Abstract
Because of growing levels of antibiotic resistance, new methods to combat bacteria are needed. We hypothesized that because bacteria evolved to survive in specific environments, the addition of compounds, including nutrient type compounds, to an environment, might result in a modification of that environment that will disrupt bacterial growth or in maladaptive bacterial behavior, i.e., gene expression. As a proof of concept, we focused on the egg white environment and the pathogen Salmonella. Despite egg white’s antibacterial nature, Salmonella is able to survive and grow in egg white, and this ability of Salmonella leads to infection of chicks and humans. Here, the 20 L-amino-acids were screened for their ability to affect the growth of Salmonella in egg white. L-arginine and L-cysteine were found to reduce growth in egg white in physiologically relevant concentrations. To determine the mechanism behind L-arginine inhibition TnSeq was utilized. TnSeq identified many Salmonella genes required for survival in egg white including genes required for iron import, biotin synthesis, stress responses, cell integrity, and DNA repair. However, a comparison of Salmonella in egg white with and without L-arginine identified only a few differences in the frequency of transposon insertions, including the possible contribution of perturbations in the cell envelope to the inhibition mechanism. Finally, both D-arginine and D-cysteine were found to inhibit Salmonella in egg white. This implied that the effect of arginine and cysteine in egg white is chemical rather than biological, likely on the egg white environment or on the bacterial outer membrane. To conclude, these results show that this approach of addition of compounds, including nutrient type compounds, to an environment can be used to limit bacterial growth. Importantly, these compounds have no inherent anti-bacterial properties, are used as nutrients by animals and bacteria, and only become anti-bacterial in a specific environmental context. Future research screening for the effects of compounds in relevant environments might uncover new ways to reduce pathogen levels in the poultry industry and beyond.
Keywords: anti-bacterial compounds, Salmonella, egg white, antibiotic resistance, tnseq
Introduction
Fueled by raising rates of antibiotic resistance, an extensive search for new antibiotics or approaches to fight bacterial pathogens has been ongoing for the last decades (Frieri et al., 2017). Here we present a proof of concept for such an approach to inhibit bacterial growth or survival: the addition of compounds, including nutrient type compounds, at levels not typical for the environment. The idea for this approach is based on the fact that bacteria evolved for very specific environments and that adding compounds to an environment, might either modulate the environment such that bacterial evolutionary programming is no longer appropriate for the modified environment, or through activation of inappropriate bacterial programming, causing the bacteria to behave, i.e., express genes, in a way that is maladaptive in that environment.
To test this idea, the egg white environment and the pathogen Salmonella were chosen. Egg white is a restrictive environment containing mainly water and proteins but with almost no sugar or fat. Furthermore, chelating proteins limit the available iron (Legros et al., 2021), and the presence of avidin limits biotin availability (Pugliese et al., 1993). Finally, several proteins in the egg white are anti-microbial enzymes. However, some Salmonella serovars are known to grow in egg white (Baron et al., 1997). Importantly, Salmonella’s ability to survive and grow in egg white is one of the mechanisms contributing to its spread to other chickens and food derived gastroenteritis (Baron et al., 2016). Indeed, Salmonella remains a health problem for humans and chickens and an economic problem for the poultry industry (Gast et al., 2022).
The 20 L-amino acids were chosen for this proof of concept as these molecules are used by all lifeforms and are a nutrient source for both Salmonella as well as chickens. Thus, to provide a proof of concept for this new anti-bacterial approach, the 20 L-amino acids were screened for their effect on Salmonella’s survival and growth in egg white.
Materials and methods
Bacterial strains
Salmonella enterica serovar Typhimurium strain 14028s was used as wild-type in this study. The following knockout mutants derived from this wild type strain were also used: ΔbcsA:kan (Mills et al., 2015), ΔSTM14_1614::cam, ΔSTM14_5387::cam, ΔSTM14_5436::cam (ΔhsdS:cam), ΔSTM14_0418::cam, ΔSTM14_0191::cam, ΔSTM14_5316::cam, ΔSTM14_5327::cam, ΔSTM14_5319::cam, and ΔcsgB:kan (Porwollik et al., 2014). Finally, two Escherichia strains were used: Escherichia coli strain MG1655 and Escherichia fergusonii strain Yara015 (Poudel et al., 2022). Bacteria were grown overnight in Lysogeny Broth (LB) at 37°C shaking. OD600 of overnight cultures was measured before dilution of bacteria into egg white.
Egg white
The experimental setup of all the experiments is described in Figure 1. Unfertilized eggs were obtained from the hen house at the Robert H. Smith Faculty of Agriculture, Food and Environment on the day of laying and stored at 4°C for up to 3 days. The egg white was separated from the egg yolk into a test tube under sterile conditions and mixed in a stirrer for half an hour to homogenize the media. This was directly used for the experiment described in Figure 2. In all other experiments, egg whites were then diluted with autoclaved saline solution (PBS) in a 1:1 ratio and stirred for an additional half hour. This facilitated better mixing of the egg white layers, resulting in a more homogeneous and pipette-able solution. This produced 50% egg white that was used in all other experiments.
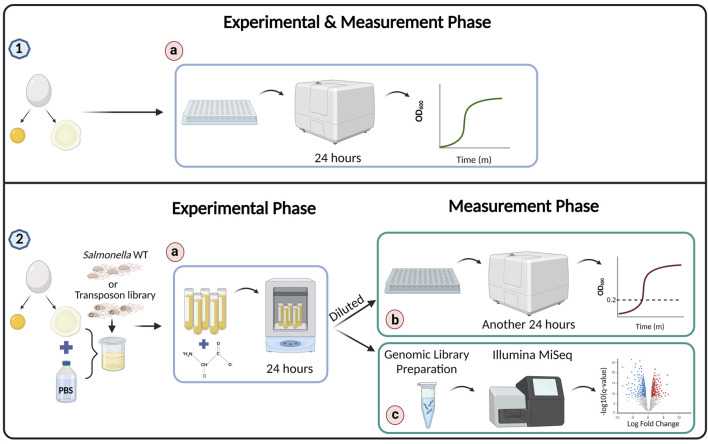
FIGURE 1.
Experiment workflow. Preparation for the experiment–egg white was separated from the yolk of a freshly laid egg in all experiments. (1a) For the experiment described in Figure 2, bacteria were mixed with egg white and bacterial growth was directly monitored using a plate reader set to 37°C, with measurements taken continuously. (2) For experiments described in Figures 3–6 and Tables 1 and 2, the egg white was mixed with PBS to obtain a homogeneous solution. Subsequently, Salmonella wild-type or the transposon library was mixed into the diluted egg white, along with a compound or DDW as a control as described in the specific experiments. The test tubes were incubated for 24 h in a shaker incubator. (2b) To measure surviving bacteria, a sample from each test tube was diluted in 96-well plate with LB and monitored in the plate reader for 24 h to obtain a growth curve. To determine relative bacterial survival in the original samples, time to OD600 = 0.2 was measured. (2c) In the transposon library experiment, at the end of the experimental phase, the samples were prepared for genomic sequencing using Illumina MiSeq.
FIGURE 2.
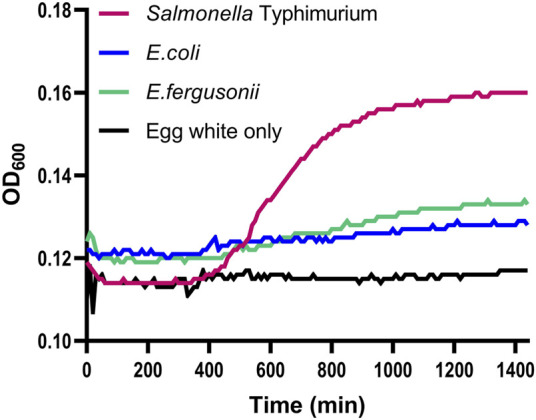
Growth of Salmonella and Escherichia strains in egg white. Wild-type Salmonella enterica serovar Typhimurium strain 14028s (Salmonella Typhimurium), Escherichia coli strain MG1655 (E. coli), and Escherichia fergusonii strain Yara15 (E. fergusonii) were grown overnight and diluted to a concentration of 8*103 cells per 1 mL in 100% egg white. Growth was measured in a plate reader. The background control contained egg white without any added bacteria (Egg white only). Shown are the averages of three technical repeats per strain of a representative experiment out of 2 separate experiments.
Experiment setting–growth in egg white
In most experiments, bacteria were diluted into egg white or 50% egg white at a concentration of 8*103 cells per 1 mL. Only for the Transposon sequencing (TnSeq) experiment a concentration of 8*104 cells per 1 mL was used. Compound stocks were prepared at 500 µM. Compounds were mixed at a 1:3 ratio with the bacteria pre-diluted into egg white, such that the final concentration of compounds was 125 µM. For the experiment described in Figure 2, growth in egg white was directly monitored in a plate reader set to 37°C. For all other experiments, samples were incubated with egg white for 24 h at 37°C. 37°C was used, as this is the optimal temperature for Salmonella growth. To determine the relative numbers of bacteria at the end of the 24 h incubation, 20 µL of sample was diluted into 180 µL of LB, and growth at 37°C was monitored by plate reader. To facilitate comparisons between samples, for some experiments the time in minutes till OD600 = 0.2 was determined.
Calibration curve
To convert differences in the time it took for a sample to reach OD600 = 0.2 given in minutes to fold differences in the relative numbers of bacteria in the original sample, we performed a calibration curve experiment. One reason for this experiment is that while samples were diluted into LB it was possible that residual egg white was slowing bacterial growth. Thus, Salmonella wild type was grown over night, and 10-fold dilutions starting at OD600 = 1.0 were created in a mixture containing egg white at 5% in LB which represents the final concentration of 50% egg white after the 1:10 dilution into LB performed for the other experiments. This calibration curve experiment was repeated three times. Because lag time is unknown, instead we calculated generation time using the time in minutes to OD600 = 0.2 reached by the different 10-fold dilutions. We found that the average generation time in LB containing 5% egg white to be 25.589 ± 3.77 min. We used this to convert differences in the time till OD600 = 0.2 in minutes to relative bacterial numbers in the original samples. For example, two samples which reached OD600 = 0.2 51 min apart have a difference of two duplications or a 4-fold difference between them in the numbers of bacteria at the end of the 24 h incubation in egg white. Finally, it should be noted that differences in the time till OD600 = 0.2 can either reflect differences in bacterial numbers in the original samples or differences in the lag time. For example, if the presence of an amino acid causes membrane damage it might take longer for bacteria to recover and start growing when diluted into LB.
Preparation of a Salmonella transposon library
Because a preliminary attempt to create a transposon library on the wild-type background resulted in a low number of transformants, we decided to use as a background a mutant in the host restriction system. A mutation in the hsdRMS system is known to result in high efficiency transformation (O’Callaghan and Charbit, 1990) and commercially available electrocompetent cells are often optimized by introducing mutations in the hsdRMS genes. Plasmid pBAD24 DNA was utilized for a preliminary electroporation experiment comparing several Salmonella mutants in known and putative host restriction systems, which included the following strains; ΔSTM14_1614 (putative DNA/RNA non-specific endonuclease), ΔSTM14_5387 (putative restriction endonuclease), ΔSTM14_5436 (type I restriction enzyme specificity protein, hsdS), ΔSTM14_0418 (DNA restriction enzyme, res), ΔSTM14_0191 (putative restriction endonuclease), ΔSTM14_5316 (putative endonuclease), ΔSTM14_5327 (putative endonuclease), and ΔSTM14_5319 (putative endonuclease). This analysis found the mutant in hsdS was by a large difference the most electrocompetent. Notably, a knockout mutation in hsdS gene increased transformation efficiency up to 220,000 transformants per reaction, compared to only 7,000 transformants obtained in the wild-type background. Of note, the growth inhibition in egg white upon arginine addition phenotype was confirmed for this hsdS mutant strain (data not shown).
To generate the transposon library the ΔhsdS::cam strain was grown overnight in 2 mL LB and then diluted 1:100 into 200 mL 2 x YT broth [16.0 g/L Tryptone, 10.0 g/L Yeast Extract, and 5.0 g/L NaCl]. The culture was incubated at 37°C with shaking at 250 rpm to an OD600 of 0.4–0.5. Cells were harvested by centrifugation at 1,000 ×g for 8 min at 4°C and washed 3 times in ½ vol cold 10% glycerol. The final electrocompetent cells pellet was resuspended in 60 µL of cold 10% glycerol and used immediately. Cells were mixed with 0.5 µL of EZ-Tn5™ <KAN-2>Tnp Transposome™ Kit (Lucigen, Cat. No. TSM99K2). After incubation in ice for 10 min, the cells were electroporated in a 1 mm electrode gap cuvette using a Bio-Rad MicroPulser set to 2.5 kV. Electroporated cells were immediately resuspended in 500 µL of pre-warmed Super Optimal broth with Catabolite repression (SOC) medium [0.5% Yeast Extract, 2% Tryptone, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, 20 mM Glucose]. After incubation for 90 min at 37°C with shaking, the volume was adjusted by a dilution factor of 20 in SOC medium to create 10 mL. 87 LB agar plates with kanamycin (100 μg/μL) were plated with 100 µL each. After 18 h of incubation, estimated colony yield was calculated. 1 mL of LB was added to each plate and colonies were scraped and pooled together into 50 mL tubes. Bacterial suspension from all tubes was vortexed, and transferred to a sterile bottle. 80% glycerol was added to a final concentration of 25%. Finally, the Tn5 transposon library was aliquoted to 1 mL microcentrifuge tubes and stored at −80°C. The library was estimated to contain 260,000 mutants.
TnSeq experiment in egg white with and without L-arginine
The protocol was similar to the basic experiment described above with the following modifications. In short, a tube of the Tn5 library was thawed on ice and 100 µL were diluted into 10 mL LB with kanamycin (50 μg/mL) to a final bacterial concentration of 108 CFU/mL. The library was incubated at 37°C shaking at 250 rpm for 1 h to initiate log phase. To prepare the library input stock, OD600 was measured and the library was diluted to OD600 = 1. The library was centrifuged and the pellet was resuspended in PBS and two 700 µL samples were taken into separate tubes and immediately placed in −80°C for safe keeping. These two samples are the T0 input - the initial population of the Tn5 library (before selection). Aliquots of 2 µL from the PBS resuspended library input stock were added to 20 mL PBS to create a library dilution of 8*104 CFU/mL for 2 biological repeats. Four sterile flasks were prepared, representing 2 biological replicates for 2 treatments: DDW control 1, DDW control 2, L-arginine treatment 1 and L-arginine treatment 2. Each flask contained a 100 mL mixture composed of: (a) 7.5 mL Tn5 library–from library dilution 1 or 2. (b) 67.5 mL egg white + PBS (1:1) mixture. (c) 25 mL L-arginine for a final concentration of 125 µM or DDW carrier as a control. Flasks were aliquoted into separate 5 mL tubes with 1 mL mixture in each tube. The first replicate consisted of 100 tubes for the L-arginine treatment and another 100 for the control. In the second replicate, the pool consisted of 50 culture tubes, rather than 100, due to technical complications. A total of 300 tubes were incubated for 24 h at 37°C shaking at 250 rpm. This large number of tubes was required to allow testing of a broad number of the transposon mutants and avoid a bottleneck. After incubation, tubes originating from the same flask were pooled into 50 mL tubes and centrifuged at 7197 RCF (maximum speed) for 5 min to eliminate the egg white mixture. The pellet of each tube was resuspended in 10 mL of LB and incubated for 1 h at 37°C shaking. This step was required in order to differentiate live bacteria from dead bacteria. By allowing live bacteria to replicate their representation compared to the DNA of dead cells increased. This step also improved DNA yield. Finally, samples were washed, resuspended in 700 µL LB and stored at −80°C for DNA extraction.
Creating genomic amplicon libraries and sequencing
The DNA extraction protocol was adapted from Stevenson and Weimer (2007). DNA was extracted from 6 samples: control DDW 1, control DDW 2, L-arginine treatment 1, L-arginine treatment 2, input T0 1 and input T0 2. Samples were disrupted with 0.1 mm glass beads in the presence of Tris-saturated phenol following phenol-chloroform extraction. DNA concentration was measured using a Qubit 2.0 fluorometer and dsDNA Quantification Assay Kit and samples were kept at 4°C.
The protocol for amplification of transposon junctions using the semi-arbitrary PCR strategy was adapted from Christen et al. (2011) with a few modifications. In a first-round of PCR, a transposon specific primer (Ez tn5 <KAN2> FP, Table 1) was used with a semi-arbitrary primer (Rnd1-PE-arb1). The semi-arbitrary primer contains a defined 3′ penta-nucleotide sequence that anneals on average every 233 bp in the Salmonella S14028 genome, an interspace of 10 bp arbitrary sequence and a 5’ PE 2.0 sequence that serves as a binding site for the subsequent PCR. The first round of PCR was performed with the following program: (1) 94°C for 3 min, (2) 94°C for 30 s, (3) 42°C for 30 s, slope −1°C per cycle, (4) 72°C for 1 min, (5) go to step 2, 6 times, (6) 94°C for 30 s, (7) 58°C for 30 s, (8) 72°C for 1 min, (9) go to step 6, 25 times, (10) 72°C for 3 min. The 50 µL PCR reaction consisted of 25 µL Platinum™ Hot Start PCR Master Mix (Invitrogen, Carlsbad, CA, United States), 1 µL Ez tn5 <KAN2> FP primer (10 µM), 1 µL Rnd1-PE-arb1 (10 µM), 10 µL Template DNA (50–80 ng), and 13 µL PCR grade dH2O. PCR products were purified using Zymo Clean and Concentrator-5 kit (Zymo Research, Irvine, CA) and eluted with 12 µL DDW. Products served as a template for a second round PCR reaction using Illumina paired-end primers.
TABLE 1.
Primers used for TnSeq.
| Name | Sequence (5'→3′) | Sample barcode | Description | Source |
|---|---|---|---|---|
| P5-IR2-1 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNAGgatcagTCAGGGTTGAGATGTGTATAAGAGACAG | DDW 1 | Semi-arbitrary PCR method | Mandal et al. (2021) |
| P5-IR2-2 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNAGcttgtaTCAGGGTTGAGATGTGTATAAGAGACAG | L-Arginine 1 | Semi-arbitrary PCR method | Mandal et al. (2021) |
| P5-IR2-3 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNAGataacgTCAGGGTTGAGATGTGTATAAGAGACAG | DDW 2 | Semi-arbitrary PCR method | Mandal et al. (2021) |
| P5-IR2-4 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNAGggctagTCAGGGTTGAGATGTGTATAAGAGACAG | L-Arginine 2 | Semi-arbitrary PCR method | Mandal et al. (2021) |
| P5-IR2-5 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNAGttcggcTCAGGGTTGAGATGTGTATAAGAGACAG | T0 1 | Semi-arbitrary PCR method | Mandal et al. (2021) |
| P5-IR2-6 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNAGcagatcTCAGGGTTGAGATGTGTATAAGAGACAG | T0 2 | Semi-arbitrary PCR method | Mandal et al. (2021) |
| Ez tn5 <KAN2> FP | ACCTACAACAAAGCTCTCATCAACC | Semi-arbitrary PCR method | EZ-Tn5™ <KAN-2>Tnp Transposome™ Kit | |
| Rnd1-PE-arb1 | CTCGGCATTCCTGCTGAACCGCTCTTCCGATCTNNNNNNNNNNTGCTG | Semi-arbitrary PCR method | Custom primer based on (Christen et al., 2011) primer | |
| round2-PE-P7 | CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATCT | Semi-arbitrary PCR method | Christen et al. (2011) |
The second reaction was composed of a flanking P5-Tn5 barcoded primer specific to the transposon, downstream to the transposon specific primer used in the 1st PCR reaction to limit amplification of non-specific products, and a PE P7 primer 2.0. To determine the optimal cycle number for amplification at the second PCR reaction and by that minimize bias introduction due to over amplification of specific fragments (Gallagher, 2019), suggests tracking amplification by using real-time PCR. Thus, prior to the second PCR reaction, a real-time PCR was carried using the following parameters: (1) 94°C for 1 min, (2) 94°C for 30 s, (3) 64°C for 30 s, (4) 72°C for 1 min, scan, (5) go to step 2, 20 times, followed by a dissociation curve reaction. The 25 µL qPCR reaction consisted of 12.5 µL 2x qPCRBIO Fast qPCR SyGreen Blue Mix (PCR Biosystems, London, UK), 0.5 µL P5 barcoded primer (10 µM), 0.5 µL Round2-PE-P7 primer (10 µM), 1 µL 1st round PCR product, and 10.5 µL PCR grade dH2O. As a result of this qPCR the optimal number of cycles was determined as 14 for the DDW control and L-arginine treatment samples and 9 for the T0 input samples. Second-round PCR was preformed using the following parameters: (1) 94°C for 3 min, (2) 94°C for 30 s, (3) 64°C for 30 s, (4) 72°C for 1 min, (5) go to step 2, 9 or 14 times (as determined for each sample), (6) 72°C for 3 min. The 50 µL PCR reaction consisted of 25 µL Platinum™ Hot Start PCR Master Mix, 1 µL P5 barcoded primer (10 µM), 1 µL Round2-PE-P7 primer (10 µM), 10 µL 1st round PCR product, and 13 µL PCR grade dH2O. Finally, second round PCR products were purified using Zymo Clean and Concentrator-5 kit and eluted in 25 µL DDW. Library molarity was calculated using Qubit measurements and average fragment size reported by TapeStation (Agilent, Santa Clara, United States). Equimolar amounts from each sample were pooled together to 4 nM and sent for single-read sequencing with 150 cycles on an Illumina MiSeq platform (Genomic Technologies Facility of the Hebrew University of Jerusalem, Givat Ram, Jerusalem).
Analysis of TnSeq data
Salmonella enterica subsp. Enterica serovar Typhimurium str. 14028S, assembly ASM2216v1, was downloaded from ENA database (Accession: GCA_000022165) and used as a reference genome. Raw fastq files were demultiplexed using Cutadapt (Martin, 2011) based on the sample inline barcode and 6 nucleotides from the transposon 5’ end sequence (TCAGGG), allowing no mismatches. Fastq files were processed through the BioTradis analysis pipeline (Barquist et al., 2016). The pipeline was executed on Ubuntu 20.04. Reads were filtered based on the presence of the transposon tag (TTGAGATGTGTA) in the beginning of the reads, allowing for 1 mismatch. Three comparisons were carried separately: (1) input T0 vs. control DDW, (2) input T0 vs. L-arginine treatment, and (3) DDW control vs. L-arginine treatment. Genes with low read counts were filtered using a threshold of 10. p-values were corrected for multiple testing using Benjamini–Hochberg method. Genes with corrected p-value (Q-value) ≤ 0.05 were considered significant.
Statistics
To statistically evaluate the outcomes, we employed paired T-tests using R software (version 4.3.2), and designed the graphs with GraphPad Prism version 5.03 (GraphPad Software, San Diego California United States, www.graphpad.com). Statistical tests with a p-value < 0.05 were considered significant.
Results
Salmonella enterica serovar Typhimurium strain 14028s grows in egg white
As a first step, we confirmed that the strain used, S. enterica serovar Typhimurium strain 14028s (Salmonella), is able to survive and grow in the egg white environment. As a comparison to Salmonella growth, the growth in egg white of additional phylogenetically closely related bacteria, E. coli strain MG1655 and a poultry derived E. fergusonii (Yara15) strain (Poudel et al., 2022), was examined. Overnight cultures of all three bacteria were diluted into egg white at a concentration of 8*103 cells per 1 mL. This low concentration was used so that the bacteria themselves would not substantially modulate the egg white environment for the duration of the experiment, i.e., by degrading egg white proteins or changing the pH through their own metabolism. Growth in egg white was measured overnight. Compared to egg white-alone background readings, it seemed that the two Escherichia species did display some growth (Figure 2). This limited growth might be a result of residual media carried over from the overnight culture. However, Salmonella displayed much more significant growth. This shows that Salmonella encodes systems enabling it to overcome at least some of the stressors that this environment uses to inhibit bacterial growth. That being said, compared to growth in LB which can reach OD600 = 0.5 in the plate reader (results not shown) this represents limited growth. To conclude, the S. enterica serovar Typhimurium strain utilized here not only survives in the egg white environment but can also grow at least to a limited degree.
L-arginine and L-cysteine inhibit Salmonella growth in egg white
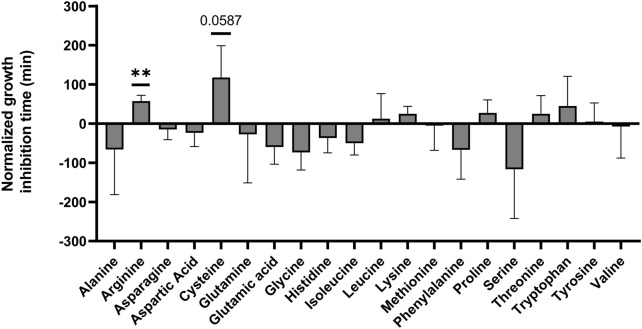
To test the hypothesis that adding compounds to an environment might limit bacterial growth, we chose the 20 L-amino acids. This is because L-amino acids are found in all living cells, and are taken up as nutrients by both animals and bacteria including Salmonella. For this set of experiments, Salmonella was incubated for 24 h in egg white at 37°C, each time with a different L-amino acid at a final concentration of 125 µM. After incubation, samples were diluted into LB and growth was recorded to determine how many bacteria survived the 24 h treatment. We chose OD600 = 0.2 because at this OD Salmonella were in logarithmic growth. Differences in the time to reach OD600 = 0.2 compared to the control are therefore indicative of differences in the numbers of surviving bacteria after egg white incubation. Among the amino acids, only L-arginine was found to delay Salmonella growth in a statistically significant manner (Figure 3). L-arginine exposure resulted in a 57.5 min delay in the time to reach OD600 = 0.2 compared to the double distilled water (DDW) control. Considering our measured generation time of 25.589 min, this equals a 4.75-fold reduction in the number of bacteria surviving in egg white. Interestingly, L-cysteine displayed a much bigger inhibition, 117.5 min, which is equivalent to a 24.11-fold reduction. However, because of a large variation in L-cysteine measurements, this was not statistically significant (p = 0.0587). To conclude, these results show that the addition of L-arginine and likely also L-cysteine inhibits the growth of Salmonella in the egg white environment, even though these are nutrient type compounds.
FIGURE 3.
Screen for L-amino acids which limit Salmonella growth in egg white. Salmonella was grown overnight and diluted to a concentration of 8*103 cells per 1 mL in 50% egg white. Each diluted sample was mixed with one of the amino acids at a 1:3 ratio to a final concentration of 125 µM of the amino acid. Samples were incubated for 24 h at 37°C. Finally, samples were diluted into LB and growth was monitored by a plate reader to determine relative bacterial levels at the end of the 24 h incubation. To compare samples, the time to reach OD600 = 0.2 was recorded. Results were normalized to the time a double distilled water (DDW) control reached OD600 = 0.2. Shown is the average of 3 separate experiments. Standard deviation is shown. ** p-value<0.01.
Growth inhibition is not mediated through biofilm formation mechanisms
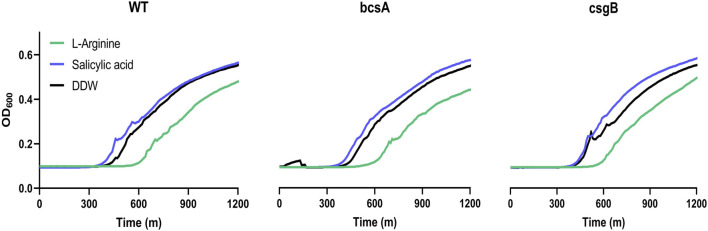
Of note, environmental L-arginine is known as a signal used by Salmonella to modulate cellular cyclic-di-GMP (CDG) levels and downstream cellulose synthesis and secretion (Mills et al., 2015). CDG is a bacterial second messenger considered the switch controlling biofilm formation versus motility (Mills et al., 2011). Furthermore, cellulose is the major exopolysaccharide of Salmonella and a major component of the Salmonella biofilm. Thus, we hypothesized it was possible that biofilm formation was involved in the inhibition of Salmonella growth upon L-arginine addition. Because salicylic acid is known to effect CDG and cellulose levels in the opposite direction to L-arginine (Mills et al., 2015), we tested if salicylic acid would increase Salmonella’s growth in egg white. In addition, two knockout mutants of Salmonella were tested for growth and survival in the presence of L-arginine, a bcsA knockout strain unable to produce cellulose and a csgB knockout strain unable to produce curli fibers that are also a major component of the Salmonella biofilm (Jonas et al., 2007). On the one hand, the presence of salicylic acid did seem to have a small positive effect on the growth of Salmonella in egg white (Figure 4). However, L-arginine inhibition still occurred in the bcsA knockout strain background in a statistically significant manner. Due to large variation in measurements when using the csgB knockout strain background, only a trend was observed (Figure 4). To conclude, it seems that cellulose, but likely also curli and CDG, are not part of the mechanism of action of L-arginine inhibition on Salmonella growth in egg white.
FIGURE 4.
Cyclic-di-GMP, cellulose and curli fibers are not involved in arginine inhibition of Salmonella in egg white. Salmonella wild-type, or knockout mutants ΔbcsA and ΔcsgB were grown overnight, diluted to a concentration of 8*103 cells per 1 mL in 50% egg white, mixed with 125 μM L-arginine, 125 µM salicylic acid, or carrier DDW as a control, and incubated for 24 h at 37°C. Finally, samples were diluted into LB and growth was monitored by a plate reader to determine relative bacterial levels at the end of the 24 h incubation. Shown are the full growth curves for a representative experiment out of 3 separate experiments. For both wild-type and the ΔbcsA mutant the difference between exposure to L-arginine to the DDW control at OD600 = 0.2 was statistically significant (p-value<0.05), while for the ΔcsgB mutant it was not.
Addition of arginine has a much smaller effect on Salmonella transposon insertions frequencies than the egg white environment itself
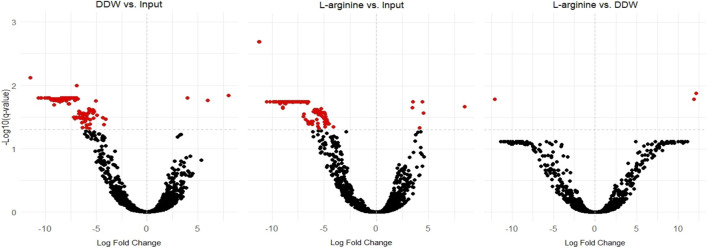
We hypothesized that arginine was possibly transported into Salmonella cells where it was dysregulating gene expression causing decreased bacterial survival. Thus, a TnSeq analysis might identify genes which are required for arginine inhibition of Salmonella in the egg white environment. Furthermore, TnSeq might identify genes which are involved in whatever stress response is produced due to the presence of arginine. Therefore, a TnSeq analysis of Salmonella exposed to egg white either in the presence or absence of arginine was performed. Furthermore, both of these were compared to the input Salmonella transposon library. A comparison of transposon insertions after exposure to egg white or to egg white with arginine, to the input library, identified 226 and 220 genes respectively which were significantly differentially represented (Figure 5). Furthermore, the vast majority of these genes had a lower representation than in the input. This supports the notion that egg white is a harsh environment for Salmonella, and that many transposon insertions which are tolerated in rich media become essential in egg white. Importantly, a direct comparison of gene insertions between Salmonella exposed to egg white with arginine to those exposed only to egg white identified just three genes which were differentially represented in a statistically significant manner (Figure 5). This implies that either the inhibition by arginine was not a large stress on Salmonella in comparison to egg white itself, or that arginine inhibition could not be countered by knocking out Salmonella genes by transposon insertions or both.
FIGURE 5.
The effect of egg white and arginine on the fitness of transposon insertions into Salmonella genes. Volcano plots showing changes in the abundances of transposon mutants comparing (middle) arginine exposed samples to input samples, (left) DDW control samples to input samples, and (right) arginine exposed samples to DDW control samples. All statistically significant genes are marked in red. Note, that the majority of statistically significant genes were negatively affected by the egg white treatment.
TnSeq identifies multiple genes required for egg white survival
As a first step in analyzing TnSeq data, genes required for Salmonella survival in egg white were characterized. To stringently identify transposon insertions into genes that effect survival in egg white the data from both arginine and control, DDW treated, samples were used. A gene was considered affected by egg white if it was significantly affected (q < 0.05) in both the arginine vs. input comparison as well as the control DDW vs. the input. This resulted in the identification of 82 high confidence genes. To expand the analysis a bit more, genes that were significant in one comparison, either arginine vs. input or control vs. input, and had a significant p < 0.05 (statistical significance before multiple comparison correction) in the other comparison were also included. This resulted in the identification of another 44 genes with lower confidence. We also verified that the effect was in the same direction, i.e., increased in both data sets in comparison to the input, or reduced in both data sets in comparison to the input. All of the high confidence genes were affected in the same direction. However, eight genes of the lower confidence genes were not affected in the same direction by egg white and were removed. This produced a set of 118 genes that were differentially represented after egg white treatment compared to the input library (Table 2). Notably, only one gene, stm14_4543, had a higher representation after egg white treatment compared to the input library whereas all the other 117 genes had a lower representation in egg white compared to the input. Among the genes identified as required for Salmonella growth are genes required during iron starvation, biotin metabolism genes, genes required for the transport and metabolism of amino acids, purine and other nutrients, stress response genes, genes required for outer membrane maintenance, and genes required for DNA repair and recombination. This further supported the notion that egg white is a harsh environment requiring the function of many genes.
TABLE 2.
Transposon mutants’ abundance in egg white.
| Egg white with DDW control vs. input | Egg white with arginine vs. input | |||||||
|---|---|---|---|---|---|---|---|---|
| Locus tag | Gene name | Function | Log Fold Change | P-value | Q-value | Log Fold Change | P-value | Q-value |
| Iron restriction response | ||||||||
| STM14_0231 | fhuB | iron-hydroxamate transporter permease subunit | −6.65 | 0.00216 | 0.02588 | −5.50 | 0.00485 | 0.04709 |
| STM14_0228 | fhuA | ferrichrome outer membrane transporter | −5.12 | 0.01320 | 0.10880 | −6.51 | 0.00363 | 0.03784 |
| STM14_3825 | exbB | biopolymer transport protein ExbB - iron transport | −5.82 | 0.00746 | 0.06534 | −9.01 | 0.00121 | 0.01780 |
| STM14_0894 | STM14_0894 | iron ABC transporter permease | −6.96 | 0.00071 | 0.01585 | −6.73 | 0.00108 | 0.01780 |
| STM14_3117 | nifU | scaffold protein for iron-sulfur cluster assembly | −7.93 | 0.00060 | 0.01585 | −7.70 | 0.00049 | 0.01780 |
| Biotin metabolism | ||||||||
| STM14_0920 | bioB | biotin synthetase | −7.02 | 0.00065 | 0.01585 | −6.80 | 0.00103 | 0.01780 |
| STM14_0922 | bioC | biotin biosynthesis protein BioC | −9.15 | 0.00108 | 0.01627 | −8.93 | 0.00121 | 0.01780 |
| Metabolism and transport of amino acids, purine and other nutirents | ||||||||
| STM14_0133 | leuB | 3-isopropylmalate dehydrogenase - Valine, leucine and isoleucine biosynthesis | −7.26 | 0.00068 | 0.01585 | −7.04 | 0.00095 | 0.01780 |
| STM14_5358 | STM14_5358 | ornithine carbamoyltransferase - Arginine biosynthesis | −9.84 | 0.00048 | 0.01585 | −3.70 | 0.04635 | 0.28689 |
| STM14_0077 | carA | carbamoyl phosphate synthase small subunit - Pyrimidine metabolism | −6.94 | 0.00075 | 0.01585 | −6.72 | 0.00103 | 0.01780 |
| STM14_3133 | glyA | serine hydroxymethyltransferase - Glycine, serine and threonine metabolism | −7.48 | 0.00043 | 0.01585 | −7.25 | 0.00049 | 0.01780 |
| STM14_3931 | garL | alpha-dehydro-beta-deoxy-D-glucarate aldolase - Ascorbate and aldarate metabolism | −8.61 | 0.00102 | 0.01627 | −4.90 | 0.00912 | 0.08004 |
| STM14_3019 | eutG | ethanolamine utilization protein - Glycolysis/Gluconeogenesis | −5.27 | 0.00321 | 0.03284 | −4.14 | 0.01051 | 0.08951 |
| STM14_4543 | STM14_4543 | Pentose phosphate pathway - glycolysis | 5.97 | 0.00129 | 0.01736 | 4.58 | 0.01740 | 0.13379 |
| STM14_0510 | phnW | 2-aminoethylphosphonate--pyruvate transaminase | −7.72 | 0.00054 | 0.01585 | −7.50 | 0.00045 | 0.01780 |
| STM14_2683 | STM14_2683 | putative glutathione S-transferase - Tyrosine metabolism | −5.70 | 0.00188 | 0.02355 | −7.58 | 0.00051 | 0.01780 |
| STM14_2875 | STM14_2875 | 5'-deoxynucleotidase - Purine metabolism | −5.33 | 0.00197 | 0.02439 | −4.12 | 0.00459 | 0.04503 |
| STM14_3065 | purN | phosphoribosylglycinamide formyltransferase - Purine metabolism | −7.67 | 0.00042 | 0.01585 | −3.27 | 0.03169 | 0.21774 |
| STM14_3067 | ppx | exopolyphosphatase - Purine metabolism | −3.58 | 0.01769 | 0.13909 | −7.17 | 0.00061 | 0.01780 |
| STM14_3098 | ndk | nucleoside diphosphate kinase - Purine metabolism | −3.30 | 0.02444 | 0.17845 | −7.24 | 0.00080 | 0.01780 |
| STM14_5248 | purA | adenylosuccinate synthetase - Purine metabolism | −9.50 | 0.00086 | 0.01627 | −9.28 | 0.00092 | 0.01780 |
| STM14_1726 | ydhB | regulates a gene involved in the purine transport process - Purine metabolism | −7.94 | 0.00094 | 0.01627 | −7.70 | 0.00077 | 0.01780 |
| STM14_2106 | oppF | oligopeptide transport protein | −6.64 | 0.00305 | 0.03249 | −6.19 | 0.00369 | 0.03830 |
| STM14_1100 | focA | formate transporter | −7.89 | 0.00064 | 0.01585 | −7.67 | 0.00051 | 0.01780 |
| STM14_1781 | ydgI | putative arginine:ornithine antiporter | −8.88 | 0.00113 | 0.01666 | −6.34 | 0.00374 | 0.03858 |
| STM14_3252 | kgtP | alpha-ketoglutarate transporter | −7.99 | 0.00080 | 0.01585 | −3.35 | 0.04547 | 0.28272 |
| STM14_4792 | trkH | potassium transporter | −8.06 | 0.00090 | 0.01627 | −4.09 | 0.01640 | 0.12740 |
| STM14_2694 | yeiA | dihydropyrimidine dehydrogenase - Pyrimidine metabolism | −3.05 | 0.04936 | 0.30628 | −5.35 | 0.00259 | 0.02896 |
The red color in the table indicates statistical significance in Q-value and/or p-value <0.05.
The effect of arginine presence on Salmonella fitness in the egg white environment
An analysis of transposon insertions deferentially distributed between egg white with L-arginine and egg white control, identified only three genes that were statistically significant (Table 3). The three genes were citB, stm14_0668, and stm14_0670. However, the known functions of these genes, mannose transport, and citrate utilization do not seem to relate to arginine transport and metabolism nor to a possible stress acting on the bacterial cell.
TABLE 3.
Transposon mutants’ abundance in egg white with and without arginine.
| Locus tag | Gene name | Function | Log fold change | P-value | Q-value |
|---|---|---|---|---|---|
| Statistically significant | |||||
| STM14_0668-0670 area | |||||
| STM14_0668 | STM14_0668 | putative phosphosugar isomerase | 11.93 | 0.00003 | 0.01636 |
| STM14_0670 | STM14_0670 | putative PTS system mannose-specific enzyme IID | 12.16 | 0.00001 | 0.01329 |
| citAB operon | |||||
| STM14_0805 | citB | citrate utilization protein b | −12.11 | 0.00003 | 0.01636 |
| Near statistical significance | |||||
| Arginine transport | |||||
| STM14_2901 | argT | lysine/arginine/ornithine transport protein | 4.87 | 0.02455 | 0.21751 |
| Arginine metabolism related | |||||
| STM14_4955 | argE | acetylornithine deacetylase | −5.89 | 0.00097 | 0.07678 |
| STM14_0817 | potE | putrescine transporter | 8.07 | 0.00056 | 0.07678 |
| STM14_1402 | potB | spermidine/putrescine ABC transporter membrane protein | −9.94 | 0.00416 | 0.08413 |
| Stress related | |||||
| STM14_0286 | rcsF | outer membrane lipoprotein | −3.34 | 0.01142 | 0.13268 |
| Systems which were not affected by arginine exposure | |||||
| Arginine transport | |||||
| STM14_2897 | hisP | histidine/lysine/arginine/ornithine transporter subunit | 1.19 | 0.58936 | 0.97854 |
| STM14_2898 | hisM | histidine/lysine/arginine/ornithine transport protein | −1.50 | 0.44912 | 0.93284 |
| STM14_2899 | hisQ | histidine/lysine/arginine/ornithine transport protein | 0.76 | 0.59059 | 0.97854 |
| STM14_5166 | yjdE, adiC | arginine:agmatin antiporter | 1.00 | 0.58828 | 0.97854 |
| Arginine metabolism | |||||
| STM14_5357 | STM14_5357 | putative arginine repressor | −2.97 | 0.14279 | 0.63919 |
| STM14_5169 | adi | catabolic arginine decarboxylase | 2.68 | 0.17457 | 0.69005 |
| STM14_3729 | speA | arginine decarboxylase | 1.01 | 0.37472 | 0.90177 |
| STM14_4175 | argD_2 | bifunctional N-succinyldiaminopimelate-aminotransferase | 1.13 | 0.46895 | 0.93932 |
| STM14_4957 | argB | acetylglutamate kinase | −1.32 | 0.56035 | 0.96410 |
| STM14_3973 | argG | argininosuccinate synthase | −0.72 | 0.59484 | 0.97854 |
| STM14_4958 | argH | argininosuccinate lyase | 0.44 | 0.74602 | 0.99384 |
| STM14_5361 | STM14_5361 | arginine deiminase | −0.16 | 0.88496 | 0.99682 |
| STM14_0818 | speF | ornithine decarboxylase | 2.30 | 0.08999 | 0.51049 |
| STM14_1584 | astD | succinylglutamic semialdehyde dehydrogenase | 0.56 | 0.76601 | 0.99384 |
| STM14_0379 | proA | gamma-glutamyl phosphate reductase | −0.28 | 0.88428 | 0.99682 |
| STM14_3781 | STM14_3781 | putative oxidoreductase | 0.01 | 0.99512 | 1.00000 |
| STM14_2874 | yfbQ | aminotransferase AlaT | 1.05 | 0.60874 | 0.97854 |
| Cellulose synthase | |||||
| STM14_4355 | yhjL | cellulose synthase subunit BcsC | 0.04 | 0.97922 | 1.00000 |
| STM14_4356 | STM14_4356 | endo-1,4-D-glucanase | 0.79 | 0.56134 | 0.96410 |
| STM14_4357 | yhjN | cellulose synthase regulator protein | 0.77 | 0.57711 | 0.97552 |
| STM14_4358 | bcsA | cellulose synthase catalytic subunit | 0.70 | 0.53302 | 0.96040 |
| Cationic antimicrobial peptide (CAMP) resistance | |||||
| STM14_5163 | basS | sensor protein BasS/PmrB | 1.99 | 0.31336 | 0.86032 |
| STM14_0249 | htrA | serine endoprotease | 1.73 | 0.34431 | 0.89041 |
| STM14_5239 | amiB | N-acetylmuramoyl-l-alanine amidase II | −1.30 | 0.55898 | 0.96410 |
| STM14_4179 | ppiA | peptidyl-prolyl cis-trans isomerase A (rotamase A) | −1.18 | 0.58848 | 0.97854 |
| Lipopolysaccharide biosynthesis | |||||
| STM14_0271 | lpxB | lipid-A-disaccharide synthase | 3.35 | 0.12774 | 0.61594 |
| STM14_4484 | rfaQ | lipopolysaccharide core biosynthesis protein | −3.14 | 0.15095 | 0.65309 |
| STM14_4479 | rfaI | lipopolysaccharide-alpha-1, 3-D-galactosyltransferase | −2.72 | 0.16157 | 0.67025 |
| STM14_4476 | rfaZ | lipopolysaccharide core biosynthesis protein | 1.90 | 0.36038 | 0.89810 |
| STM14_4480 | rfaB | UDP-D-galactose: (glucosyl)lipopolysaccharide-1, 6-D-galactosyltransferase | −1.50 | 0.41286 | 0.91438 |
| STM14_4475 | rfaK | putative hexose transferase | 0.84 | 0.49112 | 0.95099 |
| STM14_4467 | yibD | glycosyl transferase | 1.03 | 0.57084 | 0.96959 |
| STM14_4952 | yijP | hypothetical protein | −0.38 | 0.74784 | 0.99384 |
| STM14_2839 | arnT | 4-amino-4-deoxy-L-arabinose transferase | 0.29 | 0.82939 | 0.99682 |
| STM14_1612 | STM14_1612 | putative outer membrane protein | −0.32 | 0.83375 | 0.99682 |
| STM14_4474 | rfaL | O-antigen ligase | 0.24 | 0.89773 | 0.99969 |
| STM14_4382 | yhjW | phosphoethanolamine transferase | 0.01 | 0.99312 | 1.00000 |
| Peptidoglycan biosynthesis | |||||
| STM14_2218 | STM14_2218 | putative penicillin-binding protein 3 | −3.34 | 0.14718 | 0.64860 |
| STM14_2324 | STM14_2324 | putative penicillin-binding protein | 1.99 | 0.29085 | 0.84136 |
| STM14_4205 | mrcA | peptidoglycan synthetase | 1.80 | 0.38562 | 0.90446 |
| STM14_3986 | dacB | D-alanyl-D-alanine carboxypeptidase/endopeptidase | 1.37 | 0.47282 | 0.94216 |
| STM14_3039 | STM14_3039 | hypothetical protein | 1.27 | 0.55045 | 0.96410 |
| STM14_3104 | pbpC | penicillin-binding protein 1C | −0.88 | 0.67231 | 0.98169 |
| STM14_3881 | uppP | undecaprenyl pyrophosphate phosphatase | 0.29 | 0.82391 | 0.99682 |
| STM14_4014 | mtgA | monofunctional biosynthetic peptidoglycan transglycosylase | −0.31 | 0.86142 | 0.99682 |
| Exopolysaccharide biosynthesis | |||||
| STM14_2597 | wcaJ | putative UDP-glucose lipid carrier transferase | 4.67 | 0.03986 | 0.30244 |
| STM14_2595 | wcaK | putative pyruvyl transferase | 2.06 | 0.22404 | 0.75851 |
| STM14_4727 | wecG | putative UDP-N-acetyl-D-mannosaminuronic acid transferase | 0.98 | 0.55181 | 0.96410 |
| STM14_2605 | wcaF | putative colanic acid biosynthesis acetyltransferase WcaF | 1.10 | 0.59739 | 0.97854 |
| STM14_2608 | wcaC | glycosyl transferase | −0.11 | 0.95541 | 1.00000 |
The red color in the table indicates statistical significance in Q-value and/or p-value <0.05.
Next, specific molecular functions were examined to better characterize the effect of arginine on Salmonella in egg white. One such function is arginine transport. It can be hypothesized that if arginine inhibition functions by causing dysregulation of bacterial gene expression, transposon insertions into genes involved in arginine transport should confer protection. One such insertion, into the gene argT was expended in the arginine treatment samples compared to the control but was not statistical significance. Moreover, insertions into other genes of the same transport machinery, hisQMP, seems to not have been affected by the presence of arginine (Table 3). Another function analyzed is genes involved in arginine metabolism. It was hypothesized that uptake and utilization of arginine as a nutrient source might affect insertions in genes involved in arginine metabolism. Three genes involved in arginine metabolism were affected by the presence of arginine, but below statistical significance. Other genes involved in arginine metabolism did not seem to have been affected by the presence of arginine (Table 3). To conclude, it was unclear if arginine was transported into Salmonella cells for its inhibitory function.
Next, the database was searched for gene insertions that might allow the characterization of the molecular mechanism of arginine inhibition. Interestingly, insertions in rcsF were deleterious in the presence of arginine, but not statistically significantly so. A search of other systems involved in cell integrity, such as lipopolysaccharide biosynthesis, peptidoglycan biosynthesis, exopolysaccharide biosynthesis, and cationic antimicrobial peptide (CAMP) resistance found that most genes were not affected by arginine presence (Table 3). Finally, it was confirmed that insertions into genes of the cellulose pathway were not affected by arginine. To conclude, apart from a possible involvement of rcsF, no stress related pathways were identified.
D-arginine and D-cysteine also inhibit Salmonella growth in egg white
To determine if the mechanism of action of L-arginine and L-cysteine includes interaction with Salmonella biological systems or not, we turned to their D-isomers, D-arginine and D-cysteine. This is because most enzymes, receptors, and transporters have evolved to bind the L-isomers but not D-isomers of the 20 amino acids (Genchi, 2017). For example, proteins are made with L-isomers and not D-isomers. Both D-isomers, D-arginine and D-cysteine, were found to inhibit Salmonella growth in 50% egg white (Figure 6). These results imply but do not prove that the inhibition of Salmonella growth by cysteine and arginine do not include bacterial metabolism or signaling as part of their mechanism of action but is connected to chemical interactions of arginine and cysteine with the bacterial membrane or egg white environment itself.
FIGURE 6.
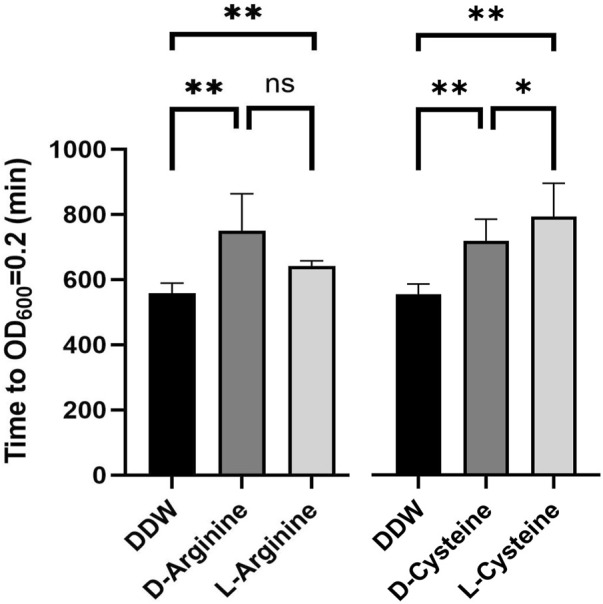
D-isoforms of arginine and cysteine also inhibit Salmonella in egg white. Salmonella wild-type was grown overnight, diluted to a concentration of 8*103 cells per 1 mL in 50% egg white, mixed with L-arginine, D-arginine, L-cysteine, or D-cysteine to a final concentration of 125 μM, or carrier DDW as a control, and incubated for 24 h at 37°C. Finally, samples were diluted into LB and growth was monitored by a plate reader to determine relative bacterial levels at the end of the 24 h incubation. Shown is the time in minutes till samples reached OD600 = 0.2. Shown are the averages of six sperate experiments. Standard deviation is shown. * p-value<0.05, ** p-value<0.01.
Discussion
The main result of our work is that addition of a nutrient type compound to an environment at levels in which it is not normally found (as a free amino acid) can negatively impact bacterial growth and survival. Importantly, arginine and cysteine were added at physiological levels (125 µM) that are within the range of what can be found in human plasma (Lüneburg et al., 2011) and at levels in which in other environments lead to uptake and assimilation into bacterial metabolism. Thus, a compound which is usually benign and even helpful as a nutrient source becomes anti-bacterial when added to a specific environment in which it is not normally found. These results represent a new approach to the development of anti-bacterials, which to our knowledge has not been considered before. This approach has several inherit advantages. One, is that the use of nutrient type compounds means that toxicity to animals should not pose a problem. Indeed, these compounds are not toxic even to bacteria in other contexts. Second, the chance that bacteria would develop resistance to treatment is low. If compound internalization, metabolism, or resultant internal signaling are required for the inhibitory effect, any mutations which disrupt these processes are likely to be detrimental to the bacteria in other environments in which the compound acts as a nutrient source. Finally, this approach adds a large number of compounds which have not been considered to date as potential candidates for anti-bacterial development. To conclude, environment modulation by the addition of nutrient type compounds has a number of advantages and opens up a new avenue for development.
The results shown in this report indicate that arginine and cysteine can be used to limit Salmonella growth in egg white. While this result provides a proof of concept for the use of nutrient type compounds as anti-bacterials, it is not clear that this specific result will have an applied use. One possibility is to add arginine to egg powder so that upon hydration, arginine would inhibit possible Salmonella growth. However, because dehydrated egg powder might be chemically different than actual egg white, more experiments are needed to determine if arginine would inhibit Salmonella growth in that environment. Another possibility is to add arginine when performing artificial insemination of chickens. This might have an impact on Salmonella levels in eggs and vertical transmission of Salmonella as the reproductive tract is where egg white is produced and is also where Salmonella contamination can pass to developing eggs (Baron et al., 2016).
A number of potential mechanisms of action might explain the ability of arginine and cysteine to inhibit the growth of Salmonella in egg white. The first is that these compounds somehow chemically modulate the egg white environment, making it less suitable for Salmonella growth. Another option is that these compounds chemically interact with the outer membrane or outer envelope of Salmonella. It is important to note, that in both of these cases, it is possible that arginine and cysteine somehow make Salmonella more sensitive to the natural anti-bacterial substances found in the egg white environment. Finally, it is possible that these compounds modulate the biology of Salmonella. For example, it is possible that arginine and cysteine modulate the behavior of Salmonella, i.e., modifying gene expression, resulting in behavior that is no longer optimal for survival and growth in egg white. A number of observations support a mechanism of action that does not require interaction with the biological systems of Salmonella, apart from the external membrane. The first is the fact that the D-isomers inhibit Salmonella as well as the L-isomers. Because D-isomers are less likely to interact with transporters, sensors and enzymes, which are adapted to the L-forms, the fact that they still inhibit Salmonella, implies a mechanism of action which includes effecting the external environment or the outer membrane. However, it should be noted that Salmonella does encode a D-cysteine desulfhydrase which is able to incorporate D-cysteine into Salmonella metabolism. Salmonella also encodes an amino acid racemase, YgeA, reported to have a broad substrate specificity, but low catalytic activity, which could potentially convert D-isomers into L-isomers (Miyamoto et al., 2017). It is unknown if these conversion mechanisms can work with such an efficiency as to allow D-isomers to inhibit Salmonella growth at the same levels of L-isomers. The second observation is that if transport into the cell was required for the inhibitory effect, transposon insertions into transport genes should have been identified. Insertions into argT did seem protective but were not statistically significant, and other components of the same transporter system, hisPMQ, were not affected. Thus, it did not seem that transposon insertions into arginine transporter genes were protective. Finally, TnSeq analysis identified rcsF as a required gene during arginine exposure, but not in a statistically significant manner. RcsF, is an outer membrane sensor which detects perturbations in the cell envelope (Laubacher and Ades, 2008). Transposon insertions into this gene might eliminate required stress responses needed to limit arginine derived damage. To conclude, it is likely, but not proven, that arginine and cysteine effect either the external egg white environment or the external membrane of Salmonella.
When considering the mechanism of action, it is interesting to note that the results show specificity as the other L-amino acids tested were not found to inhibit Salmonella growth in egg white. It should also be noted that arginine and cysteine might have different mechanisms of action. For example, it can be speculated that the positive charge of arginine might interact with the Salmonella envelope in a way that makes it more sensitive to the anti-bacterial properties of egg white. However, cysteine is not positively charged. To conclude, the mechanism of action by which arginine and cysteine inhibit Salmonella in egg white might differ.
Apart from the effects of arginine and cysteine on Salmonella inhibition, this study also identified genes required for Salmonella growth in egg white. Transposon insertions into 118 genes were affected differentially when comparing the input library to growth in egg white. Notably, insertions into 117 genes which were tolerated during LB growth were found to be essential in egg white. Only insertions into one gene, stm14_4543, were expended in egg white compared to the input. STM14_4543 is a 2-dehydro-3-deoxy-phosphogluconate aldolase, in the D-glucosaminate degradation pathway. It is not clear why a mutation in this function should be protective in the egg white environment. An analysis of the functions represented by the 117 genes which are required for survival in egg white was mostly as expected. A large number of genes were required for egg white survival due to the fact that egg white was a poor media for Salmonella. For example, a number of genes involved in iron uptake were identified as required for egg white survival. This is because of the presence of ovotransferrin which binds and sequesters iron in egg white (Legros et al., 2021). Another example, is two of the biotin metabolism genes which were significantly reduced as well, emphasizing the importance of biotin metabolism in this environment. This again is expected as avidin a biotin binding protein is abundant in egg white (Pugliese et al., 1993). It is hypothesized that the function of both ovotransferrin and avidin in egg white is to restrict the availability of iron and biotin respectively making it hard for microorganisms to grow in egg white. Finally, other genes involved in the metabolism or transport of amino acids, purine and other nutrients were also identified. Again, this implies that egg white is a poor nutrient environment requiring Salmonella to create all of these compounds by itself. Insertions in genes required for lipopolysaccharide, peptidoglycan and outer membrane maintenance were also found to be required for egg white survival. This is also expected, as egg white includes a number of anti-bacterial proteins and peptides and any malfunction in cell integrity is likely to result in cell death. Interestingly, three genes which are involved in cell division, as well as a number of genes involved in the transport of proteins to the outer membrane, were also found to be required for egg white survival. This is likely because the absence of these genes causes envelope defects which might be tolerated during growth in rich media but are deleterious in egg white due to its antibacterial properties. A number of genes involved in stress responses were also identified as required. This again is expected, as egg white is a stress inducing environment and the absence of these genes abolishes required stress responses. Three virulence associated genes were identified as required for egg white survival. As it is hard to imagine they have a direct role in egg white survival, it is possible that they have regulatory roles which have been dysregulated by their absence. Finally, four genes involved in DNA repair and recombination were also identified as required for egg white survival. This might imply that egg white functions to damage Salmonella DNA, or possibly, like in the case of virulence factors, this represents dysregulation of regulatory roles rather than the absence of their enzymatic DNA repair functions. Some of the genes identified in this experiment, bioB, rfe, and rfbK, were also identified in a screen performed to identify genes required for Salmonella Enteritidis survival in egg white (Raspoet et al., 2014). However, other than these three genes the two analyses did not overlap. This is possibly because two different Salmonella serovars, Typhimurium and Enteritidis, were used in the two screens, two different transposon systems, T5 and T7, were used, and finally the fact that Raspoet et al used a more stringent screen using exposure to egg white at 41°C while this screen was conducted at 37°C. Interestingly, some of the current results also mirror gene expression analysis. Baron et al analyzed differences in gene expression of Salmonella upon exposure to egg white (Baron et al., 2017). They found changes in the expression of genes involved in biotin metabolism, iron restriction response, and membrane stress. To conclude, as egg white is a harsh medium many more genes were required for Salmonella survival than those required for survival in rich LB medium.
In conclusion, this report serves as a proof of concept showing that nutrient type compounds can be used as anti-bacterials by adding them to environments in which they are not normally found. Considering the rise in antibiotic resistance and the large hurdles slowing the development of new antibiotics, this approach may offer additional tools in the fight against microbial pathogens.
Acknowledgments
We would like to thank BioRender for providing the platform to create Figure 1. This figure was created with BioRender.com.
Funding Statement
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the US-Israel Binational Agricultural Research and Development Fund (grant No. IS-5242-20) and by the Israeli Science Foundation (grant No. 1272/20). EM is chair of the Vigevani Senior Lectureship in Animal Sciences.
Data availability statement
Tn-Seq reads are available in the SRA database under the BioProject number PRJNA1089541.
Author contributions
NP: Writing–review and editing, Writing–original draft, Methodology, Investigation, Formal Analysis, Conceptualization. AO: Writing–review and editing, Methodology, Investigation, Formal Analysis. TR: Writing–review and editing, Supervision, Methodology. AM: Writing–review and editing, Formal Analysis. EP: Writing–review and editing, Methodology, Formal Analysis. EM: Writing–review and editing, Writing–original draft, Supervision, Project administration, Funding acquisition, Conceptualization.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Baron F., Bonnassie S., Alabdeh M., Cochet M.-F., Nau F., Guérin-Dubiard C., et al. (2017). Global gene-expression analysis of the response of Salmonella enteritidis to egg white exposure reveals multiple egg white-imposed stress responses. Front. Microbiol. 8, 829. 10.3389/fmicb.2017.00829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron F., Gautier M., Brule G. (1997). Factors involved in the inhibition of growth of Salmonella enteritidis in liquid egg white. J. Food Prot. 60, 1318–1323. 10.4315/0362-028X-60.11.1318 [DOI] [PubMed] [Google Scholar]
- Baron F., Nau F., Guérin-Dubiard C., Bonnassie S., Gautier M., Andrews S. C., et al. (2016). Egg white versus Salmonella Enteritidis! A harsh medium meets a resilient pathogen. Food Microbiol. 53, 82–93. 10.1016/j.fm.2015.09.009 [DOI] [PubMed] [Google Scholar]
- Barquist L., Mayho M., Cummins C., Cain A. K., Boinett C. J., Page A. J., et al. (2016). The TraDIS toolkit: sequencing and analysis for dense transposon mutant libraries. Bioinformatics 32, 1109–1111. 10.1093/bioinformatics/btw022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen B., Abeliuk E., Collier J. M., Kalogeraki V. S., Passarelli B., Coller J. A., et al. (2011). The essential genome of a bacterium. Mol. Syst. Biol. 7, 528. 10.1038/msb.2011.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieri M., Kumar K., Boutin A. (2017). Antibiotic resistance. J. Infect. Public Health 10, 369–378. 10.1016/j.jiph.2016.08.007 [DOI] [PubMed] [Google Scholar]
- Gallagher L. A. (2019). Methods for tn-seq analysis in Acinetobacter baumannii . Methods Mol. Biol. Clifton N. J. 1946, 115–134. 10.1007/978-1-4939-9118-1_12 [DOI] [PubMed] [Google Scholar]
- Gast R. K., Dittoe D. K., Ricke S. C. (2022). Salmonella in eggs and egg-laying chickens: pathways to effective control. Crit. Rev. Microbiol. 0, 39–63. 10.1080/1040841X.2022.2156772 [DOI] [PubMed] [Google Scholar]
- Genchi G. (2017). An overview on D-amino acids. Amino Acids 49, 1521–1533. 10.1007/s00726-017-2459-5 [DOI] [PubMed] [Google Scholar]
- Jonas K., Tomenius H., Kader A., Normark S., Römling U., Belova L. M., et al. (2007). Roles of curli, cellulose and BapA in Salmonella biofilm morphology studied by atomic force microscopy. BMC Microbiol. 7, 70. 10.1186/1471-2180-7-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubacher M. E., Ades S. E. (2008). The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J. Bacteriol. 190, 2065–2074. 10.1128/JB.01740-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros J., Jan S., Bonnassie S., Gautier M., Croguennec T., Pezennec S., et al. (2021). The role of ovotransferrin in egg-white antimicrobial activity: a review. Foods 10, 823. 10.3390/foods10040823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüneburg N., Xanthakis V., Schwedhelm E., Sullivan L. M., Maas R., Anderssohn M., et al. (2011). Reference intervals for plasma L-arginine and the L-arginine:asymmetric dimethylarginine ratio in the framingham offspring cohort. J. Nutr. 141, 2186–2190. 10.3945/jn.111.148197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal R. K., Jiang T., Kwon Y. M. (2021). Genetic determinants in Salmonella enterica serotype typhimurium required for overcoming in vitro stressors in the mimicking host environment. Microbiol. Spectr. 9, e0015521–21. 10.1128/Spectrum.00155-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 17, 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Mills E., Petersen E., Kulasekara B. R., Miller S. I. (2015). A direct screen for c-di-GMP modulators reveals a Salmonella Typhimurium periplasmic ʟ-arginine–sensing pathway. Sci. Signal 8, ra57. 10.1126/scisignal.aaa1796 [DOI] [PubMed] [Google Scholar]
- Mills E., Pultz I. S., Kulasekara H. D., Miller S. I. (2011). The bacterial second messenger c-di-GMP: mechanisms of signalling. Cell. Microbiol. 13, 1122–1129. 10.1111/j.1462-5822.2011.01619.x [DOI] [PubMed] [Google Scholar]
- Miyamoto T., Katane M., Saitoh Y., Sekine M., Homma H. (2017). Identification and characterization of novel broad-spectrum amino acid racemases from Escherichia coli and Bacillus subtilis . Amino Acids 49, 1885–1894. 10.1007/s00726-017-2486-2 [DOI] [PubMed] [Google Scholar]
- O’Callaghan D., Charbit A. (1990). High efficiency transformation of Salmonella typhimurium and Salmonella typhi by electroporation. Mol. Gen. Genet. MGG 223, 156–158. 10.1007/BF00315809 [DOI] [PubMed] [Google Scholar]
- Porwollik S., Santiviago C. A., Cheng P., Long F., Desai P., Fredlund J., et al. (2014). Defined single-gene and multi-gene deletion mutant collections in Salmonella enterica sv typhimurium. PLOS ONE 9, e99820. 10.1371/journal.pone.0099820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poudel B., Shterzer N., Sbehat Y., Ben-Porat N., Rakover M., Tovy-Sharon R., et al. (2022). Characterizing the chicken gut colonization ability of a diverse group of bacteria. Poult. Sci. 101, 102136. 10.1016/j.psj.2022.102136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugliese L., Coda A., Malcovati M., Bolognesi M. (1993). Three-dimensional structure of the tetragonal crystal form of egg-white avidin in its functional complex with biotin at 2·7 Å resolution. J. Mol. Biol. 231, 698–710. 10.1006/jmbi.1993.1321 [DOI] [PubMed] [Google Scholar]
- Raspoet R., Shearer N., Appia-Ayme C., Haesebrouck F., Ducatelle R., Thompson A., et al. (2014). A genome-wide screen identifies Salmonella Enteritidis lipopolysaccharide biosynthesis and the HtrA heat shock protein as crucial factors involved in egg white persistence at chicken body temperature. Poult. Sci. 93, 1263–1269. 10.3382/ps.2013-03711 [DOI] [PubMed] [Google Scholar]
- Stevenson D. M., Weimer P. J. (2007). Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 75, 165–174. 10.1007/s00253-006-0802-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Tn-Seq reads are available in the SRA database under the BioProject number PRJNA1089541.