Abstract
The bovine papillomavirus type 1 (BPV-1) E2 protein is the master regulator of papillomavirus replication and transcription. We have raised a panel of monoclonal antibodies (MAbs) against the BPV-1 E2 protein and used them to probe the structure and function of the protein. Five MAbs reacted with linear epitopes, and four MAbs recognized conformation-dependent epitopes which mapped within the C-terminal DNA-binding and dimerization domain. MAb 1E2 was able to recognize the replication- and transactivation-defective but not the competent conformation of the transactivation domain of the E2 protein. MAb 5H4 prevented efficiently the formation of E2-DNA as well as E2-dependent E1-E2-origin complexes and also dissociated preformed complexes in a concentration-dependent manner. Cotransfection of several MAbs with the BPV-1 minimal origin plasmid pUCAlu into CHO4.15 cells resulted in a dose-dependent inhibition of replication. Inhibition of replication by MAb 5H4 and the Fab′ fragment of 5H4 correlated with their ability to dissociate the E2 protein from the DNA. MAb 3F12 and MAbs 1H10 and 1E4, directed against the hinge region, were also capable of inhibiting BPV-1 origin replication in CHO4.15 cells. However, the Fab′ fragments of 1H10 and 3F12 had no effect in the transient replication assay. These data suggest that MAbs directed against the hinge region sterically hinder the inter- or intramolecular interactions required for the replication activity of the E2 protein.
Bovine papillomavirus type 1 (BPV-1) has been studied extensively as a model for papillomavirus replication and transcription. The viral E2 protein is the master regulator of the viral life cycle—this protein modulates the transcription of viral genes (41) and is responsible for the initiation of DNA replication (43, 44, 48) and for the stable maintenance of the viral genome (31), which is achieved presumably through facilitation of the association of the viral genome with chromatin (19a, 23, 40). E2 is a sequence-specific DNA-binding protein, and it interacts with the components of the cellular transcription (33, 49) and replication (24) machinery. The viral E2 and E1 proteins interact with each other (2, 4, 30, 35) during the initiation of replication, resulting in cooperative binding of E1 and E2 on the BPV-1 replication origin (25–27, 35–39).
The BPV-1 E2 protein, like other transcription factors, is composed of relatively well-defined function-specific modules. Structural and mutational analyses have revealed three distinct domains. The amino-terminal part (residues 1 to 210) is an activation domain for transcription (12, 28) and replication (43). It is followed by the unstructured hinge region and the carboxy-terminal DNA-binding and dimerization domain (residues 310 to 410) (29). Deletion analysis of the E2 protein has shown that the transactivation domain and the DNA-binding and dimerization domains are necessary for both replication and transcription, while large deletions in the hinge region affect replication preferentially and transcription less (46). The structure of the carboxy-terminal DNA-binding and dimerization domain has been solved by X-ray analysis and has revealed a dimeric DNA-binding and dimerization motif (15, 16). Most of the information about structural and functional determinants in the amino-terminal activation domain of the E2 protein has been obtained by mutational analysis (7, 12, 14, 46). These data confirm that the E2 amino-terminal domain, like the C-terminal domain, has a highly organized structure and that even a single point mutation can inactivate the function of the E2 protein in the activation of transcription, replication, or both (1, 5, 9, 13, 34).
Antibodies are efficient and highly specific tools for identifying the structural determinants of macromolecules and/or for studying the role of a protein in functional assays (18, 19, 21, 42, 45). Antibodies have been used for the characterization of the human papillomavirus (HPV) E2 protein. For example, polyclonal antibodies against overlapping synthetic peptides that cover the HPV type 16 (HPV-16) E2 protein have been used to test the structure of this protein (10), and the interaction of the HPV-16 E2 protein with the E1 protein could be blocked by a monoclonal antibody (MAb) that bound E2 in the region of amino acids 18 to 41 (17).
In this study, we describe the production of a set of MAbs against the BPV-1 E2 protein and characterize their ability to interfere with the functions of the E2 protein in vivo and in vitro in biochemical and functional assays.
MATERIALS AND METHODS
Production of the BPV-1 E2 protein.
E2 protein was expressed in the pET11c-based system in Escherichia coli and was purified to homogeneity by conventional methods (37) with modifications. First, we precipitated nucleic acids from clarified cell lysates by the slow addition of polyethylenimine (Polymin P; Sigma) to a final concentration of 0.6%. Precipitation was carried out on ice for 30 min, and the pellet was collected by centrifugation. Proteins were recovered from the supernatant by precipitation with 35% ammonium sulfate and purified to homogeneity by conventional chromatography.
Production of MAbs.
Female BALB/C mice were injected with 50 μg of purified BPV-1 E2 protein five times at 3- to 4-week intervals. The injections were intraperitoneal, with E2 suspended initially in Freund’s complete adjuvant and subsequently in phosphate-buffered saline (PBS). Following the final injection, mice were allowed to rest for 5 weeks and then were injected with 100 μg of antigen. One week later, final boosts with 100, 200, and 200 μg of protein in PBS at 4, 3, and 2 days before fusion, respectively, were performed. Sp2/0 myeloma cells and cells from one third of the spleen were washed three times with sterile PBS. The final pellet was mixed by tapping the tube, and 1 ml of 50% polyethylene glycol (PEG) 4000 (Merck) was added over 1 min with gentle shaking. The cells were centrifuged at 100 × g for 5 min, the PEG solution was removed, and the resuspended cells were plated on five 96-well microtiter plates containing hypoxanthine-aminopterin-thymidine medium. Supernatants were tested 10 days after fusion as described below by a direct enzyme-linked immunosorbent assay (ELISA).
Screening of hybridomas.
Wells of ELISA plates (Maxisorp; Nunc, Roskilde, Denmark) were coated with 100 μl of E2 (2 μg/ml in PBS) overnight at 4°C. After the coating step, the plates were washed three times with PBS containing 0.05% Tween 20 (PBS/T) and blocked with PBS/T containing 0.05% casein for 30 min. Then, 80 μl of PBS/T and 20 μl of hybridoma supernatants were added to the wells, and the plates were incubated for 60 min on a shaker at room temperature. The plates were washed with PBS/T, followed by the addition of 100 μl of peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) (LabAS Ltd., Tartu, Estonia) diluted 1:2,500 in PBS/T supplemented with 2% PEG 6000 (Merck). The plates were incubated for 15 min and then washed with PBS/T. Then, 100 μl of TMB substrate solution (3,3′,5,5′-tetramethylbenzidine–H2O2 in 0.1 M acetate-citrate buffer [pH 4.5]) was added.
The antibody subclasses were determined by an ELISA as described above with peroxidase-labelled goat anti-mouse isotype antibodies (LabAS).
Purification of MAbs.
The MAbs were purified from ascitic fluid by ammonium sulfate precipitation and ion-exchange chromatography on Blue DEAE-Toyopearl 650S with a Pharmacia standard fast protein liquid chromatography system (20). The IgG concentration was estimated at 280 nm by use of an extinction coefficient of 14. The purified MAbs were stored in PBS containing 50% glycerol at −20°C.
Preparation of Fab′ fragments.
Fab′ fragments were prepared from the MAbs as described by Porter (32) with modifications. The MAbs were dialyzed against 0.1 M sodium acetate buffer (pH 5.5) containing 1 mM EDTA and 25 mM 2-mercaptoethanol. The antibodies were digested with papain at an enzyme/antibody ratio of 1:10 (wt/wt) for 24 h at 37°C. The reaction was stopped by the addition of iodoacetamide to a final concentration of 30 mM. The digested antibodies were dialyzed against 20 mM Tris-HCl buffer (pH 7.2), and Fab′ fragments were purified on a Mono Q column.
Peptide synthesis.
Peptides were assembled in a stepwise manner on a solid support with a model 431A peptide synthesizer (Applied Biosystems) by the standard NMP/HOBt solvent activation strategy on a 0.1-mmol scale (22).
ELISA with peptides.
The surfaces of the microtiter wells were activated with 0.25% glutaraldehyde in PBS for 30 min at 60°C. The plates were washed three times with PBS, and a peptide solution at a concentration of 20 μg/ml in PBS was added to the wells. The plates were incubated overnight at room temperature, washed with PBS/T, and blocked with 1% nonfat dry milk in PBS/T for 2 h. The plates were washed with PBS/T, and antibodies diluted in PBS/T were added to the wells, incubated, and processed as described above.
Plasmids.
The pET-E2 vector used for the expression of E2 in E. coli was generated by PCR amplification with specific primers and was cloned between the NdeI and BamHI sites of plasmid pET11c. The E2 expression constructs pCGE2, pCGE2C, pCGE8/E2, and pCGE2(D92-161) have been described previously (43, 44). Plasmid VP16:E2 (24) contains 80 C-terminal amino acids from VP16 fused to the C terminus of E2 (starting from amino acid 250) in the context of pCG. The E2 N-terminal deletion mutants E2(D1-23), E2(D1-85), E2(D1-112), and E2(D1-183) were generated by PCR with appropriate oligonucleotide primers containing an initiation methionine codon in the optimal Kozak context and were cloned into the pCGE2 expression vector at the XbaI-BamHI sites. For the E2 C-terminal deletion mutants E2(D219-410), E2(D284-410), and E2(D310-410), PCR primer pairs were designed with terminal recognition sequences for KpnI-BclI and were cloned into the corresponding sites in pCGE2. The replication reporter plasmid pUCAlu has been described previously (43). pHookAlu was made by cloning the Alu fragment from pUCAlu at the HindIII-BamHI sites of pHook-2 (Invitrogen) and deleting the cytomegalovirus promoter with restriction enzymes HindIII and BssHI.
Cells and transfections.
E2 expression constructs (100 ng) were electroporated at 180 V into COS-7 cells (2 × 106 to 6 × 106) in 250 μl of Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (FCS) and 50 μg of denatured salmon sperm DNA at room temperature (43). For replication assays, CHO4.15 cells were trypsinized, centrifuged, and resuspended in F12 medium containing 10% FCS at a density of 107 cells/ml. The cell suspension (250 μl) was mixed with 100 ng of pUCAlu DNA, 50 μg of salmon sperm DNA, and various concentrations of MAbs or Fab′ fragments in a disposable electroporation cuvette and subjected to an electric discharge of 230 V from an Invitrogen Gene Pulser. After the discharge, the cell suspension was left at room temperature for 15 min, and then the cells were washed and plated in F12 medium supplemented with 10% FCS. The extraction of episomal DNA from cells and its analysis by Southern blotting were performed as described previously (43). For Western blot analysis, 500 ng of pHookAlu was cotransfected with MAbs (80 μg/ml), and transiently transfected cells were separated from the total population of CHO4.15 cells with magnetic beads (Invitrogen) according to the manufacturer’s recommendations.
Immunoblotting of E2.
COS-7 cells transfected with E2 proteins in 60-mm diameter dishes were lysed 36 h after electroporation in 200 μl of Laemmli sample buffer. Transfected CHO4.15 cells were separated from the magnetic beads by boiling in 100 μl of Laemmli sample buffer. Proteins were separated by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (PAGE). After transfer, the nitrocellulose membranes were incubated with mouse E2-specific MAbs and a secondary horseradish peroxidase-conjugated antibody by use of an ECL detection kit (Amersham) according to the manufacturer’s recommendations. To analyze the E2 protein level in CHO4.15 cells transfected with MAbs, rabbit anti-E2 polyclonal antibody was used.
Mobility shift assays.
For preparation of COS-7 cell extracts, transfected cells were removed from semiconfluent growth on 100-mm-diameter plates with a rubber policeman, washed, and lysed in 100 μl of lysis buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 30 mM KCl, 0.1 mM EDTA, 0.35% Nonidet P-40, 10 mM dithiothreitol, protease inhibitors) on ice for 30 min. Cell debris was removed by centrifugation, glycerol was added to a final concentration of 20%, and the extracts were divided into aliquots and stored at −70°C. For gel shift assays, 2 μl of cell extract was incubated in 10 μl of binding buffer (10 mM Tris-HCl [pH 7.5], 100 mM KCl, 2 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 15% glycerol, 5 mg of bovine serum albumin per ml, 1 μg of leupeptin per ml, 1 μg of aprotinin per ml) at room temperature for 15 min in the presence of 1 μg of sonicated salmon sperm DNA and 0.2 ng of 32P-labelled oligonucleotide containing the E2 binding site. For bacterially expressed E2, 2 ng of protein was used per reaction. Double-stranded high-affinity binding site 9 of BPV-1 (5′-ACAAAGTACCGTTGCCGGTCGAA-3′) was used as a probe. Protein-DNA complexes were resolved by 6% PAGE (acrylamide–N,N-methylene-bisacrylamide, 80:1) with 0.25× Tris-borate-EDTA. Gels were dried and exposed to X-ray film. MAbs (1 to 10 ng/μl) were added either before or after DNA binding and were incubated for an additional 20 min. For protease digestion experiments, bacterially expressed E2 protein was incubated with 2 μg of pronase for 5 min. The E1-E2-origin complex formation assay was performed as described by Sedman and Stenlund (35). The effect of antibodies on E1-E2-origin complex formation was tested either before or after the assembly of the complex.
RESULTS
Generation of E2-specific MAbs.
The soluble E2 protein was purified to apparent homogeneity from lysates of isopropyl-β-D-thiogalactopyranoside (IPTG)-induced E. coli overexpressing BPV-1 E2 from the pET11c expression vector by conventional chromatography (37). BALB/c mice were immunized with the purified functionally active E2 protein as described in Materials and Methods. We obtained nearly 200 hybridoma cell lines, 17 of which were positive for the E2 protein in both ELISAs and immunoblot assays, while 5 hybridomas were positive in ELISAs only. Nine MAbs that were deemed most useful were purified from the ascitic fluids of the respective hybridoma cell lines and studied in various assays as described below (Table 1). All studied antibodies belonged to the IgG1 subtype, with the exception of 3C1, which belonged to the IgG2a subtype. All of these MAbs had high affinities for and fast kinetics of binding to their respective epitopes, as found by the concentration dependence of antibody binding in ELISAs (data not shown).
TABLE 1.
MAbs recognizing the BPV-1 E2 proteina
| Antibody | Location of epitope (residues) | Type of epitope | Binding to E2-DNA complex | Inhibition of E2 binding to oligo | Binding to E1-E2-origin complex |
|---|---|---|---|---|---|
| 1E2 | 184–190 | Linear | − | − | − |
| 3F12 | 199–206 | Linear | + | − | + |
| 1H10 | 208–218 | Linear | + | − | + |
| 1E4 | 250–280 | Linear | + | − | + |
| 3C1 | 280–309 | Linear | + | − | + |
| 3E8 | 310–410 | Conformational | + | − | + |
| 3H5 | 310–410 | Conformational | + | − | + |
| 5F10 | 162–410 | Conformational | + | − | + |
| 5H4 | 310–410 | Conformational | − | + | − |
+, positive result; −, negative result.
Epitope mapping.
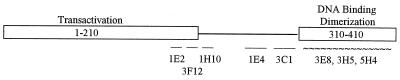
To define the continuous epitopes recognized by the antibodies, the reactivity of each MAb to full-length and truncated E2 proteins expressed in COS-7 cells was determined by Western blot analysis. The linear epitopes for the initially isolated 17 MAbs were mapped in the region between amino acids 184 and 309; for 12, the epitopes were found within the region between residues 184 and 218 (data not shown). We concluded from these results that the sequence of the 34 amino acids within the region from residues 184 to 218 is the major immunodominant determinant of the BPV-1 E2 protein. The results of immunoblot analysis for five selected purified antibodies with the linear epitopes are shown in Fig. 1A. To map the epitopes for the 1E2, 3F12, and 1H10 antibodies more precisely, we synthesized four overlapping peptides covering the region between amino acids 162 and 210. The sequences of the synthesized peptides and the ability of the MAbs to bind to these peptides in an ELISA are shown in Fig. 1B. Peptides P2 (residues 171 to 192) and P3 (residues 184 to 201) were recognized by 1E2, while peptide P4 was recognized by 3F12. To narrow down the sizes of the epitopes, two additional peptides, P5 (residues 179 to 190) and P6 (residues 197 to 208), were synthesized and confirmed by an ELISA to contain the recognition sequences for the 1E2 and 3F12 antibodies, respectively. 1H10 did not recognize any of the synthesized peptides and was therefore mapped by the Western blot analysis to the region between amino acids 208 and 218.
FIG. 1.
Epitope mapping of E2-specific MAbs. (A) Schematic representation of the truncated E2 proteins used and the results of the immunoblot analysis of the E2 proteins (diagram at right). wt, wild type; conf., MAbs with discontinuous epitopes. (B) Reactivity of MAbs to synthetic peptides (P1 to P6) covering the region from amino acids 162 to 210 of E2 in an ELISA. +, positive result; −, negative result.
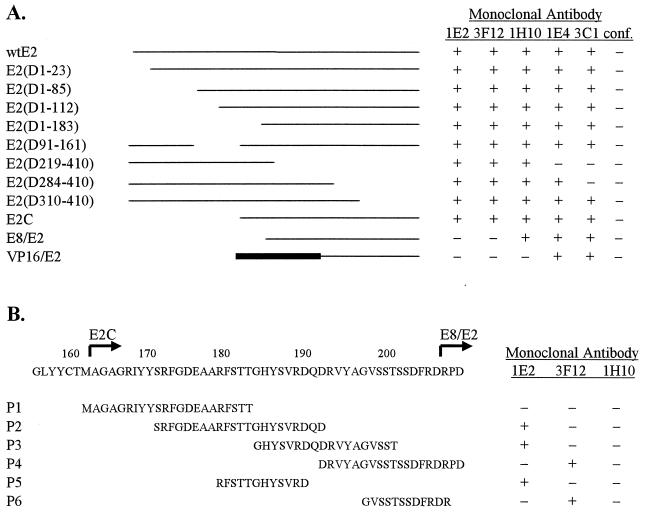
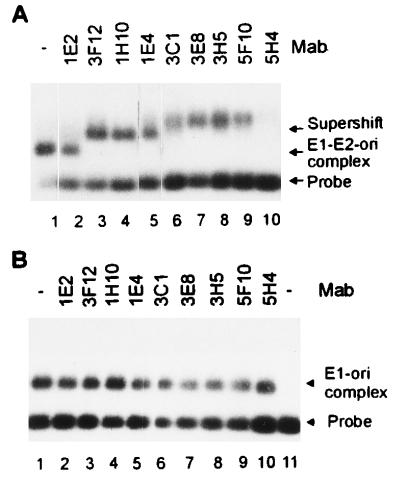
To test the ability of the antibodies to recognize their respective linear epitopes on the E2 protein in the E2-DNA complex, a mobility shift assay was used. All MAbs against the linear epitopes, with the exception of 1E2, were able to induce a supershift (Fig. 2A), indicating that their epitopes are exposed on the surface of the DNA-bound E2 molecule.
FIG. 2.
Characterization of E2-specific MAbs. (A) Reactivity of E2-specific MAbs with the native E2 protein. The mobility shift assay was carried out with 2 ng of bacterially expressed and purified E2 protein and 0.2 ng of radiolabelled E2 binding site for 15 min at room temperature. (B) Lanes 1 to 10 show reactivity of MAbs with discontinuous epitopes with truncated E2 proteins expressed in COS-7 cells. Band shift assays were performed with 2 μl of cell extract. Lanes 11 to 16 show reactivity of MAbs to the E2 DNA-binding domain (DBD). Bacterially expressed E2 protein was treated with 2 μg of pronase for 10 min at room temperature, and then reactivity was determined. (C) Reactivity of MAb 1E2 with truncated E2 proteins expressed in COS-7 cells. MAbs were added after E2 was mixed with its DNA target. neg., cells transfected with carrior only; wt, wild type. Protein-DNA complexes were resolved by 6% PAGE with 0.25× Tris-borate-EDTA.
Accessibility of the MAb 1E2 epitope in the E2 protein.
The epitope for MAb 3F12 (residues 199 to 206) is efficiently exposed in the DNA-bound E2 protein, while the epitope for MAb 1E2, located 8 residues upstream, is poorly recognized by the respective antibody in the full-length E2 protein in complex with DNA (Fig. 2A, compare lanes 2 and 3 with lanes 4 and 5). However, the epitope for MAb 1E2 was readily exposed in truncated E2 proteins bound to DNA—E2(D1-23), E2(D1-85), E2(D1-112), E2(D1-183), and E2C (Fig. 2C)—as well as in the internal deletion mutant E2(D92-161) (data not shown). All of these deletion mutants were inactive for the activation of transcription and replication (21a). Therefore, the epitope for MAb 1E2 could be identified as a transactivation domain denaturation-specific epitope of the E2 protein; the exposure of the 1E2 epitope indicates that the E2 protein transactivation domain has an inactive conformation for transcription and replication.
Antibodies against the C-terminal domain of the E2 protein.
MAbs 3E8, 3H5, 5F10, and 5H4 did not react with the E2 protein on immunoblots, indicating that the epitopes of these MAbs are sensitive to denaturation. To define further the epitopes for these MAbs, the reactivity of each MAb to the full-length or truncated E2 protein expressed in COS-7 cells was determined by a gel shift assay. All four studied antibodies were able to react with both the full-length E2 protein (data not shown) and E2C expressed in COS-7 cells (Fig. 2B, lanes 1 to 5). MAbs 3E8, 3H5, and 5F10 were able to induce a supershift, and MAb 5H4 prevented the formation of the E2-DNA or E2C-DNA complex (Fig. 2B, lane 5; see also Fig. 5A, lanes 2 to 5). MAb 5H4 not only prevented the formation of the E2-DNA complex but also dissociated the preformed E2-DNA complex, and this effect was dependent on the concentration of the antibody. The dissociation of the preformed complex required concentrations of MAb 5H4 higher than those required to block complex formation. The chimeric protein VP16-E2, which contains amino acids 250 to 410 of the E2 protein, was recognized by MAbs 3E8, 3H5, and 5H4 but poorly, if at all, by MAb 5F10 (Fig. 2B, lanes 6 to 10).
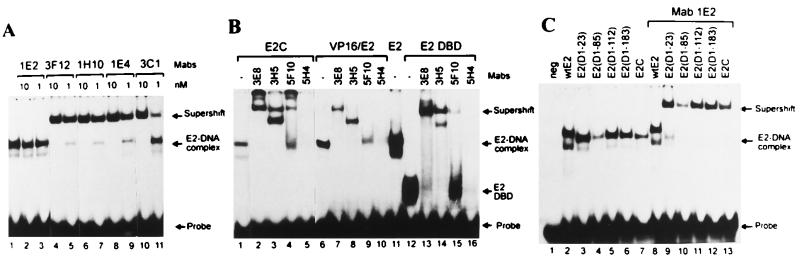
FIG. 5.
Effect of E2-specific Fab′ fragments on DNA replication. (A) Ability of MAb 5H4 and its Fab′ fragment to inhibit the formation of the E2-DNA complex at various antibody concentrations. The mobility shift assay was carried out with 2 ng of bacterially expressed and purified E2 protein and 0.2 ng of radiolabelled E2 binding site for 15 min at room temperature. MAb 5H4 or its Fab′ fragment was added after E2 was mixed with its DNA target, and incubation was carried out for an additional 20 min. (B) Inhibition of DNA replication by E2-specific Fab′ fragments at various concentrations. Reporter plasmid pUCAlu (100 ng) was cotransfected together with the Fab′ fragment of 3F12 (⧫), the Fab′ fragment of 1H10 (■), or the Fab′ fragment of 5H4 (▴) into cell line CHO4.15. At 72 h after electroporation, cells were harvested, and episomal DNA was digested with DpnI and linearizing enzyme HindIII and analyzed by Southern blotting. The replication signals of three independent experiments were quantified with a PhosphorImager, and signals from cells transfected with the origin-containing plasmid only were used as a control to normalize the results. (C) Southern blot analysis of transient replication of the BPV-1 origin-containing plasmid pUCAlu in the CHO4.15 cell line in the presence of Fab′ fragments at a concentration of 0.3 mg/ml. Episomal DNA was extracted from cells either 48 or 72 h ( ) after transfection. Filters were probed with radiolabelled plasmid pUCAlu.
) after transfection. Filters were probed with radiolabelled plasmid pUCAlu.
The carboxy-terminal DNA-binding and dimerization domain of the E2 protein forms a protease-resistant core (8). When the E2 protein was incubated with pronase prior to the addition of antibodies, the DNA-binding domain of the E2 protein was still able to interact with MAbs 3E8, 3H5, and 5H4 and weakly with MAb 5F10 (Fig. 2B, lanes 11 to 16), resulting in a supershift or dissociation of the E2-DNA complex. These data allowed us to map the epitopes for the conformational MAbs 3E8, 3H5, 5F10, and 5H4 within the carboxy-terminal ∼100 residues of the E2 protein. Immunoprecipitation studies with truncated E2 proteins mapped the epitopes for these MAbs within amino acids 310 to 410 (data not shown). The epitope mapping for MAb 5F10 was less definitive because the accessibility of the epitope for this MAb was dependent on the context of the protein. Although this MAb recognized an epitope in the E2-DNA and E2C-DNA complexes, the same epitope in VP16-E2 and the E2 DNA-binding and dimerization domain (Fig. 2B, compare lanes 4, 9, and 15) was poorly recognized.
Effect of MAbs on E1-E2-origin complex formation.
The BPV-1 minimal origin of replication comprises the E1 binding site, the A/T-rich region, and the E2 binding site (44). The E1 and E2 proteins bind cooperatively to the origin and form an E1-E2-origin complex (27, 35, 39). It has been shown that the ability of the E2 protein to form a complex with the E1 protein on DNA correlates with the efficiency of initiation of replication in vivo (11, 36). We studied the effect of the antibodies on E2-dependent E1-E2-origin complex formation. Antibodies 3F12, 1H10, 1E4, 3C1, 3E8, 3H5, and 5F10 all recognized their respective E2 epitopes in the E1-E2-origin complex and supershifted this complex (Fig. 3A, lanes 3 to 9). Interestingly, MAbs which recognize the C-terminal part of the E2 molecule resulted in an E1-E2-origin MAb complex which migrated much more slowly than complexes in which the MAbs bound to epitopes closer to the center of the E2 molecule. The epitope for MAb 1E2 is masked in the DNA-bound E2 protein and remains nonaccessible to this antibody in the E1-E2-DNA complex (Fig. 3A, lane 2). MAb 5H4, which specifically dissociated E2 from DNA, prevented the formation of the E1-E2-origin complex (Fig. 3A, lane 10). None of the antibodies had any effect on the mobility of the E1-origin complex formed at a high E1 protein concentration (Fig. 3B). These results showed that the epitopes for the most studied MAbs, with the exception of 1E2, are exposed in the E1-E2-origin complex and that MAb 5H4 is the only antibody which interferes with E2-dependent E1-E2-origin complex formation.
FIG. 3.
Effect of E2-specific MAbs on the formation of the E1-E2-origin (ori) complex. (A) A gel mobility shift assay was performed with 2 ng of E1 protein and 5 ng of E2 protein for 20 min. MAbs were added to a final concentration of 10 ng/μl and incubated for an additional 20 min. (B) A gel mobility shift assay was used to analyze the complex formed in the presence of E1 only. The resulting complexes were treated with 0.4% glutaraldehyde and separated on agarose gels.
Effect of anti-E2 MAbs on BPV-1 origin replication in cells.
The observation that antibodies recognized their respective epitopes in the DNA-bound E2 protein raised the possibility that some of these antibodies could interfere with some functions of the E2 protein in the initiation of DNA replication in vivo. Therefore, the purified antibodies were tested in a transient replication assay in vivo for their ability to block E2 protein functions in BPV-1 origin replication. Transfer of the MAbs into mammalian cells was carried out by electroporation (6). The transfection conditions were optimized to a level which allowed the uptake of both DNA and protein into CHO4.15 cells, which constitutively express viral E1 and E2 proteins (31) (see Materials and Methods). The presence of the relatively high concentrations of antibodies in the transfection mixture had no effect on CHO4.15 cell growth. We estimated that 1 to 2% of the input antibody was taken up by the cells under these conditions, as determined by Western blot analysis. Analysis of the cells immediately and 24 and 48 h after transfection showed structurally intact immunoglobulin heavy chains within the cells, indicating that no active degradation of the transfected antibodies took place in the cells (data not shown).
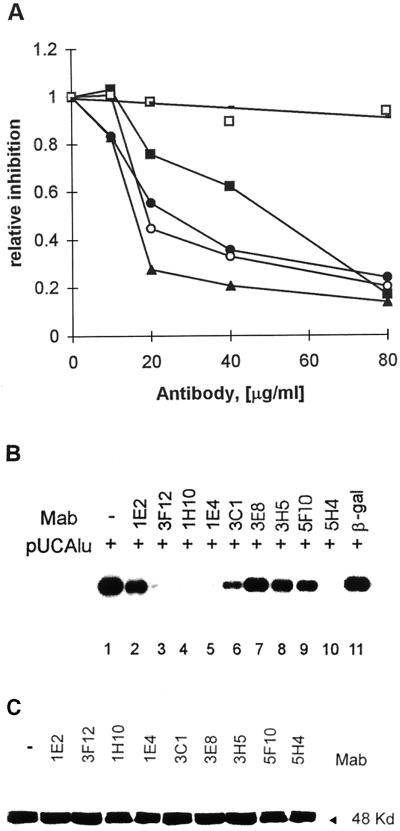
Different amounts of E2-specific MAbs were cotransfected with 100 ng of origin-containing plasmid pUCAlu by electroporation into CHO4.15 cells. Episomal DNA was extracted by alkaline lysis at 2 or 3 days after transfection, purified, digested with DpnI and linearizing enzyme HindIII, and analyzed by Southern blotting as described earlier (43). The effect of the E2-specific antibodies on the replication of the BPV-1 origin was dependent on the antibody concentration used (Fig. 4A). At a low concentration (20 μg/ml), MAb 1H10 strongly inhibited and MAbs 3F12 and 1E4 moderately inhibited the replication of origin-containing plasmid pUCAlu in CHO4.15 cells. The inhibition of replication by MAbs 3F12, 1H10, and 1E4 became almost complete when the antibody concentration in the cell suspension was increased to 80 μg/ml (Fig. 4A and B, lanes 3 to 5). MAb 5H4, which efficiently inhibited the formation of both E2-DNA and E1-E2-origin complexes in vitro, exhibited only a weak inhibitory effect in the transient replication assay at a low concentration. However, at a higher concentration (80 μg/ml), strong inhibition of replication was achieved with MAb 5H4 (Fig. 4A and B, lane 10). MAbs 1E2, 3C1, 3E8, 3H5, and 5F10 exhibited only weak inhibition, and a nonrelated anti-β-galactosidase MAb had no effect on origin-containing plasmid pUCAlu replication at all concentrations tested. The differences in the abilities of the MAbs to inhibit replication were not caused by differential uptake of MAbs by cells, since equivalent concentrations of intracellular MAbs were used in the cell suspension during electroporation and comparable amounts of the antibodies were detected in the cells by Western blotting (data not shown). The affinities of all of these antibodies were similar, as indicated by the concentration-dependent binding of the antibodies in the ELISA (data not shown).
FIG. 4.
Effect of E2-specific MAbs on papillomavirus replication. (A) CHO4.15 cells constitutively expressing BPV-1 E1 and E2 proteins were electroporated with 100 ng of reporter plasmid pUCAlu and various concentrations of MAbs. Cells were harvested 72 h after electroporation. Episomal DNA was digested with DpnI and linearizing enzyme HindIII and analyzed by Southern blotting. The replication signals of three independent experiments were quantified with a PhosphorImager, and signals from cells transfected with the origin-containing plasmid only were used as a control to normalize the results. Symbols: ●, MAb 3F12; ○, MAb 1E4; ▴, MAb 1H10; ■, MAb 5H4; □, nonspecific anti-β-galactosidase (β-gal) MAb. (B) Southern blot analysis of transient replication of the BPV-1 origin-containing plasmid pUCAlu in the CHO4.15 cell line in the presence of MAbs at a concentration of 80 μg/ml. Episomal DNA was extracted from cells 72 h after transfection. Filters were probed with radiolabelled plasmid pUCAlu. (C) Western blot analysis of E2 protein levels in transfected CHO4.15 cells with rabbit anti-E2 polyclonal antibody.
We next studied the effect of the E2-specific MAbs on the steady-state level of the E2 protein and on the localization of this protein in transfected cells. To select and isolate transiently transfected cells from the total population of CHO4.15 cells, a Capture-Tec Hook-2 kit (Invitrogen) was used. Briefly, MAbs were cotransfected with 500 ng of the origin-containing plasmid pHookAlu, which expresses a fusion protein comprised of the PDGFR transmembrane domain fused to the variable region of the antibody capable of recognizing phOx (4-etoxymethylene-2-phenyl-2-oxazolin-5-one), into CHO4.15 cells. At 24 h after electroporation, transfected cells were selected with magnetic beads carrying immobilized phOx and analyzed for the level of the E2 protein by Western blotting as well as for the localization of the E2 protein by direct immunofluorescence analysis with rabbit anti-E2 polyclonal antibody. We did not find any effect of the cotransfected E2-specific MAbs on the localization (data not shown) or steady-state level of the E2 protein in CHO4.15 cells (Fig. 4C).
Effect of Fab′ fragments on DNA replication.
Our results showed that MAbs 1H10, 1E4, 3F12, and 5H4 suppressed BPV-1 origin replication in a dose-dependent fashion (Fig. 4A). At the same time, MAbs 1H10, 3F12, and 1E4 did not influence the formation of the E1-E2-origin complex and supershifted this complex efficiently (Fig. 3, lanes 3 to 5). These data suggest that MAb 3F12 (epitope at residues 199 to 206) and MAbs 1H10 and 1E4, directed against the hinge region of the E2 protein, would not interfere directly with E1 and E2 interactions with DNA; however, these antibodies can sterically interfere with the inter- or intramolecular interactions required for the replication activity of the E2 protein. In order to study the possibility that antibodies would have an effect on replication due to steric hindrance of the formation of the replication initiation complexes, the Fab′ fragments of MAbs 3F12, 1H10, and 5H4 were prepared by a modified procedure (see Materials and Methods). An ELISA with E2-coated microtiter plates showed that all of the Fab′ fragments were active in binding to the E2 protein. The affinities of the Fab′ fragments of 1H10 and 5H4 were similar, while the Fab′ fragment of 3F12 had a lower affinity, as determined by titration on the ELISA plates (data not shown).
The produced Fab′ fragments were tested in biochemical assays as well. The ability of the MAb 5H4 Fab′ fragment to inhibit the formation of the E2-DNA complex is shown in Fig. 5A. Concentrations of MAb 5H4 and its Fab′ fragment of 1 ng/μl and at least 10 ng/μl, respectively, were required to prevent the formation of the E2-DNA complex (Fig. 5A). The MAb 5H4 Fab′ fragment was also capable of dissociating the preformed E2-DNA complex (data not shown).
We transfected 100 ng of origin-containing plasmid pUCAlu in the presence of increasing concentrations of Fab′ fragments into CHO4.15 cells by electroporation. A representative replication assay is shown in Fig. 5B and C. The Fab′ fragment of MAb 1H10 had no significant inhibitory effect on DNA replication at any concentration tested (Fig. 5B and 5C, lanes 5 and 6). The Fab′ fragment of MAb 3F12 activated rather than inhibited replication (Fig. 5B and C, lanes 3 and 4), and the Fab′ fragment of MAb 5H4 inhibited BPV-1 origin replication (Fig. 5B and 5C, lanes 7 and 8) in a dose-dependent fashion.
DISCUSSION
The crystal structure of the DNA-binding domain of the E2 protein with and without DNA has been solved (15, 16). Until the crystal structure of the full-length E2 protein is determined, we will have to rely on other methods to examine the structural organization of the whole protein and the molecular interactions that must occur to accomplish the replication and/or transcription activity of the protein. Even if the crystal structure were known, information about possible interactions should be gathered by other methods. In this study, we have produced and characterized a panel of MAbs as probes and tools for studying the structure and function of the BPV-1 E2 protein.
A total of 22 MAbs that were reactive to the E2 protein in an enzyme immunoassay were isolated. Seventeen of these MAbs were directed against linear epitopes that were mapped within the region between amino acids 180 and 309 of E2. In fact, the last part of the amino-terminal transactivation domain and the first 10 amino acids of the hinge region, residues 180 to 218, appear to constitute a highly immunogenic “hot spot,” since epitopes for 12 of these 17 MAbs were found to be localized within this region. The reason for the highly immunogenic properties of the region between residues 180 and 218 is unknown. Epitopes for 5 of the 22 MAbs were mapped within the C-terminal DNA-binding and dimerization domain. Interestingly, all of these antibodies recognized the composite epitopes and did not react with the denatured E2 protein. None of the epitopes for the MAbs tested were mapped to the first 180 residues of the E2 protein.
When only a purified transactivation domain, residues 1 to 218, of E2 was used for immunization, four MAbs against the region between amino acids 1 and 180 of E2 were obtained; however, none of them was able to recognize the E2-DNA complex in a mobility shift assay (21a). In contrast, Hibma and coworkers (17) raised antibodies against the N-terminal part of the HPV-16 E2 protein, indicating that the HPV-16 and BPV-1 E2 proteins are considerably different in terms of structure and epitope presentation. The most antigenic regions are usually the less ordered regions of the protein without packed internal side chains. From this point of view, the differential antigenicity may be a reflection of the differences in the structures of the HPV-16 and BPV-1 E2 proteins. Gauthier and coworkers (10) probed the structure of HPV-16 E2 with polyclonal antibodies raised against synthetic peptides that cover the whole region of the HPV-16 E2 protein. They found that antipeptide antibodies against the hinge region but not against the transactivation domain or the DNA-binding and dimerization domain were able to recognize the native form of the HPV-16 E2 protein.
In our study, MAb 1E2 (epitope within residues 184 to 190) was able to recognize neither E2-DNA nor E1-E2-origin complexes in a mobility shift assay. Curiously, deletion of the first alpha helix from the BPV-1 E2 protein revealed the epitope for MAb 1E2, and the protein in the protein-DNA complex was recognized by the antibody. Thus, the epitope for this MAb is probably buried within the compact structure of the N-terminal domain and is not accessible unless the structure of the molecule is distorted in some fashion. These data suggest that the transactivation domain of the E2 protein, unlike many other transactivation domains, has remarkable structural integrity. As shown by X-ray analysis, the C-terminal DNA-binding and dimerization domain has a compact structure (15). Deletion of the last 13 C-terminal residues of E2 resulted in an inactive protein unable to bind DNA and support replication (21a). So, our data confirm that in a native context, both the transactivation domain and the DNA-binding and dimerization domain of BPV-1 E2 have a complex and relatively rigid structure, while the central, hinge region is highly mobile and flexible.
MAb 5H4 and its Fab′ fragment efficiently inhibited the formation of both E2-DNA and E1-E2-origin complexes. They not only competed with DNA for binding but also were able to dissociate the preformed E2-DNA complex. In a transient replication assay, MAb 5H4 and its Fab′ fragment efficiently suppressed BPV-1 DNA replication. This assay is another way to demonstrate that the BPV-1 E2 protein interaction with the specific recognition sequence within an origin of replication is essential for the initiation of viral DNA replication. In addition, the results indicate that it is possible to target the E2 protein interaction with DNA for therapeutic purposes by using this specific MAb or Fab′ fragment to block the replication of papillomaviruses.
In our study, MAb 3F12 (epitope at residues 199 to 206), directed against the last 10 amino acids of the transactivation domain, and MAbs that bind the hinge region, 1H10 (epitope at residues 208 to 218) and 1E4 (epitope at residues 250 to 280), efficiently suppressed BPV-1 DNA replication. However, the Fab′ fragments of 1H10 and 3F12 had no inhibitory effect on and even activated replication. None of these antibodies interfered with the formation of the E1-E2-origin complex. These data suggest that antibodies 3F12 and 1H10 sterically hindered the inter- or intramolecular interactions required for the replication activity of the E2 protein. On the other hand, our results demonstrate the importance of the hinge region for the replication activity of the E2 protein. This hypothesis is based on results from several laboratories. E2 proteins containing large internal in-frame deletions of the hinge region (from amino acids 195 to 309, 213 to 309, and 220 to 309) were not able to support DNA replication (or had a decreased efficiency) but could efficiently enhance the binding of E1 to the replication origin (46, 47). A fusion protein which contained the transactivation domain together with the hinge region of BPV-1 E2 linked to the GCN4 DNA-binding domain supported replication much more efficiently than a fusion protein in which only the transactivation domain of E2 was linked to the GCN4 DNA-binding domain (3). These data suggest that the conformational freedom of the E2 protein is important for its role in replication and that the inhibitory effect of the antibodies against the epitopes in the hinge region as well as in the very last part of the transactivation domain may be explained by interference with conformational freedom, which would not allow E2 to assume the proper conformation required for its replication activity. However, another possibility is that this region is important for interactions with replication factors, so that antibodies that bind to the first part of the hinge region can but Fab′ fragments cannot prevent the binding of replication factors to the same region of the E2 protein.
ACKNOWLEDGMENTS
We are grateful to Aire Allikas and Saul Kivimae for providing the E2 deletion mutants, Marko Piirsoo for providing the pHookAlu construct, and Anne Kalling for technical assistance.
This work was supported by grants 2496 and 2497 from the Estonian Science Foundation, grant HHMI 75195-541301 from the Howard Hughes Medical Institute, grant CIPA-CT94-0154 from the EU, and a grant from the Citrina Foundation.
REFERENCES
- 1.Abroi A, Kurg R, Ustav M. Transcriptional and replicational activation functions in the bovine papillomavirus type 1 E2 protein are encoded by different structural determinants. J Virol. 1996;70:6169–6179. doi: 10.1128/jvi.70.9.6169-6179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson J D, Howley P M. Amino-terminal domains of the bovine papillomavirus type 1 E1 and E2 proteins participate in complex formation. J Virol. 1995;69:4364–4372. doi: 10.1128/jvi.69.7.4364-4372.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg M, Stenlund A. Functional interactions between papillomavirus E1 and E2 proteins. J Virol. 1997;71:3853–3863. doi: 10.1128/jvi.71.5.3853-3863.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blitz I, Laimins L A. The 68-kilodalton E1 protein of bovine papillomavirus is a DNA-binding phosphoprotein which associates with the E2 transcriptional activator in vitro. J Virol. 1991;65:649–656. doi: 10.1128/jvi.65.2.649-656.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brokaw J L, Blanco M, McBride A A. Amino acids critical for the function of the bovine papillomavirus type 1 E2 transactivator. J Virol. 1996;70:23–29. doi: 10.1128/jvi.70.1.23-29.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakrabarti R, Wylie D E, Schuster S M. Transfer of monoclonal antibodies into mammalian cells by electroporation. J Biol Chem. 1989;264:15494–15500. [PubMed] [Google Scholar]
- 7.DiMaio D, Settleman J. Bovine papillomavirus mutant temperature sensitive for transformation, replication and transactivation. EMBO J. 1988;7:1197–1204. doi: 10.1002/j.1460-2075.1988.tb02931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dostatni N, Thierry F, Yaniv M. A dimer of BPV-1 E2 containing a protease resistant core interacts with its DNA target. EMBO J. 1988;7:3807–3816. doi: 10.1002/j.1460-2075.1988.tb03265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson M, Botchan M. Genetic analysis of the activation domain of bovine papillomavirus protein E2: its role in transcription and replication. J Virol. 1996;70:4193–4199. doi: 10.1128/jvi.70.7.4193-4199.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gauthier J-M, Dillner J, Yaniv M. Structural analysis of the human papillomavirus type 16-E2 transactivator with antipeptide antibodies reveals a high mobility region linking the transactivation and the DNA-binding domains. Nucleic Acids Res. 1991;19:7073–7079. doi: 10.1093/nar/19.25.7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillette T, Lusky M, Borowiec J. Induction of structural changes in the bovine papillomavirus type 1 origin of replication by the viral E1 and E2 proteins. Proc Natl Acad Sci USA. 1994;91:8846–8850. doi: 10.1073/pnas.91.19.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giri I, Yaniv M. Structural and mutational analysis of E2 trans-activating proteins of papillomaviruses reveals three distinct functional domains. EMBO J. 1988;7:2823–2829. doi: 10.1002/j.1460-2075.1988.tb03138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grossel M J, Sverdrup F, Breiding D E, Androphy E J. Transcriptional activation function is not required for stimulation of DNA replication by bovine papillomavirus type 1 E2. J Virol. 1996;70:7264–7269. doi: 10.1128/jvi.70.10.7264-7269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haugen T H, Turek L P, Mercurio F M, Cripe T P, Olson B J, Anderson R D, Seidl D, Karin M, Schiller J. Sequence-specific and general transcriptional activation by the bovine papillomavirus-1 E2 transactivator requires an N-terminal amphipathic helix-containing E2 domain. EMBO J. 1988;7:4245–4253. doi: 10.1002/j.1460-2075.1988.tb03322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hegde R, Grossman S R, Laimins L A, Sigler P. Crystal structure at 1.7Å of the bovine papillomavirus-1 E2 DNA binding domain bound to its DNA target. Nature. 1992;359:505–512. doi: 10.1038/359505a0. [DOI] [PubMed] [Google Scholar]
- 16.Hegde R, Wang A, Kim S, Schapira M. Subunit rearrangement accompanies sequence-specific DNA binding by the bovine papillomavirus-1 E2 protein. J Mol Biol. 1998;276:797–808. doi: 10.1006/jmbi.1997.1587. [DOI] [PubMed] [Google Scholar]
- 17.Hibma M, Raj K, Ely S J, Stanley M, Crawford L. The interaction between human papillomavirus type 16 E1 and E2 proteins is blocked by an antibody to the N-terminal region of E2. Eur J Biochem. 1995;229:517–525. doi: 10.1111/j.1432-1033.1995.0517k.x. [DOI] [PubMed] [Google Scholar]
- 18.Iguchi-Ariga S M M, Itani T, Kiji Y, Ariga H. Possible function of the c-myc product: promotion of cellular DNA replication. EMBO J. 1987;6:2365–2371. doi: 10.1002/j.1460-2075.1987.tb02513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iguchi-Ariga S M M, Itani T, Yamaguchi M, Ariga H. c-myc protein can be substituted for SV40 T antigen in SV40 DNA replication. Nucleic Acids Res. 1987;15:4889–4899. doi: 10.1093/nar/15.12.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Ives I, Kivi S, Ustav M. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J Virol. 1999;73:4404–4412. doi: 10.1128/jvi.73.5.4404-4412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juronen E, Parik J, Toomik P. FPLC purification of mouse monoclonal antibodies from ascitic fluid using blue DEAE and thiophilic sorbents. J Immunol Methods. 1991;136:103–109. doi: 10.1016/0022-1759(91)90255-e. [DOI] [PubMed] [Google Scholar]
- 21.Kaczmarek L, Miller M R, Hammond R A, Mercer W E. A microinjected monoclonal antibody against human DNA polymerase-α inhibits DNA replication in human, hamster and mouse cell lines. J Biol Chem. 1986;261:10802–10807. [PubMed] [Google Scholar]
- 21a.Kurg, R., and M. Ustav. Unpublished data.
- 22.Langel U, Land T, Bartfai T. Design of chimeric peptide ligands to galanin receptors and substance P receptors. Int J Peptide Protein Res. 1992;39:516–522. doi: 10.1111/j.1399-3011.1992.tb00282.x. [DOI] [PubMed] [Google Scholar]
- 23.Lehman C, Botchan M. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc Natl Acad Sci USA. 1998;95:4338–4343. doi: 10.1073/pnas.95.8.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li R, Botchan M R. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell. 1993;73:1207–1221. doi: 10.1016/0092-8674(93)90649-b. [DOI] [PubMed] [Google Scholar]
- 25.Lusky M, Fontane E. Formation of the complex of bovine papillomavirus E1 and E2 proteins is modulated by E2 phosphorylation and depends upon sequences within the carboxyl terminus of E1. Proc Natl Acad Sci USA. 1991;88:6363–6367. doi: 10.1073/pnas.88.14.6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lusky M, Hurwitz J, Seo Y. Cooperative assembly of the bovine papilloma virus E1 and E2 proteins on the replication origin requires an intact E2 binding site. J Biol Chem. 1993;268:15795–15803. [PubMed] [Google Scholar]
- 27.Lusky M, Hurwitz J, Seo Y. The bovine papillomavirus E2 protein modulates the assembly of but is not stably maintained in a replication-competent multimeric E1-replication origin complex. Proc Natl Acad Sci USA. 1994;91:8895–8899. doi: 10.1073/pnas.91.19.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBride A A, Byrne J C, Howley P. E2 polypeptides encoded by bovine papillomavirus type 1 form dimers through the common carboxyl-terminal domain: transactivation is mediated by the conserved amino-terminal domain. Proc Natl Acad Sci USA. 1989;86:510–514. doi: 10.1073/pnas.86.2.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McBride A A, Schlegel R, Howley P. The carboxy-terminal domain shared by the bovine papillomavirus E2 transactivator and repressor proteins contains a specific DNA binding activity. EMBO J. 1988;7:533–539. doi: 10.1002/j.1460-2075.1988.tb02842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohr I J, Clark S, Sun S, Androphy E J, MacPherson P, Botchan M. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science. 1990;250:1694–1699. doi: 10.1126/science.2176744. [DOI] [PubMed] [Google Scholar]
- 31.Piirsoo M, Ustav E, Mandel T, Stenlund A, Ustav M. cis- and trans-requirements for stable episomal maintenance of the BPV-1 replicator. EMBO J. 1996;15:1–11. [PMC free article] [PubMed] [Google Scholar]
- 32.Porter R R. The hydrolysis of rabbit γ-globulin and antibodies with crystalline papain. Biochem J. 1959;73:119–126. doi: 10.1042/bj0730119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rank N, Lambert P. Bovine papillomavirus type 1 E2 transcriptional regulators directly bind two cellular transcription factors, TFIID and TBIIB. J Virol. 1995;69:6323–6334. doi: 10.1128/jvi.69.10.6323-6334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakai H, Yasugi T, Benson J, Dowhanick J, Howley P. Targeted mutagenesis of the human papillomavirus type 16 E2 transactivation domain reveals separable transcriptional activation and DNA replication functions. J Virol. 1996;70:1602–1611. doi: 10.1128/jvi.70.3.1602-1611.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sedman J, Stenlund A. Co-operative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 1995;14:6218–6228. doi: 10.1002/j.1460-2075.1995.tb00312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sedman J, Stenlund A. The initiator protein E1 binds to the bovine papillomavirus origin of replication as a trimeric ring-like structure. EMBO J. 1996;15:5085–5092. [PMC free article] [PubMed] [Google Scholar]
- 37.Sedman T, Sedman J, Stenlund A. Binding of the E1 and E2 proteins to the origin of replication of bovine papillomavirus. J Virol. 1997;71:2887–2896. doi: 10.1128/jvi.71.4.2887-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seo Y, Müller F, Lusky M, Hurwitz J. Bovine papilloma virus (BPV)-encoded E1 protein contains multiple activities required for BPV DNA replication. Proc Natl Acad Sci USA. 1993;90:702–706. doi: 10.1073/pnas.90.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seo Y-S, Müller F, Lusky M, Gibbs E, Kim H-Y, Phillips B, Hurwitz J. Bovine papilloma virus (BPV)-encoded E2 protein enhances binding of E1 protein to the BPV replication origin. Proc Natl Acad Sci USA. 1993;90:2865–2869. doi: 10.1073/pnas.90.7.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skiadopoulus M, McBride A. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J Virol. 1998;72:2079–2088. doi: 10.1128/jvi.72.3.2079-2088.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spalholz B, Yang Y, Howley P. Transactivation of a bovine papilloma virus transcriptional regulatory element by the E2 gene product. Cell. 1985;42:183–191. doi: 10.1016/s0092-8674(85)80114-8. [DOI] [PubMed] [Google Scholar]
- 42.Thompson N E, Strasheim L A, Nolan K M, Burgess R R. Accessibility of epitopes on human transcription factor IIB in the native protein and in the complex with DNA. J Biol Chem. 1995;270:4735–4740. doi: 10.1074/jbc.270.9.4735. [DOI] [PubMed] [Google Scholar]
- 43.Ustav M, Stenlund A. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 1991;10:449–457. doi: 10.1002/j.1460-2075.1991.tb07967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ustav M, Ustav E, Szymanski P, Stenlund A. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 1991;10:4321–4329. doi: 10.1002/j.1460-2075.1991.tb05010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wietkowski M, Broge P, Stahl H. Monoclonal antibodies as probes for a function of large T antigen during the elongation process of simian virus 40 DNA replication. J Virol. 1987;61:411–418. doi: 10.1128/jvi.61.2.411-418.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winokur P L, McBride A A. Separation of the transcriptional activation and replication functions of the bovine papillomavirus-1 E2 protein. EMBO J. 1992;11:4111–4118. doi: 10.1002/j.1460-2075.1992.tb05504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winokur P L, McBride A A. The transactivation and DNA binding domains of the BPV-1 E2 protein have different roles in cooperative origin binding with the E1 protein. Virology. 1996;221:44–53. doi: 10.1006/viro.1996.0351. [DOI] [PubMed] [Google Scholar]
- 48.Yang L, Li R, Mohr I, Clark R, Botchan M. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature. 1991;353:628–632. doi: 10.1038/353628a0. [DOI] [PubMed] [Google Scholar]
- 49.Yao J M, Breiding D E, Androphy E J. Functional interaction of the bovine papillomavirus E2 transactivation domain with TFIIB. J Virol. 1998;72:1013–1019. doi: 10.1128/jvi.72.2.1013-1019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]