Abstract
A xenograft model of the human disease Langerhans cell histiocytosis (LCH) was investigated with severe combined immunodeficiency (SCID) mice. Transplantation of human LCH biopsy material into SCID mice resulted in the generation of mouse tumors resembling lymphomas. A thymoma cell line (ThyE1M6) was generated from one of these mice and found to display significant levels of Mg2+-dependent reverse transcriptase activity. Electron microscopy revealed particles with type D retroviral morphology budding from ThyE1M6 cells at a high frequency, whereas control cultures were negative. Reverse transcription-PCR of virion RNA with degenerate primers for conserved regions of various mouse, human, and primate retroviruses amplified novel sequences related to primate type D retroviruses, murine intracisternal A particles, Jaagsiekte sheep retrovirus, and murine long interspersed nuclear elements but not other retroviral classes. We demonstrate that these sequences represent a novel group of endogenous retroviruses expressed at low levels in mice but expressed at high levels in the ThyE1M6 cell line. Furthermore, we propose that the activation of endogenous retroviral elements may be associated with a high incidence of thymomas in SCID mice.
Langerhans cell histiocytosis (LCH) is a human disease of unknown etiology characterized by the accumulation of clonally derived Langerhans cells (65, 69). In addition, there is an accumulation of inflammatory cells, including T cells, macrophages, eosinophils, neutrophils, giant cells, and plasma cells (24, 66). The clinical spectrum of the disease varies and includes isolated, benign lesions of bone called eosinophilic granuloma (33, 44), multifocal disease (32, 58), and severe, life-threatening disseminated disease (7, 20, 28, 55).
A study of the etiology of LCH has been hindered by the limited availability of disease material, particularly progressive disease material. Consequently, sporadic biopsy samples are commonly used for research, and progress in defining the mechanisms of pathogenesis is complicated by the lack of consistent materials. In order to further characterize LCH, we xenografted human LCH biopsy material into severe combined immunodeficiency (SCID) mice with a view to observing the induction of LCH-type pathogenesis. SCID mice lack the B and T lymphocytes required for an immune response to allo- or xenografts (5, 10, 11) and have been used to establish successful long-term engraftment of human tissues (23, 46). A SCID mouse injected with LCH biopsy material developed a lymphoma. A cell line, ThyE1M6, was established from this lymphoma. Subsequent passage of this cell line in SCID mice resulted in disseminated cellular infiltrates distinct from lymphoma but similar to the multifocal LCH observed in human patients.
Here we examine the ThyE1M6 cell line and consider the possibility that xenotransplantation of human LCH tissue transmitted the LCH disease phenotype from humans to mice. During our analysis, we observed novel viral particles, resembling the type D retroviruses of primates, budding from the ThyE1M6 cell line. Since there are no type D retroviruses yet characterized from mice and many enigmatic accounts of type D retroviruses from human tumor cell lines (2, 16, 27, 41, 48, 64) and an immunocompromised patient (4), we characterized these particles at the molecular level.
MATERIALS AND METHODS
Xenografting of SCID mice.
Six- to 8-week-old female SCID mice of the original C.B-17 strain background were used in all experiments. A thymus biopsy sample from a 13-year-old female patient diagnosed with LCH (LCH patient B) was teased into a single-cell suspension and continuously cultured in the presence of 25 ng of the inflammatory cytokines tumor necrosis factor alpha and granulocyte-macrophage colony-stimulating factor (Boehringer Mannheim Biochemica, Mannheim, Germany) per ml for 35 days. Live cells were purified on a Ficoll gradient, and 6.7 × 104 cells in saline containing tumor necrosis factor alpha and granulocyte-macrophage colony-stimulating factor were injected subcutaneously into each of three SCID mice. Cytokine injections were repeated daily for 5 days only. Three additional SCID mice were injected with cytokines only as a control. Organs were harvested from the mice after 9 weeks for histopathologic examination. One of the xenotransplanted mice developed a thymic mass, with widespread involvement of lymphomatous infiltrates of liver, spleen, lymph nodes, lungs, and bone marrow. The lymphoid cells from the enlarged thymus were placed into a culture, and a nonclonal cell line, ThyE1M6, was obtained. A different xenotransplanted mouse developed an ovarian tumor with the histological appearance of a lymphoma.
Cell cultures.
All cells were grown in Iscove’s modified Dulbecco’s medium (Gibco BRL, Gaithersburg, Md.) supplemented with 10% fetal calf serum (Gibco BRL) at 37°C. Biopsy samples derived from two patients diagnosed with LCH were cultured to establish the cell lines LCH A and LCH B used in this study. A control cell line, STh1a, from a spontaneous thymoma from an aging SCID mouse was cultured and cloned twice in agar (cell line kindly provided by Ian Radford, Peter McCallum Cancer Institute, Melbourne, Victoria, Australia). The Ann-1 cell line (9) is chronically infected with Abelson murine leukemia virus (A-MuLV) and Moloney murine leukemia virus (Mo-MuLV) and was used as a type C retrovirus control. The MiCl1 (S+ L−) mink (Mustela vison) lung cell line (infected with Moloney murine sarcoma virus) was grown in RPMI medium (Gibco BRL) supplemented with 10% fetal calf serum.
FACS analysis of the ThyE1M6 cell line.
The surface antigen phenotype of ThyE1M6 cells was determined by staining with monoclonal antibodies to murine antigens CD4, CD8, H2b, Ly-2, Thy-1, F4/80, N418, Mac-1, and NLDC145 as well as to several human leukocyte antigens (including CD4, CD8, CD45, CD19, and CD14) (all listed antibodies were supplied by Becton Dickinson/Pharminogen, San Jose, Calif.). Fluorescence-activated cell sorter (FACS) analysis was performed with a FACStar II apparatus (Becton Dickinson Immunocytometry Systems, San Jose, Calif.).
PERT assay.
Culture supernatants from ThyE1M6 cells were centrifuged at 16,000 × g for 10 min to remove cellular debris and then were filtered through a 0.2-μm-pore-size filter (Millipore, Bedford, Mass.). Virions were pelleted from 5 ml by ultracentrifugation in an SW56.1 rotor (Beckman, Fullerton, Calif.) at 70,000 × g for 90 min. The pellet was resuspended in 30 μl of lysis buffer, consisting of 100 mM KCl, 25 mM Tris-HCl (7.8), 10 mM dithiothreitol, 0.25 mM EDTA, 0.6% Triton X-100, and 50% glycerol. Viral reverse transcriptase (RT) activity was assayed by the ability to generate cDNA from an MS-2 phage RNA template primed with an MS-2-specific primer. A 112-bp portion of the MS-2 cDNA was amplified by PCR (25 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C), quantified by Southern blot hybridization with 5 ng of biotinylated RT-3 (59), an internal MS-2-specific probe, and then a streptavidin-horseradish peroxidase conjugate (DAKO, Carpinteria, Calif.), and detected with chemiluminescence (ECL kit; Amersham, Buckinghamshire, England). In addition to being assayed by the highly sensitive PCR-enhanced RT (PERT) assay, RT activity was assayed by measuring the incorporation of [32P]TTP with an oligo(dT) · poly(rA) template in the presence of 0.6 mM Mn2+ or 5 mM Mg2+ as the divalent cation as previously described (43, 53).
Electron microscopy.
Pellets of ThyE1M6 cells were fixed with glutaraldehyde and osmium tetroxide and analyzed by transmission electron microscopy as described by Lee et al. (30).
Virion RNA and cDNA preparation.
Virions were pelleted from 180 ml of supernatant from early passages of ThyE1M6 cells by ultracentrifugation for 1 h at 100,000 × g in an SW28 rotor (Beckman, Buckinghamshire, England). The RNA was extracted from the pellet by the method of Chomczynski and Sacchi (6). First-strand cDNA was prepared from viral RNA by use of 2 U of avian myeloblastosis virus RT (Boehringer) per μl in the supplied buffer, which was supplemented with 1 U of RNasin (Promega, Madison, Wis.) per μl, 5 pmol of random hexamers (Promega) per μl, and 1 mM deoxynucleoside triphosphate (dNTP) mix (Gibco BRL); the mixture was incubated at 42°C for 1 h, and inactivated at 70°C for 5 min.
Degenerate PCR of novel retroviral sequences.
Degenerate primers for the human retroviruses human immunodeficiency virus type 1 (HIV-1) (45) and human T-cell leukemia virus (HTLV) type 1 (HTLV-1) (29) and for human (56) and mouse (51) type C retroviruses were used for PCR. In addition, primers for amplifying all existing type D viruses were identified from multiple alignments of simian retrovirus (SRV) type 1 (SRV-1) and type 2 (SRV-2), Mason-Pfizer monkey virus (MPMV), and squirrel monkey retrovirus (SMRV). Conserved regions were used to design degenerate type D virus PCR primers for gag [gag1, 5′ AGGGGCCAGCCCCAGG(C/G)CCC; gag2, 5′ GAGGTCCA(A/G)TCCTGCACT] and pro (pro1, 5′ GG(A/C)AGTGCAGGA(T/C)TGGACCTC(T/A)GT; pro2, 5′ AGT(A/GA/G)CATC(A/T/G)GC(T/C)CC(C/A/T)GTATC], which were used to amplify the respective genes from the viral cDNA; nucleotides in parentheses represent alternative nucleotides incorporated during oligonucleotide synthesis to account for sequence variations at these positions. The predicted PCR product sizes for the gag1-gag2 primer pair are 119 bp for SRV-1, SRV-2, and MPMV and 157 bp for SMRV. The predicted PCR product size for the pro1-pro2 primer pair is 443 bp. The PCR products generated with the gag1-pro2 primer pair are 544 bp for SRV1, SRV2, and MPMV and 583 bp for SMRV. Hot-start PCR amplification was performed with Taq DNA polymerase (Perkin-Elmer/Roche, Branchburg, N.J.) in a 1.5 mM Mg2+ reaction buffer with 25 pmol of each primer. Forty amplification cycles at 94°C for 1 min, 45°C for 1 min, and 72°C for 1 min and a final extension at 72°C for 7 min were performed with a Perkin-Elmer 9600 Thermocycler. PCR products were subsequently treated with 1.25 U of Pfu polymerase (Stratagene, La Jolla, Calif.) per reaction for 30 min at 72°C to create blunt ends, gel purified, and ligated into pCR-Script [Stratagene pCR-Script SK(+) cloning kit] according to the manufacturer’s instructions.
Plasmid sequencing.
Plasmid sequencing was performed by use of an ABI Prism Dye Terminator Cycle Sequencing Ready Reaction Kit with Amplitaq DNA polymerase, and sequences were analyzed by use of an Applied Biosystems 373 DNA sequencer (Perkin-Elmer/Applied Biosystems).
Southern analysis.
Genomic DNA was prepared from cultured cells and mouse tissues by the methods of Maniatis et al. (37) and digested with Boehringer restriction enzymes. Digestion products were separated on a 1% agarose–Tris-acetate gel and transferred to a Hybond N+ nylon membrane (Amersham) in 0.4 M NaOH. The membrane was prehybridized with 1 M NaCl–10% dextran sulfate–1% sodium dodecyl sulfate (SDS)–100 μg of salmon sperm DNA per ml for 1 h at 42°C and then hybridized overnight under the same conditions with a degenerate gag-pro PCR product labeled by four extra cycles of PCR with [α-32P]dCTP. The membrane was washed at 65°C under the following conditions: once in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 30 min, once in 1× SSC for 30 min, and once in 0.2× SSC for 30 min; the membrane was then exposed to Kodak X-Omat film.
Northern analysis.
Total cellular RNA was extracted from cell culture pellets by the method of Chomczynski and Sacchi (6). Ten micrograms of each sample was loaded onto a 1.6% agarose–formamide gel as described by Maniatis et al. (37) and, following separation, RNA was transferred to a Hybond N+ membrane in 10× SSC. The membrane was prehybridized at 65°C for 30 min with 0.5 M sodium phosphate buffer (pH 7.0)–1 mM EDTA–0.1% bovine serum albumin–7% SDS and then hybridized overnight under the same conditions with a degenerate gag-pro PCR product labeled by four extra cycles of PCR with [α-32P]dCTP. The membrane was washed twice for 30 min each time at 65°C in 40 mM sodium phosphate (pH 7.0)–1 mM EDTA–1% SDS and exposed to Kodak X-Omat film.
RESULTS
Generation of a murine thymocytic lymphoma cell line, ThyE1M6, from SCID mice.
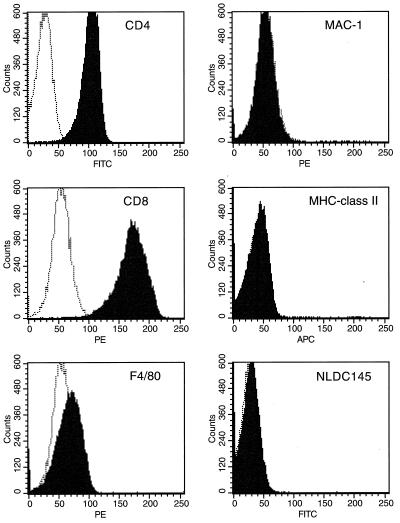
A thymic mass removed at biopsy from a human patient with LCH was continuously cultured in the presence of TNF-α and GM-CSF and, after 35 days, was injected subcutaneously into SCID mice. Two of three mice injected with the cultured cells developed a lymphoma, whereas mice receiving only cytokines were normal. The role that the injection of LCH biopsy material played in the induction of these tumors is unclear. A cell line, ThyE1M6, was generated from one of these mice. Immunophenotyping of the ThyE1M6 cell line demonstrated that it was a mouse-derived pre-T-lymphocyte thymoma bearing murine CD4inter and CD8hi surface markers and showing low expression of the F4/80low antigen (Fig. 1). Antibodies to mouse dendritic cells (NLDC145; major histocompatibility complex class II) and macrophages (Mac-1) (Fig. 1) and to human antigens (CD4, CD8, CD45, CD19, CD14) (data not shown) were negative. The first passage of the ThyE1M6 cell line into SCID mice resulted in inflammatory infiltrates in the liver, spleen, bone marrow, intestines, and mesenteric rudimentary lymph nodes and at the site of injection. Histopathologic examination of the infiltrated tissues demonstrated the presence of cell types associated with LCH, such as granulocytes, eosinophils, macrophages, giant cells, mast cells, and lymphocyte-like cells (data not shown).
FIG. 1.
FACS analysis of the ThyE1M6 cell line. The shaded area represents the test antibody, and the dotted line represents the isotype-matched negative control. MHC, major histocompatibility complex; FITC, fluorescein isothiocyanate; PE, phycoerythrin; APC,
The ThyE1M6 cell line has a high level of Mg2+-dependent RT activity.
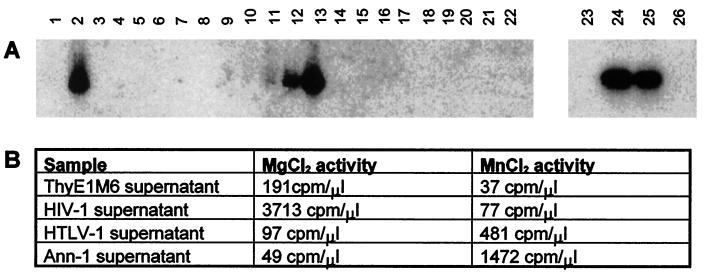
The ThyE1M6 cell line was tested for RT activity with the PERT assay (52). Cell culture supernatants were harvested from the ThyE1M6 cell line and a virus-infected control cell line, Ann-1, an A-MuLV-transformed NIH 3T3 clone superinfected with cloned Mo-MuLV (9). After ultracentrifugation to concentrate virus particles, pellets were tested for RT activity with the PERT assay, as shown in Fig. 2A. In this assay, the presence of RT enzyme is detected by PCR amplification of cDNA reverse transcribed from an MS-2 RNA template. The expected 112-bp PCR product was obtained from both the ThyE1M6 and the Ann-1 cell supernatants but not from supernatants from cultured LCH biopsy material or from human osteosarcoma, rhabdomyosarcoma, and retinoblastoma primary cultures. Furthermore, there was an increase in RT activity with continuous culturing (3 to 144 h) of the ThyE1M6 cell line, suggesting an accumulation of viral particles with culturing (Fig. 2A). Since the cell culture supernatant derived from LCH B (which was used in the genesis of the ThyE1M6 cell line) (Fig. 2A, lanes 14 to 18) was negative, it is unlikely that the particle-associated RT activity was transmitted from the biopsy material to SCID mice. Since the PERT assay is a highly sensitive, nonquantitative PCR-based method, we carried out RT assays which involved measuring the incorporation of [32P]TTP with an oligo(dT) · poly(rA) template in the presence of either Mn2+ or Mg2+ as the divalent cation to confirm the presence of RT activity in ThyE1M6 cells, as shown in Fig. 2B. This assay confirmed RT activity with a preference for Mg2+, which is commonly associated with the retroviruses of primates, whereas most murine type C viruses have a preference for Mn2+ (61).
FIG. 2.
RT activity in the ThyE1M6 cell line culture supernatant, as demonstrated by the PERT assay. In all cases, culture supernatants were used to prepare concentrated extracts used in the PERT assay. Lanes 1 and 23 are blank (H2O) PCR controls, lanes 19 and 26 are culture medium controls, lane 24 represents RT from the positive control Ann-1, and lanes 2 and 25 represent RT activity detected in supernatant preparations from confluent ThyE1M6 cells. Lanes 3 to 8 represent LCH A, and lanes 14 to 18 represent LCH B. Lanes 9 to 13 represent an RT activity time course for ThyE1M6 cells in which the supernatant was removed after 3, 24, 48, 120, and 144 h of continuous cell culturing and virion preparations were assayed for RT activity. Lanes 4 to 8 and 14 to 18 represent equivalent time courses for LCH A and LCH B, respectively. Lane 3 represents a confluent culture of the LCH A cell line. Human osteosarcoma, rhabdomyosarcoma, and retinoblastoma cell lines are represented in lanes 20 to 22, respectively. (B) RT activity in the ThyE1M6 cell culture supernatant, as determined by measurement of [32P]TTP incorporation with an oligo(dT) · poly(rA) template in the presence of either Mg2+ or Mn2+ as the divalent cation. HIV type 1 (HIV-1) and HTLV-1 supernatants were used for comparison. A background value was determined and was subtracted to give the values shown.
Electron microscopic analysis reveals a high frequency of budding type D retrovirus particles.
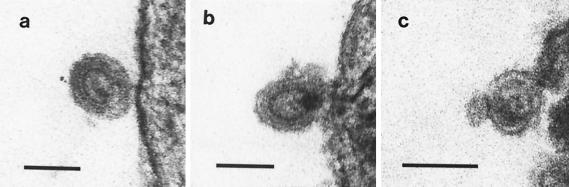
We carried out electron microscopy of the ThyE1M6, Ann-1, and STh1a (a SCID-derived spontaneous thymoma) cell lines, spleen and bone marrow derived from normal SCID mice, skin biopsy material from LCH patient B, and cultured thymus cells from LCH patient B. We were able to detect budding and mature virus particles with different morphologies and frequencies from all three cell lines, but no particles were observed from the mouse or human tissues (Fig. 3 and Table 1). Novel particles were evident from the ThyE1M6 cell line and would best be classified as type D retrovirus like on the basis of their size and rod-like core morphology (17); however, unlike the results for simian type D retroviruses, no intracellular precursor particles were observed. The ThyE1M6 particles were observed at a frequency of 1 per 24 sectioned cells examined, formed at the plasma membrane, and were round or ovoid, with a tubular core. Two matrix layers (shells) surrounded the core, a thin inner layer, and a thicker outer layer. The frequency of these particles decreased with increasing passage number. The particles observed from the STh1a thymoma cell line that spontaneously arose in SCID mice would also best be classified as type D retrovirus like. Compared to the ThyE1M6 particles, the STh1a particles had similar dimensions and morphology, including the presence of two matrix layers, although core structures could not be discerned. Therefore, the type D virus observed in the ThyE1M6 cell line was morphologically similar to but distinct from the virus observed in the STh1a cell line. As expected, type C retrovirus-like particles were observed budding from Ann-1 (infected with A-MuLV and Mo-MuLV), and these virions were clearly distinguishable from the particles budding from the SCID thymoma cell lines. Compared to the type C retrovirus-like Ann-1 particles, the type D retrovirus-like ThyE1M6 and STh1a particles were larger and had an additional matrix layer surrounding the core (Fig. 3 and Table 1).
FIG. 3.
Electron microscopy analysis of virus particles produced by ThyE1M6, STh1a, and Ann-1 cell lines. Mature type D retrovirus particles were produced by ThyE1M6 cells (a) and by STh1a cells (b). Mature type C retrovirus particles were produced by Ann-1 cells (c). Bar, 100 nm.
TABLE 1.
Characterization of the viral particles produced by the ThyE1M6, STh1a, and Ann-1 cell lines
| Cell line or tissue examined | Particles detected | Particle measurements, in mean ± SD nm (n)a
|
Core structure | |||||
|---|---|---|---|---|---|---|---|---|
| Diam
|
Core tubule
|
Shell width
|
||||||
| Total | Core | Width | Length | Inner | Outer | |||
| ThyE1M6 | Type D retrovirus (1 particle/24 cells examined) | 103.8 ± 7.4 (10) | 50.0 ± 5.7 (5) | 14.6 (2) | 43.8 (2) | 3.9 ± 1.3 (4) | 19.1 ± 2.2 (4) | Tubular |
| STh1a | Type D retrovirus | 105.1 ± 7.1 (8) | 47.4 ± 6.4 (4) | 4.9 ± 1.0 (5) | 20.9 ± 1.2 (5) | Not observed | ||
| Ann-1 | Type C retrovirus | 98.2 ± 8.1 (12) | 68.3 ± 7.5 (12) | Spherical | ||||
| SCID spleen | Not detected in 170 separate cell sections examined | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| SCID bone marrow | Not detected in 170 separate cell sections examined | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| LCH patient B skin biopsy material | Not detected in a tissue section area covering 8,500 μm2 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| LCH patient B thymic mass culture | Not detected in 40 separate cell sections examined | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
n, number of particles measured in order to derive a mean particle size. N/A, not applicable.
We were unable to detect mature virus particles in SCID mouse spleen or bone marrow samples. We cannot exclude the possibility that these particles are present at a frequency too low for detection by electron microscopy. Low-level RT activity from cultured SCID spleen and bone marrow in the PERT assay would support the presence of infrequent particles (data not shown). In addition, we were unable to detect mature virus particles in LCH patient B skin biopsy material or in cells cultured from a thymic mass from this patient. The LCH B cell line was also negative for RT activity, as determined by the PERT assay (Fig. 2A, lanes 14 to 18). Therefore, it is likely that the novel type D retrovirus particles are associated with a SCID mouse-derived thymocytic lymphoma, and we were interested in testing the possibility that these particles were of endogenous origin.
Degenerate PCR of virus preparations from ThyE1M6 cells reveals the expression of novel type D retrovirus sequences.
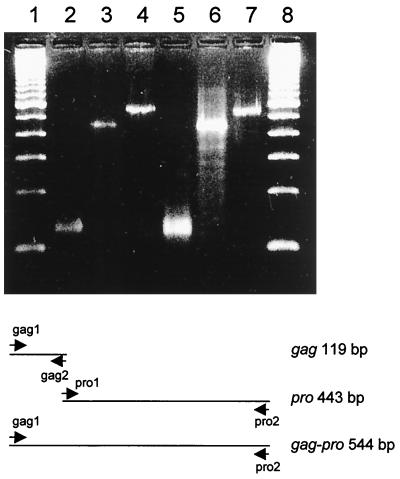
In order to identify the type D particles produced by the ThyE1M6 cell line at the molecular level, we carried out degenerate PCR cloning. Since type D virus morphology and a preference for Mg2+-dependent RT activity were observed, we designed PCR primers for highly conserved sequences of SRV-1, SRV-2, MPMV, and SMRV. Primers were designed for several regions, including the gag and pro genes, and were used to yield three overlapping PCR products of the predicted sizes. A gag region product of 119 bp, a pro region product of 443 bp, and a continuous gag-pro product of 544 bp were observed. Particles were pelleted from ThyE1M6 and STh1a cell culture supernatants. RNA was extracted from virions, reverse transcribed to cDNA, and subjected to PCR with the gag, pro, and gag-pro primer pairs. Bands of the predicted sizes were observed in both ThyE1M6 and STh1a samples, as shown in Fig. 4. All PCR products were cloned, sequenced, and summarized according to their sequence similarities (Table 2). These results showed that the PCR products contained several groups of novel sequences related to the simian type D retroviruses, murine intracisternal alpha-particle elements (IAPE) (1, 28, 36, 42), Jaagsiekte sheep retrovirus (JSRV) (67, 68), and murine long interspersed nuclear elements (LINE) (14). A number of sequence variants were identified in both ThyE1M6 and STh1a samples: those related to type D simian retroviruses (represented by clones 3.12, 3.21, 3.23, and 2.53) and those related to the IAPE (represented by clones 2.27 and 2.29). Each of these sequences was found to be present in both the ThyE1M6 and the STh1a cell lines.
FIG. 4.
RT-PCR cloning of the gag-pro genes of type D retrovirus expressed by the ThyE1M6 and STh1a cell lines. Lanes 1 and 8, molecular size markers (100-bp ladder); lanes 2 to 4, gag, pro, and gag-pro PCR products, respectively, derived from the virus from ThyE1M6 cells; lanes 5 to 7, gag, pro, and gag-pro PCR products, respectively, derived from the virus from STh1a cells. The predicted sizes were 119 bp for gag, 443 bp for pro, and 544 bp for gag-pro. The primers used to amplify these sequences are indicated at the bottom of the figure.
TABLE 2.
BLAST analysis of gag-pro PCR products generated by RT-PCR cloning from the ThyE1M6 cell line
| Group | Clone (bp) | GenBank accession no. | BLAST sequence analysis search resultsa |
|---|---|---|---|
| Primate type D retrovirus (nine clones sequenced) | 3.12 (551) | AF093700 | 64% nt identity with SRV-2 and MPMV; significant identity with SRV-1, JSRV, SMRV, and LINE |
| 3.21 (527) | AF093701 | 62% nt identity with SRV-2 and MPMV; significant identity with SRV-1, SMRV, JSRV, and LINE | |
| 3.23 (450) | AF093699 | 60% nt identity with SRV-1 and JSRV; significant identity with SRV-2 and SMRV | |
| 2.53 (403) | AF093698 | 61% nt identity with SRV-1, MPMV, and SRV-2; significant identity with SMRV, JSRV, and LINE | |
| Retrovirus-like IAPE (seven clones sequenced) | 2.27 (524) | AF093697 | 85% nt identity with Mus musculus IAPE-Y retroviral genome; significant nt identity with M. musculus type IIB IAPE; significant nt identity with SRV-1, JSRV, SRV-2, SMRV, and MPMV |
| 2.29 (545) | AF093696 | 92% nt identity with M. musculus LEC-A retroviral genome; significant nt identity with M. musculus type IIB IAPE; significant nt identity with SRV-1, SRV-2, JSRV, SMRV, and MPMV |
nt, nucleotide.
A number of other PCR primers were designed for characterized retroviral sequences, such as MPMV gag, MPMV env, degenerate SRV env, degenerate SRV pol, HTLV-1, murine leukemia virus, type C virus pol, degenerate pol, VL30, MLV env, and primate foamy virus. However, these primers did not generate PCR products of the expected sizes relative to the known retroviruses (data not shown).
The type D retrovirus-related genes are endogenous genetic elements expressed in mice.
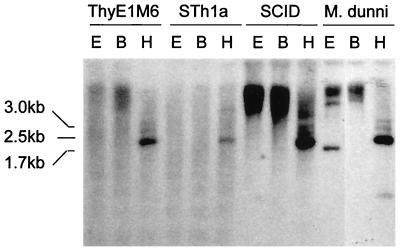
In order to investigate if the viral sequence elements that we identified by PCR were derived from endogenous mouse sequences, Southern blot analysis was performed. The type D retrovirus-related sequence gag-pro product (generated by degenerate PCR from a ThyE1M6 cell culture supernatant [Fig. 4]) was used to screen a panel of mouse-derived genomic DNAs from ThyE1M6 cells, STh1a cells, SCID mice, and Mus dunni mice and is shown in Fig. 5. A band of approximately 1.7 kb and two less prominent bands of approximately 2 and 3 kb were seen in all mouse-derived genomic DNAs digested with HindIII, including that from the wild mouse M. dunni. No cross-hybridizing bands of any size were detected in rat, hamster (CHO cells), human (HeLa cells and LCH patient tissues), and mink (MiCl1 [S+ L−]) genomic DNAs (data not shown). Therefore, the gag-pro PCR product was likely to have been derived from RNA expressed from an endogenous mouse element that is widespread across strains.
FIG. 5.
Southern analysis of mouse-derived genomic DNAs extracted from ThyE1M6 and STh1a cells and from SCID mouse and M. dunni tissues and probed with gag-pro. Restriction endonuclease digests with EcoRI (E), BamHI (B), and HindIII (H) are shown. A prominent band of approximately 1.7 kb and two less prominent bands of approximately 2 and 3 kb were present in all samples digested with HindIII.
The endogenous murine type D retrovirus-related elements are expressed as large RNA molecules.
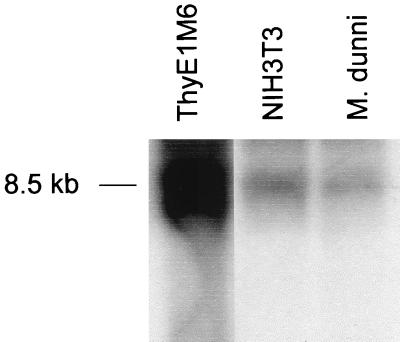
The gag-pro PCR product amplified from ThyE1M6 cDNA was radiolabelled and used as a probe in Northern blot experiments. Total RNAs from ThyE1M6, NIH 3T3, and M. dunni cell cultures were tested, and a band of approximately 8.5 kb was observed in the mouse-derived cells, with the highest abundance in the ThyE1M6 cell line (in which several bands were observed) (Fig. 6). No gag-pro-hybridizing bands were observed in normal human, LCH patient, and rat RNA samples (data not shown). Thus, the type D retrovirus-related endogenous elements are abundantly expressed as large RNA molecules that may represent a complete viral genome. This type D retrovirus-related genome may account for the type D retrovirus particles observed budding from the SCID-derived cell lines ThyE1M6 and STh1a.
FIG. 6.
Northern analysis of total cellular RNAs extracted from the mouse-derived cell lines ThyE1M6, NIH 3T3, and M. dunni. A band of approximately 8.5 kb was detected in all cells when probed with the ThyE1M6 gag-pro PCR products.
DISCUSSION
In this study, during the process of attempting to establish a xenograft model for the human disease LCH in SCID mice, a thymocytic lymphoma cell line, ThyE1M6, was generated. The ThyE1M6 cell line was shown to be of mouse origin and to express budding type D retrovirus particles containing RT activity. Degenerate PCR amplification and sequencing of the predominant encapsidated viral RNAs demonstrated a significant relationship of most PCR products to the type D retroviruses at the level of gene sequence and arrangement as well as homology to IAPE, LINE, and JSRV sequences. We have shown by hybridization analysis that the genome for these viral RNAs contains endogenous elements common to all murine strains, including the wild mouse M. dunni.
Further analysis of the full genome sequence of the virus from ThyE1M6 cells will be required to clarify whether LCH patient cells have contributed any sequences to the newly identified mouse type D retrovirus. The data presented here do not support genetic recombination with human LCH-derived genetic elements contributing to the gag-pro region of the novel virus but do support the activation of a novel endogenous mouse retrovirus. Human DNA, including LCH patient DNA, does not contain type D virus gag-pro-related sequences; furthermore, human tissues were also negative for RT activity. However, it will be interesting to address this question with further analysis of the viral genome.
The type D class of SRVs has been extensively characterized. These viruses were initially identified in studies of simian acquired immunodeficiency syndrome (38, 39, 49, 59, 62). In addition, type D retrovirus particles have been observed in a variety of transformed human cell lines (2, 16, 27, 41, 48, 64). These are presumed to be a culture contaminant (2, 48), although the activation of endogenous elements has not been ruled out. Furthermore, type D retrovirus particles have been observed in the serum of a human AIDS patient (4). A novel exogenous sequence related to type B and type D retroviruses has been identified in patients with the autoimmune condition Sjogren’s syndrome (19). In addition, the suspected etiological agent of ovine pulmonary carcinoma, JSRV, is an exogenous and endogenous type D and type B retrovirus of the ovine and caprine species (47, 67, 68). It has been speculated that JSRV may be a helper virus for an oncogenic replication-defective retrovirus, although this hypothesis is difficult to test, since JSRV cannot be grown in vitro. To our knowledge, this is the first report of the observation in mice of type D retrovirus particles, which are present as endogenous elements and are active in a mouse-derived thymocytic lymphoma.
The expression of type D retrovirus particles may be a common feature of SCID mouse-derived thymocytic lymphoma, and we speculate whether it is associated with the generation of lymphoma in the SCID mouse genetic background. SCID mice are susceptible to spontaneous thymoma, the incidence of which can be significantly increased by breeding with strains such as the NOD (nonobese diabetic) mouse strain, which is not usually susceptible to spontaneous thymoma. However, SCID/NOD mice have a 67% incidence compared to a 15 to 20% incidence for mice with the original SCID mutation background (C.B-17) (50). Differential endogenous proviral genes often account for strain-specific susceptibility or resistance to spontaneous lymphomagenesis; for SCID/NOD mice, thymomagenesis was associated with the expression of a NOD mouse-unique endogenous ecotropic murine leukemia provirus locus (Emv-30) (50). Recombination between an ecotropic virus and one or more endogenous nonecotropic proviral sequences is likely a causative agent for the high incidence of spontaneous lymphoma in strains such as AKR/J (8, 13, 18, 21, 35, 60).
Molecular genetic analysis indicates a strong association between a high spontaneous incidence of hemopoietic neoplasms and the activation of endogenous murine leukemia viruses. This hypothesis is supported by cross-strain breeding experiments (such as those with SCID/NOD mice), in which murine strains carrying different endogenous elements are brought together in one genome, facilitating potential cross-activation and recombination.
It has been demonstrated that the DNA-dependent kinase (p350) gene is the candidate gene for the SCID phenotype, resulting in an impairment of the double-strand break recombination repair pathway (15, 26). Therefore, in addition to a deficiency in B and T lymphocytes, SCID mice are highly susceptible to DNA damage, as demonstrated by hypersensitivity to ionizing radiation (3, 34). This impairment confers a high potential for mutagenesis, genomic instability, and tumorigenesis and may be linked to the occurrence of spontaneous thymomas in SCID mice. Therefore, there may be an increased susceptibility to retroviral activation in these mice.
The homology observed between the virus from ThyE1M6 cells and LINE is intriguing in the context of recent reports of the FHIT gene. It has been demonstrated that the human FHIT gene encompasses the common human fragile site FRA3B and is often inactivated or deleted in tumor cells (57). FRA3B contains a high incidence of LINE insertions (22). LINE are capable of retrotransposition (54) and have been shown to be involved in the insertional mutation of a number of genes (12, 25, 40, 63). It has been proposed that carcinogen damage to DNA results in rearrangement of the FHIT gene by homologous recombination with LINE sequences. FHIT functions as a tumor suppressor and thus may act early in tumor development. The induced expression of LINE sequences may lead to increased recombination across the SCID mouse genome and may be an early event leading to the generation of thymomas.
We have identified a novel endogenous type D retrovirus of mice which is expressed at high levels by the ThyE1M6 cell line. The results presented in this paper suggest that the activation of this endogenous virus might have been associated with the genesis of a thymocytic lymphoma in a SCID mouse. It will be interesting to determine whether the virus identified here is involved in the pathogenesis of spontaneous thymomas in other SCID mice. We aim to further characterize this endogenous murine retrovirus by cloning and sequencing of the full viral genome.
ACKNOWLEDGMENTS
We thank Wendy Cook for the Ann-1 cell line and Ian Radford for the STh1a cell line. We thank Len C. Harrison and Kaku Nakagawa for helpful discussions and Ken Shortman and Frank Battey for FACS analysis of the ThyE1M6 cell line. We thank Thulasi Murughiah, Rodney Daly, Soong Ling, and Thuy Diem for excellent assistance with SCID mice, animal experimentation, and technical assistance and Anthea Ramos for technical assistance.
D.F.J.P. was supported by the NHMRC of Australia and by Macfarlane Burnet Centre for Medical Research funds. This work was supported by grants to G.K. from the Histiocytosis Association of America, the NHMRC of Australia, CICA (Ballarat, Victoria, Australia), and St. John of God Hospital (Ballarat, Victoria, Australia).
REFERENCES
- 1.Aota S-I, Gojobori T, Shigesada K, Ozeki H, Ikemura T. Nucleotide sequence and molecular evolution of mouse retrovirus-like IAP elements. Gene. 1987;56:1–12. doi: 10.1016/0378-1119(87)90153-3. [DOI] [PubMed] [Google Scholar]
- 2.Asikainen K, Vesanen M, Kuittinen T, Vaheri A. Identification of human type D retrovirus as a contaminant in a neuroblastoma cell line. Arch Virol. 1993;129:357–361. doi: 10.1007/BF01316912. [DOI] [PubMed] [Google Scholar]
- 3.Biedermann K A, Sun J, Giaccia A J, Tosto L M, Brown J M. scid mutation in mice confers hypersensitivity to ionizing radiation and a deficiency in DNA double-strand break repair. Proc Natl Acad Sci USA. 1991;88:1394–1397. doi: 10.1073/pnas.88.4.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bohannon R C, Donehower L A, Ford R J. Isolation of a type D retrovirus from B-cell lymphomas of a patient with AIDS. J Virol. 1991;65:5663–5672. doi: 10.1128/jvi.65.11.5663-5672.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosma G C, Custer R P, Bosma M J. A severe combined immunodeficiency mutation in the mouse. Nature. 1983;301:527–530. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- 6.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 7.Christian H A. Defects in membranous bones, exophthalmos and diabetes insipidus; an unusual syndrome of dyspituitarism: a clinical study. Med Clin North Am. 1920;3:849–871. [Google Scholar]
- 8.Cloyd M W, Hartley J W, Rowe W P. Lymphomagenicity of recombinant mink cell focus-inducing murine leukemia viruses. J Exp Med. 1980;151:542–552. doi: 10.1084/jem.151.3.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook W D. Rapid thymomas induced by Abelson murine leukemia virus. Proc Natl Acad Sci USA. 1982;79:2917–2921. doi: 10.1073/pnas.79.9.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Custer R P, Bosma G C, Bosma M J. Severe combined immunodeficiency (SCID) in the mouse. Pathology, reconstitution, neoplasms. Am J Pathol. 1985;93:464–477. [PMC free article] [PubMed] [Google Scholar]
- 11.Dorshkind K, Keller G M, Phillips R A, Miller R G, Bosma G C, O’Toole M, Bosma M J. Functional status of cells from lymphoid and myeloid tissues in mice with severe combined immunodeficiency disease. J Immunol. 1984;132:1804–1808. [PubMed] [Google Scholar]
- 12.Drechsler M, Royer-Pokora B. A LINE element is present at the site of a 300-kb deletion starting in intron 10 of the PAX6 gene in a case of familial aniridia. Hum Genet. 1996;98:297–303. doi: 10.1007/s004390050210. [DOI] [PubMed] [Google Scholar]
- 13.Elder J H, Gautsch J W, Jensen F C, Lerner R A, Hartley J W, Rowe W P. Biochemical evidence that MCF murine leukemia viruses are envelope (env) gene recombinants. Proc Natl Acad Sci USA. 1997;74:4676–4680. doi: 10.1073/pnas.74.10.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fanning T G, Singer M F. LINE-1: a mammalian transposable element. Biochim Biophys Acta. 1987;910:203–212. doi: 10.1016/0167-4781(87)90112-6. [DOI] [PubMed] [Google Scholar]
- 15.Fulop G M, Phillips R A. The scid mutation in mice causes a general defect in DNA repair. Nature. 1990;347:479–482. doi: 10.1038/347479a0. [DOI] [PubMed] [Google Scholar]
- 16.Gelderblom H, Bauer H, Ogura H, Wigand R, Fischer A B. Detection of oncornavirus-like particles in HeLa cells. I. Fine structure and comparative morphological classification. Int J Cancer. 1974;13:246–253. doi: 10.1002/ijc.2910130212. [DOI] [PubMed] [Google Scholar]
- 17.Gelderblom H. Oncoviridae: type D oncovirus. In: Nermut M V, Steven A C, editors. Animal virus structure. Amsterdam, The Netherlands: Elsevier Science Publishers; 1987. pp. 289–293. [Google Scholar]
- 18.Green N, Hiai H, Elder J H, Schwartz R S, Khiroya R H, Thomas C Y, Tsichlis P N, Coffin J M. Expression of leukemogenic recombinant viruses associated with a recessive gene in HRS/J mice. J Exp Med. 1980;152:249–264. doi: 10.1084/jem.152.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths D J, Venables P J W, Weiss R A, Boyd M T. A novel exogenous retrovirus sequence identified in humans. J Virol. 1997;71:2866–2872. doi: 10.1128/jvi.71.4.2866-2872.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hand A. Defects in membranous bones, exophthalmos, and polyuria in childhood: is it dyspituitarism? Am J Med Sci. 1921;162:509. [Google Scholar]
- 21.Hartley J W, Wolford N K, Old L J, Rowe W P. A new class of murine leukemia virus associated with the development of spontaneous lymphomas. Proc Natl Acad Sci USA. 1977;74:789–792. doi: 10.1073/pnas.74.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inoue H, Ishii H, Alder H, Snyder E, Druck T, Huebner K, Croce C M. Sequence of the FRA3B common fragile region: implications for the mechanism of FHIT deletion. Proc Natl Acad Sci USA. 1997;94:14584–14589. doi: 10.1073/pnas.94.26.14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamel-Reid S, Dick J E. Engraftment of immune-deficient mice with human hematopoietic stem cells. Science. 1988;242:1706–1709. doi: 10.1126/science.2904703. [DOI] [PubMed] [Google Scholar]
- 24.Kannourakis G, Abbas A. The role of cytokines in the pathogenesis of Langerhans cell histiocytosis. Br J Cancer. 1994;70(Suppl. XXIII):S37–S40. [PMC free article] [PubMed] [Google Scholar]
- 25.Kingsmore S F, Giros B, Suh D, Bieniarz M, Caron M G, Seldin M F. Glycine receptor β-subunit gene mutation in spastic mouse associated with LINE-1 element insertion. Nat Genet. 1994;7:136–141. doi: 10.1038/ng0694-136. [DOI] [PubMed] [Google Scholar]
- 26.Kirchgessner C U, Patil C K, Evans J W, Cuomo C A, Fried L M, Carter T, Oettinger M A, Brown J M. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science. 1995;267:1178–1183. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- 27.Krause H, Wunderlich V, Uckert W. Molecular cloning of a type D retrovirus from human cells (PMFV) and its homology to simian acquired immunodeficiency type D retroviruses. Virology. 1989;173:214–222. doi: 10.1016/0042-6822(89)90237-7. [DOI] [PubMed] [Google Scholar]
- 28.Kuff E L, Feenstra A, Lueders K, Smith L, Hawlay R, Hozumi N, Shulman M. Intracisternal A-particle genes as movable elements in the mouse genome. Proc Natl Acad Sci USA. 1983;80:1992–1996. doi: 10.1073/pnas.80.7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwok S, Ehrlich G, Poiesz B, Kalish R, Sninsky J J. Enzymatic amplification of HTLV-1 viral sequences from peripheral blood mononuclear cells and infected tissues. Blood. 1988;72:1117–1123. [PubMed] [Google Scholar]
- 30.Lee J Y, Bowden D S, Marshall J A. Membrane junctions associated with rubella virus infected cells. J Submicrosc Cytol Pathol. 1996;28:101–108. [PubMed] [Google Scholar]
- 31.Leibold D M, Swergold G D, Singer M F, Thayer R E, Dombroski B A, Fanning T G. Translation of LINE-1 DNA elements in vitro and in human cells. Proc Natl Acad Sci USA. 1990;87:6990–6994. doi: 10.1073/pnas.87.18.6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Letterer E. Aleukamische retikulose (ein Beitrag zu den proliferativen Erkrankungen des Retikuendosthelisalapparates) Frankf Z Pathol. 1924;30:377–394. [Google Scholar]
- 33.Lichenstein L, Jaffe H L. Eosinophilic granuloma of bone. Am J Pathol. 1940;16:595–604. [PMC free article] [PubMed] [Google Scholar]
- 34.Lieberman M, Hansteen G A, Waller E K, Weissman I L, Sen-Majumdar A. Unexpected effects of the severe combined immunodeficiency mutation on murine lymphomagenesis. J Exp Med. 1992;176:399–405. doi: 10.1084/jem.176.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lilly F, Duran-Reynals M L, Rowe W P. Correlation of early murine leukemia virus titer and H-2 type with spontaneous leukemia in mice of the BALB/c × AKR cross: a genetic analysis. J Exp Med. 1975;141:882–889. [PMC free article] [PubMed] [Google Scholar]
- 36.Lueders K K, Kuff E L. Sequences associated with intracisternal A particles are reiterated in the mouse genome. Cell. 1977;12:963–972. doi: 10.1016/0092-8674(77)90161-1. [DOI] [PubMed] [Google Scholar]
- 37.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 38.Marx P A, Maul D H, Osborn K G, Lerche N W, Moody P, Lowenstine L J, Henrickson R V, Arthur L O, Gilden R V, Gravell M, London W T, Sever J L, Levy J A, Munn R J, Gardner M B. Simian AIDS: isolation of a type-D retrovirus and transmission of the disease. Science. 1984;223:1083–1086. doi: 10.1126/science.6695196. [DOI] [PubMed] [Google Scholar]
- 39.Marx P A, Bryant M L, Osborn K G, Maul D H, Lerche N W, Lowenstine L J, Kluge J D, Zaiss C P, Henrickson R V, Shiigi S M, Wilson B J, Malley A, Olson L C, McNulty W P, Arthur L O, Gilden R V, Barker C S, Hunter E, Munn R J, Heidecker G, Gardner M B. Isolation of a new serotype of simian acquired immune deficiency syndrome type D retrovirus from Celebes black macaques (Macaca nigra) with immune deficiency and retroperitoneal fibromatosis. J Virol. 1985;56:571–578. doi: 10.1128/jvi.56.2.571-578.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McNaughton J C, Hughes G, Jones W A, Stockwell P A, Klamut H J, Petersen G B. The evolution of an intron: analysis of a long, deletion-prone intron in the human dystrophin gene. Genomics. 1997;40:294–304. doi: 10.1006/geno.1996.4543. [DOI] [PubMed] [Google Scholar]
- 41.Oda T, Ikeda S, Watanabe S, Hatsushika M, Akiyama K, Mitsunobu F. Molecular cloning, complete nucleotide sequence, and gene structure of the provirus genome of a retrovirus produced in a human lymphoblastoid cell line. Virology. 1988;167:468–478. [PubMed] [Google Scholar]
- 42.Ono M, Cole M D, White A T, Huang R C C. Sequence organization of cloned intracisternal A particle genes. Cell. 1980;21:465–473. doi: 10.1016/0092-8674(80)90483-3. [DOI] [PubMed] [Google Scholar]
- 43.Oroszlan S, Barbacid M, Copeland T D, Aaronson S A, Gilden R V. Chemical and immunological characterization of the major structural protein (p28) of MMC-1, a rhesus monkey endogenous type C virus: homology with the major structural protein of avian reticuloendotheliosis virus. J Virol. 1981;39:845–854. doi: 10.1128/jvi.39.3.845-854.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otani S, Ehrlich J C. Solitary granuloma of bone. Am J Pathol. 1940;16:595–604. [PMC free article] [PubMed] [Google Scholar]
- 45.Ou C Y, Kwok S, Mitchell S W, Mack D H, Sninsky J J, Krebs J W, Feorino P, Warfield D, Schochetman G. DNA amplification for direct detection of HIV-1 in DNA of peripheral blood mononuclear cells. Science. 1988;239:295–297. doi: 10.1126/science.3336784. [DOI] [PubMed] [Google Scholar]
- 46.Paine-Murrieta G D, Taylor C W, Curtis R A, Lopez M H A, Dorr R T, Johnson C S, Funk C Y, Thompson F, Hersh E M. Human tumor models in the severe combined immune deficient (scid) mouse. Cancer Chemother Pharmacol. 1997;40:209–214. doi: 10.1007/s002800050648. [DOI] [PubMed] [Google Scholar]
- 47.Payne A-L, Verwoerd D W, Garnett H M. The morphology and morphogenesis of Jaagsiekte retrovirus (JSRV) Onderstepoort J Vet Res. 1983;50:317–322. [PubMed] [Google Scholar]
- 48.Popovic M, Kalyanaraman V S, Reitz M S, Sarngadharan M G. Identification of the RPMI 8226 retrovirus and its dissemination as a significant contaminant of some widely used human and marmoset cell lines. Int J Cancer. 1982;30:93–99. doi: 10.1002/ijc.2910300116. [DOI] [PubMed] [Google Scholar]
- 49.Power M D, Marx P A, Bryant M L, Gardner M B, Barr P J, Luciw P A. Nucleotide sequence of SRV-1, a type D simian acquired immune deficiency syndrome retrovirus. Science. 1986;231:1567–1572. doi: 10.1126/science.3006247. [DOI] [PubMed] [Google Scholar]
- 50.Prochazka M, Gaskins H R, Shultz L D, Leiter E H. The nonobese diabetic scid mouse: model for spontaneous thymomagenesis associated with immunodeficiency. Proc Natl Acad Sci USA. 1992;89:3290–3294. doi: 10.1073/pnas.89.8.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Purcell D F J, Broscius C M, Vanin E F, Buckler C E, Nienhuis A W, Martin M A. An array of murine leukemia virus-related elements is transmitted and expressed in a primate recipient of retroviral gene transfer. J Virol. 1996;70:887–897. doi: 10.1128/jvi.70.2.887-897.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pyra H, Boni J, Schupbach J. Ultrasensitive retrovirus detection by a reverse transcriptase assay based on product enhancement. Proc Natl Acad Sci USA. 1994;91:1544–1548. doi: 10.1073/pnas.91.4.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rabin H, Benton C V, Tainsky M A, Rice N R, Gilden R V. Isolation and characterization of an endogenous type C virus of rhesus monkeys. Science. 1979;204:841–842. doi: 10.1126/science.87013. [DOI] [PubMed] [Google Scholar]
- 54.Sassaman D M, Dombroski B A, Moran J V, Kimberland M L, Naas T P, DeBerardinis R J, Gabriel A, Swergold G D, Kazazian H H. Many human L1 elements are capable of retrotransposition. Nat Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 55.Schüller A. Über eigenartige Schadeldefekte im Jugendalter. Fortschr Rontgenstr. 1915;23:12–18. [Google Scholar]
- 56.Shih A, Misra R, Rush M G. Detection of multiple, novel reverse transcriptase coding sequences in human nucleic acids: relation to primate retroviruses. J Virol. 1989;63:64–75. doi: 10.1128/jvi.63.1.64-75.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siprashvili Z, Sozzi G, Barnes L D, McCue P, Robinson A K, Eryomin V, Sard L, Tagliabue E, Greco A, Fusetti L, Schwartz G, Pierotti M, Croce C M, Huebner K. Replacement of Fhit in cancer cells suppresses tumorigenicity. Proc Natl Acad Sci USA. 1997;94:13771–13776. doi: 10.1073/pnas.94.25.13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siwe S A. Die Reticuloendotheliose—ein neues Krankheitsbild unter den Hepatosplenomegalien. Z Kinderheilkd. 1933;55:212–247. [Google Scholar]
- 59.Sonigo P, Barker C, Hunter E, Wain-Hobson S. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell. 1986;45:375–385. doi: 10.1016/0092-8674(86)90323-5. [DOI] [PubMed] [Google Scholar]
- 60.Stoye J P, Moroni C, Coffin J M. Virological events leading to spontaneous AKR thymomas. J Virol. 1991;65:1273–1285. doi: 10.1128/jvi.65.3.1273-1285.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teich N. Taxonomy of retroviruses. In: Weiss R, Teich N, Varmus H, Coffin J, editors. RNA tumor viruses: molecular biology of tumor viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 25–208. [Google Scholar]
- 62.Thayer R M, Power M D, Bryant M L, Gardner M B, Barr P J, Luciw P A. Sequence relationships of type D retroviruses which cause simian acquired immunodeficiency syndrome. Virology. 1987;157:317–329. doi: 10.1016/0042-6822(87)90274-1. [DOI] [PubMed] [Google Scholar]
- 63.Toriello H V, Glover T W, Takahara K, Byers P H, Miller D E, Higgins J V, Greenspan D S. A translocation interrupts the COL5A1 gene in a patient with Ehlers-Danlos syndrome and hypomelanosis of Ito. Nat Genet. 1996;13:361–365. doi: 10.1038/ng0796-361. [DOI] [PubMed] [Google Scholar]
- 64.Uckert W, Fleischhacker M, Kettmann R. Isolation and characterisation of covalently closed circular proviral DNA molecules of several type D retroviruses isolated from human cell lines. Virology. 1986;155:742–746. doi: 10.1016/0042-6822(86)90236-9. [DOI] [PubMed] [Google Scholar]
- 65.Willman C L, Busque L, Griffin B B, Favara B E, McKlain K L, Duncan M H, Gilliland D G. Langerhans cell histiocytosis (histiocytosis X): a clonal proliferative disease. N Engl J Med. 1994;331:154–160. doi: 10.1056/NEJM199407213310303. [DOI] [PubMed] [Google Scholar]
- 66.Writing Group of the Histiocyte Society. Histiocytosis syndromes in children. Lancet. 1987;i:208–209. [PubMed] [Google Scholar]
- 67.York D F, Vigne R, Verwoerd D W, Querat G. Isolation, identification, and partial cDNA cloning of genomic RNA of Jaagsiekte retrovirus, the etiological agent of sheep pulmonary adenomatosis. J Virol. 1991;65:5061–5067. doi: 10.1128/jvi.65.9.5061-5067.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.York D F, Vigne R, Verwoerd D W, Querat G. Nucleotide sequence of the Jaagsiekte retrovirus, an exogenous and endogenous type D and B retrovirus of sheep and goats. J Virol. 1992;66:4930–4939. doi: 10.1128/jvi.66.8.4930-4939.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu R C, Chu C, Bulewela L, Chu A C. Clonal proliferation of Langerhans cell histiocytosis. Lancet. 1994;343:767–768. doi: 10.1016/s0140-6736(94)91842-2. [DOI] [PubMed] [Google Scholar]