Abstract
Radiomics analysis of [18F]-fluorodeoxyglucose ([18F]-FDG) PET images could be leveraged for personalised cancer medicine. However, the inherent sensitivity of radiomic features to intensity discretisation and voxel interpolation complicates its clinical translation. In this work, we evaluated the robustness of tumour [18F]-FDG-PET radiomic features to 174 different variations in intensity resolution or voxel size, and determined whether implementing parameter range conditions or dependency corrections could improve their robustness. Using 485 patient images spanning three cancer types: non-small cell lung cancer (NSCLC), melanoma, and lymphoma, we observed features were more sensitive to intensity discretisation than voxel interpolation, especially texture features. In most of our investigations, the majority of non-robust features could be made robust by applying parameter range conditions. Correctable features, which were generally fewer than conditionally robust, showed systematic dependence on bin configuration or voxel size that could be minimised by applying corrections based on simple mathematical equations. Melanoma images exhibited limited robustness and correctability relative to NSCLC and lymphoma. Our study provides an in-depth characterisation of the sensitivity of [18F]-FDG-PET features to image processing variations and reinforces the need for careful selection of imaging biomarkers prior to any clinical application.
Subject terms: Cancer imaging, Computational science, Biomarkers, Molecular imaging
Introduction
Radiological images harbour clinically imperceptible information about disease, which can be revealed through radiomics, the high-throughput extraction of quantitative features from these images1. Radiomics research involving positron emission tomography (PET), in particular with fluorine-18-labelled fluorodeoxyglucose ([18F]-FDG) as the imaging agent, is rapidly growing2 and holds promise in the prediction of disease progression3, metastasis4, survival5,6, and treatment response7—for a wide variety of cancer patients2. However, radiomic features can be sensitive to variations in imaging protocol and processing pipeline within and across institutions, ultimately challenging its translation into the clinic8,9.
The lack of reproducibility underscores the importance of feature robustness testing to develop reliable imaging biomarkers. Various parameters can affect radiomic measurements, including intensity resolution and voxel size, which are typically modified through discretisation and interpolation, respectively, to serve different purposes: noise reduction10, data compression11, image fusion with other modalities12,13, and facilitating multi-centre comparisons14. Several reports have interrogated the robustness of [18F]-FDG-PET radiomic features as a function of these two image processing parameters10,15–18. However, many of these investigations fall short of testing strategies to mitigate feature sensitivity to variations in these parameters, which would help enhance our understanding on whether datasets can be corrected and pooled for quantitative analyses19.
Non-robustness of features can be addressed via image-based solutions, which are applied directly to the images before the radiomics extraction process, or feature-based methods, applied to the downstream extracted feature values20. Previous reports contend that feature dependencies on voxel size or intensity bin configuration can be minimised either by modifying heterogeneously acquired images to utilise a fixed value for these parameters16,21,22, or through correction of radiomic features using pre-defined equations that account for these dependencies21–24. A correction factor based on the percentage change of feature values between interpolated and reconstructed PET images has also been suggested17.
However, improving feature robustness by introducing a shared imaging parameter across datasets poses the question of what parameter value(s) would be suitable for different sets of features, images, or investigations. There are currently no guidelines to help choose optimal intensity bin configurations and voxel sizes, and a comprehensive characterisation of the behaviour of radiomic features against these two parameters is necessary to help establish this25. Second, knowledge of feature definitions is usually required to reap suitable equations for correction, which is challenging as definitions vary between radiomics extraction software platforms26. While work to standardise feature definitions by the Imaging Biomarker Standardisation Initiative (IBSI) is ongoing27, a recent study contests that these efforts still do not guarantee feature reproducibility between softwares28. Third, use of reconstructed voxel size as the ground truth17 cannot be easily generalised to other images where the reconstructed voxel sizes may be different. Thus, a correction technique that is generalisable across platforms and does not require any form of ground truth is desirable.
The objectives of this work were four-fold: (i) to characterise the robustness of [18F]-FDG-PET radiomic features against varying intensity discretisation and voxel interpolation parameters; (ii) to investigate whether feature robustness could be improved by a novel solution based on imposing range conditions on these parameters; (iii) to test a software-agnostic feature correction method that minimises the sensitivity of radiomic features to variations of these parameters; and (iv) to explore feature robustness across different cancer types, i.e., non-small cell lung cancer (NSCLC), lymphoma, and melanoma.
Materials and methods
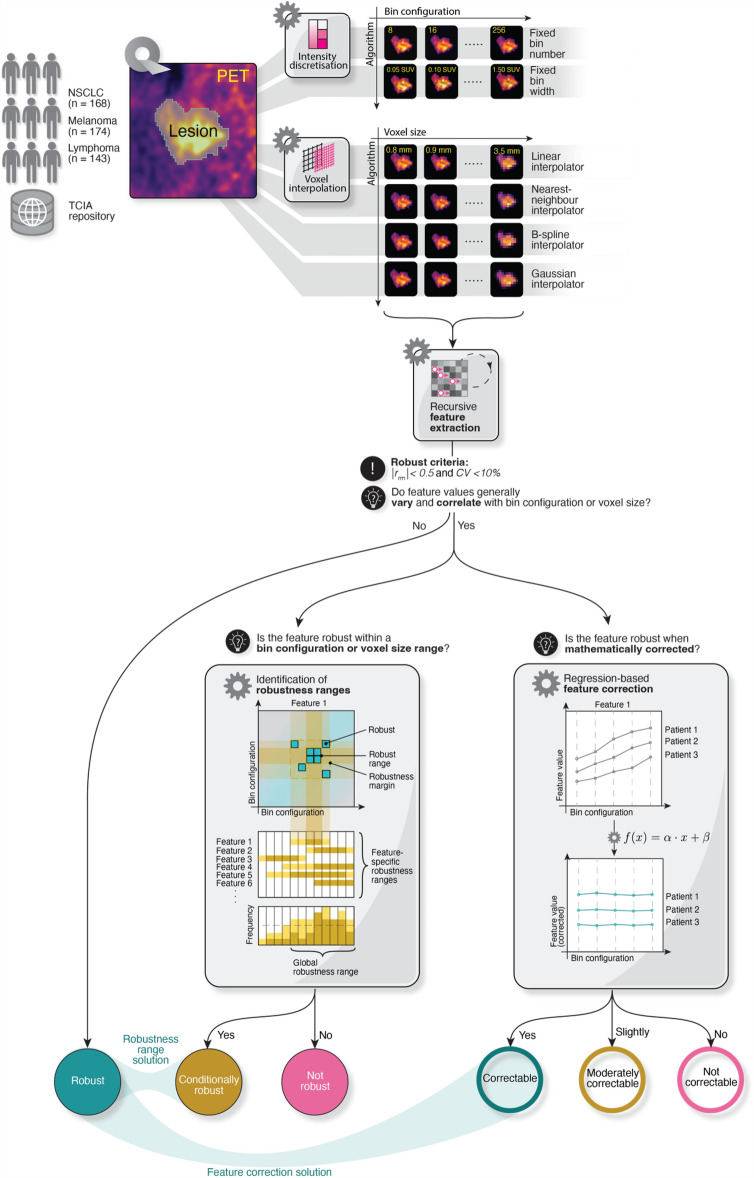
A graphic summary of the study design is illustrated in Fig. 1.
Figure 1.
Schematic representation of the study workflow.
Dataset
Nine hundred anonymised whole-body [18F]-FDG PET/CT images together with manually segmented tumour volumes were obtained from a highly curated imaging dataset publicly available at The Cancer Imaging Archive (TCIA)29–31. Patients without annotated tumour volumes, or with volumes too small (< 15 voxels) to extract valid radiomic features or too large (> 200,000 voxels) to overcome computational memory limitations were excluded. We considered one examination per patient and treated each patient segmentation mask as a single volume. After exclusions, a total of 485 patients with [18F]-FDG-avid lesions were included in this study, which spanned three cancer types: NSCLC (n = 168), melanoma (n = 174), and lymphoma (n = 143). The mean (range) of analysed tumour voxels was 21,189 (58–178,944), 8926 (31–128,920), and 24,238 (98–199,437), respectively. Corresponding tumour volumes were 264.5 (0.72–2234.1) cm3, 111.4 (0.44–1609.5) cm3, and 302.6 (1.22–2489.9) cm3.
Acquisition and reconstruction protocols for this dataset have previously been described30. In brief, images were acquired on a Biograph mCT scanner (Siemens Healthineers) at 60 min post [18F]-FDG injection, and reconstructed into a 400 × 400 matrix with 2.04 × 2.04 × 3 mm3 voxels, using ordered-subset expectation maximisation (21 subsets, 2 iterations), and a 2-mm FWHM Gaussian filter post-reconstruction. As a pre-processing step, PET images were converted from Digital Imaging and Communications in Medicine (DICOM) to Neuroimaging Informatics Technology Initiative (NIfTI) format, and subsequently normalised for injected activity and body weight to generate standardised uptake value (SUV) maps using scripts provided with the dataset29.
Image processing
Radiomic features were tested for robustness against parameter variations in two image processing schemes: intensity discretisation and voxel interpolation. Intensity discretisation was studied either by fixing bin widths (FBW) or fixing bin numbers (FBN). FBW discretisation utilised intensity bin widths ranging from 0.05 to 1.50 SUV at 0.05 SUV step intervals, while the number of bins for FBN discretisation ranged from 8 to 256 bins at 8-bin step intervals—thus, yielding 30 and 32 parameters for each discretisation scheme, respectively. All other image parameters were kept at the original values.
The effect of voxel interpolation on radiomic features was tested by employing four common interpolators: linear, nearest-neighbour, B-spline, and Gaussian. Voxels were resampled to isotropic sizes ranging from 0.8 to 3.5 mm at step intervals of 0.1 mm, yielding 28 values to test for each interpolation method. Image intensities were discretised at 64 bins, as commonly used in the literature, while all other parameters were kept at the original values.
In total, 174 image processing parameters were considered in this paper.
Recursive feature extractions
Using PyRadiomics32, version 3.0.1, a total of 107 IBSI-compliant radiomic features from the following families were extracted: shape-based (n = 14), first-order statistics (n = 18), grey-level co-occurrence matrix (GLCM) (n = 24), grey-level dependence matrix (GLDM) (n = 14), grey-level run-length matrix (GLRLM) (n = 16), grey-level size zone matrix (GLSZM) (n = 16), and neighbouring grey-tone dependence matrix (NGTDM) (n = 5). GLCM and GLRLM features were calculated by computing the corresponding matrices over 13 spatial directions in 3D (26-connectivity), with a single voxel offset for the former, after which the values were averaged to make them rotationally invariant. Feature families pre-processed with mathematical filters (higher-order features) were not evaluated. Following a recursive extraction of radiomic features from each of the 485 patient images across the 174 image processing parameters, a total of 9,029,730 feature values were obtained and analysed. Extractions were performed on Python, version 3.10.8 (Python Software Foundation, https://www.python.org).
Assessment of robustness of features
Robustness of radiomic features was assessed in terms of both the agreement and reliability of measurements33,34. The agreement of feature values against parameter variations (i.e., feature variability) was measured using the within-subject percentage coefficient of variation, which was subsequently averaged across patients to generate a mean value (CVmean) per feature. Feature reliability was assessed by means of feature correlation, using the magnitude of the repeated measures correlation coefficient (|rrm|). This correlation coefficient was chosen over other measures (e.g., Pearson, Spearman) as it addresses the non-independence of feature values across parameters35. Empirical thresholds of CVmean < 10% and |rrm|< 0.5 were used to define the robust criteria22,23,36–38 and, in turn, determine robust features.
Derivation of feature-specific robustness ranges
For features not meeting the robust criteria, CVmean and |rrm| were systematically evaluated for all possible parameter ranges. For each interpolator and a range of voxel sizes between 0.8 and 3.5 mm (i.e., 28 voxel sizes), (28C2 =) 378 distinct ranges could be interrogated for conditional robustness (e.g., 0.8 to 0.9 mm, 0.8 to 1.0 mm, etc.). This number was (30C2 =) 435 and (32C2 =) 496 for FBW and FBN discretisation, respectively. Parameter ranges were represented as tiles on a square correlogram or matrix (Fig. 1), and a robustness matrix was generated for each feature under different scenarios (image processing algorithm, cancer type). Excluding those on the diagonal, robust tiles, i.e., matrix elements for which the robust criteria were met (CVmean < 10% and |rrm|< 0.5), defined a range for which a feature was deemed conditionally robust. Features for which no robust tiles could be identified were categorised as non-robust.
To determine range conditions from the robustness matrix for conditionally robust features, two nested square regions, symmetrical about the diagonal, were delineated on the matrix, defining the robust range and robustness margin, respectively. The robust range, representing the limits of the largest closed square region containing a contiguous cluster of robust tiles, was defined via run length encoding in the horizontal and vertical directions. The contiguity constraint for the robust range was imposed to exclude tiles with spurious CVmean or |rrm| values. The robustness margin was determined by creating a non-contiguous square region using the two outermost parameter values where at least one robust tile exists, excluding tiles on the diagonal. The robust range and robustness margin, together, make up the robustness range for each feature.
Derivation of global robustness ranges
Given that a typical radiomics study would use the same discretisation and interpolation settings for the collective extraction of radiomic features, a “global” parameter range was determined by aggregating robustness ranges of conditionally robust features into a histogram, and applying a cut-off recovering at least half of these features per scenario.
Identification of correctable features
As an alternative to imposing range conditions, we tested radiomic feature correction as a potential strategy to improve the robustness of features initially not meeting the robust criteria. The relationship between feature values and bin configuration or interpolated voxel size was modelled using eight regression functions, f(x), as first utilised in23: f(x) = α · x + β, f(x) = α · x2 + β, f(x) = α · x3 + β, f(x) = α/x + β, f(x) = α/x2 + β, f(x) = α/x3 + β, f(x) = α · log(x) + β, and f(x) = α/log(x) + β where α and β are fit parameters respectively, and x is the parameter under investigation (i.e., bin configuration or voxel size).
Functions were fitted using robust regression with an iteratively reweighted least squares algorithm, applying intrinsic weights in the form of the reciprocal of feature variance values computed across patients. The function achieving the lowest value for the Akaike information criterion (AIC) amongst the ones tested was selected as best describing the dependencies of feature values on bin configuration or voxel size. Feature corrections for each patient were implemented by using a rearranged form of the best-fitting function23, e.g., f(x) = α · log(x) + β, fcorrected(x) = (f(x) − β)/log(x). When computing regressions for logarithmic functions, bin widths and voxel sizes were multiplied by 100 and 10, respectively, to circumvent division by zero errors when the x-variable equalled 1.
Wilcoxon signed-rank test was employed to compare CV values before and after correction, whereas the overlap between the 95% confidence intervals (CI) of the original and corrected |rrm| was examined. Corrections were deemed statistically significant when the p-value was less than 0.05 for the Wilcoxon signed-rank test, and the 95% CI of the |rrm| between original and corrected did not overlap. Features with a statistically significant reduction in CVmean and |rrm| upon correction were segregated into two categories: correctable, if the robust criteria (CVmean, corrected < 10 and |rrm, corrected|< 0.5) were satisfied; and moderately correctable if these criteria were not met. Features were otherwise classified as not correctable.
Statistical analysis
The effect of discretisation scheme, interpolator, or cancer type on feature robustness or correctability was investigated using mixed-effects logistic regression with per-feature random intercepts, and results were reported as odds ratios (OR) with 95% CI, where appropriate. p-values from post-hoc analyses were adjusted for multiple comparisons using the Bonferroni method. Statistical significance was defined as p < 0.05. All statistical analyses were performed using R, version 4.2.2 (R Foundation for Statistical Computing, Vienna, Austria) (https://www.R-project.org/) or jamovi version 2.3 (https://www.jamovi.org)39.
Results
Robust features
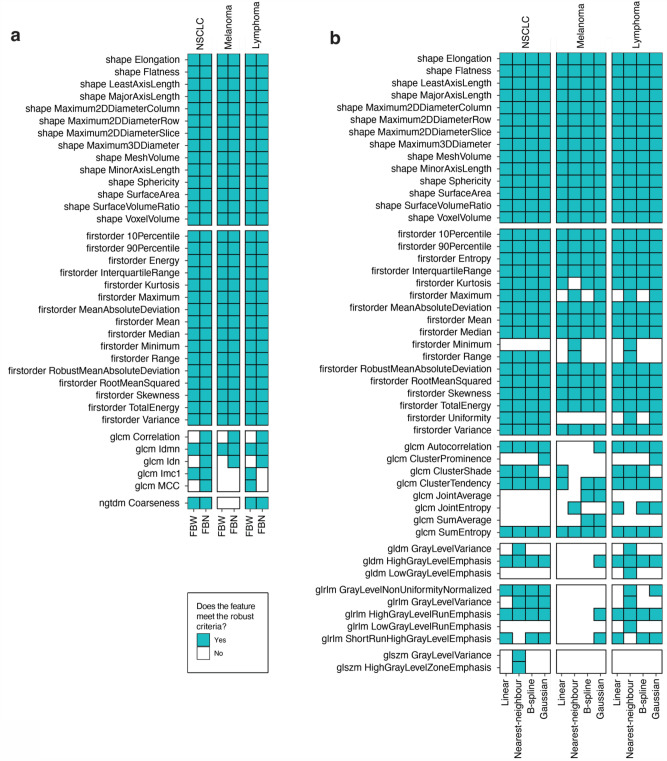
Figure 2 illustrates [18F]-FDG-PET radiomic features that meet the robust criteria (CVmean < 10 and |rrm|< 0.5), stratified by image processing scheme, algorithm, and cancer type.
Figure 2.
[18F]-FDG-PET radiomic features robust to intensity discretisation (a) or voxel interpolation (b) stratified by image processing algorithm used and cancer type.
Overall, in mixed effects logistic regression models including cancer type and image processing scheme as fixed effects with per-feature random intercepts, radiomic features were more sensitive to intensity discretisation than voxel interpolation (OR 0.57 [0.36, 0.29], p = 0.01), as also evidenced in Fig. 2. Specifically, a median 34/107 (32%) and 36/107 (34%) features were classified as robust following discretisation and interpolation, respectively (Supplementary Table ST1).
Between families, shape and first-order features were more robust than second-order ones (p < 0.001) across discretisation and interpolation schemes. Shape features were unaffected by the image processing schemes considered irrespective of cancer type. Entropy and Uniformity for discretisation, and Energy for interpolation, were the only first-order features not meeting the robust criteria in all tested instances. When considering second-order families, 5/24 (21%) GLCM, 1/5 (20%) NGTDM features, and none from the remaining families satisfied the robust criteria in at least one scenario of discretisation. These proportions were generally larger for interpolation with 8/24 (33%) GLCM, 3/14 (21%) GLDM, 5/16 (31%) GLRLM, 2/16 (13%) GLSZM, and no NGTDM features identified to be robust. Of these second-order features, only GLCM idmn (inverse difference moment normalised) or GLCM SumEntropy were robust across all tested instances of discretisation or interpolation, respectively.
FBW discretisation produced less robust features than FBN (OR 0.24 [0.05, 1.20]). However, the effect of the discretisation algorithm on robustness was not statistically significant (p = 0.08). Similarly, differences in robust categorisations between cancer types were not significant (p = 0.1), with melanoma producing the least number of robust features regardless of discretisation algorithm used (Supplementary Table ST1). The largest proportion of robust features was observed for FBN discretisation in NSCLC (36/107; 34%), and the smallest for FBW discretisation in melanoma (31/107; 29%).
In contrast, the choice of interpolator significantly affected feature robustness (p = 0.03). Use of nearest-neighbour interpolation was more likely to result in robust features than linear (OR 3.24 [1.30, 8.22]), B-spline (OR 2.59 [1.03, 6.49]), or Gaussian (OR 1.11 [0.46, 2.70]). However, upon Bonferroni correction for multiple comparisons, these differences were not statistically significant (nearest-neighbour vs. linear: p = 0.08; vs. B-spline: p = 0.25; vs. Gaussian: p = 1). Robust categorisations were significantly different between cancer types (p < 0.001), with melanoma showing greater sensitivity to interpolation than lymphoma (OR 0.16 [0.07, 0.39]) or NSCLC (OR 0.1 [0.04, 0.24]). Use of nearest-neighbour interpolation in lymphoma and linear interpolation in melanoma yielded the highest (42/107; 39%) and lowest number of robust features (30/107; 28%), respectively.
Conditionally robust features
For those features initially not fulfilling the robust criteria, we tested the possibility of making them more robust by imposing parameter range conditions. CVmean and |rrm| were evaluated across all intensity resolutions or voxel sizes considered, in a pairwise manner, and findings were compiled into a square matrix, which were subsequently used to compute the robustness ranges for each feature.
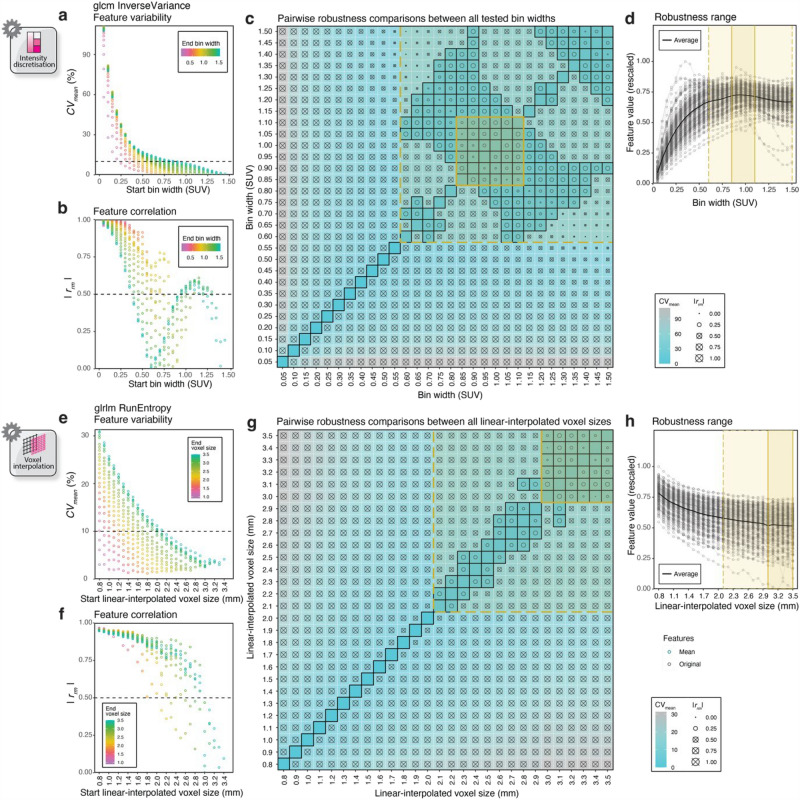
Examples of two conditionally robust features from NSCLC cases are given in Fig. 3, which examine the response to FBW discretisation for GLCM InverseVariance (Fig. 3a–d) and linear interpolation for GLRLM RunEntropy (Fig. 3e–h). The process described in the examples was repeated to identify conditionally robust features for all discretisation schemes and interpolators across cancer types. Where no robustness range could be identified, features were classified as non-robust. Scatter and correlogram plots for both conditionally robust and non-robust features have been made publicly available in https://github.com/syafiqramlee/robust-radiomics-img-processing. For each of the 107 features, robustness categorisations throughout the scenarios considered in this study are presented in Supplementary Fig. SF1, with their corresponding proportions tabulated in Supplementary Table ST1.
Figure 3.
Scatter plots (left) and square correlograms (centre) for determining the robustness range conditions for GLCM InverseVariance as a function of bin width (a–d), and GLRLM RunEntropy as a function of linear-interpolated voxel size (e–h), for the NSCLC cases, as examples. In the scatter plots, CVmean and |rrm| varied as a function of bin width (a,b) or linear-interpolated voxel size (e,f), and were dependent on the difference between the start and end bin width or linear-interpolated voxel size. By applying thresholds on these robustness metrics, start and end bin width or voxel size combinations satisfying the robust criteria were selected (represented as tiles with a black border on the square correlograms) as seen in (c) and (g). Two nested square regions that correspond to a contiguous and non-contiguous cluster of robust tiles were delineated on each correlogram, the limits of which indicate the robust range and robustness margin (outlined in yellow), respectively. Care was taken to exclude tiles on the diagonal, denoting combinations of either bin width or voxel size values with itself. The robustness ranges, wherein feature values were relatively stable (robustness margin) or stable (robust region), were overlaid onto plots showing feature response to bin width (d) or voxel size (h) variations.
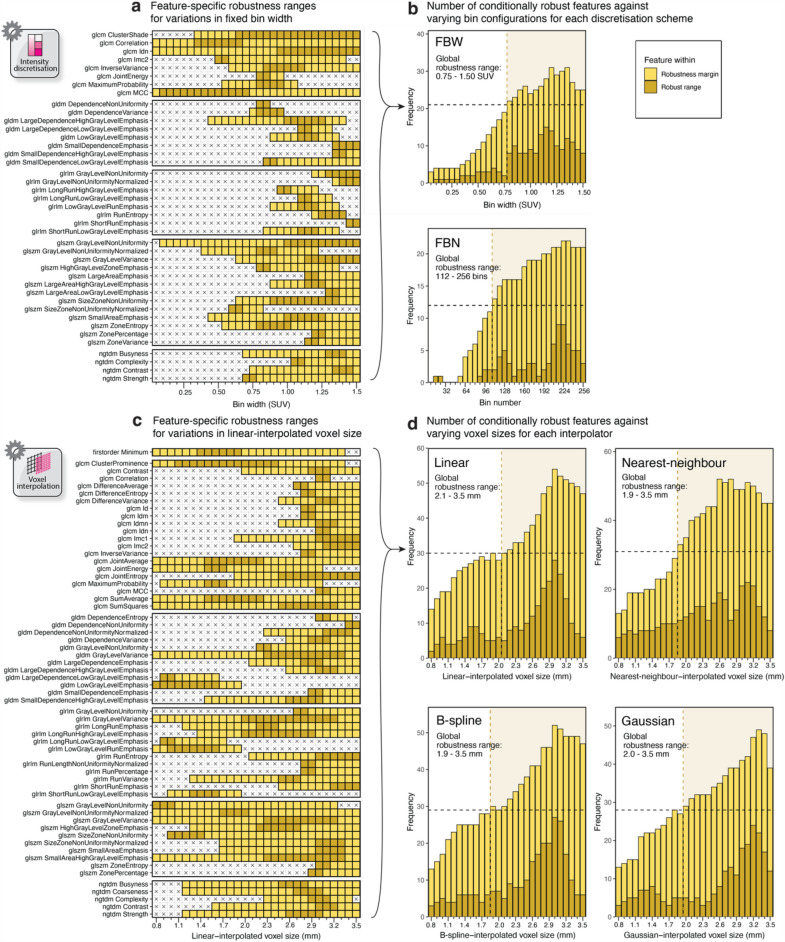
Figure 4 provides a summary of the ensemble of [18F]-FDG-PET radiomic features determined to be conditionally robust in NSCLC cases, when FBW discretisation or linear interpolation was used. Summary figures for other methods and cancer types are given in Supplementary Fig. SF2–4.
Figure 4.
Feature-specific robustness ranges when applying FBW discretisation (a), and histograms showing the number of conditionally robust features across different bin configurations for FBW and FBN discretisation (b), when NSCLC cases were analysed. Similarly, feature-specific robustness ranges for linear interpolation (c), and histograms showing the distribution of conditionally robust features against interpolated voxel size for the four interpolators: linear, nearest-neighbour, B-spline, and Gaussian (d). A histogram cut-off recovering at least half of the conditionally robust features (indicated by the horizontal black dashed line) was applied, generating the corresponding global robustness ranges as indicated by the vertical yellow dashed line and annotated in the histograms (b,d).
For NSCLC, 41/107 (38%) radiomic features were found to be robust within certain intensity resolutions introduced by FBW discretisation (Fig. 4a). These comprised 8/24 (33%) GLCM, 8/14 (57%) GLDM, 8/16 (50%) GLRLM, 13/16 (81%), GLSZM, and 4/5 NGTDM features. Across families, the robustness ranges exhibited a tendency to encompass larger bin widths, as observed in the frequency histogram in Fig. 4b (top row). A cut-off recovering at least half of the conditionally robust radiomic features (≥ 21 and ≥ 12 features for FBW and FBN, respectively) from the histogram yielded a global robustness range between 0.75 and 1.50 SUV for FBW, and 112 and 256 bins for FBN discretisation (Fig. 4b). Similarly, for the linear interpolation example in Fig. 4c, 60/107 (56%) radiomic features were robust for certain voxel sizes, with more than half (≥ 30) of these features being robust between 2.1 and 3.5 mm (Fig. 4d; top-left). Figure 4d shows the histogram analyses for other interpolators.
The histogram analyses of all discretisation or interpolation schemes across the cancer types considered in this work are provided in Supplementary Fig. SF2–3, and a summary of the resulting global robustness ranges is tabulated in Table 1. In general, there was a substantial overlap in robustness range between cancer types for both discretisation and interpolation, with melanoma exhibiting the widest ranges for the latter. Across interpolators, global robustness ranges tended to encompass larger voxel sizes, with the number of robust features peaking close to the slice thickness of the original voxels (3 mm).
Table 1.
Global robustness ranges stratified by discretisation or interpolation algorithm and cancer type. The bin configuration(s) or voxel size(s) achieving the highest number of features within the robust or robustness range are given in parentheses.
| NSCLC | Melanoma | Lymphoma | |
|---|---|---|---|
| Discretisation | |||
| Fixed bin width (SUV) | 0.75–1.50 (1.15|1.20 or 1.35) | 0.50–1.50 (1.05|1.05) | 0.50–1.50 (1.05|1.05) |
| Fixed bin number (bins) | 112–256 (216 or 224|224 or 232) | 112–256 (224|216, 224, or 240) | 112–256 (248|248) |
| Interpolation | |||
| Linear (mm) | 2.1–3.5 (3.0|3.0) | 1.5–3.5 (3.0|2.8) | 1.8–3.5 (2.4 or 3.0|3.0) |
| Nearest-neighbour (mm) | 1.9–3.5 (3.1|2.6 or 2.8) | 1.3–3.5 (2.2|3.0) | 1.9–3.5 (2.2, 3.0, or 3.2|3.0 or 3.1) |
| B-spline (mm) | 1.9–3.5 (3.0|3.0) | 1.5–3.5 (3.1|2.9 or 3.0) | 2.0–3.5 (3.0|2.7, 3.0 or 3.1) |
| Gaussian (mm) | 2.0–3.5 (3.2|3.3) | 1.5–3.5 (3.1 or 3.2|2.9, 3.0, or 3.1) | 2.2–3.5 (3.2|3.2) |
Correctable features
We also explored the feasibility of parameter dependency correction as an alternative strategy to improve the robustness of features initially not meeting the robust criteria.
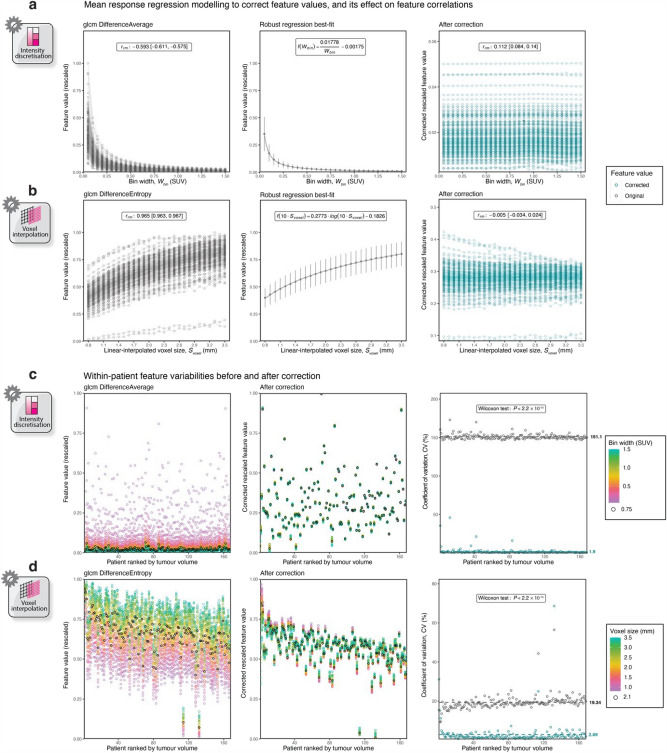
As examples, Fig. 5a,b provide the response to variations in FBW-discretised intensity resolution for GLCM DifferenceAverage and linear-interpolated voxel size for GLCM DifferenceEntropy, respectively. In these instances, the effect of bin width could be modelled by an inverse function, whereas a logarithmic function was used to model variations in voxel size. These dependencies were correctable by applying the rearranged form of their respective best-fit models, leading to a significant reduction in feature correlation after correction (i.e., |rrm|< 0.5, as annotated in the figures on the right).
Figure 5.
Feature values for GLCM DifferenceAverage and DifferenceEntropy against bin width (a) or linear-interpolated voxel size (b), respectively, tracked for every NSCLC patient. Here, original (uncorrected) values are plotted on the left; the best-fit model describing the relationship of the mean feature values as a function of bin width or linear-interpolation voxel size is presented in the centre; and feature values corrected using the inverse function of the best fit are given on the right. Uncorrected and corrected values for the same features are plotted from lowest to highest patient tumour volume, with coloured dots indicating variations in bin width (c) or linear-interpolated voxel size (d) used during feature extraction. The corresponding per-patient coefficient of variations before and after correction are provided on the right. All feature values have been rescaled using min–max normalisation.
Figure 5c,d (left) show original feature values for GLCM DifferenceAverage and DifferenceEntropy, respectively, for each patient ranked by tumour volume. Following correction (Fig. 5c,d; middle), we observed a statistically significant reduction in feature variability per patient, as evidenced by a decrease in the dispersion of coloured points in the figures. Mean feature variability for the two example cases was < 10%, satisfying the robust criterion for CVmean. As such, these features were deemed correctable.
The original and corrected scatter plots for all features with respect to image processing method and cancer type are deposited in https://github.com/syafiqramlee/robust-radiomics-img-processing. A snapshot of the correctability categorisations has been presented in Supplementary Fig. SF5, with the proportions tabulated in Supplementary Table ST2. For correctable features, the best-fit invertible function utilised for correction in each scenario is provided in Supplementary Fig. SF6.
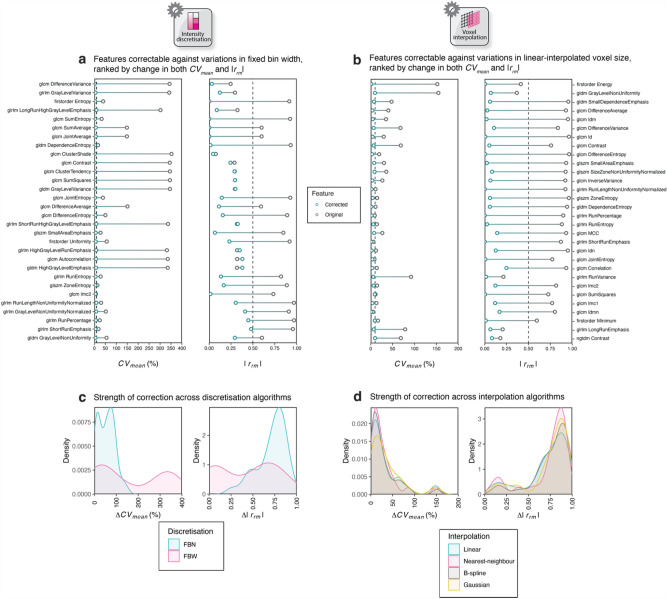
Figure 6a,b summarise all the features that were correctable in FBW discretisation and linear interpolation scenarios for the NSCLC cohort, ranked by relative change in CVmean (ΔCV) and |rrm| (Δ|rrm|). FBN discretisation resulted in a homogeneous reduction in both feature variability and correlation, with a greater reduction in the latter, than FBW (Fig. 6c). The strength of correction was similar across interpolation methods (Fig. 6d). Summary figures for other methods and cancer types can be found in Supplementary Fig. SF7–10, with the most correctable features presented in Supplementary Table ST3.
Figure 6.
Dumbbell plots showing the change in CVmean and |rrm| for correctable features when considering FBW discretisation (a) and linear interpolation (b) in NSCLC. Features are ranked by the change in feature variability and correlation upon correction. Density plots quantifying the overall effect of correction on feature variability and correlation for correctable features with respect to discretisation (c) and interpolation (d) algorithms.
Feature categorisations across methods and cancer types
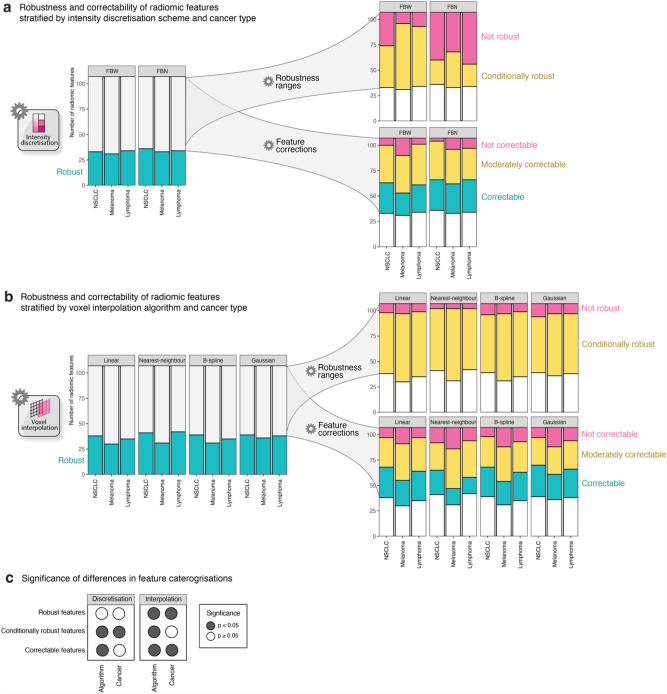
Figure 7 aggregates robustness and correctability categorisations of [18F]-FDG-PET radiomic features stratified by image processing method and cancer type.
Figure 7.
[18F]-FDG-PET radiomic feature robustness and correctability categorisations stratified by algorithm and cancer type for intensity discretisation (a) and voxel interpolation (b), and their corresponding significance of differences (c).
FBW discretisation tended to produce more conditionally robust radiomic features than FBN (p < 0.001), especially for melanoma (p < 0.001), lymphoma (p < 0.001), and less so for NSCLC (p = 0.05) (Fig. 7a). The effect of cancer type on conditional feature robustness for both discretisation methods was overall significant (p < 0.001), with melanoma exhibiting the highest number of conditionally robust features (Supplementary Table ST1). Nearest-neighbour interpolation produced more conditionally robust features than linear (p = 0.015), Gaussian (p < 0.001), and B-spline (p < 0.004), after correction for multiple comparisons. The impact of cancer type on conditional feature robustness for interpolation was not statistically significant (p = 0.3), with melanoma resulting in the highest number of conditionally robust features (Supplementary Table ST1).
Feature correctability significantly depended on the discretisation algorithm (p = 0.045), with FBN resulting in more correctable features than FBW discretisation (Supplementary Table ST2). While melanoma yielded the fewest correctable features for both discretisation schemes (Supplementary Table ST2), the effect of cancer type on correctability was not significant (p = 0.09). In contrast, feature correctability was impacted by both interpolation algorithm (p < 0.001) and cancer type (p < 0.001), with nearest-neighbour interpolation or melanoma resulting in the least number of correctable features (Supplementary Table ST2).
Discussion
Radiomic features may convey clinically meaningful information about tumours but can be susceptible to variations in factors pertaining to image acquisition, reconstruction, and processing19,40. In this work, we performed an in-depth characterisation of the robustness of [18F]-FDG-PET radiomic features to variations in bin configuration or voxel size introduced by discretisation and interpolation, respectively, across a cohort of three cancer types (NSCLC, melanoma, and lymphoma). Features not meeting our robust criteria were analysed to ascertain if their robustness could be improved by imposing parameter range conditions or dependency corrections. To our knowledge, our study is the first to collectively evaluate the sensitivity, robustness ranges, and mathematical correctability of tumour [18F]-FDG-PET radiomic features against variations in bin configuration or voxel size.
Impact of intensity discretisation and voxel interpolation on [18F]-FDG-PET radiomic feature robustness
We showed that radiomic features were significantly more sensitive to intensity discretisation than voxel interpolation, with second-order features manifesting the least stability to variations in intensity bin configuration or voxel size. These observations concur with prior reports9,16,41, as these textural features take into account the spatial relationship between voxel intensities. This is further exemplified by our findings that GLCM idmn and GLCM SumEntropy were the only second-order features found to be robust across all our discretisation and interpolation investigations, respectively. Such features were also highlighted as robust in other studies10,17,42, potentiating their utility as suitable radiomic candidates for reliable comparison between images for which different bin configurations or voxel sizes are used.
When comparing intensity discretisation algorithms used in radiomics studies, we found feature values displayed a higher tolerance to changes in bin width than bin number, as reflected by the greater number of conditionally robust features for the former. This agrees with existing studies supporting the use of FBW discretisation in radiomic interrogations10,27,43–45, given this scheme preserves the relationship between SUV units and discretised intensity levels46. However, it should be noted that FBW discretisation may introduce variations in the number of bins between tumour volumes, and should not be advocated in all use-cases as features may become non-informative or incomparable where significant inter-lesion heterogeneity is observed23,43. To this end, some authors have found the use of FBN discretisation to be advantageous as it shows little dependence on the SUV, reconstruction parameters, and noise47.
The choice of interpolation algorithm on [18F]-FDG-PET radiomics analyses should be carefully considered as we observed a significant dependency of feature robustness on the type of interpolator. Images resampled using nearest-neighbour interpolation generated the greatest number of robust features than other interpolators considered in this work (i.e., linear, B-spline, and Gaussian). This observation is concordant with an earlier study48, and was also found when we compared the conditional robustness of features. A plausible explanation for the high tolerance of radiomic features to variable voxel sizes when considering nearest-neighbour interpolation could be that such an interpolation method preserves the intensity spectrum of the original data49.
Mitigating the impact of image processing variations on radiomic features using robustness ranges
Given that features not satisfying the robust criteria may exhibit varying sensitivity across the image processing parameters tested, we hypothesised that these sensitivities could be attenuated by restricting radiomic interrogations to within certain parameter ranges. This hypothesis was confirmed for most of our investigations, where the majority of these radiomic features could be recovered by applying parameter range conditions for bin configuration or voxel size.
Robustness range conditions were derived on a per-feature or all-features (global) basis for each algorithm and cancer type. Our results indicated robustness ranges for the former varied considerably, even in the same feature family, suggesting that the feature-specific response to discretisation or interpolation is an important aspect to consider when selecting features for radiomics analyses50,51. Nevertheless, we envisage that global robustness ranges could be helpful in choosing an optimal bin configuration or interpolated voxel size, which are usually required as input parameters for radiomic feature computations.
Global robustness ranges for the discretisation methods tested (FBW: 0.5 to 1.50 SUV; FBN: 112 to 256 bins) both favoured the use of larger values for improved feature robustness, with the upper limits being the largest values tested. This indicates that although the width and number of bins are inversely related properties, their overall impact on robustness across tumours may not always be inversely related. For FBW discretisation, this may be explained by a reduction in image noise, increased clustering of similar intensity levels or, more severely, potential loss of information as larger bin widths are used23,43. Therefore, to preserve meaningful comparisons while still recovering the overall robustness of sensitive features, smaller bin widths in the global robustness range should take precedence (e.g., 0.5 SUV). On the other hand, the improvement of feature robustness as we increase the number of bins in FBN discretisation could be attributed to the large voxel sizes commonly found in PET images, where the effect of adding bins on the appearance of tumour volumes may be constrained by the limited number of voxel intensities assessed.
Across interpolators, global robustness ranges for voxel size were markedly similar, and widest for nearest-neighbour interpolation in melanoma (1.3–3.5 mm). Analysed images had an original voxel dimension of 2.03 × 2.03 × 3 mm3, which we resampled into a range of isotropic voxel sizes to ensure rotationally invariant feature values, as recommended by IBSI26. We observed that the number of features robust within certain voxel sizes peaked when voxels were interpolated to an isotropic dimension equal, or close to, the slice thickness. This may highlight the critical role of the largest voxel dimension on the interpolated value of voxels and robustness of features, and echoes previous findings in a magnetic resonance imaging study52.
Improving feature robustness to image processing variations using feature correction
Aside from determining robustness ranges, which may be circumscribed by the fact that only a subset of bin configurations or voxel sizes could be used, we explored feature correction as an alternative strategy to achieve robustness. To this end, the underlying dependency of [18F]-FDG-PET radiomic features non-robust to variations in bin configuration and voxel sizes was modelled and corrected using a library of eight invertible functions, as utilised in previous work23. This approach has been shown to be software-agnostic and feature definition non-specific23.
While we found that not all features were correctable to discretisation and interpolation, approximately a third of second-order non-robust features could be corrected. As prime examples, Entropy for discretisation and Energy for interpolation could not meet the robust criteria in all tested instances but were ranked amongst the most correctable using basic mathematical functions in our results. In a conventional radiomics modelling study, these potentially meaningful but non-robust features would have been pruned during feature selection; yet, applying corrections with simple equations has demonstrated that these features could have easily been preserved. However, we highlight that feature corrections exhibited limited generalisability, as there was a notable dependency on the image processing method used, with a lower number of features being correctable than conditionally robust.
Still, our results encourage the possibility of incorporating bin configurations or voxel sizes into existing definitions of radiomic features27, as also asserted in earlier investigations17,21,22. An alternative to the use of explicitly defined mathematical equations, which we considered in this work, would have been to include feature-specific variations to discretisation or interpolation within the construct of a mixed-effects model. To this end, a similar approach has been proposed for reducing the variability of radiomic features before pooling data from different sites53.
Effect of cancer type on feature robustness and correctability
When comparing cancer types, melanoma images consistently generated the fewest robust or correctable [18F]-FDG-PET radiomic features, with the effect of cancer type on robustness being significant when considering variations in voxel size. We postulate these observations could be owed to the considerably smaller analysed tumour volumes for melanoma cases, given prior evidence that radiomic features can exhibit volume dependence23,24,54. Nonetheless, melanoma cases demonstrated a significantly greater number of features conditionally robust to bin configurations, and the widest global robustness ranges for resampled voxel sizes. Therefore, robustness can still be achieved by using common bin configurations or resampling images to appropriate voxel sizes.
Limitations
This study carries several limitations. First, while we investigated image processing parameters covering those found in the literature, these parameters are not exhaustive (e.g., voxel sizes smaller than 0.8 mm were not investigated). However, we used an extensive set of image processing parameter configurations (e.g., intensity bin numbers as multiples of 8 instead of powers of 2) to thoroughly explore the robustness of features extracted from [18F]-FDG-PET images. Second, for feature correction, we tested the fits of only eight functions, and more reliable fits and better correction could possibly be achieved by considering a larger library of functions or implementing these equations in a piecewise manner. Third, tumour segmentations for each patient were treated as a single volumetric mask, even in cases where multiple lesions were present. While this approach is not atypical for radiomics studies55–58, it should be noted that the goal of this work was not to correlate these features with lesion-specific clinicopathological characteristics. Furthermore, lesion-by-lesion analyses would have increased the complexity of radiomic computations because of the inherently smaller volumes. Finally, the generalisability of the findings from this work warrants further investigation using data involving other cancer types and imaging protocols, ideally acquired in a prospective manner. These studies may also consider examining the combined effect of intensity discretisation and voxel interpolation on [18F]-FDG-PET features, alongside variations in other factors known to influence radiomics, such as lesion segmentation (e.g., intra- and inter-reader variability) and acquisition or reconstruction settings19,40,59. Nevertheless, our use of a large, multi-cancer, and homogeneously acquired imaging dataset allowed us to examine the aims of this study with better statistical power.
Conclusions
In conclusion, [18F]-FDG-PET radiomic features were more sensitive to intensity discretisation than voxel interpolation, especially for second-order families, with feature robustness also exhibiting a potential cancer type dependence. Non-robustness of features could be addressed by implementing range conditions or dependency corrections on bin configuration or voxel size, with both approaches significantly affected by the choice of discretisation or interpolation algorithm used. Robustness ranges were suitable for most features tested and may inform prospective study designs on which bin configurations or voxel sizes would be optimal; however, this approach could be restrictive as only parameters within this range can be selected. In contrast, feature corrections using mathematical functions work reasonably well across the entire spectrum of parameters tested and may be valuable when pooling multi-centric heterogeneous datasets, albeit this approach is appropriate for less features. Taken together, the results of this work offer a more nuanced understanding of the impact of image processing variations on [18F]-FDG-PET radiomic features, and underscore the importance of selecting reliable imaging biomarkers for clinical use.
Supplementary Information
Acknowledgements
S.R. is supported by the Brunei “Sultan’s Scholar” scholarship from the Sultan Haji Hassanal Bolkiah Foundation, and the Turing-Roche Strategic Partnership as part of the Community Scholar Scheme. R.M. is supported by the National Institute for Health and Care Research (NIHR) Cambridge Biomedical Research Centre (BRC-1215-20014). L.E.S was supported by the Cancer Research UK (CRUK) National Cancer Imaging Translational Accelerator (NCITA) (C42780/A27066), and is supported by the CRUK Cambridge Centre (C9685/A25177) and the NIHR Cambridge Biomedical Research Centre (BRC-1215-20014; NIHR203312). L.A. is supported by the Cancer Research UK Cambridge Centre (CTRQQR-2021\100012). The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care.
Author contributions
Conceptualisation, S.R. and R.M.; methodology, S.R. and R.M.; investigation, formal analysis, S.R. and R.M.; software, S.R. and L.E.S.; data interpretation, all authors; writing—original draft preparation, S.R.; visualisation, S.R.; writing—review and editing, R.M., L.E.S., and L.A.; supervision, L.A., R.M., and L.E.S. All authors reviewed and approved the final version of the manuscript.
Data availability
The imaging dataset used for this study are publicly available at The Cancer Imaging Archive, as referenced in the article. All other data generated and/or analysed during this study are included in this article, its supplementary materials, and a GitHub repository at https://github.com/syafiqramlee/robust-radiomics-img-processing.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-67239-8.
References
- 1.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pineiro-Fiel M, et al. A systematic review of PET textural analysis and radiomics in cancer. Diagnostics (Basel). 2021 doi: 10.3390/diagnostics11020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polverari G, et al. (18)F-FDG PET parameters and radiomics features analysis in advanced Nsclc treated with immunotherapy as predictors of therapy response and survival. Cancers (Basel) 2020 doi: 10.3390/cancers12051163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirienko M, et al. Ability of FDG PET and CT radiomics features to differentiate between primary and metastatic lung lesions. Eur. J. Nucl. Med. Mol. Imaging. 2018;45:1649–1660. doi: 10.1007/s00259-018-3987-2. [DOI] [PubMed] [Google Scholar]
- 5.Ahn HK, Lee H, Kim SG, Hyun SH. Pre-treatment (18)F-FDG PET-based radiomics predict survival in resected non-small cell lung cancer. Clin. Radiol. 2019;74:467–473. doi: 10.1016/j.crad.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Lovinfosse P, et al. FDG PET/CT radiomics for predicting the outcome of locally advanced rectal cancer. Eur. J. Nucl. Med. Mol. Imaging. 2018;45:365–375. doi: 10.1007/s00259-017-3855-5. [DOI] [PubMed] [Google Scholar]
- 7.Carles M, et al. FDG-PET radiomics for response monitoring in non-small-cell lung cancer treated with radiation therapy. Cancers (Basel) 2021 doi: 10.3390/cancers13040814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging-"how-to" guide and critical reflection. Insights Imaging. 2020;11:91. doi: 10.1186/s13244-020-00887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Traverso A, Wee L, Dekker A, Gillies R. Repeatability and reproducibility of radiomic features: A systematic review. Int. J. Radiat. Oncol. Biol. Phys. 2018;102:1143–1158. doi: 10.1016/j.ijrobp.2018.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leijenaar RT, et al. The effect of SUV discretization in quantitative FDG-PET radiomics: The need for standardized methodology in tumor texture analysis. Sci. Rep. 2015;5:11075. doi: 10.1038/srep11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehmann TM, Gonner C, Spitzer K. Survey: Interpolation methods in medical image processing. IEEE Trans. Med. Imaging. 1999;18:1049–1075. doi: 10.1109/42.816070. [DOI] [PubMed] [Google Scholar]
- 12.El-Gamal FE-ZA, Elmogy M, Atwan A. Current trends in medical image registration and fusion. Egypt. Inform. J. 2016;17:99–124. doi: 10.1016/j.eij.2015.09.002. [DOI] [Google Scholar]
- 13.Leng J, Xu G, Zhang Y. Medical image interpolation based on multi-resolution registration. Comput. Math. Appl. 2013;66:1–18. doi: 10.1016/j.camwa.2013.04.026. [DOI] [Google Scholar]
- 14.Bailly C, et al. Revisiting the robustness of PET-based textural features in the context of multi-centric trials. PLoS One. 2016;11:e0159984. doi: 10.1371/journal.pone.0159984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu L, et al. Robustness of radiomic features in [(11)C]Choline and [(18)F]FDG PET/CT imaging of nasopharyngeal carcinoma: Impact of segmentation and discretization. Mol. Imaging Biol. 2016;18:935–945. doi: 10.1007/s11307-016-0973-6. [DOI] [PubMed] [Google Scholar]
- 16.Altazi BA, et al. Reproducibility of F18-FDG PET radiomic features for different cervical tumor segmentation methods, gray-level discretization, and reconstruction algorithms. J. Appl. Clin. Med. Phys. 2017;18:32–48. doi: 10.1002/acm2.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whybra P, Parkinson C, Foley K, Staffurth J, Spezi E. Assessing radiomic feature robustness to interpolation in (18)F-FDG PET imaging. Sci. Rep. 2019;9:9649. doi: 10.1038/s41598-019-46030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang P, et al. Extracting and selecting robust radiomic features from PET/MR images in nasopharyngeal carcinoma. Mol. Imaging Biol. 2020;22:1581–1591. doi: 10.1007/s11307-020-01507-7. [DOI] [PubMed] [Google Scholar]
- 19.Mali SA, et al. Making radiomics more reproducible across scanner and imaging protocol variations: A review of harmonization methods. J. Pers. Med. 2021 doi: 10.3390/jpm11090842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stamoulou E, et al. Harmonization strategies in multicenter MRI-based radiomics. J. Imaging. 2022 doi: 10.3390/jimaging8110303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shafiq-Ul-Hassan M, et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med. Phys. 2017;44:1050–1062. doi: 10.1002/mp.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shafiq-Ul-Hassan M, et al. Voxel size and gray level normalization of CT radiomic features in lung cancer. Sci. Rep. 2018;8:10545. doi: 10.1038/s41598-018-28895-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Escudero Sanchez L, et al. Robustness of radiomic features in CT images with different slice thickness, comparing liver tumour and muscle. Sci. Rep. 2021;11:8262. doi: 10.1038/s41598-021-87598-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy S, et al. Optimal co-clinical radiomics: Sensitivity of radiomic features to tumour volume, image noise and resolution in co-clinical T1-weighted and T2-weighted magnetic resonance imaging. EBioMedicine. 2020;59:102963. doi: 10.1016/j.ebiom.2020.102963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yip SSF, et al. Impact of experimental design on PET radiomics in predicting somatic mutation status. Eur. J. Radiol. 2017;97:8–15. doi: 10.1016/j.ejrad.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 26.Zhao B. Understanding sources of variation to improve the reproducibility of radiomics. Front. Oncol. 2021;11:633176. doi: 10.3389/fonc.2021.633176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zwanenburg A, et al. The image biomarker standardization initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295:328–338. doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paquier Z, et al. Radiomics software comparison using digital phantom and patient data: IBSI-compliance does not guarantee concordance of feature values. Biomed. Phys. Eng. Expr. 2022;8:065008. doi: 10.1088/2057-1976/ac8e6f. [DOI] [PubMed] [Google Scholar]
- 29.Gatidis, S. K. T. A whole-body FDG-PET/CT dataset with manually annotated tumor lesions (FDG-PET-CT-Lesions). (2022). 10.7937/gkr0-xv29. [DOI] [PMC free article] [PubMed]
- 30.Gatidis S, et al. A whole-body FDG-PET/CT dataset with manually annotated Tumor Lesions. Sci. Data. 2022;9:601. doi: 10.1038/s41597-022-01718-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark K, et al. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging. 2013;26:1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Griethuysen JJM, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berchtold A. Test–retest: Agreement or reliability? Methodol. Innov. 2016;9:2059799116672875. doi: 10.1177/2059799116672875. [DOI] [Google Scholar]
- 34.de Vet HC, Terwee CB, Knol DL, Bouter LM. When to use agreement versus reliability measures. J. Clin. Epidemiol. 2006;59:1033–1039. doi: 10.1016/j.jclinepi.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 35.Bakdash JZ, Marusich LR. Repeated measures correlation. Front. Psychol. 2017;8:456. doi: 10.3389/fpsyg.2017.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNitt-Gray, M. et al. Standardization in quantitative imaging: A multicenter comparison of radiomic features from different software packages on digital reference objects and patient data sets. Tomography6, 118–128 (2020). 10.18383/j.tom.2019.00031. [DOI] [PMC free article] [PubMed]
- 37.Antonopoulos AS, et al. Machine learning of native T1 mapping radiomics for classification of hypertrophic cardiomyopathy phenotypes. Sci. Rep. 2021;11:23596. doi: 10.1038/s41598-021-02971-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ge G, Zhang J. Uniqueness of radiomic features in non-small cell lung cancer. J. Appl. Clin. Med. Phys. 2022;23:e13787. doi: 10.1002/acm2.13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The jamovi project. jamovi. Version 2.3 [Computer software]. (2023). https://www.jamovi.org
- 40.Nyflot MJ, et al. Quantitative radiomics: Impact of stochastic effects on textural feature analysis implies the need for standards. J. Med. Imaging (Bellingham) 2015;2:041002. doi: 10.1117/1.JMI.2.4.041002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hosseini SA, et al. The impact of preprocessing on the PET-CT radiomics features in non-small cell lung cancer. Front. Biomed. Technol. 2021;8:261–272. [Google Scholar]
- 42.Meijer, K. M. Accuracy and Stability of Radiomic Features for Characterising Tumour Heterogeneity Using Multimodality Imaging: A Phantom Study (University of Twente, 2019).
- 43.Pfaehler E, et al. Repeatability of (18) F-FDG PET radiomic features: A phantom study to explore sensitivity to image reconstruction settings, noise, and delineation method. Med. Phys. 2019;46:665–678. doi: 10.1002/mp.13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kolinger GD, et al. Effects of tracer uptake time in non-small cell lung cancer (18)F-FDG PET radiomics. J. Nucl. Med. 2022;63:919–924. doi: 10.2967/jnumed.121.262660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Branchini M, et al. Impact of acquisition count statistics reduction and SUV discretization on PET radiomic features in pediatric 18F-FDG-PET/MRI examinations. Phys. Med. 2019;59:117–126. doi: 10.1016/j.ejmp.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 46.Ferreira M, et al. [(18)F]FDG PET radiomics to predict disease-free survival in cervical cancer: A multi-scanner/center study with external validation. Eur. J. Nucl. Med. Mol. Imaging. 2021;48:3432–3443. doi: 10.1007/s00259-021-05303-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Presotto L, et al. PET textural features stability and pattern discrimination power for radiomics analysis: An "ad-hoc" phantoms study. Phys. Med. 2018;50:66–74. doi: 10.1016/j.ejmp.2018.05.024. [DOI] [PubMed] [Google Scholar]
- 48.Bleker J, Roest C, Yakar D, Huisman H, Kwee TC. The effect of image resampling on the performance of radiomics-based artificial intelligence in multicenter prostate MRI. J. Magn. Reson. Imaging. 2023 doi: 10.1002/jmri.28935. [DOI] [PubMed] [Google Scholar]
- 49.Parker J, Kenyon RV, Troxel DE. Comparison of interpolating methods for image resampling. IEEE Trans. Med. Imaging. 1983;2:31–39. doi: 10.1109/TMI.1983.4307610. [DOI] [PubMed] [Google Scholar]
- 50.Papanikolaou N, Matos C, Koh DM. How to develop a meaningful radiomic signature for clinical use in oncologic patients. Cancer Imaging. 2020;20:33. doi: 10.1186/s40644-020-00311-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCague C, et al. Introduction to radiomics for a clinical audience. Clin. Radiol. 2023;78:83–98. doi: 10.1016/j.crad.2022.08.149. [DOI] [PubMed] [Google Scholar]
- 52.Park S-H, et al. Robustness of magnetic resonance radiomic features to pixel size resampling and interpolation in patients with cervical cancer. Cancer Imaging. 2021;21:19. doi: 10.1186/s40644-021-00388-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orlhac F, et al. A guide to ComBat harmonization of imaging biomarkers in multicenter studies. J. Nucl. Med. 2022;63:172–179. doi: 10.2967/jnumed.121.262464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Traverso A, et al. Machine learning helps identifying volume-confounding effects in radiomics. Phys. Med. 2020;71:24–30. doi: 10.1016/j.ejmp.2020.02.010. [DOI] [PubMed] [Google Scholar]
- 55.Jakoba JE, et al. Quantitative radiomics features in diffuse large B-cell lymphoma: Does segmentation method matter? J. Nucl. Med. 2022;63:389. doi: 10.2967/jnumed.121.262117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images are more than pictures, they are data . Radiology. 2015;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Helden EJ, et al. Radiomics analysis of pre-treatment [(18)F]FDG PET/CT for patients with metastatic colorectal cancer undergoing palliative systemic treatment. Eur. J. Nucl. Med. Mol. Imaging. 2018;45:2307–2317. doi: 10.1007/s00259-018-4100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henry T, et al. Investigation of radiomics based intra-patient inter-tumor heterogeneity and the impact of tumor subsampling strategies. Sci. Rep. 2022;12:17244. doi: 10.1038/s41598-022-20931-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang F, et al. Impact of contouring variability on oncological PET radiomics features in the lung. Sci. Rep. 2020;10:369. doi: 10.1038/s41598-019-57171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The imaging dataset used for this study are publicly available at The Cancer Imaging Archive, as referenced in the article. All other data generated and/or analysed during this study are included in this article, its supplementary materials, and a GitHub repository at https://github.com/syafiqramlee/robust-radiomics-img-processing.