Abstract
By comparative analysis of the hemagglutinin-esterase (HE) protein of mouse hepatitis virus strain S (MHV-S) and the HE protein of influenza C virus, we found major differences in substrate specificities. In striking contrast to the influenza C virus enzyme, the MHV-S esterase was unable to release acetate from bovine submandibulary gland mucin. Furthermore, MHV-S could not remove influenza C virus receptors from erythrocytes. Analysis with free sialic acid derivatives revealed that the MHV-S HE protein specifically de-O-acetylates 5-N-acetyl-4-O-acetyl sialic acid (Neu4,5Ac2) but not 5-N-acetyl-9-O-acetyl sialic acid (Neu5,9Ac2), which is the major substrate for esterases of influenza C virus and bovine coronaviruses. In addition, the MHV-S esterase converted glycosidically bound Neu4,5Ac2 of guinea pig serum glycoproteins to Neu5Ac. By expression of the MHV esterase with recombinant vaccinia virus and incubation with guinea pig serum, we demonstrated that the viral HE possesses sialate-4-O-acetylesterase activity. In addition to observed enzymatic activity, MHV-S exhibited affinity to guinea pig and horse serum glycoproteins. Binding required sialate-4-O-acetyl groups and was abolished by chemical de-O-acetylation. Since Neu4,5Ac2 has not been identified in mice, the nature of potential substrates and/or secondary receptors for MHV-S in the natural host remains to be determined. The esterase of MHV-S is the first example of a viral enzyme with high specificity and affinity toward 4-O-acetylated sialic acids.
Mouse hepatitis virus (MHV) is a positive-strand RNA virus belonging to the family Coronaviridae. Several viruses have been classified as members of this family, which can be subdivided into three antigenic clusters (3, 28). MHV belongs to the same cluster as bovine coronavirus (BCV) and human coronavirus OC43. A major characteristic of this cluster is the presence of a hemagglutinin-esterase (HE) surface glycoprotein in addition to the viral spike protein. The latter is present in all coronaviruses, while the HE protein may be present or absent in viruses of the MHV cluster.
The HE protein of MHV is encoded by a gene located immediately upstream of the spike gene. It is expressed from mRNA 2-1; the molecular mass is approximately 60 to 69 kDa. In virions, it is found as a dimer anchored in the viral membrane by a C-terminal transmembrane region. Expression of the HE gene is highly variable between MHV strains. Functional HE proteins have been detected in MHV-JHM (27), MHV-S (38), and MHV-DVIM (31, 32). In MHV-S, large levels of the HE protein are found, while MHV-JHM expresses relatively low amounts (27, 38). Different MHV-JHM isolates express variable levels of HE, depending on the number of UCUAA repeats at the 3′ end of the leader RNA (18).
The presence of HE is not strictly required for MHV replication. MHV-A59 and several other MHV strains do not express HE (17, 27, 38). In MHV-A59, it is not expressed due to a missing initiation codon (17). In addition, the upstream promoter determining synthesis of mRNA 2-1 is destroyed in this strain. In other MHV strains, mutations and deletions at the 3′ end of the HE gene have been detected; as a result, HE proteins without a transmembrane anchor are encoded. Biosynthesis of such truncated forms could be detected neither in lysates of infected cells nor in culture supernatants (38). Interaction of HE alone with target cells is apparently not sufficient for infectivity. MHV-DVIM replication is inhibited by a monoclonal antibody specific for the MHV receptor, indicating that interaction of the viral spike protein with cellular receptor molecules is mandatory for infection (6). The presence of HE may, however, modulate tissue tropism particularly within the central nervous system. MHV strains expressing an HE protein exhibit some preference for infecting neurons (for a review, see reference 1). Differences in neuropathogenicity of viruses with or without HE expression are at least partially derived from immune responses against HE. Passive immunization of mice with HE-specific monoclonal antibodies resulted in protection from a lethal infection, possibly by inhibition of virus spread through the central nervous system (39). MHV variants with mutations in the HE gene were isolated from such animals at late stages of infection (41). In a recent study, mice were infected with a chimeric MHV-A59 strain containing an HE protein derived from cells transfected with a defective interfering vector expressing the HE gene of MHV-JHM. Data obtained in this study indicated an enhanced early innate response caused by transient expression of HE (42).
In addition to MHV strains, several other viruses have been shown to express HE proteins. Among these, BCV and influenza C virus have been studied most extensively. HE proteins of these viruses are receptor-destroying enzymes, removing 9-O-acetyl groups from sialic acid-containing cellular receptor glycoproteins (11, 23, 25, 34, 35). In contrast, data on substrate specificities of MHV esterases are limited. Enzymatic activity was mostly determined with p-nitrophenylacetate (pNPA) as the substrate (6, 21, 40). Recently, the esterase of MHV-DVIM was found to remove acetyl groups from the natural substrate bovine mandibulary gland mucin (BSM) at very low levels (33).
We recently characterized the HE protein of puffinosis virus (PV), a coronavirus closely related to MHV (14). In that study, we compared substrate specificities of PV and influenza C virus. Results obtained from this comparison led us to propose that compounds different from 5-N-acetyl-9-O-acetyl sialic acid (Neu5,9Ac2) may be natural substrates for the PV HE. Because of the high amino acid sequence similarity between the HE proteins of PV and MHV, we have now extended our investigation on the substrate specificity of the MHV esterase. In this report, we provide evidence that 4-O-acetylated sialic acid (Neu4,5Ac2), but not Neu5,9Ac2, is a natural substrate for the HE protein of MHV-S.
MATERIALS AND METHODS
Viruses and cells.
MHV-S was kindly supplied by M. Buchmeier (Scripps Research Institute, La Jolla, Calif.). MHV-A59 and MHV-S were grown in mouse L cells. Influenza C/JJ/50 virus was isolated from embryonated eggs as described elsewhere (34).
Recombinant vaccinia virus.
RNA derived from L cells infected with MHV-S was isolated as described by Spaan et al. (29). Purified RNA was reverse transcribed with Superscript II reverse transcriptase (Gibco), using oligonucleotide C171 (5′ AGGCGAATTCGTTATGCCTCATGCAATCTAACAC 3′) as the primer. The underlined segment is complementary to the 3′ region of the HE gene in addition, an EcoRI site was added to allow cloning. Then the HE gene was amplified by PCR, using oligonucleotide C171 and upstream primer oligonucleotide C170 (5′ AGTCGAATTCGGTACCGTGTGTAGAATGAAGGG 3′). The resulting PCR product was digested with EcoRI and cloned into pUC21. Cloning of the authentic gene was verified by sequencing of the resultant recombinant plasmid. For expression in vaccinia virus, the cloned gene was amplified by PCR, using oligonucleotides MHV-S-forward (5′ CGCGAATTCATGTGCATAGCTATGGCTCCTCGC 3′) and oligonucleotide MHV-S-reverse (5′ CCACAATCTAACGTACTCCGTATTGGGCCCCCT 3′). The PCR product was digested with EcoRI and SmaI and cloned into the EcoRI/SmaI fragment of pATA gpt stop3. This plasmid is a derivative of pATA-18 (30), modified by the addition of the Escherichia coli xanthine guanine phosphoribosyltransferase gene (gpt) under the control of the vaccinia virus early/immediate promoter I3 and insertion of stop codons in all three reading frames into the SalI/SphI fragment of the polylinker. Homologous recombination was performed by infection of human TK− cells with wild-type vaccinia virus strain WR and subsequent transfection of 100 ng of plasmid pATA-S-HE. Recombinant vaccinia viruses were isolated by TK− selection (30), followed by threefold plaque purification in RK13 cells with gpt selection (4).
HA assay.
Hemagglutinin (HA) assays were performed as described previously (36) with 0.5% human type O erythrocytes obtained from the local blood bank or 0.5% murine erythrocytes. HA titers were expressed as the reciprocal of highest virus dilution resulting in full agglutination of erythrocytes.
Esterase assays.
Acetylesterase activity was determined with pNPA as described previously (34). One unit of viral esterase was defined as the amount of enzymatic activity resulting in cleavage of 1 μmol of pNPA per min. Release of acetate from glycoconjugates was determined with a commercial test kit as described previously (36). BSM types I and I-S were obtained from Sigma-Aldrich. Sialic acids were either chemically O-acetylated Neu5Ac according to the method of Ogura et al. (20) or prepared from BSM, equine submandibulary gland mucin (ESM), or guinea pig serum by acid hydrolysis (22). To detect esterase activity with free sialic acids, 1.5 nmol-aliquots of O-acetylated sialic acids were incubated for 45 min at 37°C with 2.5 mU of MHV-S or influenza C/JJ/50 virus in phosphate-buffered saline (PBS) containing 0.5% bovine albumin. For assays involving glycosidically bound sialic acids, 2.5 mU of virus was incubated with guinea pig serum (8.4 μg of sialic acid) under the same conditions. Alternatively, guinea pig serum was incubated with membrane fractions derived from HeLa cells infected with recombinant vaccinia virus for 4 h at 37°C. For control, heat-inactivated virus or membrane fractions of cells infected with wild-type vaccinia virus were used. Reactions were stopped by heating for 10 min at 96°C.
Fluorimetric high-pressure liquid chromatography (HPLC) analysis.
Samples containing glycosidically bound sialic acids were first hydrolyzed with 2 M propionic acid for 4 h at 80°C (19). The hydrolyzed mixtures were centrifuged at 100,000 × g, and the supernatants, as well as those derived from assays with free sialic acids, were lyophilized. Samples were then incubated with 20 μl of 2 M acetic acid and 49 μl of 1,2-diamino-4,5-methylenedioxybenzene reagent for 1 h at 56°C (9). After centrifugation at 100,000 × g, 20 μl of the supernatant was injected on an RP-18 column (Lichrospher 100; particle size, 5 μm; 4 mm [inside diameter] by 250 mm; Merck, Darmstadt, Germany) and eluted isocratically by water-methanol-acetonitrile (86/7/9 by vol) at a flow rate of 1 ml/min and compared with authentic standard sialic acids (see above). Fluorimetric detection occurred at an excitation wavelength of 373 nm and emission wavelength of 448 nm.
Solid-phase binding assay.
Virus binding assays were performed on coated 96-well microtiter plates as described elsewhere (44). Glycoproteins were dissolved in PBS and allowed to bind at 4°C overnight (50 μl/well). Wells were then washed with PBS, and remaining binding sites were blocked with 3% bovine serum albumin in PBS for 2 h at room temperature. For saponification of O-acetyl esters, coated wells were incubated with 200 mM NaOH for 30 min at room temperature. The wells were then washed with PBS, and virus suspensions were added (1 mU of esterase/well) and incubated for 2 h at 4°C. After virus was removed, wells were washed three times with PBS. Bound virus was detected by incubation with 4-methylumbelliferyl acetate (4-MUAc). Hydrolysis of substrate was monitored at an excitation wavelength of 365 nm.
RESULTS
Comparison of enzymatic activities of MHV-S and influenza C/JJ/50 virus esterases.
In a recent study, we obtained data on major differences in substrate specificities between the esterases of PV and those of influenza C virus and BCV. Because of high sequence similarities of the HE proteins of PV and MHV esterases (85% identical amino acid sequence), we assumed that MHV may exhibit substrate requirements similar to those of PV (14). Esterase activity of MHV strains with an expressed HE protein has been demonstrated in the past with a synthetic low-molecular-weight substrate, pNPA. Compared to assays determining acetate release from natural substrates as mucin, esterase activity can be more easily determined with pNPA (34). With this assay, esterase activities of MHV-JHM (40), MHV-DVIM (6), and HE derived from the cloned gene of MHV-JHM Wb1 expressed by vaccinia virus (21) have been determined. Although this type of assay allows rapid determination of esterase activity, it does not provide evidence for sialate-O-acetylesterase activity of MHV. When we used a purified MHV-S preparation and compared its esterase activity with that of influenza C/JJ/50 virus, we found acetylesterase activity associated with both viruses in a pNPA assay. For further experiments, we defined 1 U of viral esterase as the amount required to hydrolyze 1 μmol of pNPA/min. Other p-nitrophenyl esters, like pNP-propionate, -butyrate, and -valerate, were not hydrolyzed to a significant extent by the viruses tested, indicating a high specificity of both esterases towards acetyl esters (data not shown). In contrast, when we used glycoconjugates resembling natural substrates, major differences between MHV-S and C/JJ/50 virus were observed. The latter, as well as BCV, specifically removes acetyl groups at position 9 of sialic acids from BSM (10, 11, 34, 36, 37). When we used 30 mU of esterase of influenza C/JJ/50 virus, release of 3.7 and 3.6 μg of acetate/mg of substrate/h from two different BSM preparations was observed. In contrast, after incubation of these substrates with 30 mU of MHV-S, we detected no free acetate.
The MHV HE is unable to destroy influenza C virus receptors on erythrocytes.
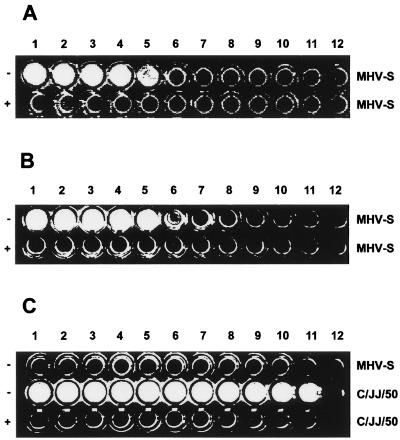
We then tried to remove influenza C virus receptors from erythrocytes by preincubation with MHV-S. If the MHV esterase cleaved these receptors, we expected a drop in HA titers of influenza C virus similar to data obtained with BCV esterase (36). We tested potential effects of the MHV esterase with human type O erythrocytes as well as murine erythrocytes from 6-week- and 6-month-old animals. In these assays, we used influenza C/JJ/50 virus, because it agglutinates human and murine erythrocytes. Mock-treated human erythrocytes were agglutinated by C/JJ/50 virus with the same titer as MHV-S-treated cells. As a control, we treated these cells with influenza C/JJ/50 virus, rendering them unagglutinable by C/JJ/50 virus (Table 1). From this control assay, we concluded that influenza C virus receptors can be removed from erythrocytes, provided that an enzyme with the correct substrate specificity is used. Similar data were obtained for murine erythrocytes. Since young mice are more susceptible to infection with MHV than adult animals, we tested cells obtained from approximately 6-week-old as well as 6-month-old mice. Erythrocytes from young animals were agglutinated by influenza C/JJ/50 virus with the same titers, regardless of whether cells were preincubated with buffer or MHV-S. There was a slight increase in HA titers with erythrocytes obtained from adult animals after incubation with MHV-S. This may indicate that the MHV esterase unmasks some additional influenza C virus receptors. However, given the only twofold increase, it appears more likely that this result can be explained by small experimental variations. Unfortunately, we were unable to do the reverse experiment using HA titration of MHV, because we could not detect any HA activity of MHV-S with the erythrocytes used.
TABLE 1.
Effects of erythrocyte pretreatments with viral esterase on hemagglutination by influenza C/JJ/50 virus
| Treatment | HA titera
|
||
|---|---|---|---|
| Human | Mouse
|
||
| 6 wk old | 6 mo old | ||
| PBS | 128 | 256 | 256 |
| C/JJ/50b | <2 | ND | ND |
| MHV-Sb | 128 | 256 | 512 |
Reciprocal of the highest virus dilution resulting in full agglutination. All experiments were performed three times. ND, not determined.
Erythrocytes were preincubated with 30 mU of viral esterase for 3 h at 37°C.
Identification of Neu4,5Ac2 as substrate for the MHV esterase.
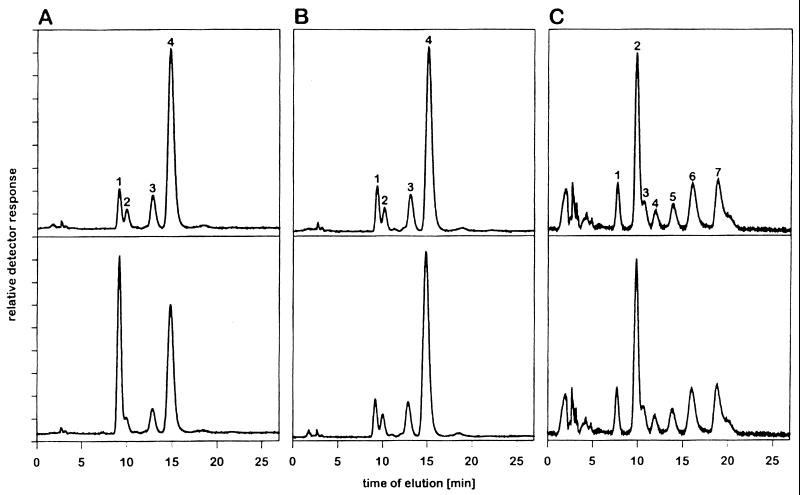
First, to determine whether the differences observed were attributable to the type of linkage of terminal sialic acids to underlying sugars, we tested the esterase activity of MHV-S with free sialic acid derivatives. We first incubated chemically prepared Neu5,9Ac2 containing small amounts of Neu5Ac, Neu5,7Ac2, and Neu5,8Ac2 with purified MHV-S. As a control, heat-inactivated virus was used. For a positive control, we incubated sialic acids with influenza C/JJ/50 virus, which was able to convert Neu5,9Ac2 to Neu5Ac (Fig. 1A). Analysis of sialic acids after incubation with MHV-S revealed no detectable de-O-acetylation of Neu5,9Ac2 (Fig. 1B). These data strongly indicate that 9-O-acetylated sialic acids are not hydrolyzed by the acetylesterase of MHV-S. Next, sialic acids isolated from BSM were incubated with active or heat-inactivated MHV-S. In addition to the above-mentioned sialic acid derivatives, Neu5Gc, Neu5Gc9Ac, and Neu5,8,9Ac3 were present in this preparation. Again, the esterase of MHV-S was unable to cleave any of these O-acetylated sialic acids (Fig. 1C).
FIG. 1.
Influenza C virus HE, but not MHV-S esterase, is able to hydrolyze acetate esters at the glycerol side chain of sialic acids. The reversed-phase C18 HPLC chromatograms show fluorescent derivatives of free sialic acids. (A and B) Chemically O-acetylated sialic acids, free Neu5Ac (peak 1), Neu5,7Ac2 (peak 2), Neu5,8Ac2 (peak 3), and Neu5,9Ac2 (peak 4), were incubated with influenza C virus (A) or with MHV-S (B). (C) Sialic acids, Neu5Gc (peak 1), Neu5Ac (peak 2), Neu5,7Ac2 (peak 3), Neu5Gc9Ac (peak 4), Neu5,8Ac2 (peak 5), Neu5,9Ac2 (peak 6), and Neu5,8,9Ac3 (peak 7), released from BSM, were incubated with MHV-S. Samples in the upper and lower chromatograms were treated with heat-inactivated and with active virus, respectively.
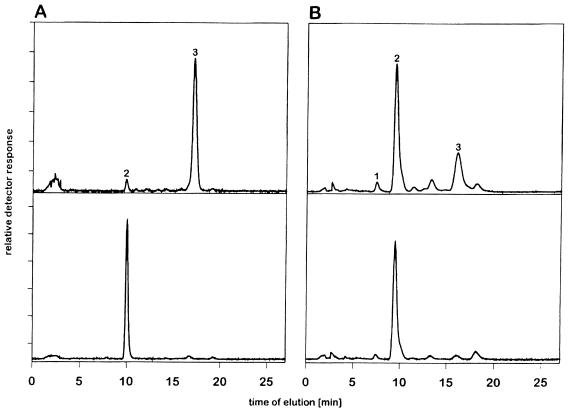
We then tested whether sialic acid with an O-acetyl group in position 4 could serve as an alternative substrate for MHV-S. When we incubated free Neu4,5Ac2 with MHV-S, we observed an almost complete (99%) loss of the sialate-4-O-acetyl ester and a corresponding increase of the Neu5Ac peak (Fig. 2A). In control incubations with heat-inactivated virus, no cleavage occurred, which indicates that this conversion was due to the viral esterase. To date, glycoproteins containing this sialic acid derivative have been identified in only a few animals (e.g., guinea pigs [for a review, see reference 24]). Therefore, to clarify whether the MHV-S HE protein also recognizes glycosidically linked Neu4,5Ac2 on glycoproteins, we used guinea pig serum proteins. These contain sialic acids, of which approximately 25 to 29% represent Neu4,5Ac2 (13). Again, an almost quantitative conversion of Neu4,5Ac2 to Neu5Ac was observed within 45 min at 37°C (Fig. 2B).
FIG. 2.
MHV-S esterase is able to de-O-acetylate free and glycosidically bound Neu4,5Ac2. Samples in the upper and lower reversed-phase C18 HPLC chromatograms of fluorescent derivatives of free sialic acids were treated with heat-inactivated and active MHV-S, respectively. (A) Free Neu4,5Ac2 (peak 3). Peak 2 represents Neu5Ac. (B) Sialic acids released from guinea pig serum glycoconjugates after treatment with MHV-S. Peak 1, Neu5Gc; peak 2, Neu5Ac; and peak 3, Neu4,5Ac2.
Expression of the HE protein by recombinant vaccinia virus.
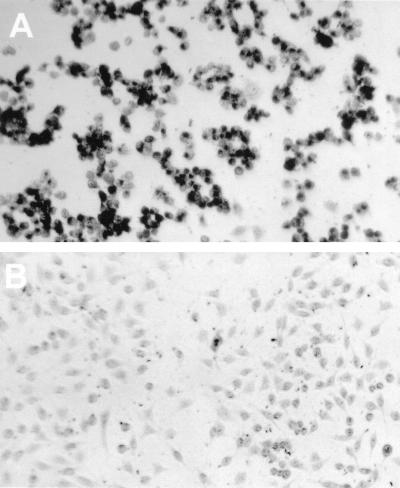
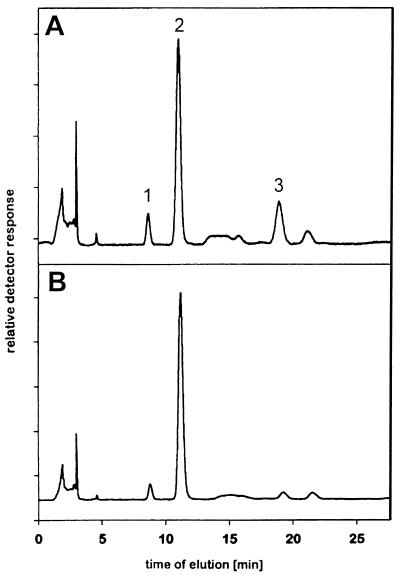
To determine whether the observed sialate-4-O-acetylesterase activity is an intrinsic property of the viral HE protein, we cloned the corresponding gene and inserted it into the thymidine kinase gene of vaccinia virus by targeted recombination. Vaccinia virus expressing the MHV esterase, termed VV-S-HE, was used to infect HeLa cells. Expression of esterase activity was monitored by incubating infected cells with α-naphthyl acetate, which results in precipitation of an insoluble dye on cells expressing the recombinant enzyme (Fig. 3). Mock-infected cells were not stained by this procedure. We then isolated membranes from infected cells and incubated them with guinea pig serum. Degradation of Neu4,5Ac2 and a corresponding increase of the Neu5Ac peak were observed after incubation with membranes from cells infected with VV-S-HE. Control incubations with membranes of cells infected with wild-type vaccinia virus revealed no change in the Neu4,5Ac2 content of guinea pig serum (Fig. 4). These data confirm that specific O-acetylesterase activity is encoded by the HE gene of MHV-S. We additionally observed a small but reproducible decrease in the amount of Neu5Gc after incubation of guinea pig serum with membranes from HeLa cells infected with recombinant VV-S-HE.
FIG. 3.
Identification of cells expressing recombinant MHV-S HE by staining with α-naphthyl acetate. L cells were infected with recombinant vaccinia virus VV-S-HE (multiplicity of infection of 5) (A) or mock infected (B); 16 h postinfection, cells were fixed with citrate-acetone-formaldehyde and stained with α-naphthyl acetate–fast blue BB for 15 min.
FIG. 4.
De-O-acetylation of Neu4,5Ac2 by recombinant MHV-S esterase. HeLa cells were infected (multiplicity of infection of 0.1) with wild-type vaccinia virus (A) or recombinant vaccinia virus VV-S-HE (B); 48 h postinfection, cells were lysed by freeze-thawing, and plasma membrane fractions were prepared. Membranes were incubated with guinea pig serum for 4 h at 37°C, and sialic acids were analyzed by HPLC. Peak 1, Neu5Gc; peak 2, Neu5Ac; peak 3, Neu4,5Ac2.
MHV-S exhibits binding activity toward glycoproteins with Neu4,5Ac2.
Next we examined if MHV-S is able to bind to Neu4,5Ac2 on glycoproteins. We used a solid-phase binding assay with immobilized proteins coated in microtiter plates. Since viral esterases commonly can cleave fluorogenic substrates (7, 25), we first investigated if the MHV esterase exhibits enzymatic activity with fluorescein diacetate or 4-MUAc. Incubation of MHV-S with these substrates clearly resulted in cleavage of substrates (data not shown), comparable to rates observed with influenza C/JJ/50 and PV (14). In the assay, we used 4-MUAc as the substrate to detect virus binding. Since guinea pig serum glycoproteins were a substrate for the MHV esterase, we also used them in the assay. MHV-S exhibited binding activity with these immobilized glycoproteins in a concentration-dependent manner (Fig. 5). Binding was abolished by saponification of acetyl esters, indicating that this binding activity is specific for 4-O-acetyl esters present on guinea pig serum proteins. Similar results were obtained when we used horse serum, which is also a source of Neu4,5Ac2 (8). Again, MHV-S was able to bind to these immobilized proteins, and no binding was observed after removal of O-acetyl groups by mild alkali treatment. In contrast, no reactivity of MHV-S was observed when we used plates coated with BSM. These data suggest that the HE protein of MHV-S binds to Neu4,5Ac2 but not to other O-acetylated sialic acids.
FIG. 5.
Binding of MHV-S to immobilized 4-O-acetylated glycoproteins. Serially diluted glycoproteins were immobilized in microtiter wells. Then proteins were either treated with 0.2 M NaOH (+) or mock treated with PBS (−) at room temperature for 30 min, washed with PBS, and incubated with MHV-S or influenza C/JJ/50 virus, exhibiting 1 mU of esterase activity, for 2 h at 4°C as indicated. Bound virus was detected with 4-MUAc. (A) Guinea pig serum; (B) horse serum; (C) BSM. Proteins used for coating in lanes 1 through 12: (A and C) 125 μg, 12.5 μg, 1.25 μg, 500 ng, 250 ng, 125 ng, 50 ng, 25 ng, 12 ng, 5 ng, 1 ng, and 0 ng, respectively, per well; (B) 25 μg, 12.5 μg, 6.25 μg, 3.12 μg, 1.6 μg, 800 ng, 400 ng, 200 ng, 100 ng, 50 ng, 25 ng, and 0 ng, respectively, per well.
DISCUSSION
In this study, we investigated the substrate specificity of the HE protein of MHV-S. Other viruses known to express evolutionarily related proteins are influenza C viruses (5, 11, 34), BCV (35, 36), human coronavirus OC43 (12, 43), hemagglutinating encephalomyelitis virus (26), and bovine torovirus (2). Several of these viral esterases have been characterized in terms of their substrate specificities and in all instances tested have been shown to recognize 9-O-acetylated sialic acids. Enzymatic activities of HE proteins in MCV strains have been described, but few data on their substrates and binding activities have been published (for a review, see reference 1). Recently, more data became available. First, Sugiyama et al. (33) reported significant differences on cleavage of a natural substrate known to contain high amounts of O-acetylated sialic acids. They found that MHV-DVIM, the only MHV strain exhibiting hemagglutinating activity, can hydrolyze O-acetylated sialic acids present on the natural substrate BSM, but at limited rates compared to other viral esterases. Furthermore, data published in this work indicated that MHV-S was essentially unable to liberate acetic acid from BSM (33). We have recently investigated another coronavirus, PV, and found similar differences regarding acetate release from Neu5,9Ac2. Particularly, BSM was found to be no substrate for PV, a virus closely related to MHV (14). These data had prompted us to hypothesize that other, unidentified O-acetylated compounds may be substrates for PV and closely related coronaviruses.
In this study we used MHV-S, a strain expressing high levels of HE protein (38). In contrast to BCV and influenza C virus, MHV-S exhibited no esterase activity with BSM and was in addition unable to remove influenza C virus receptors from erythrocytes. To clarify the reasons for these differences, we wanted to gain further information on the enzymatic activity of the MHV-S HE protein. We used either chemically synthesized sialic acid derivatives or sialic acids prepared from BSM, ESM, or guinea pig serum glycoproteins to characterize substrate specificities of the MHV-S esterase. We identified Neu4,5Ac2 as the only sialic acid derivative hydrolyzed by MHV-S. Other sialic acids with O-acetylation on the glycerol side chain were not de-O-acetylated at detectable amounts. MHV-S was able to hydrolyze acetyl esters from free as well as glycosidically linked Neu4,5Ac2. In addition, we have demonstrated that this novel substrate specificity of MHV-S is a property of the viral HE protein. HE expressed by recombinant vaccinia virus exhibited the same reactivity with Neu4,5Ac2 as observed with MHV-S.
Recently, Sugiyama et al. reported acetate release by MHV-DVIM from isolated murine brush border membranes (33). However, in the case of MHV-DVIM, it remains to be determined whether this MHV strain also exhibits 4-O-acetylesterase or the more classical 9-O-acetylesterase activity. Taking into consideration the close relationship between amino acid sequences of MHV esterases, it appears likely that all MHV HE proteins are specific for Neu4,5Ac2. On the other hand, the exclusive specificity observed for the HE protein of MHV-S may be the result of subtle changes in the three-dimensional configuration of the viral enzyme during evolution. Possibly there exist viral esterases that recognize O-acetyl esters on sialic acids in positions 4 and 9. The possibility arises that in addition to MHV, other viruses with an esterase specific for Neu4,5Ac2 exist, infecting particularly animals which are known to possess such sialic acid derivatives, e.g., horses or guinea pigs (13). However, to our knowledge there is no evidence that MHV-S itself causes infections in these animals. It will be interesting to test specificities of other coronaviruses with HE proteins.
Binding assays revealed that MHV-S exhibits a concentration-dependent affinity to glycoproteins with Neu4,5Ac2. We provide two forms of evidence that 4-O-acetylation is required for binding. First, saponification of O-acetyl groups on guinea pig and horse serum glycoproteins resulted in a complete loss of affinity. Second, BSM, which possesses sialic acids O-acetylated in the glycerol side chain but not in position 4, was not a binding substrate for MHV-S. Thus, one may speculate that 4-O-acetylated sialic acids on glycoconjugates at the surface of cells can serve as additional viral receptors.
Current evidence suggests that infection by MHV strictly depends on the interaction of the viral spike protein with the MHV receptor present at the surface of target cells (6). This was concluded from experiments designed to infect cells expressing influenza C virus receptors containing Neu5,9Ac2. Such cells were not infected by MHV-DVIM unless the MHV receptor was expressed from the transfected gene. Since we now provide evidence that influenza C virus receptors are not bound by the HE protein of MHV-S, it remains to be determined if this also applies to MHV-DVIM. In the future, we will test whether MHV-S can infect cells expressing Neu4,5Ac2 but lacking the MHV receptor. Such experiments may shed light on whether the presence of the MHV receptor is a prerequisite for infection by MHV-S. Neu4,5Ac2 may either serve as secondary receptor modulating tissue tropism of HE-expressing MHV strains or represent an alternative receptor facilitating infection of cells devoid of the MHV receptor.
Since Neu4,5Ac2 has not yet been found in mice (13), the question arises about potential substrates for the HE protein in this host. In further experiments, it may be rewarding to explore the binding activity of recombinant, soluble MHV-S HE with 4-O-acetylated sialic acid-bearing glycoconjugates. This may be a useful tool for the histochemical detection of Neu4,5Ac2 in mice. Similar approaches to detect Neu5,9Ac2 have been described for recombinant, soluble influenza C virus HE (15, 16) as well as for purified influenza C virus (10, 44, 45).
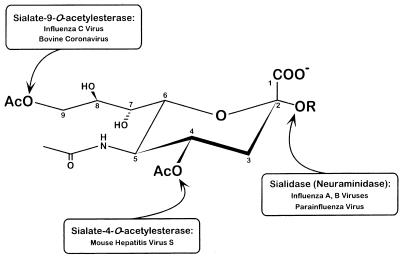
In summary, we have identified a viral enzyme exhibiting a previously unidentified specificity. In addition to the sialidases of influenza A and B viruses and paramyxoviruses and the sialate-9-O-acetylesterases of influenza C and BCV, a third type of receptor-destroying enzyme specifically cleaving 4-O-acetyl groups, has now been identified (Fig. 6).
FIG. 6.
Substrate specificities of viral sialidases and O-acetylesterases.
ACKNOWLEDGMENTS
We thank Michael Buchmeier for kindly providing MHV-S and Ulrike Hubl for the chemically O-acetylated sialic acids. The contribution of Jessica Vorgel (University of Berlin) at initial stages of the project is hereby acknowledged.
This work was partly supported by grant P-09945-Med from the Austrian Fonds zur Förderung der wissenschaftlichen Forschung, by the Fond der Chemischen Industrie, Frankfurt, and the Sialic Acid Society, Kiel, Germany, and by Commett grant 94/1/8273 (to B.S.).
REFERENCES
- 1.Brian D A, Hogue B G, Kienzle T E. The coronavirus hemagglutinin esterase glycoprotein. In: Siddell S, editor. The coronaviridae. New York, N.Y: Plenum Press Inc.; 1995. pp. 165–179. [Google Scholar]
- 2.Cornelissen L A H M, Wierda C M H, van der Meer F J, Herrewegh A A P M, Horzinek M C, Egberink H F, de Groot R J. Hemagglutinin-esterase, a novel structural protein of torovirus. J Virol. 1997;71:5277–5286. doi: 10.1128/jvi.71.7.5277-5286.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dea S, Verbeek A J, Tijssen P. Antigenic and genomic relationships among turkey and bovine enteric coronaviruses. J Virol. 1990;64:3112–3118. doi: 10.1128/jvi.64.6.3112-3118.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falkner F G, Moss B. Escherichia coli gpt gene provides dominant selection for vaccinia virus open reading frame expression vectors. J Virol. 1988;62:1849–1854. doi: 10.1128/jvi.62.6.1849-1854.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Formanowski F, Meier-Ewert H. Isolation of the influenza C virus glycoprotein in a soluble form by bromelain digestion. Virus Res. 1988;10:177–192. doi: 10.1016/0168-1702(88)90014-7. [DOI] [PubMed] [Google Scholar]
- 6.Gagneten S, Gout O, Duois-Dalcq M, Rottier P, Rossen J, Holmes K V. Interaction of mouse hepatitis virus (MHV) spike glycoprotein with receptor glycoprotein MHVR is required for interaction with an MHV strain that expresses the hemagglutinin-esterase glycoprotein. J Virol. 1995;69:889–895. doi: 10.1128/jvi.69.2.889-895.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Sastre A, Villar E, Manuguerra J C, Hannoun C, Cabezas J A. Activity of influenza C virus O-acetylesterase with O-acetyl-containing compounds. Biochem J. 1991;273:435–441. doi: 10.1042/bj2730435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanaoka K, Pritchett T J, Takasaki S, Kochibe N, Sabesan S, Paulson J C, Kobata A. 4-O-Acetyl-N-acetylneuraminic acid in the N-linked carbohydrate structures of equine and guinea pig alpha 2-macroglobulins, potent inhibitors of influenza virus infection. J Biol Chem. 1989;264:9842–9849. [PubMed] [Google Scholar]
- 9.Hara S, Yamaguchi M, Takemori Y, Furuhata K, Ogura H, Nakamura M. Determination of mono-O-acetylated N-acetylneuraminic acids in human and rat sera by fluorometric high-performance liquid chromatography. Anal Biochem. 1989;179:162–166. doi: 10.1016/0003-2697(89)90218-2. [DOI] [PubMed] [Google Scholar]
- 10.Harms G, Reuter G, Corfield A P, Schauer R. Binding specificity of influenza C virus to variably O-acetylated glycoconjugates and its use for histochemical detection of N-acetyl-9-O-acetylneuraminic acid in mammalian tissues. Glycoconj J. 1996;13:621–630. doi: 10.1007/BF00731450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrler G, Rott R, Klenk H-D, Müller H P, Shukla A K, Schauer R. The receptor-destroying enzyme of influenza C virus is neuraminate-O-acetylesterase. EMBO J. 1985;4:1503–1506. doi: 10.1002/j.1460-2075.1985.tb03809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hogue B G, Brian D A. Structural proteins of human respiratory coronavirus OC43. Virus Res. 1986;5:131–144. doi: 10.1016/0168-1702(86)90013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwersen M, Vandamme-Feldhaus V, Schauer R. Enzymatic 4-O-acetylation of N-acetylneuraminic acid in guinea-pig liver. Glycoconj J. 1998;15:895–904. doi: 10.1023/a:1006911100081. [DOI] [PubMed] [Google Scholar]
- 14.Klausegger, A., B. Strobl, G. Regl, A. Kaser, W. Luytjes, and R. Vlasak. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 15.Klein A, Krishna M, Varki N M, Varki A. 9-O-Acetylated sialic acids have a widespread but selective expression: analysis using a chimeric dual-function probe derived from influenza C hemagglutinin-esterase. Proc Natl Acad Sci USA. 1994;91:7782–7786. doi: 10.1073/pnas.91.16.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishna M, Varki A. 9-O-Acetylation of sialomucins: a novel marker of murine CD4 T cells that is regulated during maturation and activation. J Exp Med. 1997;185:1997–2013. doi: 10.1084/jem.185.11.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luytjes W, Bredenbeek P, Noten A, Horzinek M C, Spaan W. Sequence of mouse hepatitis virus A59 mRNA 2: indications for RNA recombination between coronaviruses and influenza C virus. Virology. 1988;166:415–422. doi: 10.1016/0042-6822(88)90512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makino S, Lai M M C. Evolution of the 5′-end of genomic RNA of murine coronaviruses during passages in vitro. Virology. 1989;169:227–232. doi: 10.1016/0042-6822(89)90060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mawhinney T P, Chance D L. Hydrolysis of sialic acids and O-acetylated sialic acids with propionic acid. Anal Biochem. 1994;223:164–167. doi: 10.1006/abio.1994.1564. [DOI] [PubMed] [Google Scholar]
- 20.Ogura H, Furuhata K, Sato S, Anazawa K, Itoh M, Shitori Y. Synthesis of 9-O-acyl- and 4-O-acetyl-sialic acids. Carbohydr Res. 1987;167:77–86. doi: 10.1016/0008-6215(87)80269-0. [DOI] [PubMed] [Google Scholar]
- 21.Pfleiderer M, Routledge E, Herrler G, Siddell S G. High level expression of the murine coronavirus hemagglutinin-esterase. J Gen Virol. 1991;72:1309–1315. doi: 10.1099/0022-1317-72-6-1309. [DOI] [PubMed] [Google Scholar]
- 22.Reuter G, Pfeil R, Stoll S, Schauer R, Kamerling J P, Versluis C, Vliegenthart J F G. Identification of new sialic acids derived from glycoprotein of bovine submandibular gland. Eur J Biochem. 1983;134:139–143. doi: 10.1111/j.1432-1033.1983.tb07542.x. [DOI] [PubMed] [Google Scholar]
- 23.Rogers G N, Herrler G, Paulsen J C, Klenk H-D. Influenza C virus uses 9-O-acetyl-N-acetyl-neuraminic acid as high affinity receptor determinant for attachment to cells. J Biol Chem. 1986;261:5947–5951. [PubMed] [Google Scholar]
- 24.Schauer R, Kamerling J P. Chemistry, biochemistry and biology of sialic acids. In: Montreuil J, Vliegenthart J F G, Schachter H, editors. Glycoproteins II. Amsterdam, The Netherlands: Elsevier; 1997. pp. 243–402. [Google Scholar]
- 25.Schauer R, Reuter G, Stoll S, Posadas del Rio F, Herrler G, Klenk H-D. Isolation and characterization of sialate 9(4)-O-acetylesterase from influenza C virus. Biol Chem Hoppe-Seyler. 1988;369:1121–1130. doi: 10.1515/bchm3.1988.369.2.1121. [DOI] [PubMed] [Google Scholar]
- 26.Schultze B, Wahn K, Klenk H-D, Herrler G. Isolated HE-protein from hemagglutinating encephalomyelitis virus and bovine coronavirus has receptor-destroying and receptor-binding activity. Virology. 1991;180:221–228. doi: 10.1016/0042-6822(91)90026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shieh C K, Lee H J, Yokomori K, La Monica N, Makino S, Lai M M C. Identification of a new transcriptional initiation site and the corresponding functional gene 2b in the murine coronavirus RNA genome. J Virol. 1989;63:3729–3736. doi: 10.1128/jvi.63.9.3729-3736.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spaan W J M, Cavanagh D, Horzinek M. Coronaviruses. In: Regenmortel M H V, Neurath A R, editors. Immunochemistry of viruses. 2. The basis for serodiagnosis and vaccines. Amsterdam, The Netherlands: Elsevier; 1990. pp. 359–379. [Google Scholar]
- 29.Spaan W J M, Rottier P J M, Horzinek M C, Van der Zeijst B A M. Isolation and identification of virus-specific mRNAs in cells infected with mouse hepatitis virus (MHV-A59) Virology. 1981;108:424–434. doi: 10.1016/0042-6822(81)90449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stunnenberg H G, Lange H, Philipson L, van Miltenburg R T, van der Vliet P C. High expression of functional adenovirus DNA polymerase and precursor terminal protein using recombinant vaccinia virus. Nucleic Acids Res. 1988;16:2431–2444. doi: 10.1093/nar/16.6.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugiyama K, Amano Y. Hemagglutination and structural polypeptides of a new coronavirus associated with diarrhea in infant mice. Arch Virol. 1980;66:95–105. doi: 10.1007/BF01314978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugiyama K, Ishikawa R, Fukuhara N. Structural polypeptides of the murine coronavirus DVIM. Arch Virol. 1986;89:245–254. doi: 10.1007/BF01309893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugiyama K, Kasai M, Kato S, Kasai H, Hatakeyama K. Haemagglutinin-esterase protein (HE) of murine coronavirus: DVIM (diarrhea virus of infant mice) Arch Virol. 1998;143:1523–1534. doi: 10.1007/s007050050395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vlasak R, Krystal M, Nacht M, Palese P. The influenza C virus glycoprotein (HE) exhibits receptor-binding (hemagglutinin) and receptor destroying (esterase) activities. Virology. 1987;160:419–425. doi: 10.1016/0042-6822(87)90013-4. [DOI] [PubMed] [Google Scholar]
- 35.Vlasak R, Luytjes W, Leider J, Spaan W, Palese P. The E3 protein of bovine coronavirus is a receptor-destroying enzyme with acetylesterase activity. J Virol. 1988;62:4686–4690. doi: 10.1128/jvi.62.12.4686-4690.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vlasak R, Luytjes W, Spaan W, Palese P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc Natl Acad Sci USA. 1988;85:4526–4529. doi: 10.1073/pnas.85.12.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vlasak R, Muster T, Lauro A M, Powers J C, Palese P. Influenza C virus esterase: analysis of catalytic site, inhibition, and possible function. J Virol. 1989;63:2056–2062. doi: 10.1128/jvi.63.5.2056-2062.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yokomori K, Banner L R, Lai M M C. Heterogeneity of gene expression of the hemagglutinin-esterase (HE) protein of murine coronaviruses. Virology. 1991;183:647–657. doi: 10.1016/0042-6822(91)90994-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yokomori K, Baker S C, Stohlman S A, Lai M M C. Hemagglutinin-esterase-specific monoclonal antibodies alter the neuropathogenicity of mouse hepatitis virus. J Virol. 1992;66:2865–2874. doi: 10.1128/jvi.66.5.2865-2874.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yokomori K, La Monica N, Makino S, Shieh C K, Lai M M C. Biosynthesis, structure, and biological activities of envelope protein gp65 of murine coronavirus. Virology. 1989;173:683–691. doi: 10.1016/0042-6822(89)90581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yokomori K, Stohlman S A, Lai M M C. The detection and characterization of multiple hemagglutinin-esterase (HE)-defective viruses in the mouse brain during subacute demyelination induced by mouse hepatitis virus. Virology. 1993;192:170–178. doi: 10.1006/viro.1993.1019. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X, Hinton D R, Park S, Parra B, Liao C-L, Lai M M C, Stohlman S A. Expression of hemagglutinin/esterase by a mouse hepatitis virus coronavirus defective-interfering RNA alters viral pathogenesis. Virology. 1998;242:170–183. doi: 10.1006/viro.1997.8993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Kousoulas K G, Storz J. The hemagglutinin/esterase of human coronavirus strain OC43: phylogenetic relationship to bovine and murine coronaviruses and influenza C virus. Virology. 1992;186:318–323. doi: 10.1016/0042-6822(92)90089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zimmer G, Reuter G, Schauer R. Use of influenza C virus for detection of 9-O-acetylated sialic acids on immobilized glycoconjugates by esterase activity. Eur J Biochem. 1992;204:209–215. doi: 10.1111/j.1432-1033.1992.tb16626.x. [DOI] [PubMed] [Google Scholar]
- 45.Zimmer G, Suguri T, Reuter G, Yu R K, Schauer R, Herrler G. Modification of sialic acids by 9-O-acetylation is detected in human leucocytes using the lectin property of influenza C virus. Glycobiology. 1994;4:343–349. doi: 10.1093/glycob/4.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]