Abstract
The C terminus of the HIV-1 Gag protein contains a proline-rich domain termed p6Gag. This domain has been shown to play a role in efficient virus release and incorporation of Vpr into virions. In a previous study (X. F. Yu, L. Dawson, C. J. Tian, C. Flexner, and M. Dettenhofer, J. Virol. 72:3412–3417, 1998), we observed that the removal of the p6 domain of Gag as well as drastic mutations in the PTAP motif resulted in reduced virion-associated Pol proteins from transfected COS cells. In the present study, amino acid substitutions at residues 5 and 7 of p6Gag resulted in a cell type-dependent replication of the mutant virus in CD4+ T cells; the virus was replication competent in Jurkat cells but restricted in H9 cells and primary blood-derived monocytes. Established Jurkat and H9 cell lines expressing p6Gag mutant and parental virus were used to further understand this defect. Mutant virions produced from H9 cells, which displayed no defect in extracellular virion production, showed an ∼16-fold reduction in Pol protein levels, whereas the levels of Pol proteins were only marginally reduced in mutant virions produced from Jurkat cells. The reduction in the virion-associated Pol proteins could not be accounted for by differences in the levels of intracellular p160Gag-Pol or in the interaction between p55Gag and p160Gag-Pol precursors. Electron microscopic analysis of the p6Gag mutant virions showed a predominately immature morphology in the absence of significant defects in Gag proteolytic cleavage. Taken together, these data suggest that the proline-rich motif of p6Gag is involved in the late stages of virus maturation, which include the packaging of cleaved Pol proteins in viral particles, a process which may involve cell-type-specific factors.
Common to all retroviruses are three major structural proteins, Gag, Pol, and Env (38). In human immunodeficiency virus type 1 (HIV-1), Gag is expressed in the form of a 55-kDa precursor, p55Gag, which is posttranslationally cleaved into p17MA, p24CA, p7NC, and p6Gag by the viral protease during virus assembly and maturation (38). The Pol proteins of HIV-1 (protease, reverse transcriptase (RT), and integrase) are synthesized in the form of a fusion protein that includes part of the Gag protein (MA, CA, and NC, but not p6Gag). The Gag-Pol fusion protein is the result of a −1 ribosomal frameshifting event between the p7NC and p6Gag domains of Gag which occurs with a frequency of 5 to 10% of the total Gag proteins synthesized (38). Incorporation of these Pol proteins into virions is believed to occur through the shared Gag regions of the p55Gag and p160Gag-Pol precursors (21, 36).
The expression of retroviral Gag proteins, which constitute the principal structural components of the virus, is sufficient to drive viral particle assembly and budding (38). The domains that constitute the Gag molecule play different roles in the ability of the budding particle to emerge from the plasma membrane of an infected cell. Mutagenesis studies have shown that the addition of a myristic acid at the N terminus of HIV-1 Gag (5, 18), as well as basic residues within the N terminus of the MA domain (44, 46), is critical for the accumulation of p55Gag at the plasma membrane, the site of virus budding. MA also plays another role during virus assembly, that of incorporating the viral envelope glycoproteins into the surfaces of virions (9, 12, 40) by interacting with the cytoplasmic tail of gp41 (6, 10, 12, 41).
The formation of virus particles involves the multimerization of p55Gag through protein-protein interactions (38). This process is controlled at least in part by the I domain in NC (3, 8, 34, 45). Positively charged amino acids in HIV-1 NC seem to be important for the function of the I domain (4, 8). In addition, regions in MA that are important for trimer formation (19, 28) and regions in CA that are involved in dimer formation (7, 11a, 13, 27, 29, 37) are also thought to play important roles in HIV-1 assembly. The p7NC domain also contributes to the incorporation of the viral genomic RNA into the virions (38).
In addition to the protein domains described above, retroviral Gag molecules also encode regions which have been termed late-assembly (L) domains. Mutational analysis has suggested that these domains are involved in the late stage of virus particle formation, possibly controlling the efficient release of particles from the cell surface (17, 20, 30, 32, 35, 39, 43). Motifs of the L domains in HIV-1 p6Gag (PTAPP), equine infectious anemia virus p9Gag (YXXL), and Rous sarcoma virus p2 (PPPPY) have been shown to be functionally interchangeable with regard to their effects on particle release (30). The L domain of Rous sarcoma virus p2 has also been shown to interact with WW protein binding motifs; however, the comparable domains in HIV-1 and equine infectious anemia virus fail to bind this motif (16).
In addition to its involvement in particle release, HIV-1 p6Gag is also critical for the incorporation of Vpr into particles (23, 25, 31). Mutagenesis studies have shown that the C-terminal (LXX)4 repeat in p6Gag is important for Vpr virion incorporation (25). This conclusion is supported by the incorporation of Vpr into heterologous virions when p6Gag is fused to the C terminus of the murine leukemia virus Gag protein (23). More recently, a role for HIV-1 p6Gag in the control of particle size has been reported (15).
Regions in p24CA have been implicated in the association of p55Gag and p160Gag-Pol prior to viral protease-directed cleavage of these molecules (21, 36). These may constitute some of the early intermolecular contacts to promote the recruitment of p160Gag-Pol into virions. Additionally, we have shown that the p6 domain, which is unique to p55Gag, contributes to the recruitment of Pol proteins into virions (42). In the present study, we have demonstrated that residues 5 and 7 of HIV-1 p6Gag contribute to the incorporation of the Pol proteins within the virions. This was observed to be cell type dependent, which correlated with the replication of the mutant virus, and was distinct from the role of p6Gag in virus release. These data suggest that cell-type-specific factors may be involved in the packaging of cleaved Pol proteins during HIV-1 assembly and budding.
MATERIALS AND METHODS
DNA constructs.
The parental virus used for this study was derived from the HXB2 clone (33). The EcoRI site in the cellular flanking region distal to the 3′ end of the viral genome was destroyed by digestion with XbaI, which overlaps the EcoRI site, and the DNA strands were treated with mung bean nuclease to create blunt ends and then ligated with T4 ligase. A ClaI site was created in the nef coding region (downstream from the env coding region) with a mutagenic oligonucleotide: 5′-TTTTGCTATAACATCGATGGCAAGTGGTCA-3′ (the restriction site is underlined). Mutagenesis was performed according to the manufacturer’s protocol (Bio-Rad, Richmond, Calif.). The neomycin phosphotransferase gene (neoR) was PCR amplified from pLXSN (14) with primers that incorporated ClaI and XhoI sites at the 5′ and 3′ ends of neoR, respectively. PCR was performed according to conditions described by Perkin-Elmer with primers 5′-ATGAGGATCGATCGATATGATTGAACAAGA-3′ and 5′-AACCCCAGAGCTCGAGTCAGAAGAACTCGT-3′ (restriction sites are underlined). PCR-amplified neoR was then digested with ClaI and XhoI and cloned into the modified HXB2 construct at the ClaI and XhoI sites of nef. The p6Gag mutant construct (HXB2Pro) was created as previously described (43). The env deletion (HXB2Bgl) and p6Gag mutant with the env deletion (HXB2ProBgl) vectors were created by removing nucleotides 6620 to 7200 of HXB2 by digestion with BglII and were religated with T4 ligase.
Cells, DNA transfection, and infection.
COS-7 cells were maintained in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum and antibiotics and passaged upon confluence. CEM-ss, Jurkat, and H9 cell lines were grown in RPMI 1640 with 10% fetal bovine serum and antibiotics and maintained at a density of <1 × 106/ml. Primary blood-derived mononuclear cells (PBMC) were activated with phytohemagglutinin and interleukin-2 prior to infection. Establishment of chronically infected Jurkat and H9 cell lines was performed in the presence of G418. Cell culture reagents were obtained from GIBCO/BRL.
COS-7 cells were transfected by the DEAE-dextran method. Briefly, COS-7 cells were trypsinized and seeded at 50% confluence 24 h prior to transfection. The cells (5 × 106) were then trypsinized, pelleted, and resuspended in 1 ml of TD buffer (25 mM Tris-HCl [pH 7.4], 140 mM NaCl, 5 mM KCl, 0.7 mM K2HPO4) containing 500 μg of DEAE-dextran and 5 μg of HXB2 or HXB2Pro DNA. Transfection was carried out at 37°C for 30 min, and the cells were then washed in 5 ml of complete medium and reseeded in T-75 flasks. Transfection of COS-7 cells with HXB2-derived env mutants was performed as described above, except that the cells were cotransfected with a murine leukemia virus Env expression vector, SV-A-MLV-env (24).
Cell culture supernatants containing viral particles were harvested 3 days after transfection, precleared by centrifugation in a Sorvall RT 6000B centrifuge at 3,000 rpm for 30 min, filtered through a 0.2-μm-pore-size membrane, and used for infectivity analysis. Chronically infected cell lines were established by exposing cells to virus-containing cell culture supernatants and then growing the cells in the presence of 1.2 mg of G418/ml for at least 2 weeks before analysis of the cellular and viral protein profiles.
RT assay.
Cell culture supernatants were cleared of cells and cellular debris by centrifugation in a Sorvall MC 12V centrifuge at 14,000 rpm for 2 min. For each sample, 250 μl of culture supernatant was mixed with 125 μl of 30% polyethylene glycol-8000 with 0.5 M NaCl at 4°C overnight. The samples were centrifuged at 2,500 rpm for 30 min (Sorvall RT 6000B), the viral pellets were dissolved in 25 μl of RT lysis buffer (1% Triton X-100, 20 mM Tris-HCl [pH 7.5], 60 mM KCl, 1 mM dithiothreitol, 30% glycerol), and 10 μl of each sample was used for the RT reaction. Viral lysates were combined with 90 μl of RT reaction cocktail {40 mM Tris-HCL (pH 7.8), 8 mM dithiothreitol, 10 mM MgCl2, 0.05 A260 unit of poly(rA) · poly(dT)15 (Boehringer Mannheim), and 2.5 μCi of [3H]dTTP} at 37°C for 2 h. The reaction product was precipitated with 3 ml of chilled 10% (wt/vol) trichloroacetic acid, with tRNA as a carrier. Incorporation of [3H]dTTP was determined from material bound to GF/C glass microfiber filters following five washes with chilled 5% trichloroacetic acid and quantified with a Beckman LS 6500 scintillation counter.
Immunoblotting.
Virion-associated viral proteins were prepared from cell culture supernatants by centrifugation at 3,000 rpm for 30 min in a Sorvall RT 6000B centrifuge followed by filtration through a 0.2-μm-pore-size membrane. Virus particle-containing supernatants were concentrated by centrifugation through a 20% sucrose cushion at 100,000 × g for 2 h in a Sorvall Ultra80. Viral pellets were resuspended in phosphate-buffered saline. Cell-associated viral proteins were analyzed from chronically infected Jurkat and H9 cell lines. Cells (105) were lysed in 1× loading dye (0.08 M Tris [pH 6.8], 2.0% sodium dodecyl sulfate [SDS], 10% glycerol, 0.1 M dithiothreitol, 0.2% bromophenol blue). Samples were boiled for 10 min, and proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE). The proteins were transferred onto two separate nitrocellulose membranes by passive diffusion for 48 h, producing identical mirror image blots. The membranes were probed with HIV-1-positive human serum (1:200), mouse monoclonal antibody (MAb) against RT (1:400; BTI), or rabbit polyclonal serum against integrase (1:400).
Radioimmunoprecipitation analysis.
Chronically infected Jurkat and H9 cells (5 × 106) were lysed in lysis buffer (0.15 M NaCl, 0.01 M Tris-HCl [pH 7.4], 1% Triton X-100) containing phenylmethylsulfouyl fluoride, leupeptin, aprotinin, and antipain. The cell lysates were precleared of nuclei by centrifugation at 1,500 × g for 5 min, and postnuclear supernatants were then immunoprecipitated for 3 h at 4°C with p6 antiserum (43) that had been preabsorbed with protein A-Sepharose. Cell-associated viral proteins were analyzed by SDS-PAGE, followed by Western blotting as described above.
Radioimmunoprecipitation of cell- and virion-associated proteins was performed as described above, with the following modifications. Infected H9 cells (5 × 106) were starved for 30 min in cysteine-free medium containing 5% fetal calf serum, labeled with [35S]cysteine (200 μCi/ml) for 12 h, and lysed in lysis buffer. Virions were pelleted through 20% sucrose and resuspended in lysis buffer. Postnuclear supernatants and the pelleted virions were immunoprecipitated for 3 h at 4°C with the HIV-1-positive human serum that had been preabsorbed with protein A-Sepharose. Cell and virion-associated viral proteins were analyzed by SDS–12% PAGE and autoradiography.
Ultrastructural studies.
Electron microscopy (EM) was performed as previously described (43). Briefly, H9 cells chronically infected with either HXB2Bgl or HXB2ProBgl were pelleted and fixed for 2 h at 4°C in 0.13 M sodium phosphate containing 2.5% glutaraldehyde. The fixative was then removed, and the cells were washed three times with 0.13 M sodium phosphate and stored overnight at 4°C. The cell pellets were minced, postfixed in 1.0% osmium tetroxide, and washed in H2O. The cells were stained en bloc with 2.0% aqueous uranyl acetate, dehydrated in ethanol, and infiltrated and embedded in Spurr’s plastic resin, which was polymerized overnight at 70°C. Embedded blocks from the cell samples were ultrathin sectioned with a Reichert-Jung Ultracut E ultramicrotome. Ultrathin sections 60 to 80 nm thick were collected and mounted onto copper mesh grids. The grids were poststained with Reynold’s lead citrate and examined in a Hitachi HU-12A transmission EM.
RESULTS
Mutation in HIV-1 p6Gag and replication growth curves.
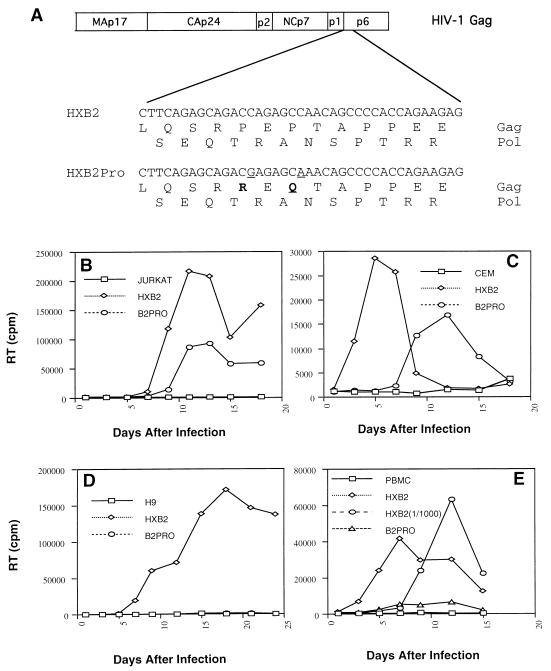
The amino acid sequence of p6Gag includes a proline-rich amino terminus which has a high degree of conservation among primate lentiviruses. In order to investigate the functional role of HIV-1 p6Gag, we introduced amino acid substitutions at two proline residues, P5R and P7Q (Fig. 1A). These mutations retain the original amino acid sequence for the Pol open reading frame.
FIG. 1.
(A). Schematic depiction of partial HIV-1 Gag and Pol sequences of the p6Gag mutant (HXB2Pro) and parental construct (HXB2) used in this study. Proline residues at positions 5 and 7 of p6Gag where changed to arginine and glutamine (boldface), respectively, without altering the Pol amino acid sequence. (B to E) Replication growth curve of HXB2 and HXB2Pro viruses as monitored by RT activity from cell-free supernatants. Virions were generated from transfected COS-7 cells and used to initiate infections in Jurkat (B), CEM (C), or H9 (D) cells or PBMC (E). PBMC were additionally infected with a 1,000-fold-reduced concentration of the HXB2 virus [HXB2(1/1000)], to check the relative degree of infectivity of the HXB2Pro virus.
In order to examine the replication capacity of the p6Gag mutant and the parental virus (Gag+ Pol+ Vif+ Vpr− Vpu− Tat+ Rev+ Env+ Nef−), supernatants derived from transfected COS-7 cells were used to initiate infection in Jurkat, CEM, and H9 cells and PBMC. Virus production was monitored by RT activity in the cultured supernatants. In Jurkat and CEM cells, the p6Gag mutant HXB2Pro was competent for replication, although it displayed a delay in its replication compared to the parental virus HXB2 (Fig. 1B and C). In contrast, HXB2Pro failed to replicate in H9 cells (Fig. 1D) and PBMC (Fig. 1E). In PBMC, HXB2Pro was at least 1,000-fold less infectious than the parental virus, HXB2 (Fig. 1E). These data demonstrate a clear cell type-dependent replication of this p6Gag mutant.
Virion production by the p6Gag mutant virus in nonpermissive H9 cells.
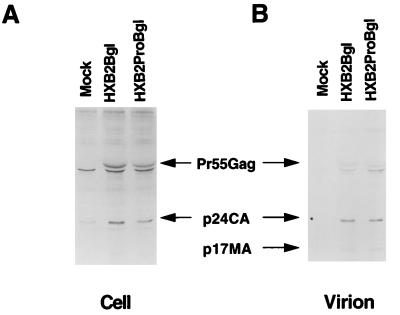
We next examined whether failure to replicate in H9 cells was caused by a defect in virus release by the p6Gag mutant virus. H9 cells expressing the parental virus or p6Gag mutant virus lacking env were established, and virus release was evaluated by radioimmunoprecipitation analysis. The parental virus (HXB2Bgl) and p6Gag mutant virus (HXB2ProBgl) expressed comparable levels of intracellular p55Gag and p24CA in H9 cells (Fig. 2A). Examination of released virions revealed similar quantities of p55, p24, and p17 Gag molecules in both the parental and p6Gag mutant viruses (Fig. 2B). These data demonstrate that mutations at residues 5 and 7 of p6Gag did not significantly influence the release of virions from H9 cells.
FIG. 2.
Radioimmunoprecipitation of viral proteins from uninfected H9 (Mock) cells, established H9 cell lines expressing p6Gag mutant (HXB2ProBgl), and parental (HXB2Bgl) constructs. The cells were metabolically labeled with [35S]cysteine for 12 h. The proteins were immunoprecipitated with HIV-1-positive patient serum either from cell lysates (A) or from particles pelleted through 20% sucrose (B). The proteins were separated by SDS-PAGE and visualized by autoradiography.
Cell-associated p6Gag mutant viral protein profiles.
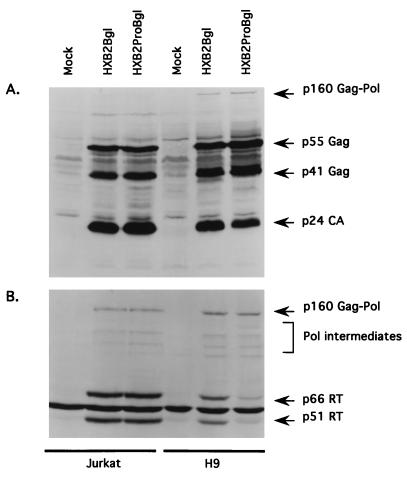
In order to determine why infection with the p6Gag mutant virus led to a cell type-dependent replication, we also established chronically infected Jurkat cells. The cell-associated viral protein composition was first examined by probing Western blots with an HIV-1-positive human serum (Fig. 3A). A comparison of the parental virus (HXB2Bgl) with the p6Gag mutant virus (HXB2ProBgl) in Jurkat cell lines revealed no significant difference in the quantity or degree of processing of Gag proteins. In nonpermissive H9 cells, the Gag protein profiles displayed no major differences, although the degree of Gag processing was subtly defective in the p6Gag mutant-infected cells. There appeared to be a slight accumulation of the Gag intermediate p41Gag and a corresponding reduction in p24CA.
FIG. 3.
Intracellular viral protein profiles derived from established Jurkat and H9 cell lines containing HXB2Bgl (parent) or HXB2ProBgl (p6Gag mutant). Proteins from mock-infected and HXB2Bgl- and HXB2ProBgl-infected cell lysates were separated by 12% (A) or 7.5% (B) polyacrylamide gels and transferred onto nitrocellulose membranes. The blots were reacted with either HIV-1-positive human serum (A) or anti-RT MAb (B).
When similar blots were probed with an anti-RT MAb, again no significant differences were apparent in Jurkat cells infected with HXB2Bgl or HXB2ProBgl (Fig. 3B). Comparable levels of p160Gag-Pol precursor, fully cleaved p66RT and p51RT, and RT-reactive Pol processing intermediates were apparent. Comparable quantities of p160Gag-Pol precursors as well as RT-reactive Pol processing intermediates were also detected in HXB2Bgl- and HXB2ProBgl-infected H9 cells (Fig. 3B). However, the p6Gag mutant had significantly (approximately 5-fold) reduced levels of mature p66RT and p51RT compared to the parental virus (Fig. 3B). The comparable levels of p55Gag and p160Gag-Pol indicate that protein synthesis was not compromised, but the reduced levels of the fully mature forms of RT in the p6Gag mutant suggest either a defect in proteolyic cleavage or a selective degradation of mature RT proteins. It should be noted that protease function itself did not seem to be drastically impaired, as indicated by the existence of the Pol processing intermediates as well as partially cleaved p41Gag and fully cleaved p24CA.
Virion-associated p6Gag mutant protein profiles.
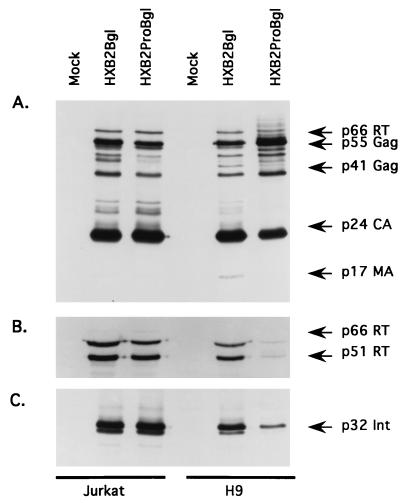
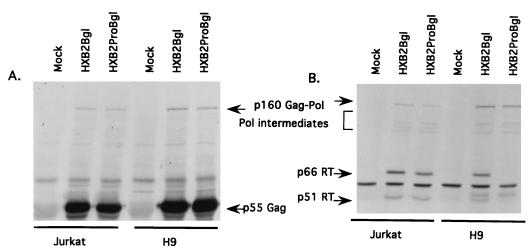
We next examined the protein composition in cell-free virions derived from Jurkat and H9 cell lines. When we compared the p6Gag mutant (HXB2ProBgl) with its parent virus (HXB2Bgl) in Jurkat cell lines, we found that the two sets of Gag, integrase, and RT proteins were indistinguishable (Fig. 4A to C). In contrast, the protein profiles of virions derived from p6Gag mutant virus- and parental virus-infected H9 cells display two noticeable differences. First, the p6Gag mutant virus-infected cells showed a partial defect in the cleavage of the Gag protein, as evidenced by the accumulation of p55Gag and p41Gag and a corresponding decrease in the mature p24CA and p17MA. Although a cleavage defect in the p6Gag mutant might suggest a defect in viral protease activity, it should be noted that this defect was slight and, therefore, that the protease may not be the functional target of p6Gag activity. A more striking difference between the parental virus and the p6Gag mutant was evident when virion-associated proteins were stained with the anti-RT MAb (Fig. 4B) or with an anti-integrase antibody (Fig. 4C). The p6Gag mutant virions contained drastically reduced levels of both RT and integrase compared with those of the parental viruses.
FIG. 4.
Virion-associated-protein profiles derived from established Jurkat and H9 cell lines containing HXB2Bgl (parent) or HXB2ProBgl (p6Gag mutant). Virion proteins from mock-infected and HXB2Bgl- and HXB2ProBgl-infected cells were separated by 12% polyacrylamide gels and transferred onto nitrocellulose membranes. The blots were reacted with either HIV-1-positive human serum (A), anti-RT MAb (B), or anti-integrase (Int) antiserum (C).
In order to get a better estimate of the degree to which the Pol proteins failed to be retained in the p6Gag mutant virions, we ran sequential twofold dilutions of the parental viral lysates on SDS–12% PAGE alongside the p6Gag mutant viral lysates. Virions derived from Jurkat cells were stained with either the HIV-1-positive human serum (Fig. 5A) or the anti-RT MAb (Fig. 5B). When the overall levels of Gag proteins were normalized, an approximately twofold-lower level of the Pol proteins was observed in the p6Gag mutant virus than in the parental virus isolated from Jurkat cells.
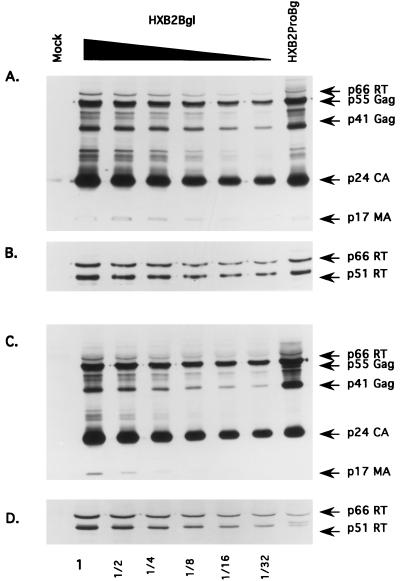
FIG. 5.
Quantitation of virion-associated protein profiles derived from Jurkat (A and B) and H9 (C and D) cell lines. Twofold dilutions of HXB2Bgl virion lysates (as indicated below each corresponding lane) were run side by side with a fixed amount of HXB2ProBgl (furthest-right lane) to compare the relative amounts of Gag proteins and RT proteins in each sample. Virion proteins from mock-infected and HXB2Bgl- and HXB2ProBgl-infected cells were separated by 12% polyacrylamide gels and transferred onto nitrocellulose membranes. The blots were reacted with either HIV-1-positive human serum (A and C) or anti-RT MAb (B and D).
When a similar analysis was performed on the H9 cell-derived virions, we observed an approximately 16-fold-lower level of the Pol proteins in the p6Gag mutant virions than in the parental virions (Fig. 5C and D). This defect could not be accounted for by a corresponding reduction in the level of the Gag-Pol precursor proteins (p160Gag-Pol) in the producer cell (Fig. 3B) or by an accumulation of uncleaved p160Gag-Pol in the p6Gag mutant virions (data not shown). It should be noted that a nonspecific, cross-reacting band with mobility similar to that of p66RT is recognized by the HIV-1-positive serum used (Fig. 4 and 5). However, this protein band is not p66RT as it was not recognized by the MAb against HIV-1 p66RT and p51RT.
Intracellular p55Gag and p160Gag-Pol association.
Since the incorporation of Pol proteins in the virions was disrupted in the case of the p6Gag mutant virus, we then studied the intracellular association between Gag and Gag-Pol precursors. These experiments involved immunoprecipitation of the Gag complex with a polyclonal antibody to p6Gag, which did not recognize the p160Gag-Pol molecule but did precipitate Gag-interacting molecules. The profiles of the immunoprecipitated proteins were then examined by Western blotting with either the HIV-1-positive serum or the RT MAb. When we compared the p6Gag mutant with its parental virus in both Jurkat and H9 cells, we found that similar quantities of p160Gag-Pol and p55Gag were coimmunoprecipitated by the p6Gag antiserum in each case (Fig. 6A). When similar blots were stained with the anti-RT MAb, we found that comparable levels of Pol-reactive proteins were coimmunoprecipitated by the p6Gag antiserum from Jurkat cells infected with the mutant virus and from cells infected with the parental virus (Fig. 6B). In contrast, lower levels of the mature p66RT and p51RT proteins were coimmunoprecipitated by the p6Gag antiserum from H9 cells infected with the p6Gag mutant virus than from parental virus-infected H9 cells. At the same time, the levels of the coimmunoprecipitated p160Gag-Pol precursor and Pol intermediates were indistinguishable (Fig. 6B).
FIG. 6.
Coimmunoprecipitation of intracellular viral complex. Analysis of p55Gag precursor association with p160Gag-Pol precursor- and Pol domain-containing proteins. Proteins from mock-infected HXB2Bgl- and HXB2ProBgl-infected establish Jurkat or H9 cell lysates were immunoprecipitated with anti-p6 antiserum as described in Materials and Methods. The anti-p6 antiserum used specifically recognizes only p55Gag-related proteins containing the p6 domain of Gag. The precipitated proteins were separated on 7.5% polyacrylamide gels and transferred onto nitrocellulose membranes. The Western blots were reacted with either HIV-1-positive human serum (A) or anti-RT MAb (B).
Ultrastructural analysis of p6Gag mutant by EM.
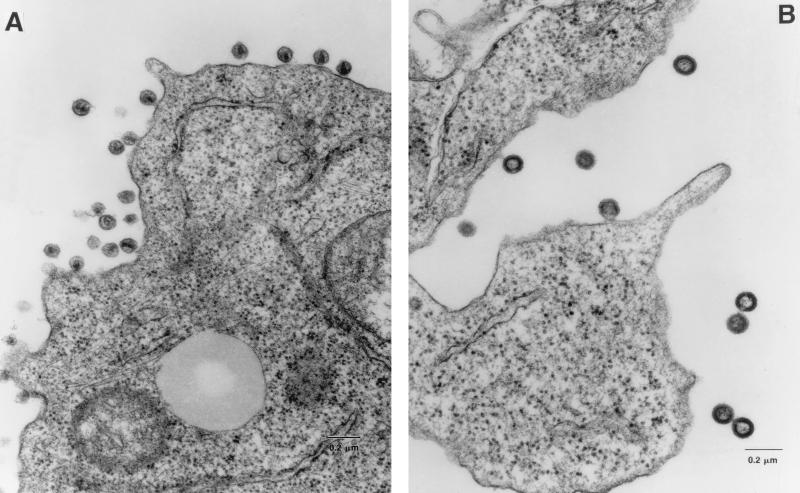
It has previously been shown that deletions of the entire p6Gag domain result in inefficient release of virions from the cell surface, with a corresponding accumulation of virions at the cell surface in intermediate stages of budding (17, 43). Since the p6Gag mutant described here displayed cell type-dependent defects, with little apparent influence on the release of virions, it seemed useful to examine the ultrastructure of these particles. H9 cells producing either the p6Gag mutant or parental virions were embedded and examined by EM. Over 200 p6Gag mutant and parental virions were examined for morphological differences (Table 1). Most of the parental virions appeared to be mature, possessing electron-dense cone-shaped cores (Fig. 7 and Table 1). In contrast, the majority of the p6Gag mutant virions were immature, as evidenced by the presence of electron-dense structural material underlying the inner leaflet of the membrane and an absence of cone-shaped cores. There was no evidence for an increased accumulation of p6Gag mutant budding particles at the plasma membrane compared to that seen for the parental virus, suggesting that release of p6Gag mutant virions was not impaired. Thus, the p6Gag mutant phenotype described here may be more subtle than those of previously reported p6Gag deletion mutants (17, 43).
TABLE 1.
Characterization of HXB2Bgl and HXB2ProBgl virions derived from H9 cellsa
| Virion morphology | HXB2Bgl | HXB2ProBgl |
|---|---|---|
| Mature particles (%) | 76 | 14 |
| Immature particles (%) | 24 | 86 |
| No. of particles examined | 322 | 234 |
Virion morphology was distinguished as either mature or immature.
FIG. 7.
Ultrastructure analysis of parental and p6 mutant virions. Viruses produced from HXB2Bgl- (A) and HXB2ProBgl-containing (B) established H9 cells were examined by EM. Representative fields are shown.
The EM data revealed that the majority of the p6 mutant virions possessed immature phenotypes and a majority of the wild-type virions possessed mature phenotypes. However, the biochemical data showed the viral protease in the p6 mutant virions to be active, albeit not at wild-type levels (Fig. 4A). It is possible that the immature-looking p6 mutant virions that were newly released but did not wash away during the EM fixation procedure may have had a more pronounced Gag cleavage defect because of reduced incorporation of viral protease than did those mutant virions analyzed by immunoblotting, which represent older virions. It is also possible that the Gag proteins must be cleaved to a certain threshold before particles may appear fully mature. Therefore, if not all of the p55Gag precursors are cleaved within a given particle, the virion may appear to be immature by EM yet look biochemically “mature.” Alternatively, it is possible that Gag processing may be necessary but not sufficient for the maturation of HIV-1 virions.
DISCUSSION
In this study, we have examined the cell type-dependent role of the HIV-1 p6Gag domain. Mutations introduced at residues 5 and 7 of p6Gag resulted in a lack of viral replication in H9 cells and PBMC, whereas the mutant virus remained replication competent in Jurkat and CEM cells despite an initial delay. Analysis of virion-associated protein profiles demonstrated a failure of the Pol proteins RT and integrase to be retained in the p6Gag mutant virus particles. This defect was correlated with the cell type-dependent replication of the p6Gag mutant virus. A previous study, using more drastic mutations in p6Gag, has suggested a role for the p6Gag domain in the packaging of the Pol proteins during virus assembly and budding (42). However, the role of the p6Gag domain in Pol protein retention could not be clearly distinguished from the domain’s proposed role in virus release (42).
During the assembly process in HIV-1, the p55Gag and p160Gag-Pol proteins are targeted to the plasma membrane, and proteolytic processing is initiated (38). Since proteolytic cleavage, budding, and release of virions are dynamic and coordinated processes, the altered timing of one of these events could in turn affect the other steps. One interpretation of our data is that the introduction of mutations within p6Gag results in slower virus release. For example, if virus budding were slowed down but proteolytic processing was not affected, the Pol domain would become separated from the upstream Gag domain, the principal domain for interaction between the Gag-Pol precursors and the Gag precursors. If interaction between Gag and Gag-Pol precursors were solely dependent on the Gag regions that are shared between the two precursors, the Pol proteins would be excluded from the assembling p6Gag mutant viruses if they had already been cleaved from the Gag-Pol precursor prior to the completion of virus budding. In the present study, the introduction of more subtle mutations in p6Gag failed to produce a significant defect in virion release yet at the same time caused a 16-fold reduction in the Pol proteins of mutant virions. Since the p6Gag mutant virions studied here showed efficient virus release, this mutation may be viewed as an intermediate between the wild-type parental virus and the more drastic p6Gag mutants, which show inefficient virus release as well as failure to retain the Pol proteins. On the basis of these data, there does not appear to be a direct link between the efficiency of virus release and the ability to package Pol proteins in released virus particles.
The principal protein-protein interaction domains of p55Gag and p160Gag-Pol precursor molecules are found within the shared Gag region, including MA, CA, and NC (38). At the C terminus of p55Gag is the p6Gag molecule, which is not present in the p160Gag-Pol precursor. The self-cleavage of the virus-encoded protease from the p160Gag-Pol precursor is thought to be necessary to initiate subsequent proteolytic cleavage events along the Gag and Pol molecules. Since the protease is situated at the N terminus of the Pol domain, its cleavage physically separates Gag from Pol so that the Pol proteins lose their Gag interaction domain and no longer associate with the virus assembly complex in the absence of other interactions. It is possible that the p6Gag domain is involved in a protein-protein interaction with the Pol domain. The data presented in this report suggest that the p6Gag domain functions to retain the Pol domain within the assembling virus after the activation of the viral protease. This idea is also consistent with studies in which mutations in either the RT (2, 26) or integrase (1, 11) molecules were associated with a reduction in the levels of Pol proteins in released mutant virions.
Since p6Gag mutations at prolines 5 and 7 were associated with a cell type-dependent process, it appears that cell-derived factors are involved in the retention of the Pol proteins during virus assembly. The fact that H9 cell-derived virions showed a more drastic defect in Pol protein incorporation than did those from Jurkat cells suggests that a cellular factor in Jurkat cells may interact with both wild-type and p6Gag mutant molecules, whereas the analogous factor in H9 cells may only interact with the wild-type p6Gag molecule. Alternatively, the putative cellular factor might be present at varying concentrations that are cell type specific. It remains to be determined whether there is indeed a factor that acts as a molecular bridge between p6Gag and a determinant within the Pol domain to ensure the recruitment of the Pol proteins into virions after the initial proteolytic processing events.
Examination of the virion protein profiles supports the notion that p6Gag may control the incorporation of Pol into virions, although analysis of the intracellular viral proteins still leaves some questions to be answered. Even though the synthesis of the Pr160 Gag-Pol appeared to be unimpaired in the p6Gag mutant cells (Fig. 3), and the degree of proteolytic cleavage of Gag was not drastically altered, a reduction in the fully cleaved forms of RT was evident. This suggests one of two possibilities. First, in the p6Gag mutant virus-infected cells, the final cleavage of the mature RT molecules may be selectively suppressed. We believe this possibility to be less likely because, in such a case, an accumulation of p160Gag-Pol would result and be detectable in virions, and this situation has not been observed. The second possibility is that the mature forms of RT are indeed produced but are degraded. This idea would have to be considered if we assume that the mature RT molecules no longer remain part of the virus assembly complex because of their lack of association with p6Gag and as a result they are not protected from digestion by cellular protease(s). These data are similar to those in previous reports in which point mutations in the RT coding region reduced RT incorporation into virions without influencing the intracellular level of p160Gag-Pol (26). Selective degradation of mature RT molecules but not of p160Gag-Pol has also been reported for certain temperature-sensitive HIV-1 RT mutants (22). In the current study we have shown that p160Gag-Pol is not only present intracellularly in similar quantities in the p6Gag mutant and the parental virus but is also found in association with p55Gag (Fig. 6). This finding argues that the introduction of point mutations within p6Gag did not interfere with the initial intracellular interactions between p55Gag and p160Gag-Pol nor did it dramatically alter the structure of p55Gag. Further studies are in progress to investigate the mechanism by which p6Gag is involved in the incorporation of the Pol proteins into the virion.
ACKNOWLEDGMENTS
We are grateful to Zene Matsuda, Tun-Hou Lee, and Max Essex for several DNA constructs; to John Birnbaum for EM assistance; and to Oliver Laeyendecker for technical assistance. The following reagent was obtained through the AIDS Research and Reference Reagents Program, Division of AIDS, NIAID, NIH: antiserum against HIV-1 integrase (catalog no. 756).
M.D. was supported in part by a training grant from NIEHS (ES07141). Support also came from a National Institutes of Health grant (AI-35525 to X.-F.Y.).
REFERENCES
- 1.Ansari-Lari M A, Donehower L A, Gibbs R A. Analysis of human immunodeficiency virus type 1 integrase mutants. Virology. 1995;213:680. [PubMed] [Google Scholar]
- 2.Ansari-Lari M A, Gibbs R A. Expression of human immunodeficiency virus type 1 reverse transcriptase in trans during virion release and after infection. J Virol. 1996;70:3870–3875. doi: 10.1128/jvi.70.6.3870-3875.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett R P, Nelle T D, Wills J W. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus gag proteins. J Virol. 1993;67:6487–6498. doi: 10.1128/jvi.67.11.6487-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowzard J B, Bennett R B, Krishna N K, Ernst S M, Rein A, Wills J W. Importance of basic residues in nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J Virol. 1998;72:9034–9044. doi: 10.1128/jvi.72.11.9034-9044.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cosson P. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 1996;15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 7.Craven R C, Leure-duPree A E, Weldon R A, Jr, Wills J W. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J Virol. 1995;69:4213–4227. doi: 10.1128/jvi.69.7.4213-4227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawson L, Yu X F. The role of nucleocapsid of HIV-1 in virus assembly. Virology. 1998;251:141–157. doi: 10.1006/viro.1998.9374. [DOI] [PubMed] [Google Scholar]
- 9.Dorfman T, Mammano F, Haseltine W A, Gottlinger H G. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1994;68:1689–1696. doi: 10.1128/jvi.68.3.1689-1696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubay J W, Roberts S J, Hahn B H, Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J Virol. 1992;66:6616–6625. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelman A, Englund G, Orenstein J M, Martin M A, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Franke E K, Yuan H E H, Bossolt K L, Goff S P, Luban J. Specificity and sequence requirements for interactions between various retroviral Gag proteins. J Virol. 1994;68:5300–5305. doi: 10.1128/jvi.68.8.5300-5305.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed E O, Martin M A. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamble T R, Yoo S, Vajdos F F, von Schwedler U K, Worthylake D K, Wang H, McCutcheon J P, Sundquist W I, Hill C P. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 14.Garcia J V, Miller A D. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature. 1991;350:508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- 15.Garnier L, Ratner L, Rovinski B, Cao S X, Wills J W. Particle size determinants in the human immunodeficiency virus type 1 Gag protein. J Virol. 1998;72:4667–4677. doi: 10.1128/jvi.72.6.4667-4677.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garnier L, Wills J W, Verderame M F, Sudol M. WW domains and retrovirus budding. Nature. 1996;381:744–745. doi: 10.1038/381744a0. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 17.Gottlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill C P, Worthylake D, Bancroft D P, Christensen A M, Sundquist W I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang M, Orenstein J M, Martin M A, Freed E O. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang M, Martin M A. Incorporation of Pr160gag-pol into virus particles requires the presence of both the major homology region and adjacent C-terminal capsid sequences within the Gag-Pol polyprotein. J Virol. 1997;71:4472–4478. doi: 10.1128/jvi.71.6.4472-4478.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang M, Zensen R, Cho M, Martin M A. Construction and characterization of a temperature-sensitive human immunodeficiency virus type 1 reverse transcriptase mutant. J Virol. 1998;72:2047–2054. doi: 10.1128/jvi.72.3.2047-2054.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondo E, Mammano F, Cohen E A, Gottlinger H G. The p6gag domain of human immunodeficiency virus type 1 is sufficient for the incorporation of Vpr into heterologous viral particles. J Virol. 1995;69:2759–2764. doi: 10.1128/jvi.69.5.2759-2764.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landau N R, Page K A, Littman D R. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J Virol. 1991;65:162–169. doi: 10.1128/jvi.65.1.162-169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Y L, Bennett R P, Wills J W, Gorelick R, Ratner L. A leucine triplet repeat sequence (LXX)4 in p6gag is important for Vpr incorporation into human immunodeficiency virus type 1 particles. J Virol. 1995;69:6873–6879. doi: 10.1128/jvi.69.11.6873-6879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mak J, Khorchid A, Cao Q, Huang Y, Lowy I, Parniak M A, Prasad V R, Wainberg M A, Kleiman L. Effects of mutations in Pr160gag-pol upon tRNA(Lys3) and Pr160gag-pol incorporation into HIV-1. J Mol Biol. 1997;265:419–431. doi: 10.1006/jmbi.1996.0742. [DOI] [PubMed] [Google Scholar]
- 27.Mammano F, Ohagen A, Hoglund S, Gottlinger H G. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J Virol. 1994;68:4927–4936. doi: 10.1128/jvi.68.8.4927-4936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morikawa Y, Zhang W, Hockley D J, Nermut M V, Jones I M. Detection of a trimeric human immunodeficiency virus type 1 Gag intermediate is dependent on sequences in the matrix protein, p17. J Virol. 1998;72:7659–7663. doi: 10.1128/jvi.72.9.7659-7663.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orlinsky K J, Gu J, Hoyt M, Sandmeyer S, Menees T M. Mutations in the Ty3 major homology region affect multiple steps in Ty3 retrotransposition. J Virol. 1996;70:3440–3448. doi: 10.1128/jvi.70.6.3440-3448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paxton W, Connor R I, Landau N R. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J Virol. 1993;67:7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puffer B A, Parent L J, Wills J W, Montelaro R C. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J Virol. 1997;71:6541–6546. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, et al. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 34.Sandefur S, Varthakavi V, Spearman P. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J Virol. 1998;72:2723–2732. doi: 10.1128/jvi.72.4.2723-2732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwartz M D, Geraghty R J, Panganiban A T. HIV-1 particle release mediated by Vpu is distinct from that mediated by p6. Virology. 1996;224:302–309. doi: 10.1006/viro.1996.0532. [DOI] [PubMed] [Google Scholar]
- 36.Srinivasakumar N, Hammarskjold M-L, Rekosh D. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J Virol. 1995;69:6106–6114. doi: 10.1128/jvi.69.10.6106-6114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strambio-de-Castillia C, Hunter E. Mutational analysis of the major homology region of Mason-Pfizer monkey virus by use of saturation mutagenesis. J Virol. 1992;66:7021–7032. doi: 10.1128/jvi.66.12.7021-7032.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swanstrom R, Wills J. Synthesis, assembly, and processing of viral proteins. In: Coffin J, Hughes S, Varmus H, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1998. pp. 263–334. [PubMed] [Google Scholar]
- 39.Yasuda J, Hunter E. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J Virol. 1998;72:4095–4103. doi: 10.1128/jvi.72.5.4095-4103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu X, Yuan X, Matsuda Z, Lee T H, Essex M. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol. 1992;66:4966–4971. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu X, Yuan X, McLane M F, Lee T H, Essex M. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of Env proteins into mature virions. J Virol. 1993;67:213–221. doi: 10.1128/jvi.67.1.213-221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu X F, Dawson L, Tian C J, Flexner C, Dettenhofer M. Mutations of the human immunodeficiency virus type 1 p6Gag domain result in reduced retention of Pol proteins during virus assembly. J Virol. 1998;72:3412–3417. doi: 10.1128/jvi.72.4.3412-3417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu X F, Matsuda Z, Yu Q C, Lee T H, Essex M. Role of the C terminus Gag protein in human immunodeficiency virus type 1 virion assembly and maturation. J Gen Virol. 1995;76:3171–3179. doi: 10.1099/0022-1317-76-12-3171. [DOI] [PubMed] [Google Scholar]
- 44.Yuan X, Yu X, Lee T H, Essex M. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J Virol. 1993;67:6387–6394. doi: 10.1128/jvi.67.11.6387-6394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Qian H, Love Z, Barklis E. Analysis of the assembly function of the human immunodeficiency virus type 1 gag protein nucleocapsid domain. J Virol. 1998;72:1782–1789. doi: 10.1128/jvi.72.3.1782-1789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]