspended in 1 ml of TNE buffer (10 mM Tris [pH 7.4], 150 mM NaCl, 1 mM EDTA). Virion RNA was extracted with TRIzol reagent (Gibco/BRL) following the instructions of the manufacturer and was used for first-strand cDNA synthesis. First-strand cDNA synthesis was carried out at 42°C for 2 h in a 20-μl reaction mixture containing viral RNA, 200 pmol of the primer (5′-GAATTCAAGCT17-3′), 200 U of Superscript RT RNase H− reverse transcriptase (RT) (Gibco/BRL), and 40 U of RNase inhibitor (Promega) in a buffer provided by the manufacturer (Gibco/BRL). The synthesized first-strand cDNA was used as the template for the subsequent PCR amplification. A primer, 5′-CTCTCCCGAATGGCAA-3′, complementary to the viral genomic sequence 5′-TTGCCATTCGGGAGAG-3′ (nt 6378 to 6491), was used in the reaction for the synthesis of first-strand cDNA covering the nonstructural protein gene region.
Construction of full-length RV M33 cDNA clone.
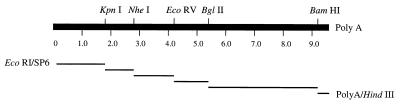
The viral cDNA was amplified in six distinct overlapping fragments by PCR with six pairs of primers in individual reactions. These six DNA fragments were cut with restriction endonucleases as shown in Fig. 1 and ligated into a full-length cDNA representing the entire viral genomic RNA. PCR amplifications were performed in separate reactions containing cDNA, 1 pmol of primers, 0.4 mM deoxynucleoside triphosphates, 10% dimethyl sulfoxide, and proofreading TaKaRa Taq polymerase (Takara Shuzo Co., Ltd.) in a buffer provided by the manufacturer. The PCR program consisted of 1 min at 98°C followed by 25 cycles of 20 s at 98°C, 1 s at 56°C, and varied extension times (depending on the length of each sequence) at 70°C. Amplification of the DNA fragment covering the 5′ terminus of the viral genome was carried out with a sense primer, 5′-TTCGAATTCATTTAGGTGACACTATAGCAATGGAAGCTATC-3′, which includes the EcoRI site (italic), the SP6 RNA polymerase promoter sequence (single underline), and the first 14 nucleotides (double underline) of the 5′ terminus of the viral genome, and with the antisense primer 5′-GGTGGGCGGGGTGGCGGTAGA-3′ (nt 1737 to 1757). For amplification of the DNA covering the 3′ terminus of the viral genome, the PCR was done with a sense primer, 5′-TACCGTCGAAATGCCCGAGT-3′ (nt 9158 to 9175), and an antisense primer, 5′-GAATTCAAGCTT15, which includes a HindIII site immediately downstream from the poly(dT15) tract. The rest of the cDNA fragments were amplified with the following pairs of primers: a KpnI-NheI fragment with 5′-AGGCTCTCCGCGCGCGCGCCG-3′ (sense primer; nt 1658 to 1679) and 5′-GCACGCGACGCGCGCCCACCG-3′ (antisense primer; nt 2819 to 2839); an NheI-EcoRV fragment with 5′-GCTGCTCGAGCGCGCCTACCG-3′ (sense primer; nt 2770 to 2790) and 5′-GTAGGTGGCGGCGTTCTTGAT-3′ (antisense primer; nt 4220 to 4240); an EcoRV-BglII fragment with 5′-GATCCAGGCCAAACTCCGCGC-3′ (sense primer; nt 4186 to 4206) and 5′-ACGCTCAGGCTCTGGGCGGTG-3′ (antisense primer; nt 5371 to 5391); and a BglII-BamHI fragment with 5′-CGCGGGAGCTCACCGACCGCT-3′ (sense primer; nt 5313 to 5333) and 5′-GTCCTCGCCATTGACGGTAAGATGGCAGTT-3′ (antisense primer; nt 9351 to 9381). The assembled full-length cDNA clone was cut with EcoRI and HindIII and inserted into a modified plasmid, pBR322, that had been digested with the same enzymes (in this vector, the NheI-NruI fragment had been removed to delete the Tet gene). The resulting construct was named pBRM33.
FIG. 1.
Construction of RV M33 virus full-length cDNA clone. The numbers on the viral genome scale refer to the distance from the 5′ end in kilobases. Six DNA fragments were amplified by proofreading TaKaRa Taq DNA polymerase with the requisite primers as described in Materials and Methods. The amplified DNA fragments were ligated into a full-length cDNA representing the whole viral genome by using the restriction sites indicated above the genome. The full-length cDNA was cut with EcoRI and HindIII and inserted into a modified pBR322 plasmid that had been cut with the same enzymes to obtain the full-length cDNA clone pBRM33.
Construction of mutants.
A series of mutations was introduced into the E1 coding region by PCR-mediated mutagenesis with appropriate primers containing the desired nucleotide changes. To facilitate mutagenesis, a silent mutation was introduced into pBRM33 to create a new SphI site by changing T to A at nt 9647. For this purpose, a fusion PCR (27) was employed with pBRM33 DNA as a template, Pfu DNA polymerase (Stratagene), and two pairs of primers: 5′-TACCGTCGAAATGCCCGAGT-3′ (sense primer) and 5′-AATGAGCGTACGACACGG-3′ (antisense primer) and 5′-AATCTCGCATGCTGTGCC-3′ (sense primer) and 5′-GAATTCAAGCTTT15-3′ (antisense primer). The PCR product was cut with BamHI and HindIII and inserted into pBRM33 (minus the BamHI/HindIII fragment). This construct was named pBRM34.
To construct mutants L464A, C467A, C470A, and L471A, PCR amplifications were performed in a series of reactions with pBRM34 DNA as a template and primers containing the desired mutations. The mutagenic primers were 5′-GGCACAGCATGCGGCTAAGCC-3′ for mutant L464A, 5′-TTACTCGCATGCGCTGCCAAATGC-3′ for mutant C467A, 5′-TTACTCGCATGCTGTGCCAAAGCCTTGTAC-3′ for mutant C470A, and 5′-TTACTCGCATGCTGTGCCAAATGCGCGTACTAC-3′ for mutant L471A (mutated nucleotides are underlined). To construct mutants C466A and C466A/C467A, the fusion PCR described above was employed. The mutation was introduced into mutant C466A with a pair of primers, 5′-TTACTCGCAGCCTGTGCC-3′ (sense primer) and 5′-GGCACAGGCTGCGAGTAA-3′ (antisense primer). For mutant C466A/C467A, a pair of primers, 5′-GGCTTACTCGCGGCCGCCGCCAAATGC-3′ (sense primer) and 5′-GCATTTGGCGGCGGCCGCGAGTAAGCC-3′ (antisense primer), was used. All of the amplified DNA fragments containing the desired mutations were reintroduced into pBRM34, and the mutations were verified by sequencing and restriction analysis.
RNA transcription and transfection.
RNA transcripts were synthesized with SP6 RNA polymerase in the presence of m7Gpp(5′)G cap analog as described previously (29). Vero cells were transfected by a Lipofectin-mediated transfection method. Briefly, 10 μl of RNA transcription reaction mixture was mixed with 10 μl of Lipofectin (Gibco/BRL) at room temperature. The mixtures were added to Vero cells that had been washed twice with MEM. After incubation at 37°C for 2 h, the mixtures were removed and replaced with culture medium (liquid or agarose medium). At day 5 posttransfection, the liquid medium was harvested as viral stock. The cells, overlaid with agarose, were stained with neutral red to visualize viral plaques. BHK cells were transfected by electroporation as described previously (27). Briefly, BHK cells were harvested by trypsin treatment and washed twice with ice-cold phosphate-buffered saline without Ca2+ and Mg2+ and resuspended at a concentration of 107/ml. RNA transcripts (20 μl) were mixed with 0.5 ml of cells, and the mixture was transferred to a 2-mm-diameter cuvette. Electroporation was done at room temperature with two consecutive 1.5-kV, 25-μF pulses with a Gene-pulser (Bio-Rad). After electroporation, the cells were diluted in 10 ml of culture medium and distributed into six-well culture plates.
RNA slot blot hybridization.
Viral RNA replication was examined by RNA slot blot hybridization. For this purpose, total RNAs were extracted with TRIzol reagent from Vero cells transfected with mutant RNAs at 12, 24, or 36 h after transfection. Total RNAs (3 μg) were denatured with formamide and formaldehyde, slot blotted into a nitrocellulose membrane, and then hybridized with minus- or plus-strand RNA probes synthesized in vitro. For synthesis of minus- or plus-strand RNA probes, a DNA fragment representing the 5′ terminus of the viral RNA genome (nt 1 to 1070) was cloned into plasmid pSPT19 (Pharmacia Biotech) at the SmaI site in both orientations. The resulting constructs were used as templates for RNA transcription. The minus- or plus-strand RNA probe was transcribed in the presence of 50 μCi of [α-32P]CTP (NEN) (3,000 Ci/mmol) with SP6 RNA polymerase. The hybridization was in a buffer (50% formamide, 6× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 1% sodium dodecyl sulfate [SDS], 0.1% Tween-20, 100 μg of tRNA/ml) containing minus- or plus-strand RNA probe at 55°C overnight. After hybridization, the membrane was washed with 1× SSC containing 0.1% SDS twice at room temperature and with 0.1× SSC containing 0.1% SDS twice at 65°C and then exposed to X-ray film.
Metabolic labeling, immunoprecipitation, and endoglycosidase H (endo H) digestion.
For analysis of viral structural protein synthesis and transport, Vero cells infected with mutant viruses or BHK cells transfected with mutant RNAs were incubated at 37°C for 40 h, washed with MEM, and starved in methionine-free medium for 30 min at 37°C. This medium was replaced with one containing [35S]methionine (NEN) (200 μCi/ml), and the cells were pulse-labeled for 80 min. After being labeled, the cells were chased for various times in chase medium containing unlabeled methionine at 10 times the usual concentration. At each chase point, the chase medium was harvested for assay of released virus particles. The cells were washed with ice-cold phosphate-buffered saline and lysed in 200 μl of lysis buffer (1% Triton X-100, 10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 100 μg of phenylmethylsulfonyl fluoride/ml). The lysates were centrifuged to remove nuclei and immunoprecipitated with human anti-rubella virus serum. For immunoprecipitation, 50 μl of lysate was diluted to 150 μl with lysis buffer and 2 μl of human anti-rubella virus serum was added. After incubation at 4°C for 1 h, 40 μl of 50% protein A-Sepharose beads (Pharmacia Biotech) was added and incubated for a further 1 h at room temperature with shaking. The beads were washed three times with lysis buffer and resuspended in sample buffer (0.1 M citrate [pH 5.5], 0.15% SDS) and boiled for 5 min. After centrifugation, the immunoprecipitates were collected and mixed with SDS gel-loading buffer (62.5 mM Tris-HCl [pH 6.0], 2% SDS, 5% 2-mercaptoethanol, 500 mM sucrose) and then analyzed by SDS–10% polyacrylamide gel electrophoresis (PAGE) under reducing conditions.
For assay of released virus particles from transfected cells, the virus particles in the chase medium were precipitated with PEG and suspended in TNE buffer as described above. Triton X-100 was added to the suspension to a final concentration of 1%, and immunoprecipitation and SDS-PAGE analysis were done as described above.
For endo H digestion, the immunoprecipitates were digested with endo H (1 mU per 10 μl of immunoprecipitates) in the presence of phenylmethylsulfonyl fluoride for 14 h at 37°C. After digestion, the immunoprecipitates were mixed with SDS gel-loading buffer and analyzed by SDS-PAGE.
Sucrose gradient centrifugation.
Cell lysates were layered onto 5 to 20% (wt/wt) sucrose gradients in TNE buffer containing 0.1% Triton X-100 and centrifuged in an SW41 rotor at 38,000 rpm for 28 h at 5°C (2). Fractionated gradients (total, 26 fractions; 25 μl per fraction) were subjected to SDS–10% PAGE under nonreducing conditions. After electrophoresis, the proteins were transferred to nitrocellulose membranes and probed with human anti-rubella virus serum as described previously (24).
RESULTS
Characterization of pBRM33 cDNA clone.
RV M33 is one of the wild-type isolates of RV. For the construction of a full-length RV M33 cDNA clone, six overlapping DNA fragments covering the entire viral genome were individually amplified by RT-PCR with TaKaRa Taq polymerase, which has been used successfully for construction of the infectious cDNA clone of wild-type RV (f-Therien strain) (21). We have used Pfu DNA polymerase in PCR amplification of RV sequence and found that this enzyme produced smaller amounts of DNA than TaKaRa Taq polymerase and also amplified DNA fragments over 3 kb poorly. The six DNA fragments were assembled into a full-length cDNA by ligation after appropriate restriction sites were used (Fig. 1). For synthesis of RNA transcripts in vitro, an SP6 RNA polymerase promoter sequence was positioned immediately upstream of the full-length cDNA by using a primer containing the promoter sequence plus the first 14 nt at the 5′ terminus of RV (M33 strain) (29). The 3′-terminal sequence of the full-length cDNA consisted of a poly(A) tract (A31) followed by a HindIII site, which was introduced in cDNA synthesis by using a primer containing poly(dT15) and a HindIII site. The assembled full-length cDNA was cut with EcoRI and HindIII and inserted into modified plasmid, pBR322, to produce recombinant plasmid pBRM33 (Fig. 1). The HindIII site was used to linearize the plasmid for in vitro transcription of RNA by SP6 RNA polymerase.
A second recombinant plasmid, pBRM34, was constructed, in which a new SphI site in the E1 transmembrane domain was created by introduction of a silent mutation (changing T to A at nt 9647) in the pBRM33 clone. This clone was used for mutant construction as described below.
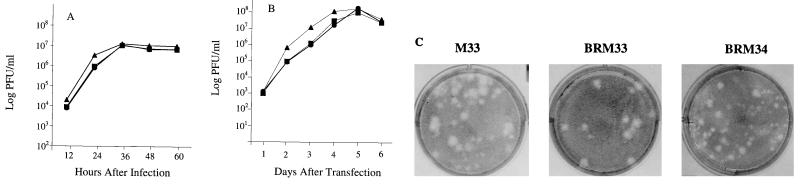
In the initial characterization of the full-length cDNA clone pBRM33, Vero cells were transfected with RNA transcripts synthesized from pBRM33. At day 5 posttransfection, the culture medium containing BRM33 virus was harvested as a viral stock. The relative growth rate of this BRM33 virus compared to that of the parental M33 virus was examined in Vero cells by viral infection at a multiplicity of infection (MOI) of 10 PFU/cell. The infected culture medium was harvested at 12-h intervals and replaced with fresh medium. The virus titers at each time point were determined and are shown in Fig. 2A. As can be seen, the growth rates for BRM33 and parental M33 virus were similar, with an exponential phase between 10 and 24 h postinfection and reaching a maximum rate at 36 h. Virus production continued at this rate up to 60 h after infection. At the maximum rate, BRM33 produced 107 PFU/ml/12 h, which was approximately the same as the parental M33 virus, although in the exponential phase the virus titer of BRM33 was twofold lower than that of M33.
FIG. 2.
Properties of BRM33, BRM34, and M33 viruses. BRM34 virus is a restriction site variant of BRM33 in which a new SphI site in the E1 transmembrane domain was created by introduction of a silent mutation (changing T to A at nt 9647) in the pBRM33 clone. (A) Vero cells were infected with virus at an MOI of 10 PFU/cell. At 12-h intervals, the medium was completely removed for titration and new medium was added. The titer was determined by plaque formation on Vero cells, and the mean titer from two experiments is shown for each time point. ●, BRM33 virus; ■, BRM34 virus; ▴, parental M33 virus. (B) Vero cells were transfected with RNA and Lipofectin and overlaid with liquid growth medium. At 24-h intervals, the medium was completely removed for titration and new medium was added. The titer was determined by plaque formation, and the mean titer from two experiments is shown. ●, pBRM33; ■, pBRM34; ▴, parental M33 RNA; The data points for the growth curves represent two independent experiments. (C) Morphology of plaques produced by BRM33 and BRM34. Vero cells were transfected with RNAs from pBRM33, pBRM34, or parental M33 by using Lipofectin, and the transfected cells were overlaid with agarose medium. After incubation for 6 days at 35°C, the cells were stained with neutral red.
The growth rate of BRM33 virus was also directly determined in primary transfected cells by titration of the culture medium. Vero cells were transfected with pBRM33 RNA and Lipofectin. The culture medium was harvested daily and replaced with fresh medium. The virus titers in the culture medium were quantitated and are shown in Fig. 2B. RNA transcripts from pBRM33 produced a large amount of infectious virus with titer and growth kinetics similar to those of the parental M33 RNA. The amount of virus produced at day 1 posttransfection was approximately 103 PFU/ml. After day 1, the rate of virus production rose rapidly until 4 days posttransfection and reached a peak at day 5, with the titer approaching 108 PFU/ml. After that, the virus production declined due to cell death. Taken together, these data indicate that RNA transcribed in vitro from pBRM33 is capable of producing infectious virus with a growth capacity similar to that of the parental M33 virus.
To determine the specific infectivity of BRM33 RNA, transfected cells were overlaid with agarose medium and viral plaques were allowed to develop. Vero cells transfected with BRM33 RNA synthesized in the presence of m7G(5′)ppp(5′)G cap analog gave rise to 3 × 103 plaques per μg of RNA (Table 1), indicating that BRM33 RNA is as infectious as the RNA transcripts from another infectious cDNA clone derived from RV, f-Therien strain (21). Plaques formed from BRM33 RNA were large (3-mm diameter), similar to those of M33 RNA (Fig. 2C). Some smaller plaques were also present in both M33 and BRM33 RNA-transfected cells. No plaques were detected with BRM33 RNA transcribed in the absence of cap analog, suggesting that the cap structure at the 5′ terminus of the RV genome is required for viral infectivity.
TABLE 1.
Specific infectivity of RV M33 full-length cDNA clone
| RNA used for transfection |
Specific infectivity (plaques/μg)a |
|---|---|
| pBRM33 transcripts | |
| No cap | 0 |
| m7G(5′)ppp(5′)G | 3 × 103 |
| pBRM34 transcriptsb | |
| m7G(5′)ppp(5′)G | 1.5 × 103 |
| M33 RNA | 2 × 104 |
Vero cells were transfected with RNA transcripts and Lipofectin. After transfection, the cells were overlaid with agarose medium and incubated at 35°C for 6 days. The plaques were visualized by neutral red stain, and the specific infectivity of the RNA was determined from the number of plaques formed per microgram of RNA.
pBRM34 is a restriction site variant of pBRM33 in which a new SphI site in the E1 transmembrane domain was created by changing T to A at nt 9647 in the pBRM33 clone.
It is notable that BRM33 virus grew more slowly than did the parental M33 virus. The RNA transcripts synthesized with SP6 RNA polymerase start with an additional G residue at their 5′ ends that is contributed by the last nucleotide of the SP6 promoter. It has been reported by Pugachev et al. (21) that in the cDNA clone derived from f-Therien strain, after one passage, 60% of 5′-terminal clones still had the extra G, but after five passages, no clones containing extra G were found. After five passages, plaques of the virus appear to be larger and the virus grows to a higher titer than does virus harvested after one passage. Data presented in Fig. 2B and C were obtained from RNA transcript transfection without subsequent virus passage. The lag in virus growth and smaller-plaque phenotype observed in BRM33 and BRM34 viruses are likely due to the extra G present at the 5′ ends of the RNA transcripts.
Construction of mutants in transmembrane and cytoplasmic domains of E1.
Possession of the infectious cDNA clone pBRM33 enabled us to introduce mutations of interest into RV. It has been suggested that RV E1 plays an important role in virus assembly, possibly by mediating the interaction with nucleocapsids (11). To investigate this, alanine substitution mutagenesis was performed in the E1 transmembrane and cytoplasmic domains with the pBRM33 cDNA clone. Three cysteine and two leucine residues in the E1 transmembrane and cytoplasmic domains (Fig. 3A) were chosen as the targets in mutagenesis. In alphaviruses, it has been reported that the cysteine residues in the cytoplasmic domain of E2 are critical in the interaction between the nucleocapsid core and the membrane-embedded E2 and E1 (8, 13, 30). To facilitate the mutagenesis process, we used the pBRM34 clone containing a created SphI site in the E1 transmembrane domain. Since the changed base is the third base of a cysteine codon, the change is silent. The characterization of the resulting construct, pBRM34, is shown in Fig. 2 and Table 1. These data clearly show that there is no distinguishable difference between pBRM33 and pBRM34 in terms of virus growth, plaque size, and specific infectivity. Thus, alteration of nucleotide T to A at position 9647 had no effect on viral replication.
FIG. 3.
Mutations in E1 transmembrane and cytoplasmic domains and growth curves of mutant viruses. (A) Amino acid sequence of E1 transmembrane and cytoplasmic domains and substitutions produced by mutagenesis. The numbering is based on the sequence published by Clarke et al. (4). (B) Growth of mutant viruses. Vero cells were transfected with RNAs and Lipofectin and overlaid with liquid growth medium. At 24-h intervals, the medium was removed for virus titration and new medium was added. ●, BRM34; □, C466A/C467A; ■, C467A; ○, C470A; ⧫, L471A; ▴, L464A; ▵, C466A.
By taking advantage of the SphI site created, we constructed six mutants based on pBRM34 by site-directed mutagenesis: three mutations in the E1 transmembrane domain and two mutations in the cytoplasmic domain. As shown in Fig. 3A, these comprised three single-amino-acid changes of Leu 464, Cys 466, and Cys 467 to Ala; a double change of Cys 466 and 467 to Ala in the E1 transmembrane domain; and two single-amino-acid substitutions of Ala for Cys 470 and Leu 471 in the E1 cytoplasmic domain.
Growth characteristics of mutant viruses.
To examine the effect of these mutations on viral growth, Vero cells were transfected with mutant RNAs by using Lipofectin and the titers of virus released into the medium were determined. The results are shown in Fig. 3B. In mutants C467A and C466A/C467A (transmembrane domain mutants), virus production was severely impaired and the virus yield was only about 1% of that of the parental BRM34 virus. Though mutant C466A also released slightly less virus than did BRM34, its virus titer was much higher than that of mutant C467A with the same mutation at the next position. Mutant L464A also produced slightly less virus, notably in the later stage of infection. In the case of cytoplasmic domain mutants, mutant C470A produced amounts of virus equal to that produced by BRM34. However, substitution of Ala for Leu 471 (next to Cys 470) had a profound effect on virus release: mutant L471A produced about 10% as much virus as the parental BRM34 virus. The plaques formed by C467A, C466A/C467A, and L471A mutants in Vero cells were found to be smaller than those of the BRM34 virus (data not shown).
RNA replication.
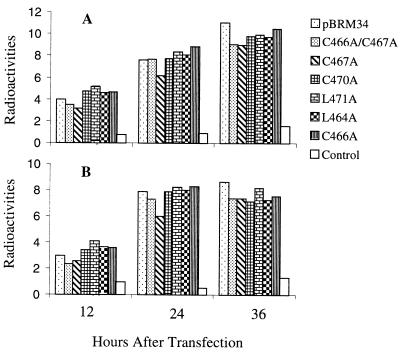
It is possible that the reduction in virus yield in mutants C467A, C466A/C467A, and L471A was due to the effect of these mutations on viral RNA replication, since the mutated nucleotides (between nt 9642 and 9665) are located near a potential stem-and-loop structure (nt 9669 to 9720). This stem-and-loop structure has been shown to associate with host cell proteins (20), although its function is unknown. To determine whether RNA replication of these mutants was affected, both minus- and plus-strand RNA synthesis at 12, 24, or 36 h postinfection were examined in transfected Vero cells by slot blot hybridization. The amount of RNA in each dot was quantitated by densitometry of autoradiographs. RNA extracted from a mock infection was used as a control. As shown in Fig. 4, synthesis of both minus- and plus-strand RNA was detected as early as 12 h after transfection. The amount of RNA was increased in both blots hybridized with either minus- or plus-strand RNA probes from 12 to 24 h posttransfection. No significant difference was observed between the mutants and parental BRM34 at 24 and 36 h in either minus- or plus-strand RNA blots except that for mutant C467A the RNA level was slightly lower than that of the parental BRM34 at 24 h posttransfection. However, the RNA synthesis of mutant C467A caught up with those of the other mutants at 36 h after transfection. These data indicate that the nucleotide changes introduced in the mutants had no significant effect on viral RNA replication, though the changes in mutant C467A may slightly impair early RNA synthesis.
FIG. 4.
RNA slot blot hybridization. Total RNA was extracted from Vero cells transfected by Lipofectin with RNAs from mutants or parental BRM34 at 12, 24, or 36 h posttransfection. RNA extracted from mock-infected Vero cells was used as a control. Total RNA (3 μg) from each culture was dotted onto nitrocellulose membranes and hybridized with 32P-labeled RNA probes as described in Materials and Methods. The amount of radiolabel in each dot was quantitated by densitometry of the autoradiographs. (A) Membrane hybridized with 32P-labeled minus-strand RNA probe. (B) Membrane hybridized with 32P-labeled plus-strand RNA probe. All dots were exposed to X-ray film for the same period. The data shown represent two separate experiments.
Viral structural protein synthesis, transport, and release as virus particles.
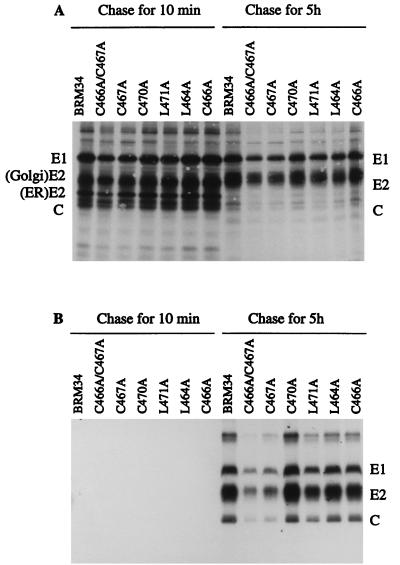
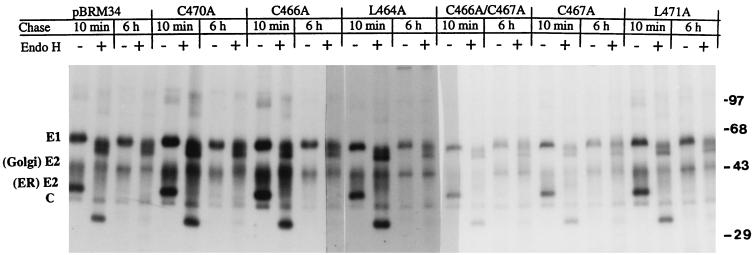
Since RNA replication in the mutants was not significantly affected by the introduced mutations, viral structural protein synthesis, transport, and release as virus particles were examined to determine which stages of viral replication were affected. Vero cells were infected with mutant viruses, and at 40 h after infection, the cells were pulse-labeled with [35S]methionine for 80 min and chased with unlabeled methionine for 10 min or 5 h. The cellular lysates and corresponding chase media containing virus particles were immunoprecipitated with human anti-rubella virus serum and analyzed by SDS-10% PAGE (Fig. 5). After 80 min of pulse-labeling and a short chase of 10 min, the structural proteins E1, E2, and capsid were observed in cells infected with all mutant and BRM34 viruses (Fig. 5A). Two E2 forms, an ER form (39 kDa), and a Golgi form (42 to 47 kDa) were observed (10). Below the 39-kDa E2 is a 33-kDa capsid protein. After 5 h of chase, the ER form of E2 in all mutant viruses was completely converted to the Golgi form of E2 (Fig. 5A) and virus particles were observed in the corresponding 5-h chase medium (Fig. 5B). These results indicate that the structural proteins in all of the mutant viruses were synthesized, cleaved, and transported out of the ER. To compare the viral structural protein synthesis and virus release of the mutants and parental BRM34 virus, the amounts of intracellular E1 after 10 min of chase and extracellular E1 after 5 h of chase in each mutant virus were quantified by densitometry. The results are shown in Table 2. The amounts of intracellular E1 synthesized by mutants C470A, L464A, and C466A were equal to that of the parental BRM34 virus, indicating that structural protein synthesis in these mutants was similar to that of the parental BRM34 virus. Slightly smaller amounts of structural proteins were observed in mutants C466A/C467A, C467A, and L471A, as they produced about 60, 70, and 80%, respectively, as much E1 protein as the parental BRM34 virus (Table 2). However, the amounts of virus released from the infected cells by these mutants were very different. Compared to the parental BRM34 virus, all mutants except C470A released smaller amounts of virus. The most significant reductions in virus release were observed in mutants C466A/C467A, C467A, and L471A, which only released about 30, 40, and 70%, respectively, of the amount of parental BRM34 virus (Table 2). After normalizing the values to those of intracellular E1, the amounts of virus released by mutants C466A/C467A, C467A, and L471A were 50, 57, and 88%, respectively, of that released by the parental BRM34 virus. The normalized values are not sufficiently low to account for the 10- to 100-fold reduction in infectivity shown in mutants C466A/467A, C467A, and L471A, suggesting that the mutant viruses which were released might have reduced infectivity.
FIG. 5.
Synthesis, transport, and release of structural proteins following transfection of mutant RNAs. Vero cells were infected with mutant or parental BRM34 viruses at an MOI of 2. At 40 h postinfection, the infected cells were pulse-labeled for 80 min with [35S]methionine and chased with medium containing unlabeled methionine for 10 min or 5 h. The chase medium was harvested, and the labeled cells were lysed with buffer containing 1% Triton X-100. (A) The lysates were immunoprecipitated with human anti-rubella virus serum, and the immunoprecipitates were collected with protein A-Sepharose CL-4B and analyzed by SDS–10% PAGE. (B) Virus particles in the chase medium were precipitated with PEG and resuspended in TNE buffer containing 1% Triton X-100. The suspension was immunoprecipitated with human anti-rubella virus serum and analyzed by SDS–10% PAGE and subsequent autoradiography. The positions of migration of RV structural proteins E1, E2, and C are shown.
TABLE 2.
Relative rates of synthesis and release of viral structural protein
| Virus | Relative rate of synthesis and release of:
|
|||
|---|---|---|---|---|
| Intracellular E1 protein
|
Extracelluar E1 protein
|
|||
| Radiolabela | Ratiob | Radiolabelc | Ratiob | |
| BRM34 | 10.6 | 1.0 | 5.2 | 1.0 |
| C466A/C467A | 6.2 | 0.6 | 1.6 | 0.3 |
| C467A | 7.8 | 0.7 | 2.3 | 0.4 |
| C470A | 10.2 | 1.0 | 5.6 | 1.0 |
| L471A | 8.6 | 0.8 | 3.5 | 0.7 |
| L464A | 10.8 | 1.0 | 4.7 | 0.9 |
| C466A | 10.9 | 1.0 | 4.3 | 0.8 |
Amounts of radiolabel (expressed in densitometry units) in intracellular E1 bands after 10 min of chase in Fig. 5A were quantitated by densitometry of autoradiographs.
Ratio of radiolabel to that in BRM34.
Amounts of radiolabel (expressed in densitometry units) in extracellular E1 bands in the virions released into medium after 5 h of chase (Fig. 5B) were quantitated by densitometry of autoradiographs.
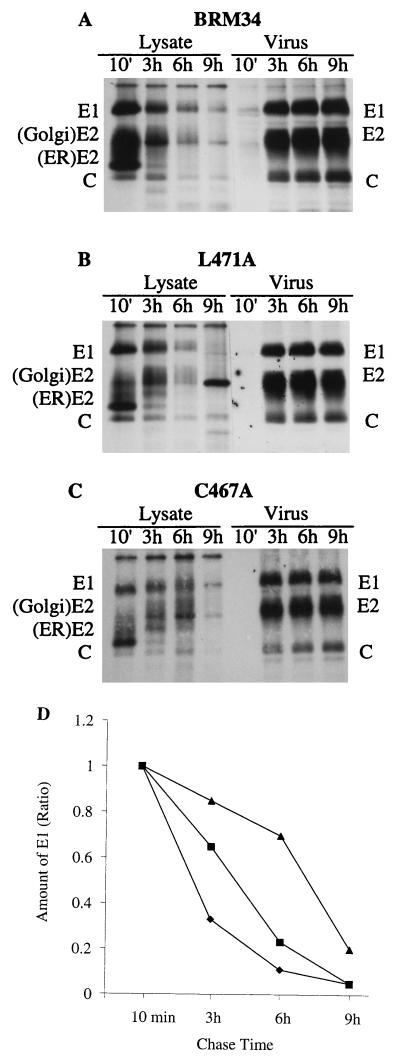
Since a reduction in virus release was observed on substitution of cysteine 467 and leucine 471, further pulse-chase experiments were performed to determine whether an intracellular accumulation of structural proteins occurred in the two mutants that would indicate a possible block in virus budding. BHK cells were transfected with mutant C467A or L471A or parental BRM34 RNA and incubated for 40 h. The transfected cells were pulse-labeled with [35S]methionine for 80 min and then incubated with excess unlabeled methionine for 10 min and 3, 6, and 9 h. At each chase point, the cell lysates and corresponding chase medium were immunoprecipitated and analyzed by SDS–10% PAGE (Fig. 6A to C). The amount of radiolabel in intracellular E1 at each point was quantitated by densitometry. The ratios of radiolabel in E1 at 3, 6, and 9 h of chase to that at 10 min of chase for mutants C467A, L471A, and parental BRM34 were plotted and are shown in Fig. 6D. In parental BRM34 after 3 h of chase, the intracellular E1 was 33% of that at 10 min of chase, whereas about 85 and 65% of E1 radiolabel in C467A and L471A, respectively, were intracellular. After a 6-h chase, most of the intracellular E1 in BRM34 and mutant L471A was released from the cells, with only 11 and 23%, respectively, of E1 remaining intracellular. However, in mutant C467A, most of the E1 (70%) remained intracellular. By comparison with the ratios of E1 at each chase point between the mutants and parental BRM34, it is clear that the E1 protein accumulated in mutants C467A and L471A during the chase period, with mutant C467A accumulating more proteins than mutant L471A. Thus, these results, consistent with the results shown in Fig. 3B and 5 and Table 2, suggest that the defect in virus release is at the late stage of virus assembly. It is worth noting that the reduction in radiolabel in intracellular E1 may not be due solely to release of E1 from the cells. Quantitation of the release of E1 in the mutants revealed that the reduction in intracellular E1 was not proportional to the increase in the release of E1 from the cells (data not shown), indicating that degradation of intracellular E1 might occur expecially after 9 h of chase. Nonetheless, less virus was released in mutants C467A and L471A at each chase point compared to the parental virus.
FIG. 6.
Pulse-chase analysis of intracellular structural proteins. BHK cells were transfected with mutant C467A, L471A, or parental BRM34 RNAs and incubated for 40 h. The transfected cells were pulse-labeled with [35S]methionine for 80 min and then chased with unlabeled methionine for 10 min or 3, 6, or 9 h. The cell lysates and corresponding chase media were immunoprecipitated with human anti-rubella virus serum and analyzed by SDS–10% PAGE. (A) Parental BRM34; (B) mutant L471A; (C) mutant C467A. (D) The amount of radiolabel in intracellular E1 at each time point was quantitated by densitometry of the autoradiographs. The ratios of radiolabel in E1 at 3, 6, and 9 h of chase to that at 10 min of chase (set to 1) are shown. ⧫, parental BRM34; ■, L471A; ▴, C467A.
To examine whether the transport of RV structural proteins was affected, the immunoprecipitates of cell lysates were treated with endo H. RV structural proteins undergo extensive glycosylation during transport from the ER and passage through the Golgi complex to the cell surface (10). The acquisition of resistance to endo H digestion by RV structural proteins is an indication that they have reached the cis-Golgi compartment (10, 23). As shown in Fig. 7, during the 10-min chase period, the ER form of E2 in all mutants was sensitive to endo H digestion, resulting in a reduction in apparent molecular mass from 39 to 31 kDa. However, treatment of E1 with endo H led to only partial digestion (Fig. 7). Complete digestion would reduce the molecular mass of E1 from 58 to 51 kDa. After 6 h of chase, the ER form of E2 in all mutants became completely endo H resistant. The majority of E1 also became resistant. E1 contains three glycosylation sites, and it appears that only one site is processed to add a complex type of glycan (10). No change in molecular mass was found with capsid after endo H digestion. These results indicate that E1 and E2 of all mutants were transported normally.
FIG. 7.
endo H treatment of structural proteins. Immunoprecipitates were isolated as described in the legend to Fig. 5, digested with endo H at 37°C for 14 h, and then analyzed by SDS–10% PAGE and autoradiography. −, not treated with endo H; +, treated with endo H. The positions of molecular mass markers are shown on the right (in kilodaltons).
E2/E1 heterodimer formation.
RV E2 and E1 form a noncovalent protein complex, not dissociated by nonionic detergents, and the formation of intracellular E2/E1 heterodimers can be detected by sedimentation analysis in sucrose gradients (1). To examine whether E2/E1 heterodimer formation was affected in mutants C467A and L471A, we performed sucrose gradient sedimentation analysis of cellular lysates treated with Triton X-100. Vero cells were infected with the mutant or the parental BRM34 viruses. At 3 days postinfection, Triton X-100 lysates were prepared and centrifuged on a 5 to 20% sucrose gradient followed by fractionation. The fractionated gradients were analyzed by SDS-PAGE under nonreducing conditions. RV antigens transferred to membranes were detected with human anti-rubella virus serum. E1 protein was found in all fractions of the gradient, whereas E2 was found in two regions (Fig. 8). According to Baron and Forsell (1), the slower-migrating E1 (fractions 16 to 20) was monomeric E1 and the faster-sedimenting peak (fractions 5 to 15) contained E2/E1 heterodimers as well as E1 oligomers (Fig. 8). Therefore, it is likely that the fractions between 11 and 15 contained mostly E2/E1 heterodimer. Comparison of the sedimentation profiles of BRM34, L471A, and C467A shows that E2 and E1 form a heterodimer in the mutants, unaffected by the introduced mutations. RV E2 protein is heavily but heterogeneously glycosylated. The heavier E2 fractions (5 to 10) contained mostly Golgi-form E2, while the lighter fractions (11 to 20) contained mostly ER-form E2 and some unglycosylated E2 with a molecular mass of 31 kDa (Fig. 8).
FIG. 8.
E2/E1 heterodimer formation. Vero cells were infected with C467A, L471A, or parental BRM34 virus. After 3 days of infection, the cells were lysed with buffer containing 1% Triton X-100. The lysates were applied to a 5 to 20% (wt/wt) sucrose gradient in TNE buffer containing 0.1% Triton X-100. After centrifugation in an SW41 rotor at 5°C and at 38,000 rpm for 28 h, the gradients were fractionated from the bottom of the tube. Samples (25 μl per fraction) were analyzed by SDS–10% PAGE under nonreducing conditions followed by immunoblotting and probing with human anti-rubella virus serum. Note that fractions 5 to 20 are shown in the figure. Sedimentation is from right to left. The capsid protein was found in the pellet fractions at the bottom of the gradient. The positions of molecular mass markers are shown on the right (in kilodaltons).
DISCUSSION
In this study, we constructed a full-length cDNA clone, pBRM33, derived from the M33 strain of RV. The RNA transcripts synthesized from pBRM33 in vitro were highly infectious, with specific infectivity approaching 104 plaques per μg of RNA, and produced viruses showing characteristics of growth rate and plaque size similar to those of the parental M33 virus. The amounts of virus produced by BRM33 were comparable to those produced by parental M33 (107 PFU/ml). The full-length cDNA in pBRM33 plasmid is also stable, without any loss of infectivity after several passages in Escherichia coli. During construction of the cDNA clone, we found that purification of the virion RNA used in RT-PCR by oligo(dT)-cellulose chromatography, as was reported previously, was not necessary (21). Interestingly, we were also able to amplify a DNA fragment that covered the region between KpnI (nt 1723) and NheI (nt 2800) previously reported not to be amplified (21). The reasons are not clear. Perhaps there are differences in RNA secondary structure between M33 and f-Therien viruses or the primers used in our experiments are more efficient.
We also demonstrated the feasibility of using an infectious cDNA clone for the analysis of RV RNA replication and gene expression. We were able to detect the synthesis of minus- or plus-strand RNA in Vero cells transfected with RNA transcripts by slot blot hybridization as early as 12 h posttransfection and to express larger amounts of structural proteins in BHK cells by electroporation of RNA transcripts without using amplified virus. This expression system is important in studying RV mutants with higher frequencies of reversion or second-site mutation, since it allows phenotypic analysis of the mutants after one round of RNA replication. The system also permits the study of null mutants from which virus cannot be rescued.
It has been proposed that E1 in the E2/E1 heterodimer may be involved in the assembly of RV, possibly through interaction with nucleocapsids by its cytoplasmic domain (11). We studied the role of E1 in viral assembly by introducing mutations in its transmembrane and cytoplasmic domains into our constructed infectious cDNA clone. Nucleotide changes introduced in the transmembrane or cytoplasmic domain did not significantly alter the rate of minus- or plus-strand RNA synthesis, and all mutant RNAs replicated normally. Biosynthesis studies showed that E2 and E1 were synthesized and efficiently cleaved in all mutants, although slightly reduced levels of protein synthesis were observed in mutants C466A/C467A and C467A. Both E2 and E1 were also transported normally, as evidenced by pulse-chase experiments in which E2 protein synthesized in the ER was completely converted to the Golgi form with a higher molecular mass of 42 to 47 kDa, and both E2 and E1 acquired endo H resistance after the chase. However, a reduction in virus release was observed with all mutants except C470A. Substitution of Ala for Leu 471 in the cytoplasmic domain and of Ala for Cys 467 in the transmembrane domain led to significant reductions in virus release. This defect in virus release was not due to a failure of E2-E1 interaction, since the mutated E1 formed heterodimers with E2 (Fig. 8). However, an accumulation of intracellular structural proteins was observed during pulse-chase experiments (Fig. 6). Thus, these results indicate that E1 plays a critical role at a very late stage of viral assembly and probably in the final virus budding process. Hobman et al. (11) have studied RV assembly by expression of RV structural proteins with an expression vector. They showed that COS cells transiently expressing the structural proteins secrete virus-like particles into the medium in a capsid-dependent manner. Deleting or replacing the E1 cytoplasmic domain was found to inhibit the secretion of RV structural proteins from the transfected cells, suggesting that the cytoplasmic domain of E1 is required for virus assembly. However, the effect of deletion of the cytoplasmic domain on E2/E1 heterodimer formation and transport was not examined. Nonetheless, the results of our studies are consistent with those of Hobman et al., indicating that the E1 cytoplasmic domain is involved in virus assembly. It is unclear how mutation of Leu 471 in the E1 cytoplasmic domain affects virus assembly. However, we found that substitution of Tyr 472 by Ala (next to Leu 471) also resulted in severe reduction in virus release (unpublished data). In contrast, no inhibition of virus production was observed with mutation of Cys 470 in the cytoplasmic domain, suggesting that the leucine residue at position 471 may represent the N terminus of the interactive region in the cytoplasmic domain. The reduction in virus release observed with the mutant L471A, therefore, is likely caused by altered interaction between its E1 cytoplasmic domain and other structural proteins, possibly nucleocapsids. Similar results were reported with alphaviruses, in which it is known that virus budding is driven by the interaction of the E2 cytoplasmic domain with nucleocapsids (25). Any changes in the amino acid residues that form the N-terminal region of the E2 cytoplasmic domain were found to be deleterious to alphavirus assembly (8, 13, 19, 28, 30).
Substitution of Cys 467 in the E1 transmembrane domain dramatically reduced virus release, indicating that the transmembrane domain of E1 is also involved in virus assembly. It is not clear at present how the alteration of Cys 467 influences virus assembly. E1 is known to undergo palmitoylation (10, 26). The Cys 467 is, therefore, one of the possible sites for attachment of palmitic acid, as suggested by others (7). Palmitoylation usually results in better anchoring of membrane proteins in cellular membranes. If this is the site for palmitoylation, mutation of Cys 467 would block fatty acid acylation in E1 and would lead to weakly anchored E1 protein. Since our experimental results showed that substitution of Ala for Cys 467 did not affect E2/E1 heterodimer formation, therefore, it is tempting to speculate that the palmitoylation of Cys 467 is required for better E1 anchoring, thereby allowing correct positioning of its cytoplasmic domain in the cytoplasm. This possibility is under investigation.
ACKNOWLEDGMENTS
This work was supported by a grant from the Medical Research Council of Canada. Shirley Gillam is an investigator of British Columbia’s Children’s Hospital Foundation.
REFERENCES
- 1.Baron M D, Forsell K. Oligomerization of the structural proteins of rubella virus. Virology. 1991;185:811–819. doi: 10.1016/0042-6822(91)90552-m. [DOI] [PubMed] [Google Scholar]
- 2.Barth B-U, Wahlberg J M, Garoff H. The oligomerization reaction of the Semiliki Forest virus membrane protein subunits. J Cell Biol. 1995;128:283–291. doi: 10.1083/jcb.128.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J-P, Strauss J H, Strauss E G, Frey T K. Characterization of the rubella virus nonstructural protease domain and its cleavage site. J Virol. 1996;70:4707–4713. doi: 10.1128/jvi.70.7.4707-4713.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarke D M, Loo T W, Hui I, Chong P, Gillam S. Nucleotide sequence and in vitro expression of rubella virus 24S subgenomic mRNA encoding the structural proteins E1, E2 and C. Nucleic Acids Res. 1987;15:3041–3057. doi: 10.1093/nar/15.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke D M, Loo T W, McDonald H, Gillam S. Expression of rubella virus cDNA coding for the structural proteins. Gene. 1988;65:23–30. doi: 10.1016/0378-1119(88)90413-1. [DOI] [PubMed] [Google Scholar]
- 6.Dominguez G, Wang C, Frey T K. Sequence of the genome RNA of rubella virus: evidence for genetic rearrangement during Togavirus evolution. Virology. 1990;177:225–238. doi: 10.1016/0042-6822(90)90476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frey T K. Molecular biology of rubella virus. Adv Virus Res. 1994;44:69–160. doi: 10.1016/S0065-3527(08)60328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaedigk-Nitschko K, Schlesinger M J. Site-directed mutations in Sindbis virus E2 glycoprotein’s cytoplasmic domain and the 6K protein lead to similar defects in virus assembly and budding. Virology. 1991;183:206–214. doi: 10.1016/0042-6822(91)90133-v. [DOI] [PubMed] [Google Scholar]
- 9.Hobman T C, Gillam S. In vitro and in vivo expression of rubella virus glycoprotein E2: the signal peptide is contained in the C-terminal region of capsid protein. Virology. 1989;173:241–250. doi: 10.1016/0042-6822(89)90240-7. [DOI] [PubMed] [Google Scholar]
- 10.Hobman T C, Lundstrom M L, Gillam S. Processing and intracellular transport of rubella virus structural proteins in COS cells. Virology. 1990;178:122–133. doi: 10.1016/0042-6822(90)90385-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hobman T C, Lundstrom M L, Mauracher C A, Woodward L, Gillam S, Farquhar M G. Assembly of rubella virus structural proteins into virus-like particles in transfected cells. Virology. 1994;202:574–585. doi: 10.1006/viro.1994.1379. [DOI] [PubMed] [Google Scholar]
- 12.Hobman T C, Shukin R, Gillam S. Translocation of rubella virus glycoprotein E1 into the endoplasmic reticulum. J Virol. 1988;62:4259–4264. doi: 10.1128/jvi.62.11.4259-4264.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivanova L, Schlesinger M J. Site-directed mutations in the Sindbis virus E2 glycoprotein identify palmitoylation sites and affect virus budding. J Virol. 1993;67:2546–2551. doi: 10.1128/jvi.67.5.2546-2551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopes S, Yao J, Kuhn R, Strauss E G, Strauss J H. The E2 glycoprotein-nucleocapsid interaction is required for alphavirus assembly. J Virol. 1994;68:1316–1323. doi: 10.1128/jvi.68.3.1316-1323.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matthews R E F. Classification and nomenclature of viruses. Intervirology. 1982;17:1–99. doi: 10.1159/000149278. [DOI] [PubMed] [Google Scholar]
- 16.McDonald H, Hobman T C, Gillam S. The influence of capsid protein cleavage on the processing of E2 and E1 glycoproteins of rubella virus. Virology. 1991;183:52–56. doi: 10.1016/0042-6822(91)90117-t. [DOI] [PubMed] [Google Scholar]
- 17.Oker-Blom C. The gene order for rubella virus structural proteins is NH2-C-E2-E1-COOH. J Virol. 1984;51:964–973. doi: 10.1128/jvi.51.2.354-358.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oker-Blom C, Ulmanen I, Kääriäinen L, Pettersson R F. Rubella virus 40S genome RNA specifies a 24S subgenomic mRNA that codes for a precursor to structural proteins. J Virol. 1984;49:403–408. doi: 10.1128/jvi.49.2.403-408.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Owen K E, Kuhn R J. Alphavirus budding is dependent on the interaction between the nucleocapsid and hydrophobic amino acid on the cytoplasmic domain of the E2 envelope glycoprotein. Virology. 1997;230:187–196. doi: 10.1006/viro.1997.8480. [DOI] [PubMed] [Google Scholar]
- 20.Pogue G P, Cao X-Q, Singh N K, Nakhasi H L. 5′ sequences of rubella virus RNA stimulate translation of chimeric RNA and specifically interact with two host encoded proteins. J Virol. 1993;67:7106–7117. doi: 10.1128/jvi.67.12.7106-7117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pugachev K V, Abernathy E S, Frey T K. Improvement of the specific infectivity of the rubella virus (RUB) infectious clone: determinants of cytopathogenicity induced by RUB map to the nonstructural proteins. J Virol. 1997;71:562–568. doi: 10.1128/jvi.71.1.562-568.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pugachev K V, Abernathy E S, Frey T K. Genomic sequence of the RA27/3 vaccine strain of rubella virus. Arch Virol. 1997;142:1165–1180. doi: 10.1007/s007050050150. [DOI] [PubMed] [Google Scholar]
- 23.Qiu Z, Hobman T C, McDonald H L, Seto N O L, Gillam S. Role of N-linked oligosaccharides in processing and intracellular transport of E2 glycoprotein of rubella virus. J Virol. 1992;66:3514–3521. doi: 10.1128/jvi.66.6.3514-3521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu Z, Ou D, Wu H, Hobman T C, Gillam S. Expression and characterization of virus-like particles containing rubella virus structural proteins. J Virol. 1994;68:4086–4091. doi: 10.1128/jvi.68.6.4086-4091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waxham M N, Wolinsky J S. A model of the structural organization of rubella virions. Rev Infect Dis. 1985;7(Suppl. 1):S133–S139. doi: 10.1093/clinids/7.supplement_1.s133. [DOI] [PubMed] [Google Scholar]
- 27.Yao J, Strauss E G, Strauss J H. Interaction between PE2, E1, and 6K required for assembly of alphaviruses studied with chimeric viruses. J Virol. 1996;70:7910–7920. doi: 10.1128/jvi.70.11.7910-7920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao J, Strauss E G, Strauss J H. Molecular genetic study of the interaction of Sindbis virus E2 with Ross River E1 for virus budding. J Virol. 1998;72:1418–1423. doi: 10.1128/jvi.72.2.1418-1423.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao J, Yang D, Chong P, Hwang D, Liang Y, Gillam S. Proteolytic processing of rubella virus non-structural proteins. Virology. 1998;246:74–82. doi: 10.1006/viro.1998.9179. [DOI] [PubMed] [Google Scholar]
- 30.Zho H, Lindqvist B, Garoff H, von Bonsdorf C H, Liljestrom P. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 1994;13:4204–4211. doi: 10.1002/j.1460-2075.1994.tb06740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]