Abstract
The effects of hepatitis C virus (HCV) proteins on anti-Fas (CD95/APO-1) antibody- and tumor necrosis factor alpha (TNF-α)-mediated apoptosis in different human cell lines were investigated by magnetic concentration of cells which transiently produced the exogenous protein. HepG2 cells, which produced whole HCV proteins, became resistant to anti-Fas-induced apoptotic cell death. Furthermore, the core protein among HCV proteins had a key role in protecting the various cells from apoptosis mediated by not only anti-Fas but also TNF-α. We also found that the core functioned in the activation of nuclear factor κB (NF-κB) in all cells examined. Deletion analysis of the core revealed that the region required for NF-κB activation was closely correlated with that for its antiapoptotic function. In addition, we revealed in some cases that the antiapoptotic effect of the core was restrained by coproduction of the inhibitor of NF-κB, IκB-α protein. These results demonstrated that the core inhibits Fas- and TNF-α-mediated apoptotic cell death via a mechanism dependent on the activation of NF-κB in particular cell lines.
Hepatitis C virus (HCV) is a major causative agent of chronic liver disease including chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma worldwide (1, 7, 17, 18, 40). The majority of individuals infected by HCV cannot resolve their infection and suffer from persistent chronic hepatitis. This chronic infection by HCV is suspected to be strongly associated with the development of hepatocellular carcinoma.
Apoptotic cell death with viral infection can be induced by the host immune response through the function of cytotoxic T lymphocytes (CTL) and natural killer cells, or by viral proteins themselves, and apoptosis has been suggested to be a common pathway of virus clearance by host organisms (25, 29). On the other hand, many virus genomes encode proteins which suppress apoptosis so as to escape from immune attack by the host (41). For example, CrmA, a cowpox virus gene product, encodes a protease inhibitor of the caspase family and prevents apoptosis caused by CTL, tumor necrosis factor alpha (TNF-α), or Fas (CD95/APO1) signaling (23, 42). Adenovirus E1B and human papillomavirus E6 proteins suppress p53-dependent apoptosis by binding and inactivating wild-type p53 (8, 31, 36, 49). In the case of HCV infection, it was suggested that apoptosis in hepatocytes, especially that mediated by Fas, plays an important role as the main mechanism of viral clearance (11, 15), which would result in the liver damage observed in chronic hepatitis.
The HCV genome encodes a polypeptide precursor consisting of about 3,010 amino acid (aa) residues, and this precursor protein is cleaved by the host and viral proteases to generate at least 10 functional protein units: the core, envelope 1 (E1), E2, p7, nonstructural protein 2 (NS2), NS3, NS4A, NS4B, NS5A, and NS5B (12–14, 19, 24). Ray et al. reported that the core suppresses apoptosis induced by cisplatin in human cervical epithelial cells, by c-myc overexpression in Chinese hamster ovary cells, and by TNF-α in human breast carcinoma cell lines (32, 33). Fujita et al. also suggested that NS3 protein inhibits actinomycin D-induced apoptosis in NIH 3T3 cells (10). In a different study, the core was proposed to sensitize HepG2 cells to apoptosis mediated by the Fas signaling pathway (34). The core was also suggested to enhance TNF-α-induced apoptosis (56). Thus, these previous studies have demonstrated controversial phenomena, and the discrepancies between these results might be partially due to the differences in cell lines or apoptosis-inducing agents used. However, an alternative explanation is that the cloned permanent transfectants differ in characteristic responses from parental cell lines, irrespective of exogenously introduced protein production, because all these results were obtained with cloned permanent transfectant cells producing the viral protein. To avoid this possibility, recent reports showing the function of the exogenous proteins in apoptosis have tended to include the results of transient-transfection experiments in which transfection-positive cells were detected by production of β-galactosidase or green fluorescent protein originating from a cotransfected expression plasmid (48, 53). Therefore, we used a magnetic concentration system for transiently DNA-transfected cells to analyze the functions of viral proteins in a population of cells expressing HCV proteins. Using this system, we examined whether HCV proteins affect apoptotic responses in various cells, especially those mediated by Fas and TNF-α, and directly analyzed the biochemical characteristics of the concentrated cells.
Our results demonstrated that the core among HCV proteins protected several types of cells from apoptotic cell death induced by anti-Fas and TNF-α and that the activation of NF-κB is an important pathway of the antiapoptotic effects of the core in certain cells.
MATERIALS AND METHODS
Plasmid constructs.
The plasmids used in this study were constructed to produce several proteins under the control of the cytomegalovirus immediate-early promoter and named the pCMV series. pCMV-3010, which expressed the whole HCV genome, was made by replacing the EcoRI-AvrII fragment of pC980 with the EcoRI-AvrII fragment of pCMV/729-3010 (14). pCMV-980, encoding the core, E1, E2, p7, and the C-terminally truncated NS2, was constructed by insertion of the EcoRI-HindIII fragment of pC980 (13) into the EcoRI-HindIII sites of the pKS+/CMV vector (14). pCMV-Core, for expression of the core gene encoding a polypeptide spanning from aa 1 to 191 of the HCV precursor polyprotein, was made by inserting the PCR product with oligonucleotide primers (5′-TGTGGATCCATGAGCACAAATCCTAAACC-3′ [named core-s] and 5′-CTCGAATTCTCAAGCGGAAGCTGGGATGGTCA-3′) into the BamHI-EcoRI sites of pKS+/CMV. pCMV-FLAG-Core, encoding the core which was N-terminally fused with FLAG epitope tag, was constructed by inserting the amplified core sequences into the BglII-SalI sites of pCMV-Tag1 (Stratagene). The expression plasmids of truncated core protein were also prepared as described above by PCR with pCMV-Core as a template. The oligonucleotides core-s and 5′-TCTGAATTCTCAAGAGCAACCGGGCAGATTCC-3′ were used as primers for pCMV-ΔCore173, and core-s and 5′-ACTGAATTCTCAC AGGGCCCTGGCAACGCCTC-3′ were used for pCMV-ΔCore151. The resultant expression plasmids, pCMV-ΔCore173 and pCMV-ΔCore151, encode C-terminally truncated core proteins in which the C-terminal 18 and 40 aa, respectively, were deleted. pCMV-E1E2, for expression of the C-terminal portion of core, E1, E2, p7, and C-terminally truncated NS2, was produced by inserting the EcoRI-HindIII fragment of pN124 (13) into EcoRI-HindIII-digested pKS+/CMV.
The cDNA fragment of human Bcl-2 was excised from pB4 Bcl-2 (kindly supplied by Y. Tsujimoto, Osaka University). The EcoRI fragment of pB4 Bcl-2 was inserted into pKS+/CMV, and the Bcl-2 expression plasmid pCMV-Bcl-2 was obtained. The expression plasmid for NF-κB-inducing kinase, pcDNA3-NIK, was kindly provided by David Wallach (Weizmann Institute of Science, Rehovot, Israel). The cDNA fragment of human IκB-α was synthesized by reverse transcription-PCR with mRNA from Jurkat cells as a template. After reverse transcription with the oligonucleotide primer (5′-CAAGTCCATGTTCTTTCAGC-3′), PCR was performed with oligonucleotides 5′-ATAGGATCCAGCTCGTCCGCGCCATGTTC-3′ and 5′-TTAGGATCCGTTCTTTCAGCCCCTTTGCA-3′ as primers. After digestion with BamHI, the PCR product was then cloned into the BamHI site of pKS+/CMV. The sequence of the resultant plasmid, pCMV-IκB, was verified by sequencing. The reporter plasmid, pNF-κB(Mut)-Luc, was constructed by inserting the synthetic oligonucleotide for the mutated element of NF-κB binding sequences (51) into the pGL3 promoter vector (Promega).
Cell culture.
HepG2, HeLa, and Saos-2 cells were cultured in Dulbecco’s modified Eagle medium (Nissui) with 10% fetal bovine serum (FBS) and l-glutamine. Jurkat cells (a generous gift from S. Yonehara, Kyoto University) were grown in RPMI 1640 (Nissui) supplemented with 10% FBS. Huh-7 cells were grown in RPMI 1640 with l-glutamine, lactalbumin, and 2.5% FBS. MCF-7 cells were grown in Eagle’s minimum essential medium with nonessential amino acids (GIBCO BRL) and 10% FBS.
Transfection of cells.
For plasmid transfection into the adherent cells, we used the FuGENE 6 transfection reagent (Boehringer Mannheim). The DNA transfection procedure for Jurkat cells was performed with SuperFect transfection reagent (Qiagen). All these experiments were performed essentially according to the manufacturer’s protocols.
Concentration of cells transiently transfected with the expression plasmids.
We utilized the MACSelect system (Miltenyi Biotec) for specific concentration of transiently DNA-transfected cells from the heterogeneous cell population. The concentration of the plasmid-transfected cells was achieved by magnetic isolation of the cells producing a trypsin resistance cell surface marker, a truncated mouse H-2k molecule, which was expressed from the cotransfected plasmid, pMacsKκ.
A total of 5 × 106 HepG2 or MCF-7 cells were cotransfected with 7.5 μg of an expression plasmid together with 2.5 μg of pMacsKk with 15 μl of FuGENE 6. After 18 h, cells were treated with 0.02% trypsin and dispersed by being pipetted into single-cell suspensions after addition of trypsin inhibitor. The cells were resuspended with 600 μl of PBE buffer (phosphate-buffered saline [PBS] supplemented with 0.5% bovine serum albumin and 2 mM EDTA) containing 80 μl of micromagnetic beads conjugated with a monoclonal antibody against mouse H-2k and incubated for 15 min at room temperature. Thus, magnetically labeled cells were recovered by the magnetic separation column. By this procedure, we collected up to 2 × 105 to 5 × 105 transiently transfected cells and used them for further analysis. In the case of Jurkat cells, we used the same procedure as described above without treatment with trypsin.
Evaluation of cell death.
Apoptotic cell death was evaluated by determining the cell viability and caspase activation and by the detection of DNA fragmentation. The cell viability was measured by scoring 200 cells in each experiment, and the score of viable cell ratio represented the average of three independent experiments. The cells transfected with several plasmids and concentrated magnetically were treated with anti-Fas antibody (CH-11; MBL, Nagoya, Japan) or recombinant human TNF-α (Sigma). Anti-Fas and TNF-α were used at final concentrations of 100 and 10 ng/ml, respectively. The minimum concentration of cycloheximide (CHX) required for anti-Fas-induced apoptosis in HepG2 cells was determined as a final concentration of 500 ng/ml, which was 1/20 less than that generally used in the same assay (50). The number of dead cells was counted as those stained with trypan blue dye within four microscopic fields after 14 or 48 h from the start of anti-Fas or TNF-α treatment, respectively. The apoptotic cell death was also measured by cell detection enzyme-linked immunosorbent assay (Boehringer Mannheim) according to the manufacturer’s protocol. This assay is based on the specific determination of mononucleosomes and oligonucleosomes in the cytoplasmic fraction of apoptosis-induced cells.
Caspase-8 activities in the anti-Fas- or TNF-α-treated cells were examined with caspase colorimetric protease assay kits for caspase-8 (MBL). The assay was based on spectrophotometric detection of the chromophore p-nitroanilide after cleavage from the labeled substrates IETD (Ile-Glu-Thr-Asp) and p-nitroanilide for caspase-8. The assays were performed according to the manufacturer’s protocol 3 or 6 h after addition of anti-Fas or TNF-α, respectively. The protein concentration of the lysates was measured by using the bicinchoninic acid protein assay reagent (Pierce, Rockford, Ill.).
Reporter plasmid assay.
The reporter pNF-κB-Luc vector contained the NF-κB binding elements upstream of the minimum promoter region driving the luciferase reporter gene (Stratagene). The luciferase activities in the cells after treatment or not with anti-Fas or TNF-α for 2 h were measured by a luminometer with a luciferase assay kit (Promega) as recommended by the manufacturer.
Immunoblotting analysis.
The preparation of cell lysates, sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, and immunoblotting analysis were performed with a polyvinylidene difluoride membrane as described previously (14). The antibodies used in this experiment were those against HCV core protein (54) (515S; a generous gift from M. Kohara, Tokyo Metropolitan Institute of Medical Science), anti-NS5A protein (14), anti-Bcl-2 (MBL), anti-caspase-8 (MBL), and anti-IκB-α (Santa Cruz). Immunocomplexes on the filters were detected by enhanced chemiluminescence assay (Renaissance; NEN, Boston, Mass.). The densitometric analysis of detected protein by immunoblotting was performed by using a Fluor-S multi-imager (Bio-Rad).
Immunofluorescence.
The indirect immunofluorescence experiment was performed as described previously (30). Briefly, HepG2 cells were fixed in 2% paraformaldehyde for 1 h at room temperature. After being washed twice with PBS, the fixed cells were permeabilized with 0.05% Triton X-100 for 15 min and washed with PBS. Then, the cells were incubated with a 1:1,000 dilution of anti-HCV core monoclonal antibody. After being washed with PBS, the cells were incubated with rhodamine-conjugated secondary antibody and 4′,6-diamidino-2-phenylindole (DAPI). After washing, the samples were mounted on glass slides and observed by fluorescence microscopy.
RESULTS
Selective concentration of plasmid DNA-transfected cells.
To assess the efficiency of the transfection and selective concentration method, two reporter plasmids were used: pEGFP-N1 (Clontech), which was designed to express a green fluorescent protein in transfected cells; and pCMV-lacZ, for expression of Escherichia coli β-galactosidase in mammalian cells, the production of which can be easily monitored by staining with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) in situ. Compared with other conventional methods, a relatively high efficiency of plasmid transfection into HepG2 and MCF-7 cells (∼10 and ∼20%, respectively) was obtained with minimum cytotoxicity with FuGENE 6 in this study. Almost the same amount of plasmid was likely to be distributed to each cell, as demonstrated by the intensity of the signals from the reporter gene product (data not shown). In addition, the expression levels of transiently transfected plasmids in a single cell obtained by our procedure were likely to be lower than those obtained by using a conventional calcium phosphate technique (data not shown). The SuperFect reagent also produced efficient transfection in Jurkat cells, although the transfection efficiency remained at 2 to 3%. After concentration by the magnetic separator, the ratio of transfected cells was markedly increased in each case. We confirmed by the observation of green fluorescent signals from transfected pEGFP-N1 that more than 80% of collected cells were usually transfection positive.
HepG2 cells that produced the whole HCV proteins showed resistance against Fas-mediated apoptosis.
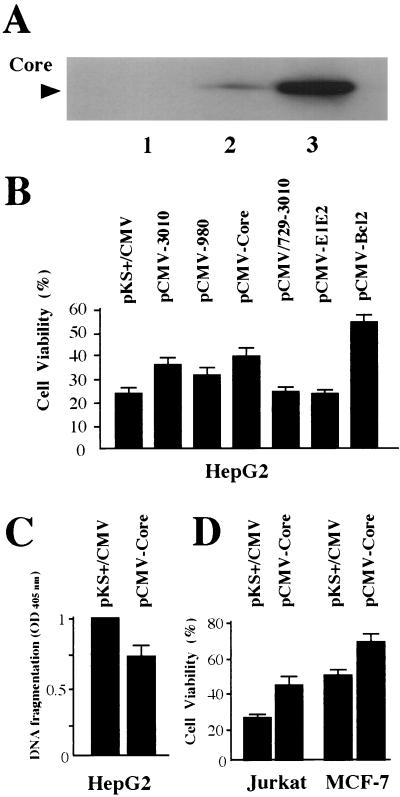
To examine whether HCV proteins alter the fate of cells in which apoptotic cell death is induced by anti-Fas, the expression plasmid pCMV-3010, encoding all HCV proteins, was transfected into HepG2 cells and the enriched fraction of transfected cells was used as a model of hepatocytes infected by HCV. The production and processing of each HCV protein in these cells were confirmed by immunoblotting analysis (Fig. 1A and data not shown). As a negative control, pKS+/CMV without any insert was used, and pCMV-Bcl-2 encoding Bcl-2 protein, a well-known inhibitor of apoptosis (43, 46, 52), was used as a positive control. Fourteen hours after treatment with anti-Fas, the numbers of viable and dead cells in the enriched population were counted after trypan blue staining, and the viability of the population was determined. As shown in Fig. 1B, cell viability of control HepG2 cells transfected with pKS+/CMV was 23.7% (±4.0% [standard error]). However, the viability of cells transfected with pCMV-3010 was 35.9% (±3.7%) under similar conditions. The HepG2 cells expressing Bcl-2 protein were significantly more resistant to anti-Fas-induced cell death (54.3% ± 3.1%) than were the HCV protein-producing cells. No change in cell viability was observed following treatment with CHX only or no treatment (data not shown). From these results, we concluded that HepG2 cells which produced all the proteins of HCV become resistant to apoptotic cell death induced by anti-Fas despite the proapoptotic effect of CHX.
FIG. 1.
Effects of HCV protein production on the sensitivity of various cell lines to Fas- or TNF-α-mediated apoptosis. (A) Production levels of the HCV core protein in HepG2 cells. HepG2 cells (4 × 105) were transiently transfected with 2.0 μg of pKS+/CMV, pCMV-3010, or pCMV-Core by using 3 μl of FuGENE 6. At 48 h posttransfection, the level of production of the core was analyzed by SDS-polyacrylamide gel electrophoresis followed by immunoblotting analysis with anticore antibody. Lane 1, pKS+/CMV; lane 2, pCMV-3010; lane 3, pCMV-Core. (B) Cell viabilities of HepG2 cells producing various HCV proteins. HepG2 cells (5 × 106) transfected with 7.5 μg of various HCV protein expression plasmids and 2.5 μg of pMacsKk were enriched magnetically at 18 h posttransfection. A total of 2 × 105 selected transfectants were propagated for 24 h and then treated with anti-Fas (100 ng/ml) in the presence of CHX (500 ng/ml) for 14 h. A total of 200 cells were counted after staining with trypan blue in randomly chosen fields. The cell viabilities are presented as percentages of living cells from three independent experiments. (C) Suppressive effect on DNA fragmentation of the core production in HepG2 cells. The enriched HepG2 cells transfected with pKS+/CMV or pCMV-Core were treated with anti-Fas and CHX for 10 h. Then, cells were lysed and the supernatant was assayed by a sandwich enzyme-linked immunosorbent assay system with antihistone and anti-DNA-peroxidase antibodies. The reaction was measured with a microplate reader at a 405-nm wavelength. (D) Viabilities of Jurkat and MCF-7 cells producing the core protein. A total of 2 × 106 Jurkat cells were transfected with 1.2 μg of pMacsKk and 3.6 μg of pKS+/CMV or pCMV-Core with 20 μl of SuperFect reagent and enriched magnetically at 18 h posttransfection. Then, the collected cells were propagated for 24 h and treated with 100 ng of anti-Fas per ml for 24 h. MCF-7 cells producing the core were also enriched according to the same protocol as that for HepG2 cells and analyzed after treatment with 10 ng of TNF-α per ml for 48 h.
HCV core protein protected the cells from Fas- and TNF-α-mediated apoptotic cell death.
To investigate which HCV protein might have this antiapoptotic effect, cells transfected with plasmids for expression of several HCV protein units were treated with anti-Fas after magnetic enrichment of the cells. First, we examined the effects of HCV structural and nonstructural protein production in HepG2 cells on the Fas-mediated apoptosis. As shown in Fig. 1B, cells transfected with pCMV-980, which is for production of core, E1, E2, p7, and C-terminally truncated NS2, showed resistance to anti-Fas-induced cytotoxicity (the mean viability was 32.0% ± 5.2%) compared with control cells. However, the viability of the cells transfected with pCMV-N729/3010, which produced the C-terminal portion of E2, p7, and all NS proteins of HCV, was the same as that of negative controls (24.4% ± 1.4%). Then, HepG2 cells transfected with pCMV-Core or pCMV-E1E2, for production of the core or E1, E2, p7, and C-terminally truncated NS2, respectively, were also analyzed to examine the ability of each protein to resist the effects of anti-Fas (Fig. 1B). Increased viability was observed in the cells with core production (39.0% ± 3.7%), whereas transfection with pCMV-E1E2 did not confer any protection against anti-Fas-induced cell death (23.4% ± 1.4%). The suppressive effect of the core on Fas-mediated apoptosis was also confirmed by measuring the cytoplasmic histone-associated DNA fragments (Fig. 1C). Taken together, the decreased sensitivity against Fas-mediated apoptosis observed in whole-HCV protein-producing HepG2 cells was assumed to be due to the effect of the core protein.
To examine whether the core would also show this antiapoptotic effect in other cell lines, Jurkat cells transfected with pCMV-Core were assayed as described above except for the absence of CHX. As shown in Fig. 1D, the Jurkat cells producing the core protein also showed higher viability than did the control cells (42.2% ± 3.6% versus 26.3% ± 1.8%, respectively). Thus, the antiapoptotic effect of the core was observed in different cell types treated with anti-Fas in either the presence or the absence of CHX. We also examined the effects of the core on the TNF-α-mediated apoptotic cell death in MCF-7 cells. The viability of MCF-7 cells transfected with pCMV-Core or pKS+/CMV was assessed after 48 h of treatment with TNF-α (Fig. 1D). As observed for HepG2 and Jurkat cells, MCF-7 cells producing the core also showed a high cell survival ratio in comparison with that of negative control cells (68.0% ± 4.1% versus 49.5% ± 2.3%, respectively) against TNF-α-induced apoptosis. We also observed that expression of other plasmids expressing the HCV open reading frame, pCMV-3010, pCMV-980, pCMV/729-3010, and pCMV-E1E2, resulted in cellular responses in MCF-7 and Jurkat cells quite similar to those found for HepG2 cells (data not shown). The apparent induction of cell death was not observed in HepG2 and MCF-7 cells after the treatment with TNF-α and anti-Fas, respectively.
From the above results, we concluded that the core was responsible for suppressing the apoptotic response mediated by either Fas or TNF-α in different cell lines.
Caspase-8 activation was suppressed in HCV core protein-producing cells.
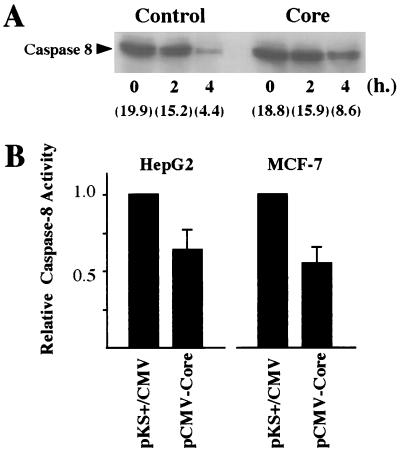
It has been suggested that the binding of Fas ligand to Fas results in the activation of a cascade of caspases including caspase-8 (FLICE) and caspase-3 (CPP32) (26, 27, 38). Therefore, to determine whether the core affects the protease cascade, we examined the activation of caspase-8 in HepG2 cells induced by anti-Fas under conditions of core production. As shown in Fig. 2B, the activity of caspase-8, which has been suggested to be located at the most upstream site in the caspase cascade activated by anti-Fas (6, 9) and TNF-α (48), was suppressed in the core-producing HepG2 cells after anti-Fas treatment (68.0% of that in negative controls). Furthermore, anti-Fas-induced processing of caspase-8 was diminished in the core-producing HepG2 cells compared with that in the control cells (Fig. 2A). As observed for HepG2 cells, the activation of caspase-8 induced by TNF-α was also down-regulated in MCF-7 cells producing the core (55% of the activity found in negative control cells, respectively). We confirmed that no activation of caspase-8 occurred after the magnetic concentration of the cells in the absence of anti-Fas or TNF-α. These results indicated that the core plays a role in the suppression of caspase activation induced by anti-Fas and TNF-α in those cell lines and that the suppression was likely to be achieved upstream of caspase-8 in the caspase cascade.
FIG. 2.
Effects of HCV core production on caspase-8 activity after treatment with anti-Fas or TNF-α. (A) The processing of caspase-8 in response to anti-Fas in HepG2 cells. HepG2 cells were transfected with pKS+/CMV or pCMV-Core in combination with pMacsKκ. After magnetic concentration, cells were treated with anti-Fas (100 ng/ml) and CHX (500 ng/ml) for the indicated time. Then, cell lysates were collected and separated by SDS-polyacrylamide gel electrophoresis and probed with anti-caspase-8 antibodies after transfer onto the polyvinylidene difluoride membrane. The relative quantitation of detected proteins by immunoblotting by chemiluminescence assay was performed with a Fluor-S multi-imager, and arbitrary units of each band were indicated. (B) HepG2 and MCF-7 cells transfected with pKS+/CMV or pCMV-Core were enriched and treated with anti-Fas (100 ng/ml) for 3 h or TNF-α (10 ng/ml) for 6 h, respectively. The relative activities of caspase-8, measured with the caspase colorimetric protease assay kit, as described in Materials and Methods, are shown.
HCV core protein activates a cellular transcriptional factor, NF-κB.
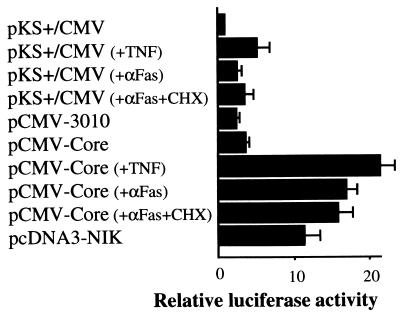
In HepG2 cells, the antiapoptotic function of the core was seen in the presence of a very low dose of CHX (500 ng/ml). However, a higher dose of CHX, 10 μg/ml, which is generally used in several apoptotic assays, eliminated that ability of the core despite no significant change in core production detected by immunoblotting analysis (data not shown). From these observations, we supposed that the core might protect the cells from apoptotic cell death in response to anti-Fas stimulation through a newly synthesized protein(s). Recently, protein synthesis-dependent protective mechanisms against apoptotic cell death have been reported to be associated with the activation of NF-κB (2, 4, 5, 45, 47, 48). To determine whether core production affects NF-κB activity in the cells, a reporter plasmid assay, in which NF-κB-dependent reporter gene expression was monitored in the presence or absence of HCV proteins, was performed with HepG2 cells cotransfected with pNF-κB-Luc and pCMV-3010, pCMV-Core, and pKS+/CMV for a negative control or pcDNA3-NIK for a positive control. As shown in Fig. 3, at 48 h after transfection, we found two- and threefold augmentation of relative luciferase activities in whole-HCV protein- and core-producing cells, respectively, compared to that in the negative control cells. In our experiment, 12-fold augmentation of the luciferase activities in the positive control cells producing NF-κB-inducing kinase was found. Furthermore, the treatment of core-producing HepG2 cells with either TNF-α or anti-Fas for 2 h resulted in 22- and 17-fold increases in luciferase activity, respectively, while the same treatments augmented the reporter activity by about 2- to 5-fold in the negative control cells. This activation of reporter gene expression was likely to be NF-κB specific for two reasons; one was that the transcription from either reporter plasmid which contained the mutated element of NF-κB binding sequence or cyclic AMP-responsive element was not affected (data not shown), and another was that exogenous production of IκB-α, a specific inhibitor of NF-κB, eliminated NF-κB activation by the core as mentioned below. When the core was produced in MCF-7 cells, both the basal and TNF-α-induced levels of NF-κB-dependent transcriptional activities were increased 5- and 15-fold, respectively, compared with that in the negative control cells (Fig. 4C and data not shown). About three- to sevenfold enhancement of NF-κB activities was also observed for Jurkat, Huh-7, Saos-2, and HeLa cells producing the core (data not shown).
FIG. 3.
Effects of HCV core production on NF-κB activity with and without treatment with anti-Fas or TNF-α. HepG2 cells were transfected with 0.5 μg of pNF-κB-Luc and 1.5 μg of various expression plasmids with 3 μl of FuGENE 6. At 46 h posttransfection, cells were treated or not with anti-Fas (100 ng/ml) or TNF-α (10 ng/ml) for 2 h as indicated. A whole-cell lysate was prepared and assayed for luciferase activity. The data represent the means of the relative luciferase activities in three independent experiments.
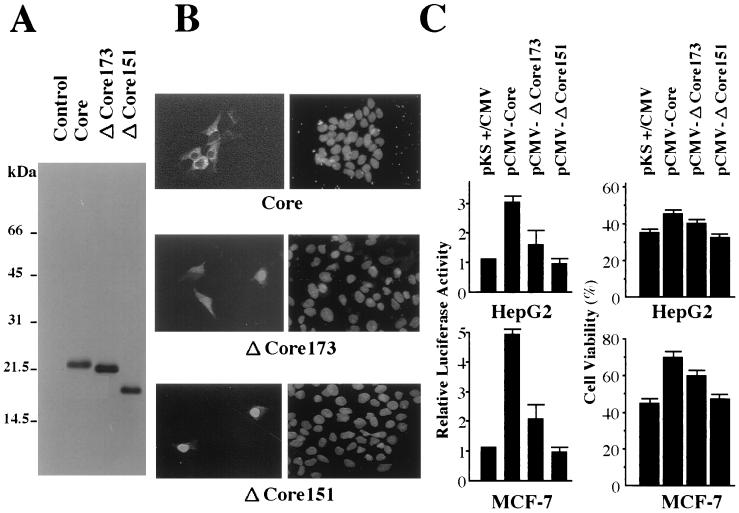
FIG. 4.
Effects of C-terminally truncated core proteins on apoptotic cell death and NF-κB activation in HepG2 and MCF-7 cells. (A) Detection of the core protein produced in HepG2 cells by immunoblotting analysis. HepG2 cells were transfected with expression plasmids for two types of the C-terminally truncated core protein as well as full-size core protein as indicated. Then, cells were harvested for immunoblotting analysis 48 h after transfection. Products expressed from pCMV-Core, pCMV-ΔCore173, and pCMV-ΔCore151 are shown. Negative control, cells transfected with pKS+/CMV. The molecular size markers are shown on the left of the panel. (B) Subcellular localization of core proteins. HepG2 cells were transfected with expression vectors for three types of core constructs, i.e., full length (191 aa), N-terminal 173 aa (ΔCore173), and N-terminal 151 aa (ΔCore151) as indicated. After 48 h, the cells were fixed and stained with anticore antibody, and the immunocomplexes were detected with rhodamine-conjugated secondary antibodies (left panels). The staining of the nucleus by DAPI in the same cells is shown in the right panels. (C) Antiapoptotic effect and NF-κB activation abilities of the C-terminally truncated core proteins. HepG2 and MCF-7 cells were transfected with 2.5 μg of pMacsKk and 7.5 μg of pKS+/CMV, pCMV-Core, pCMV-ΔCore173, or pCMV-ΔCore151. After enrichment, HepG2 and MCF-7 cells were treated with anti-Fas and TNF-α, respectively (left panels). Cell viabilities were assayed as indicated for Fig. 1. NF-κB activities in HepG2 and MCF-7 cells transfected with expression vectors for three types of core constructs were assayed as indicated for Fig. 3.
Based on this and the results described above, we concluded that the core has a function in the activation of NF-κB in all cell lines examined in this study and that this activation is synergistically enhanced in HepG2 cells by stimulation with anti-Fas and in MCF-7 cells by TNF-α, under the apoptosis-inducing conditions.
C-terminally truncated core had no effect on either NF-κB activation or suppression of apoptotic cell death.
The primary structure of the final core product after secondary processing in cells is still unknown. There have been some controversial reports suggesting that its C-terminal end is located at around 151 or 173 aa based on the results of deletion analysis (21, 35). It was also shown that the subcellular localization of the core was shifted from the cytoplasm to the nucleus by these deletions (20, 39). Therefore, to assess what type of core is functional in the suppression of apoptosis, HepG2 cells transfected with the expression plasmids for the two truncated core proteins, pCMV-ΔCore173 and pCMV-ΔCore151, were analyzed as described above. As shown in Fig. 4A, the molecular sizes of the cores from pCMV-Core, pCMV-ΔCore173, and pCMV-ΔCore151 were estimated to be similar to those previously reported (54). We also observed differences in subcellular localization of each core product by indirect immunofluorescence analysis. As shown in Fig. 4B, the core which was designed to be truncated as a polypeptide of 151 aa was detected mainly in the nucleus, in contrast to the observation that the core which was translated as a full-length protein of 191 aa was located around the perinuclear region in the cytoplasm. In contrast, the core which was produced primarily as a product of 173 aa was distributed in the nucleus and perinucleic cytoplasm.
As shown in Fig. 4C (upper panels), the maximal effects on suppression of the apoptosis induced by anti-Fas and NF-κB activation were observed for HepG2 cells transfected with pCMV-Core, whereas the cells producing the C-terminally truncated core of 151 aa showed no resistance against Fas-mediated apoptosis and no activation of NF-κB. In the HepG2 cells transfected with pCMV-ΔCore173, intermediate effects on both apoptotic cell death and NF-κB activation were observed. Similar results were obtained for MCF-7 cells transfected with each construct after stimulation with TNF-α (Fig. 4C, lower panels). These results indicated that the ability of each construct to activate NF-κB paralleled its antiapoptotic potential. Furthermore, we observed that the FLAG-core construct, in which FLAG tag was fused with the N-terminal end of the core, showed no antiapoptotic effect and no activation of NF-κB in spite of the fact that the production level and subcellular localization of this protein were similar to those of the original when it was produced in HepG2 cells (data not shown). This may suggest that a certain tertiary structure of the core which was destroyed by the N-terminal fusion is important for its biological activities.
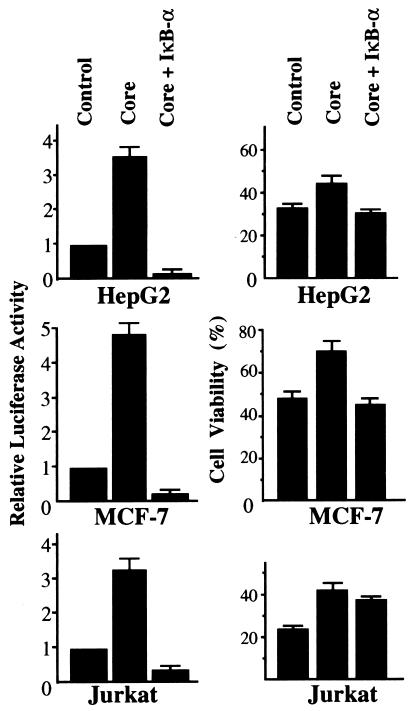
The antiapoptotic effects of the core were restrained by inhibition of NF-κB activation.
The above results suggested that at least one of the antiapoptotic effects of the core is achieved through the activation of NF-κB. Therefore, to assess this possibility, the sensitivity of MCF-7 cells cotransfected with pCMV-Core and pCMV-IκB against TNF-α-induced apoptotic cell death was evaluated after magnetic concentration as described above. As expected, the resistance against apoptosis observed for MCF-7 cells producing the core was completely abolished by coproduction of the inhibitor of NF-κB, IκB-α, in the cells (Fig. 5, middle panels). We also confirmed by reporter plasmid assay that the NF-κB activity in the cells producing the core was substantially inhibited by cotransfection of pCMV-IκB either with or without treatment with TNF-α (Fig. 5, left panels, and data not shown). When apoptotic cell death was induced by anti-Fas, this suppressive effect of IκB-α on the antiapoptotic function of the core was also observed for HepG2 cells (Fig. 5, upper panels). In contrast to these two cell lines, when IκB-α was coproduced in Jurkat cells with the core, the cell viability after treatment with anti-Fas was only partially reduced compared with that of the core-producing cells despite the complete suppression of NF-κB activities (Fig. 5, lower panels). Thus, the suppressive mechanism of apoptotic signaling by the core was believed to differ in the case of Jurkat cells from that for HepG2 and MCF-7 cells.
FIG. 5.
Coproduced IκB-α down-regulated NF-κB activities and antiapoptotic effects in core-producing cells. Combinations of 4.5 μg of pCMV-Core, 4.5 μg of pCMV-IκB, and 1.5 μg of pMacsKk and pKS+/CMV were transfected into HepG2 (upper panels) or MCF-7 (middle panels) cells with a total of 10.5 μg of expression plasmids. In Jurkat cells (lower panels), a total of 2 × 106 cells were transfected with a total of 4.9 μg of expression plasmids, including 2.1 μg of pCMV-Core, 2.1 μg of pCMV-IκB, and 0.7 μg of pMacsKk and pKS+/CMV with 20 μl of SuperFect reagent. After the concentration procedure, HepG2 and Jurkat cells were treated with anti-Fas and MCF-7 cells were treated with TNF-α (right panels). Cell viability was assayed as indicated for Fig. 1. HepG2 and MCF-7 cells were cotransfected with 0.25 μg of pNF-κB-Luc and 0.75 μg of pCMV-Core, pCMV-IκB, or pKS+/CMV with a total of 1.75 μg of expression plasmids. In Jurkat cells, a total of 4.9 μg of expression plasmids, including 0.7 μg of pNF-κB-Luc, and 2.1 μg of pCMV-Core, pCMV-IκB, or pKS+/CMV were transfected. The luciferase activity was measured as described in the legend to Fig. 3.
These results indicated that the antiapoptotic function of the core is dependent on the activation of NF-κB in HepG2 and MCF-7 cells. Moreover, our results suggested that the activation of NF-κB contributes to protection from Fas-mediated apoptosis in certain cells.
DISCUSSION
As part of the defense mechanism of host organisms, cells infected by viruses are induced to initiate apoptotic cell death by signals delivered from CTL (25). On the other hand, a number of viruses have been reported to cause infected cells to escape from this apoptosis to maintain persistent infection (29, 41). In this study, we investigated the effects of whole HCV proteins produced in HepG2 cells on Fas-mediated apoptotic cell death and found that these cells became resistant to apoptosis. These collected cells are likely to be a good model of HCV-infected cells because all HCV proteins were authentically produced from a precursor polyprotein encoded in a single open reading frame. This antiapoptotic effect of HCV proteins turned out to be a contribution of the core and was seen for different cell lines treated with anti-Fas or TNF-α. Furthermore, we demonstrated that the antiapoptotic effect of the core was exerted through enhanced activation of NF-κB, especially in HepG2 and MCF-7 cells.
Discrepancies regarding the effects of the core on the cellular apoptotic responses have been reported previously: the core functions antiapoptotically according to some papers (32, 33) and proapoptotically according to others (34, 56). The reason for the discrepancy among these reports is still unclear. It may be that the core has bipotential roles in the apoptotic signaling. This discrepancy may be, however, explained by the possibility that it was caused by use of clonally selected permanent transfectant cells in the previous studies. As cultured cell lines are likely to be mixed populations of certain cells, a clonally selected cell population cannot be certified to have characteristic features of the parental mixed population. To decrease the chance of selecting particular cells from mixed populations, we enriched the transfected cell population magnetically. Under these conditions, the cell populations which were originally sensitive to the apoptosis mediated by Fas or TNF-α gained the ability to resist such stimuli from the HCV core protein production. During preparation of the manuscript, Shrivastava et al. reported that the core suppressed TNF-mediated NF-κB activation in MCF-7 cells (37). The discrepancies between our findings and theirs might be derived from the difference in cells used in the experiments as mentioned above. The other difference between transient-transfection and permanent transfectant systems seems to be the expression levels of exogenous genes: that is, a relatively higher level of expression would be expected in the former case. Furthermore, the production of exogenous proteins, not only by transient but also by permanent transfection, may cause a kind of nonspecific stress in the cells. Therefore, to try to reduce the production level we chose the transfection method that enabled us to produce a relatively small amount of exogenous protein in a single cell but in many cells. Moreover, as shown in Fig. 1A, HepG2 cells transfected with pCMV-3010, in which only less than 1/20 of the core production was seen compared with that for pCMV-Core transfection, also showed the effects on both suppression of apoptosis and NF-κB activation. In addition, we found that the production of FLAG-core fusion protein did not show the above-reported biological effects at all despite the similarity in expression patterns to that of the wild-type core protein, including production level. Taken together, the biological effects of the core reported here should be attributable to the core-specific function irrespective of its expression levels.
We showed here that one of the antiapoptotic effects of the core was exerted through the activation of NF-κB in certain cells. The fine structure of the core required for its ability to activate NF-κB is not clear at this time. However, deletion analysis indicated that at least the C-terminal region of the core is important for that function. Although a simple explanation for this is that the C-terminal portion of the core forms the NF-κB activation domain, the real reason seems to be more complicated. Deletions of the C-terminal hydrophobic region of the core caused changes of subcellular localization of those products (Fig. 4). As this region was suggested to act as a signal peptide for E1 protein of HCV during processing of the precursor polyprotein (13), this region is likely to function as a primary topogenic signal of the core for the cytoplasmic surface of the endoplasmic reticulum. From the nuclear localization of the C-terminal deletion mutants, ΔCore173 and ΔCore151, it is assumed that the decrease in and the loss of NF-κB activation abilities of these mutants, respectively, are due to the isolation of these products from cytoplasm by translocation into the nucleus. It is well known that the regulation of NF-κB activation is based on its localization in the cell: NF-κB is present as an inactive form in the cytoplasm as a complex with IκB, but when the degradation of IκB is induced via activation of several protein kinases, for example, IKK-α and -β, NF-κB translocates into the nucleus as an active form (3, 22). Therefore, it seems reasonable that the core modulates the pathway for NF-κB activation in the cytoplasm, as do several other viral proteins (16, 28, 44, 55).
It is still unknown how NF-κB activation by the core leads to suppression of Fas- and TNF-α-mediated apoptosis. However, it was recently reported that NF-κB induces a group of gene products such as TNF receptor-associated factors 1 and 2 and inhibitor-of-apoptosis proteins 1 and 2, which suppress TNF-α-mediated apoptosis, and blocks the activation of caspase-8 (48). We have not observed the induction of those factors in the core-producing cells treated with anti-Fas or TNF-α. However, it may be possible that the core activating NF-κB acts on not only the TNF-α- but also the Fas-mediated apoptotic pathway by a similar mechanism, since we found that the activation of caspase-8 in anti-Fas- and TNF-α-treated HepG2 and MCF-7 cells, respectively, was diminished by production of the core. In contrast to HepG2 and MCF-7 cells, the mechanism of the suppressive effect on Fas-mediated apoptosis introduced by the core in Jurkat cells is unknown so far, because this effect was revealed to be independent of NF-κB activation.
We concluded from our results that HCV core protein inhibits the onset of apoptotic cell death, and at least one of the important pathways for this includes NF-κB activation by the core. This antiapoptotic effect introduced by the core might be advantageous for HCV by allowing the host hepatocytes to survive apoptosis, resulting in sustained infection. Further studies are necessary to determine the molecular mechanism by which the core enhances NF-κB activity and to find the other antiapoptotic pathway mediated by the core independently of NF-κB, because this might allow development of effective strategies for the prevention of chronic sustained viral infection.
ACKNOWLEDGMENTS
We are grateful to Y. Tsujimoto and D. Wallach for providing plasmids, S. Yonehara for Jurkat cells, M. Kohara for anticore antibody, and K. Watashi for assistance with the transfected-cell concentration procedure.
This work was supported by grants-in-aid for cancer research and for the second-term comprehensive 10-year strategy for cancer control from the Ministry of Health and Welfare and by grants-in-aid for scientific research from the Ministry of Education, Science and Culture of Japan.
REFERENCES
- 1.Alter H J, Purcell R H, Shih J W, Melpolder J C, Houghton M, Choo Q L, Kuo G. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N Engl J Med. 1989;321:1494–1500. doi: 10.1056/NEJM198911303212202. [DOI] [PubMed] [Google Scholar]
- 2.Arsura M, FitzGerald M J, Fausto N, Sonenshein G E. Nuclear factor-kappaB/Rel blocks transforming growth factor β1-induced apoptosis of murine hepatocyte cell lines. Cell Growth Differ. 1997;8:1049–1059. [PubMed] [Google Scholar]
- 3.Baeuerle P A, Baltimore D. Activation of DNA-binding activity in an apparently cytoplasmic precursor of the NF-κB transcription factor. Cell. 1988;53:211–217. doi: 10.1016/0092-8674(88)90382-0. [DOI] [PubMed] [Google Scholar]
- 4.Beg A A, Baltimore D. An essential role for NF-κB in preventing TNF-α-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 5.Bertrand F, Atfi A, Cadoret A, L’Allemain G, Robin H, Lascols O, Capeau J, Cherqui G. A role for nuclear factor kappaB in the antiapoptotic function of insulin. J Biol Chem. 1998;273:2931–2938. doi: 10.1074/jbc.273.5.2931. [DOI] [PubMed] [Google Scholar]
- 6.Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 7.Choo Q L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 8.Debbas M, White E. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 9.Fernandes-Alnemri T, Armstrong R C, Krebs J, Srinivasula S M, Wang L, Bullrich F, Fritz L C, Trapani J A, Tomaselli K J, Litwack G, Alnemri E S. In vitro activation of CPP32 and Mch3 by Mch4, a novel human apoptotic cysteine protease containing two FADD-like domains. Proc Natl Acad Sci USA. 1996;93:7464–7469. doi: 10.1073/pnas.93.15.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujita T, Ishido S, Muramatsu S, Itoh M, Hotta H. Suppression of actinomycin D-induced apoptosis by the NS3 protein of hepatitis C virus. Biochem Biophys Res Commun. 1996;229:825–831. doi: 10.1006/bbrc.1996.1887. [DOI] [PubMed] [Google Scholar]
- 11.Galle P R, Hofmann W J, Walczak H, Schaller H, Otto G, Stremmel W, Krammer P H, Runkel L. Involvement of the CD95 (APO-1/Fas) receptor and ligand in liver damage. J Exp Med. 1995;182:1223–1230. doi: 10.1084/jem.182.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grakoui A, Wychowski C, Lin C, Feinstone S M, Rice C M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci USA. 1991;88:5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hijikata M, Mizushima H, Tanji Y, Komoda Y, Hirowatari Y, Akagi T, Kato N, Kimura K, Shimotohno K. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc Natl Acad Sci USA. 1993;90:10773–10777. doi: 10.1073/pnas.90.22.10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiramatsu N, Hayashi N, Katayama K, Mochizuki K, Kawanishi Y, Kasahara A, Fusamoto H, Kamada T. Immunohistochemical detection of Fas antigen in liver tissue of patients with chronic hepatitis C. Hepatology. 1994;19:1354–1359. [PubMed] [Google Scholar]
- 16.Kaye K M, Devergne O, Harada J N, Izumi K M, Yalamanchili R, Kieff E, Mosialos G. Tumor necrosis factor receptor associated factor 2 is a mediator of NF-kappa B activation by latent infection membrane protein 1, the Epstein-Barr virus transforming protein. Proc Natl Acad Sci USA. 1996;93:11085–11090. doi: 10.1073/pnas.93.20.11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuo G, Choo Q L, Alter H J, Gitnick G L, Redeker A G, Purcell R H, Miyamura T, Dienstag J L, Alter M J, Stevens C E, Tegtmeier G, Bonino F, Colombo M, Lee W-S, Kuo C, Berger K, Shuster J R, Overby L R, Bradley D W, Houghton M. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989;244:362–364. doi: 10.1126/science.2496467. [DOI] [PubMed] [Google Scholar]
- 18.Liang T J, Jeffers L J, Reddy K R, De Medina M, Parker I T, Cheinquer H, Idrovo V, Rabassa A, Schiff E R. Viral pathogenesis of hepatocellular carcinoma in the United States. Hepatology. 1993;18:1326–1333. [PubMed] [Google Scholar]
- 19.Lin C, Lindenbach B D, Pragai B, McCourt D W, Rice C M. Processing of the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J Virol. 1994;68:5063–5073. doi: 10.1128/jvi.68.8.5063-5073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Q, Tackney C, Bhat R A, Prince A M, Zhang P. Regulated processing of hepatitis C virus core protein is linked to subcellular localization. J Virol. 1997;71:657–662. doi: 10.1128/jvi.71.1.657-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo S Y, Masiarz F, Hwang S B, Lai M M, Ou J H. Differential subcellular localization of hepatitis C virus core gene products. Virology. 1995;213:455–461. doi: 10.1006/viro.1995.0018. [DOI] [PubMed] [Google Scholar]
- 22.May M J, Ghosh S. Signal transduction through NF-κB. Immunol Today. 1998;19:80–88. doi: 10.1016/s0167-5699(97)01197-3. [DOI] [PubMed] [Google Scholar]
- 23.Miura M, Friedlander R M, Yuan J. Tumor necrosis factor-induced apoptosis is mediated by a CrmA-sensitive cell death pathway. Proc Natl Acad Sci USA. 1995;92:8318–8322. doi: 10.1073/pnas.92.18.8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizushima H, Hijikata M, Tanji Y, Kimura K, Shimotohno K. Analysis of N-terminal processing of hepatitis C virus nonstructural protein 2. J Virol. 1994;68:2731–2734. doi: 10.1128/jvi.68.4.2731-2734.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moretta A. Molecular mechanisms in cell-mediated cytotoxicity. Cell. 1997;90:13–18. doi: 10.1016/s0092-8674(00)80309-8. [DOI] [PubMed] [Google Scholar]
- 26.Muzio M, Chinnaiyan A M, Kischkel F C, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, Mann M, Krammer P H, Peter M E, Dixit V M. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 27.Nagata S, Golstein P. The Fas death factor. Science. 1995;267:1449–1456. doi: 10.1126/science.7533326. [DOI] [PubMed] [Google Scholar]
- 28.Nicot C, Tie F, Giam C Z. Cytoplasmic forms of human T-cell leukemia virus type 1 Tax induce NF-κB activation. J Virol. 1998;72:6777–6784. doi: 10.1128/jvi.72.8.6777-6784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Brien V. Viruses and apoptosis. J Gen Virol. 1998;79:1833–1845. doi: 10.1099/0022-1317-79-8-1833. [DOI] [PubMed] [Google Scholar]
- 30.Osumi T, Tsukamoto T, Hata S, Yokota S, Miura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T. Amino-terminal presequence of the precursor of peroxisomal 3-ketoacyl-CoA thiolase is a cleavable signal peptide for peroxisomal targeting. Biochem Biophys Res Commun. 1991;181:947–954. doi: 10.1016/0006-291x(91)92028-i. [DOI] [PubMed] [Google Scholar]
- 31.Rao L, Debbas M, Sabbatini P, Hockenbery D, Korsmeyer S, White E. The adenovirus E1A proteins induce apoptosis, which is inhibited by the E1B 19-kDa and Bcl-2 proteins. Proc Natl Acad Sci USA. 1992;89:7742–7746. doi: 10.1073/pnas.89.16.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ray R B, Meyer K, Ray R. Suppression of apoptotic cell death by hepatitis C virus core protein. Virology. 1996;226:176–182. doi: 10.1006/viro.1996.0644. [DOI] [PubMed] [Google Scholar]
- 33.Ray R B, Meyer K, Steele R, Shrivastava A, Aggarwal B B, Ray R. Inhibition of tumor necrosis factor (TNF-α)-mediated apoptosis by hepatitis C virus core protein. J Biol Chem. 1998;273:2256–2259. doi: 10.1074/jbc.273.4.2256. [DOI] [PubMed] [Google Scholar]
- 34.Ruggieri A, Harada T, Matsuura Y, Miyamura T. Sensitization to Fas-mediated apoptosis by hepatitis C virus core protein. Virology. 1997;229:68–76. doi: 10.1006/viro.1996.8420. [DOI] [PubMed] [Google Scholar]
- 35.Santolini E, Migliaccio E, La Monica N. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J Virol. 1994;68:3631–3641. doi: 10.1128/jvi.68.6.3631-3641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 37.Shrivastava A, Manna S K, Ray R, Aggarwal B B. Ectopic expression of hepatitis C virus core protein differentially regulates nuclear transcription factors. J Virol. 1998;72:9722–9728. doi: 10.1128/jvi.72.12.9722-9728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srinivasula S M, Ahmad M, Fernandes-Alnemri T, Litwack G, Alnemri E S. Molecular ordering of the Fas-apoptotic pathway: the Fas/APO-1 protease Mch5 is a CrmA-inhibitable protease that activates multiple Ced-3/ICE-like cysteine proteases. Proc Natl Acad Sci USA. 1996;93:14486–14491. doi: 10.1073/pnas.93.25.14486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki R, Matsuura Y, Suzuki T, Ando A, Chiba J, Harada S, Saito I, Miyamura T. Nuclear localization of the truncated hepatitis C virus core protein with its hydrophobic C terminus deleted. J Gen Virol. 1995;76:53–61. doi: 10.1099/0022-1317-76-1-53. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka K, Hirohata T, Koga S, Sugimachi K, Kanematsu T, Ohryohji F, Nawata H, Ishibashi H, Maeda Y, Kiyokawa H, Tokunaga K, Irita Y, Takeshita S, Arase Y, Nishino N. Hepatitis C and hepatitis B in the etiology of hepatocellular carcinoma in the Japanese population. Cancer Res. 1991;51:2842–2847. [PubMed] [Google Scholar]
- 41.Teodoro J G, Branton P E. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tewari M, Telford W G, Miller R A, Dixit V M. CrmA, a poxvirus-encoded serpin, inhibits cytotoxic T-lymphocyte-mediated apoptosis. J Biol Chem. 1995;270:22705–22708. doi: 10.1074/jbc.270.39.22705. [DOI] [PubMed] [Google Scholar]
- 43.Tsujimoto Y. Stress-resistance conferred by high level of bcl-2 alpha protein in human B lymphoblastoid cell. Oncogene. 1989;4:1331–1336. [PubMed] [Google Scholar]
- 44.Uhlik M, Good L, Xiao G, Harhaj E W, Zandi E, Karin M, Sun S C. NF-κB-inducing kinase and IκB kinase participate in human T-cell leukemia virus I Tax-mediated NF-κB activation. J Biol Chem. 1998;273:21132–21136. doi: 10.1074/jbc.273.33.21132. [DOI] [PubMed] [Google Scholar]
- 45.Van Antwerp D J, Martin S J, Kafri T, Green D R, Verma I M. Suppression of TNF-α-induced apoptosis by NF-κB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 46.Vaux D L, Cory S, Adams J M. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature. 1988;335:440–442. doi: 10.1038/335440a0. [DOI] [PubMed] [Google Scholar]
- 47.Wang C Y, Mayo M W, Baldwin A S., Jr TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-κB. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 48.Wang C Y, Mayo M W, Korneluk R G, Goeddel D V, Baldwin A S., Jr NF-κB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281:1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 49.Werness B A, Levine A J, Howley P M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 50.Woods K M, Chapes S K. Three distinct cell phenotypes of induced-TNF cytotoxicity and their relationship to apoptosis. J Leukoc Biol. 1993;53:37–44. doi: 10.1002/jlb.53.1.37. [DOI] [PubMed] [Google Scholar]
- 51.Yamaoka S, Inoue H, Sakurai M, Sugiyama T, Hazama M, Yamada T, Hatanaka M. Constitutive activation of NF-κB is essential for transformation of rat fibroblasts by the human T-cell leukemia virus type I Tax protein. EMBO J. 1996;15:873–887. [PMC free article] [PubMed] [Google Scholar]
- 52.Yang E, Korsmeyer S J. Molecular thanatopsis: a discourse on the Bcl2 family and cell death. Blood. 1996;88:386–401. [PubMed] [Google Scholar]
- 53.Yang X, Khosravi-Far R, Chang H Y, Baltimore D. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell. 1997;89:1067–1076. doi: 10.1016/s0092-8674(00)80294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yasui K, Wakita T, Tsukiyama-Kohara K, Funahashi S I, Ichikawa M, Kajita T, Moradpour D, Wands J R, Kohara M. The native form and maturation process of hepatitis C virus core protein. J Virol. 1998;72:6048–6055. doi: 10.1128/jvi.72.7.6048-6055.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin M J, Christerson L B, Yamamoto Y, Kwak Y T, Xu S, Mercurio F, Barbosa M, Cobb M H, Gaynor R B. HTLV-I Tax protein binds to MEKK1 to stimulate IκB kinase activity and NF-κB activation. Cell. 1998;93:875–884. doi: 10.1016/s0092-8674(00)81447-6. [DOI] [PubMed] [Google Scholar]
- 56.Zhu N, Khoshnan A, Schneider R, Matsumoto M, Dennert G, Ware C, Lai M M. Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J Virol. 1998;72:3691–3697. doi: 10.1128/jvi.72.5.3691-3697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]