Abstract
Schoenoplectus tabernaemontani (C. C. Gmelin) Palla is a typical macrophyte in diverse wetland ecosystems. This species holds great potential in decontamination applications and carbon sequestration. Previous studies have shown that this species may have experienced recent polyploidization. This would make S. tabernaemontani a unique model to study the processes and consequences of whole-genome duplications in the context of the well-documented holocentric chromosomes and dysploidy events in Cyperaceae. However, the inference was not completely solid because it lacked homology information that is essential to ascertain polyploidy. We present here the first chromosome-level genome assembly for S. tabernaemontani. By combining Oxford Nanopore Technologies (ONT) long reads and Illumina short reads, plus chromatin conformation via the Hi-C method, we assembled a genome spanning 507.96 Mb, with 99.43% of Hi-C data accurately mapped to the assembly. The assembly contig N50 value was 3.62 Mb. The overall BUSCO score was 94.40%. About 68.94% of the genome was comprised of repetitive elements. A total of 36,994 protein-coding genes were predicted and annotated. Long terminal repeat retrotransposons accounted for ∼26.99% of the genome, surpassing the content observed in most sequenced Cyperid genomes. Our well-supported haploid assembly comprised 21 pseudochromosomes, each harboring putative holocentric centromeres. Our findings corroborated a karyotype of 2n = 2X = 42. We also confirmed a recent whole-genome duplication occurring after the divergence between Schoenoplecteae and Bolboschoeneae. Our genome assembly expands the scope of sequenced genomes within the Cyperaceae family, encompassing the fifth genus. It also provides research resources on Cyperid evolution and wetland conservation.
Keywords: Schoenoplectus, karyotype, Cyperaceae, holocentric chromosome, WGD
Significance.
The soft-stem bulrush (Schoenoplectus tabernaemontani) holds promise as a valuable wetland plant. The inadequacy of accessible genetic information impedes a comprehensive understanding of its ecological significance and evolutionary uniqueness. We present the inaugural chromosome-level genome assembly for S. tabernaemontani, characterized by competent quality and detailed annotation of protein-coding genes and repeated sequences. Our genome assembly substantiates a robust karyotype inference for the sequenced individual of S. tabernaemontani (2n = 2X = 42). We validate a clade-specific whole-genome duplication occurring after the divergence between Schoenoplecteae and Bolboschoeneae, contributing an example of duplication-driven evolution within the dysploidy-prevalent Cyperaceae family.
Introduction
Schoenoplectus tabernaemontani (C. C. Gmelin) Palla, common name as soft-stem bulrush, is a flagship macrophyte in wetland ecosystems. It is a promising plant in decontamination applications. This species performs well in tolerating multiple organic pollutants, inorganic heavy metals, and nanoparticle (Zhang et al. 2009; Blanco 2018; Yan et al. 2022). However, debates exist about the biology and practical potential of S. tabernaemontani. For example, this species was reported to selectively retain arsenic and selenium in belowground tissues while conveying other heavy metals, such as lead, copper, and cadmium, to aboveground parts (Hammill et al. 2022). This selective strategy may lead to the accumulation of harmful elements among trophic levels. The notorious immunity of Schoenoplectus plants to herbicides also has negative effects on crop production (Scarabel et al. 2009). Nevertheless, Schoenoplectus plants have critical ecological significance. They typically grow fast and yield high biomass. Previous studies have shown that they are competent nontimber materials in construction practices, offering an alternative way to limit carbon emissions (Hidalgo-Cordero and García-Navarro 2018). Research on coastal wetlands also highlighted the heritable variations in the biomass allocation strategy of Schoenoplectus americanus and its relations with estuary carbon sequestration and soil surface accretion (Blum et al. 2021; Vahsen et al. 2023). However, a high-quality reference genome for S. tabernaemontani is still lacking, hindering further insights into the biological mechanisms of this promising plant.
Schoenoplectus tabernaemontani belongs to the species-rich sedge family (Cyperaceae). The prevalence of holocentric chromosomes confers evolutionary uniqueness to Cyperaceae species (Escudero et al. 2012, 2016; Hofstatter et al. 2022). The pervasive distribution of centromeres along the entire chromosome facilitates tolerance to breakages of chromosomes, which may prompt speciation through dysploidy instead of polyploidy (Lucek et al. 2022). For example, polyploidy occurrence is strikingly low in the Carex genus, despite the high volume of species diversity (∼2,000) and an exceptional chromosome number variation (2n = 10 to 132) (Márquez-Corro et al. 2021). However, it may not hold for other Cyperid species, as the chromosome number could evolve at heterogeneous rates along different clades (Márquez-Corro et al. 2019; Shafir et al. 2023). Notably, previous studies have provided some clues for polyploidization in the Schoenoplectus genus. Yano and Hoshino (2005) have examined 13 Schoenoplectus species, revealing a set of varied chromosome numbers, but individual chromosome sizes nearly hold constant, indicating a larger chance of polyploidy than dysploidy. The first record of polyploid intraindividual variation has also been found in Schoenoplectus acutus (Tena-Flores et al. 2014). Nevertheless, most of the evidence comes from chromosome counting, lacking homology information that is critical in inferring polyploidy events, especially autopolyploidy (Spoelhof et al. 2017). Thus, we present here the first chromosome-scale genome assembly of S. tabernaemontani, expanding the scale of the Cyperaceae reference genomes to the fifth genus. We aim to provide a valuable genetic resource for research on Cyperaceae evolution and wetland conservation.
Results and Discussion
Competence of the Genome Assembly
In total, we acquired 55.48 Gb (∼112×) of Oxford Nanopore Technologies (ONT) long-read data for preliminary assembly, 46.50 Gb (∼94×) of Illumina short-read data for genome profiling and back-mapping check, 45.04 Gb (∼91×) of Hi-C (all-vs.-all chromosome conformation capture) data for pseudochromosome construction, and 14.08 Gb (∼28×) of RNA-seq data for gene prediction. The average Q30 value for our short-read data was 92.76. The mean Q value for ONT data was 11.50. (supplementary fig. S1 and table S1, Supplementary Material online). Results of genome profiling showed the sequenced genome was moderately complex (∼1.3% heterozygosity). The inferred genome size was about 513 Mb, with repetitive content of ∼48.40% and GC content of ∼33.26% (supplementary fig. S2, Supplementary Material online). The estimated genome size is consistent with all four records in the comprehensive research by Elliot et al. (2022) about genome and chromosome evolution in Cyperid species, which provide essential guidance for our further assembling.
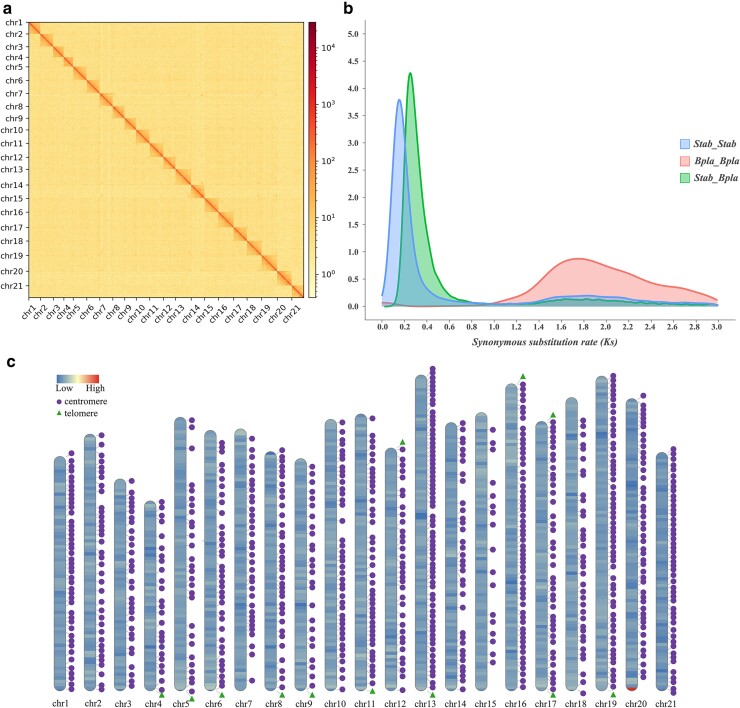
Using ONT data, we assembled a preliminary genome (supplementary table S2, Supplementary Material online). Then, we incorporated high-quality Hi-C data (supplementary table S3, Supplementary Material online) and polished this genome to chromosome level (Table 1). We successfully detected the association between most contigs. These contigs were then clustered into pseudomolecules. Eventually, we constructed a haploid assembly of 21 pseudochromosomes (Fig. 1a; supplementary table S4, Supplementary Material online). Up to 99.43% of the total bases were mapped into these pseudochromosomes. Our final assembly showed that S. tabernaemontani has a 1C genome size of 507.96 Mb (including both the well-mapped and unmapped bases). The contig N50 value is 3.62 Mb. The scaffold N50 value is 24.61 Mb. Detailed information on the assembly was listed in Table 1. We also provided a Circos graph (supplementary fig. S3, Supplementary Material online) that shows the gene density, GC content, transposable elements (TEs), and intragenome collinearity relations.
Table 1.
Statistics for the S. tabernaemontani genome assembly and BUSCO scores. “Anchored rate” refers to the proportion of bases that are well mapped into pseudochromosomes. Those unmapped bases are also included in the final assembly. “Size range” delimits the minimum and maximum size of pseudochromosomes
| Type | Statistics |
|---|---|
| Sequence | |
| Assembly size (bp) | 507,964,631 |
| GC content (%) | 33.32 |
| Number of scaffolds | 57 |
| Scaffold N50 size (bp) | 24,610,677 |
| Scaffold N90 size (bp) | 21,215,737 |
| Number of contigs | 249 |
| Contig N50 size (bp) | 3,615,529 |
| Contig N90 size (bp) | 1,429,834 |
| Pseudochromosome | |
| Number | 21 |
| Anchored rate (%) | 99.43 |
| Size range (M) | 17.35 to 28.85 |
| BUSCO score | |
| Complete BUSCOs (%) | 94.40 |
| Complete and single-copy BUSCOs (%) | 70.30 |
| Complete and duplicated BUSCOs (%) | 24.10 |
| Fragmented BUSCOs (%) | 1.90 |
| Missing BUSCOs (%) | 3.70 |
| Total groups searched | 1,614 |
Fig. 1.
Summary of the S. tabernaemontani genome assembly. a) Heatmap of Hi-C (all-vs.-all chromosome conformation capture) interactions within S. tabernaemontani pseudochromosomes. Gradients in the scale bar indicate the frequencies of Hi-C links alter from low to high. b) Density plot of the synonymous substitution rate (Ks) showing the different evolution modes for B. planiculmis (Bpla) and S. tabernaemontani (Stab). A clear peak is detected in the S. tabernaemontani genome subsequent to its divergence from B. planiculmis. c) Chromosome ideograms showing the karyotype of the assembled genome. Length of ideograms is proportional to chromosome size. Filling color scales with gene densities (300-kb window size). The putative telomeres are indicated with green triangle. The detected centromeres (purple circle) manifest pervasive distribution.
The quality of our genome assembly was supported by the following evidence: (i) The construction of pseudochromosomes was reliable. The mapping rate of Hi-C data (99.43%) was higher than formerly published genomes Bolboschoenus planiculmis (93.34%) (Ning et al. 2024) and Carex littledalei (96.28%) (Can et al. 2020). The results of our chromosome-staining experiment also supported the haploid chromosome number of 21 (supplementary fig. S4, Supplementary Material online). This value was also consistent with previous studies (2n = 42) (Elliot et al. 2022); (ii) the complete BUSCO score (94.40%) was at a comparable level to those recently reported for four Cyperid genomes (Planta et al. 2022). The back-mapping scores were good. About 98.33% Illumina short reads got projection in the genome assembly, and ∼96.74% of the whole genome was covered through back mapping; (iii) successful detection of telomeres and centromeres consolidated the high quality. Although highly repetitive in base content, centromeres and telomeres are vital components in gene regulation and cell biology (Lin et al. 2023). Both elements act as key criteria in the evaluation of telomere-to-telomere genome assembly. In our case, telomeres were detected in 11 pseudochromosomes (∼52.38% of the total), with Chr17 showing signals at both ends (Fig. 1c). Notably, our assembly supported a diffused distribution of centromeres along each chromosome, indicating that S. tabernaemontani may host holocentric chromosomes. Holocentricity has long been recognized as a critical and flexible trait in the diversification of Cyperaceae species (Escudero et al. 2016, 2012; Hofstatter et al. 2022). Our new assembly provides data resources that may benefit future research to fully ascertain the specific mechanisms of holocentricity in S. tabernaemontani.
Repetitive Elements and Gene Annotation
Repetitive elements constituted about 68.94% (∼350.19 Mb) of the S. tabernaemontani genome. Approximately 55.33% of the genome was composed of TEs. Tandem repeats consisted of ∼13.61% of the genome (see details in supplementary tables S5 and S6, Supplementary Material online). Based on the repeat-masked genome, we predicted protein-coding genes through a combination of three methods: ab initio, homology, and transcriptome-based prediction. In total, 36,994 protein-coding genes were identified in the S. tabernaemontani genome. Detailed information about gene prediction and BUSCO scores is presented in supplementary table S7, Supplementary Material online. The complete and duplicated type notably scored 22.00%, suggesting a potential large-scale duplication event. Approximately 91.76% of all the predicted genes got annotated at canonical databases (Pfam, EggNOG, Swiss-Prot, KEGG, NR, KOG, GO, and TrEMBL; see details in supplementary table S7, Supplementary Material online). We also established a computationally predicted noncoding RNA library, consisting of 550 rRNAs, 625 tRNAs, 200 miRNAs, and 464 snRNAs (supplementary table S8, Supplementary Material online).
Notably, the proportion of long terminal repeat retrotransposons (LTR-RTs) in S. tabernaemontani genome ranked high in all the available Cyperaceae genome assemblies (supplementary table S9, Supplementary Material online). Previous studies have shown that chromosomes originated from fusion (leading to large chromosome) may possess higher amounts of repetitive DNA, whereas fission (leading to small chromosome) may favor effective purge of repeat contents (Bureš and Zedek 2014; Veleba et al. 2016). Our genome assembly (26.99% LTR-RTs, mean chromosome size ∼24.19 Mb) provides preliminary clues for putative fusion events in the genome of S. tabernaemontani.
WGD and Clade-Specific Evolution Mode
Our genome assembly confirmed a clade-specific whole-genome duplication (WGD) event. The synonymous substitution rate (Ks) distribution clearly showed a burst after the divergence between S. tabernaemontani and B. planiculmis (Fig. 1b). It is also supported by the apparently higher amount of complete and duplicated BUSCOs (22.00%) compared with other species lacking genetic duplication, e.g. Cyperus esculentus (1.50%) (Zhao et al. 2023) and B. planiculmis (1.49%) (Ning et al. 2024). The strongest evidence came from the considerable amounts of collinear blocks within the S. tabernaemontani genome (supplementary fig. S5, Supplementary Material online). Previous studies have shown that intragenome collinear segments amend the possible deceiving effect of Ks plot, especially in inferring WGD events among recent divergent lineages (Zwaenepoel et al. 2019). Thus, the clade-specific WGD in S. tabernaemontani is highly possible. The prevalence of dysploidy evolution is well documented in some lineages in Cyperaceae. Our result exhibited a contrary instance. However, our result did not support polyploidy in S. tabernaemontani, as the genome profiling shows the karyotype to be 2n = 2X = 42 (supplementary figs. S2 and S4, Supplementary Material online). Furthermore, this result may provide valuable information in the transitions of evolution mode among closely related clades. Márquez-Corro et al (2019) have highlighted that, in the Fuireneae–Abildgaardieae–Eleocharideae–Cypereae clade, Cypereae showed a strikingly high rate of dysploidy events compared with the remarkably low rate of chromosome evolution in the rest lineages (Schoenoplectus included). Our inference of WGD offered a possible explanation other than chromosome number variation.
Materials and Methods
Collection and Preparation of Plant Materials
The sequenced samples were taken from a healthy individual of S. tabernaemontani at the Yongding wetland (39.887°N, 116.177°E). The sampled individual was well maintained in its original habitat for long-term research purpose. We selected vigorous leaves and treated them with caution to avoid exogenous contamination. All the field samples were swiftly transferred to lab environment and stored at −80 °C.
Genome Sequencing
We followed the cetyltrimethylammonium bromide method to extract genomic DNA. We checked the quality of DNA extraction through agarose gel electrophoresis. The SQKLSK109 ligation kit was used to generate ONT libraries. Primed R9.4 Spot-On Flow Cells were prepared following standard protocols to settle the purified libraries. We chose the PromethION platform to execute the sequencing. The raw data were treated using the Oxford Nanopore GUPPY software (v.0.3.0). Technical details could be found at https://github.com/nanoporetech. For Illumina short-read sequencing, pair-end libraries were constructed using the Nextera DNA Flex Library Prep Kit (Illumina, San Diego, CA, USA) and sequenced on the NovaSeq 6000 platform. We chose SOAPnuke (v.2.1.4) tool to clean and filter the raw reads (https://github.com/BGI-flexlab/SOAPnuke).
Transcriptome Sequencing
For gene prediction, total RNA was extracted and sequenced from four independent tissue samples (stem, tuber, spikelet, and root). The extraction of RNA was established following the manufacturer's instructions on RNA prep Pure Plant Plus Kit (Tiangen Biotech [Beijing] Co., Ltd., China). Then, the samples were pooled and sequenced on the Illumina NovaSeq 6000 platform. The library type was paired-end. The insertion size was about 350 bp on average. The generation of library followed the standard protocols of Illumina.
Genome Profiling and Draft Assembly
Genome profiling was realized using Genome Scope (v.2.0) (Ranallo-Benavidez et al. 2020) and Jellyfish (v.2.1.4) (Marçais and Kingsford 2011). The primary assembly was acquired using the NextDenovo pipeline (https://github.com/Nextomics/NextDenovo). Double rounds of error check of the primary assembly were performed using both the ONT data and the Illumina data. Heterozygous sequences were removed from the error-checking assembly using Purge_haplotigs pipeline (v.1.0.4) (Roach et al. 2018) to decrease ambiguities.
For Hi-C library construction, we followed a previously published protocol involving HindIII enzymatic digestion (Xie et al. 2015). The clean Hi-C data were then aligned with the draft assembly using Burrows–Wheeler Aligner (v.0.7.17) (Li and Durbin 2009). Only read pairs that were uniquely aligned were deemed valid-interaction reads. HiCUP (v.0.8.0) (Wingett et al. 2015) was used to screen and filter out read pairs. We clustered the contigs of the draft assembly into several groups (pseudochromosomes) using ALLHiC (v.0.9.8) (Wang and Zhang 2022). The orientation and ordination of contigs were further improved using 3D-DNA (v.180922) (Dudchenko et al. 2017) and Juciebox (v.1.11.08) (Durand et al. 2016).
Detection of Repetitive Elements
A de novo repeat library was acquired using RepeatModeler (v.2.0.1) (Flynn et al. 2020). We utilized a pipeline incorporating LTR_finder, LTR_harvester, and LTR_retriever to identify high-quality LTRs (Ou and Jiang 2017). RepeatMasker (v.4.15) and RepBase (v.20181026) were jointly used to finalize the repeat library. We utilized TRF (v.4.1.0) (Benson 1999) and MISA (v.2.1) (Beier et al. 2017) to annotate tandem repeats. Python scripts of quarTeT (Lin et al 2023) were used to detect potential centromeres and telomeres. Visualization was established using RIdeogram (v.0.2.2) (Hao et al. 2020).
Gene Prediction and Annotation
Based on the repeat-masked genome, Augustus (v 3.5.0) was utilized to generate de novo gene models (Stanke et al. 2008). The homology-based inference was achieved by using five well-annotated species as references (Arabidopsis thaliana, Oryza sativa, Triticum aestivum, Rhynchospora breviuscula, and B. planiculmis). TransDecoder (v.5.7.1) (https://github.com/TransDecoder/TransDecoder) was applied to parse the transcripts. Finally, these three types of evidence were integrated and reconciled using Maker (v.3.01) pipeline to obtain ultimate gene prediction results (https://github.com/Yandell-Lab/maker?tab). For noncoding RNA, we used tRNAscan-SE (v.1.3.1) (Lowe and Eddy 1997) to detect tRNA with eukaryote parameters. We used RNAmmer (v.1.2) to identify rRNA genes (https://services.healthtech.dtu.dk/services/RNAmmer-1.2/). We used a combination of Infernal (v.1.1.4) (Nawrocki and Eddy 2013) and Rfam (v.14.9) (Kalvari et al. 2021) to determine the miRNA, snoRNA, and snRNA in this genome. Both the Infernal and the Rfam incorporate covariance models. These models consider RNA secondary structure and primary sequence simultaneously, which greatly improves the scope of potential candidates (Kalvari et al. 2018).
Detection of Intragenome Synteny and WGD
We utilized the WGDI toolkit (Sun et al. 2022) to reveal the intragenomic synteny among pseudochromosomes and the potential WGD events. By implementing a hierarchical algorithm, WGDI has been shown to have high sensitivity and accuracy in collinearity detection. We applied the built-in functions of “-d”, “-icl”, “-ks”, “-bi”, and “-bk” to generate our inferences. Finally, we got an ideogram of pseudochromosomes to intuitively represent the multidimensional genomic information. The visualization of synonymous substitution (Ks) burst was accomplished using ggplot2 (https://github.com/tidyverse/ggplot2).
Supplementary Material
Acknowledgments
We thank Mr. Feng Y Y, Mr. Yuan Q H, and Mr Feng X for their assistance in our fieldwork. We are also grateful to anonymous reviewers whose comments helped to improve our manuscript.
Contributor Information
Yang Li, Huzhou University, Huzhou, China.
Yu Ning, Wetland Research Center, Institute of Ecological Conservation and Restoration, Chinese Academy of Forestry, Beijing, China; Sichuan Zoige Wetland Ecosystem Research Station, Prefecture of Aba, China.
Yan Chao Zheng, East China Inventory and Planning Institute, Hangzhou, China.
Xuan Yu Lou, Zhejiang Wanli University, Ningbo, China.
Zhe Pan, Sichuan Academy of Environmental Policy and Planning, Chengdu, China.
Shu Bin Dong, College of Biological Sciences and Technology, Beijing Forestry University, Beijing, China.
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online.
Author Contributions
Y.L. and Y.N. conceived and designed the research; Y.N. issued the funds and organized the related resources; Y.C.Z., X.Y.L., and Z.P. participated in the fieldwork and curated the raw data; Y.L. and Y.N. drafted the manuscript; and S.B.D., Y.C.Z., and X.Y.L. contributed in data visualization. The revision and approval of this final manuscript were established by all the authors.
Funding
The funding sources for this research are Fundamental Research Funds of the Chinese Academy of Forestry (CAFYBB2020SY042), Natural Science Foundation of Zhejiang Province (LQ21C160005), Huzhou Public Welfare Application Research Project (2022GZ21), and National Natural Science Foundation of China (NSFC31800348 and NSFC31972948).
Data Availability
The presented genome is deposited in the NCBI (Genome assembly ID CAF_SchTab_1.0, Bioproject ID PRJNA1055192, and Biosample ID SAMN38984562). The raw reads used to generate this genome assembly are stored at Sequence Read Archive (SRA) (ONT long-reads: SRR27340954; Illumina short reads: SRR27340955). The complete set of genome annotation files in gff3 format, including coding sequences, ncRNA sequences, and repeat sequences, are shared at https://figshare.com/articles/dataset/Annotation_files_for_the_chromosome-level_genome_assembly_of_soft-stem_bulrush_i_i_i_Schoenoplectus_tabernaemontani_i_/25367605.
Literature Cited
- Beier S, Thiel T, Münch T, Scholz U, Mascher M. MISA-web: a web server for microsatellite prediction. Bioinformatics. 2017:33(16):2583–2585. 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999:27(2):573–580. 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco J. Suitability of totora (Schoenoplectus californicus (C.A. Mey.) Soják) for its use in constructed wetlands in areas polluted with heavy metals. Sustainability. 2018:11(1):19. 10.3390/su11010019. [DOI] [Google Scholar]
- Blum MJ, Saunders CJ, McLachlan JS, Summers J, Craft C, Herrick JD. A century-long record of plant evolution reconstructed from a coastal marsh seed bank. Evol Lett. 2021:5(4):422–431. 10.1002/evl3.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureš P, Zedek F. Holokinetic drive: centromere drive in chromosomes without centromeres. Evolution. 2014:68(8):2412–2420. 10.1111/evo.12437. [DOI] [PubMed] [Google Scholar]
- Can M, Wei W, Zi H, Bai M, Liu Y, Gao D, Tu D, Bao Y, Wang L, Chen S, et al. Genome sequence of Kobresia littledalei, the first chromosome-level genome in the family Cyperaceae. Sci Data. 2020:7(1):175. 10.1038/s41597-020-0518-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017:356(6333):92–95. 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, Aiden EL. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 2016:3(1):99–101. 10.1016/j.cels.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott TL, Zedek F, Barrett RL, Bruhl JJ, Escudero M, Hroudová Z, Joly S, Larridon I, Luceño M, Márquez-Corro JI, et al. Chromosome size matters: genome evolution in the cyperid clade. Ann Bot. 2022:130(7):999–1014. 10.1093/aob/mcac136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero M, Hahn M, Brown BH, Lueders K, Hipp AL. Chromosomal rearrangements in holocentric organisms lead to reproductive isolation by hybrid dysfunction: the correlation between karyotype rearrangements and germination rates in sedges. Am J Bot. 2016:103(8):1529–1536. 10.3732/ajb.1600051. [DOI] [PubMed] [Google Scholar]
- Escudero M, Hipp AL, Hansen TF, Voje KL, Luceño M. Selection and inertia in the evolution of holocentric chromosomes in sedges (Carex, Cyperaceae). New Phytol. 2012:195(1):237–247. 10.1111/j.1469-8137.2012.04137.x. [DOI] [PubMed] [Google Scholar]
- Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A. 2020:117(17):9451–9457. 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammill E, Pendleton M, Brahney J, Kettenring KM, Atwood TB. Metal concentrations in wetland plant tissues influences transfer to terrestrial food webs. Ecotoxicology. 2022:31(5):836–845. 10.1007/s10646-022-02550-6. [DOI] [PubMed] [Google Scholar]
- Hao Z, Lv D, Ge Y, Shi J, Weijers D, Yu G, Chen J. RIdeogram: drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ Comput Sci. 2020:6:e251. 10.7717/peerj-cs.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo-Cordero JF, García-Navarro J. Totora (Schoenoplectus californicus (C.A. Mey.) Soják) and its potential as a construction material. Ind Crop Prod. 2018:112:467–480. 10.1016/j.indcrop.2017.12.029. [DOI] [Google Scholar]
- Hofstatter PG, Thangavel G, Lux T, Neumann P, Vondrak T, Novak P, Zhang M, Costa L, Castellani M, Scott A, et al. Repeat-based holocentromeres influence genome architecture and karyotype evolution. Cell. 2022:185(17):3153–3168.e18. 10.1016/j.cell.2022.06.045. [DOI] [PubMed] [Google Scholar]
- Kalvari I, Nawrocki EP, Argasinska J, Quinones-Olvera N, Finn RD, Bateman A, Petrov AI. Non-coding RNA analysis using the Rfam database. Curr Protoc Bioinformatics. 2018:62(1):e51. 10.1002/cpbi.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I, Nawrocki EP, Ontiveros-Palacios N, Argasinska J, Lamkiewicz K, Marz M, Griffiths-Jones S, Toffano-Nioche C, Gautheret D, Weinberg Z, et al. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 2021:49(D1):D192–D200. 10.1093/nar/gkaa1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009:25(14):1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Ye C, Li X, Chen Q, Wu Y, Zhang F, Pan R, Zhang S, Chen S, Wang X, et al. Quartet: a telomere-to-telomere toolkit for gap-free genome assembly and centromeric repeat identification. Hortic Res. 2023:10(8):uhad127. 10.1093/hr/uhad127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997:25(5):955–964. 10.1093/NAR/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucek K, Augustijnen H, Escudero M. A holocentric twist to chromosomal speciation? Trends Ecol Evol. 2022:37(8):655–662. 10.1016/j.tree.2022.04.002. [DOI] [PubMed] [Google Scholar]
- Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011:27(6):764–770. 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez-Corro JI, Martín-Bravo S, Jiménez-Mejías P, Hipp AL, Spalink D, Naczi RFC, Roalson EH, Luceño M, Escudero M. Macroevolutionary insights into sedges (Carex: Cyperaceae): the effects of rapid chromosome number evolution on lineage diversification. J Syst Evol. 2021:59(4):776–790. 10.1111/jse.12730. [DOI] [Google Scholar]
- Márquez-Corro JI, Martín-Bravo S, Spalink D, Luceño M, Escudero M. Inferring hypothesis-based transitions in clade-specific models of chromosome number evolution in sedges (Cyperaceae). Mol Phylogenet Evol. 2019:135:203–209. 10.1016/j.ympev.2019.03.006. [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013:29(22):2933–2935. 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y, Li Y, Lin HY, Kang EZ, Zhao YX, Dong SB, Li Y, Xia XF, Wang YF, Li CY. Chromosome-scale genome assembly for clubrush (Bolboschoenus planiculmis) indicates a karyotype with high chromosome number and heterogeneous centromere distribution. Genome Biol Evol. 2024:16(3):evae039. 10.1093/gbe/evae039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S, Jiang N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 2017:176(2):1410–1422. 10.1104/pp.17.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planta J, Liang Y-Y, Xin H, Chansler MT, Prather LA, Jiang N, Jiang J, Childs KL. Chromosome-scale genome assemblies and annotations for Poales species Carex cristatella, Carex scoparia, Juncus effusus, and Juncus inflexus. G3 (Bethesda). 2022:12(10):jkac211. 10.1093/g3journal/jkac211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranallo-Benavidez TR, Jaron KS, Schatz MC. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat Commun. 2020:11(1):1432. 10.1038/s41467-020-14998-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach MJ, Schmidt SA, Borneman AR. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics. 2018:19(1):460. 10.1186/s12859-018-2485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarabel L, Locascio A, Furini A, Sattin M, Varotto S. Characterisation of ALS genes in the polyploid species Schoenoplectus mucronatus and implications for resistance management. Pest Manag Sci. 2009:66(3):337–344. 10.1002/ps.1883. [DOI] [PubMed] [Google Scholar]
- Shafir A, Halabi K, Escudero M, Mayrose I. A non-homogeneous model of chromosome-number evolution to reveal shifts in the transition patterns across the phylogeny. New Phytol. 2023:238(4):1733–1744. 10.1111/nph.18805. [DOI] [PubMed] [Google Scholar]
- Spoelhof JP, Soltis PS, Soltis DE. Pure polyploidy: closing the gaps in autopolyploid research. J Sytematic Evol. 2017:55(4):340–352. 10.1111/jse.12253. [DOI] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008:24(5):637–644. 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- Sun P, Jiao B, Yang Y, Shan L, Li T, Li X, Xi Z, Wang X, Liu J. WGDI: a user-friendly toolkit for evolutionary analyses of whole-genome duplications and ancestral karyotypes. Mol Plant. 2022:15(12):1841–1851. 10.1016/j.molp.2022.10.018. [DOI] [PubMed] [Google Scholar]
- Tena-Flores JA, González-Elizondo MS, Herrera-Arrieta Y, Almaraz-Abarca N, Mayek-Pérez N, Vanzela ALL. Karyotype characterization of four Mexican species of Schoenoplectus (Cyperaceae) and first report of polyploid mixoploidy for the family. Caryologia. 2014:67(2):124–134. 10.1080/00087114.2014.931633. [DOI] [Google Scholar]
- Vahsen ML, Blum MJ, Megonigal JP, Emrich SJ, Holmquist JR, Stiller B, Todd-Brown KEO, McLachlan JS. Rapid plant trait evolution can alter coastal wetland resilience to sea level rise. Science. 2023:379(6630):393–398. 10.1126/science.abq0595. [DOI] [PubMed] [Google Scholar]
- Veleba A, Šmarda P, Zedek F, Horová L, Šmerda J, Bureš P. Evolution of genome size and genomic GC content in carnivorous holokinetics (Droseraceae). Ann Bot. 2016:119(3):409–416. 10.1093/aob/mcw229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-B, Zhang X. Chromosome scaffolding of diploid genomes using ALLHiC. Bio Protoc. 2022:12(18):1–8. 10.21769/bioprotoc.4503. [DOI] [Google Scholar]
- Wingett S, Ewels P, Furlan-Magaril M, Nagano T, Schoenfelder S, Fraser P, Andrews S. HiCUP: pipeline for mapping and processing Hi-C data. F1000Res. 2015:4:1310. 10.12688/f1000research.7334.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Zheng J-F, Liu S, Peng C, Zhou Y-M, Yang Q-Y, Zhang H-Y. De novo plant genome assembly based on chromatin interactions: a case study of Arabidopsis thaliana. Mol Plant. 2015:8(3):489–492. 10.1016/j.molp.2014.12.015. [DOI] [PubMed] [Google Scholar]
- Yan X, An J, Yin Y, Gao C, Wang B, Wei S. Heavy metals uptake and translocation of typical wetland plants and their ecological effects on the coastal soil of a contaminated bay in Northeast China. Sci Total Environ. 2022:803:149871. 10.1016/j.scitotenv.2021.149871. [DOI] [PubMed] [Google Scholar]
- Yano O, Hoshino T. Molecular phylogeny and chromosomal evolution of Japanese Schoenoplectus (Cyperaceae), based on ITS and ETS sequences. Acta Phytotax Geobot. 2005:56:183–195. 10.18942/apg.KJ00004623243. [DOI] [Google Scholar]
- Zhang Z, Rengel Z, Meney K. Kinetics of ammonium, nitrate and phosphorus uptake by Canna indica and Schoenoplectus validus. Aquat Bot. 2009:91(2):71–74. 10.1016/j.aquabot.2009.02.002. [DOI] [Google Scholar]
- Zhao X, Yi L, Ren Y, Li J, Ren W, Hou Z, Su S, Wang J, Zhang Y, Dong Q, et al. Chromosome-scale genome assembly of the yellow nutsedge (Cyperus esculentus). Genome Biol Evol. 2023:15(3):evad027. 10.1093/gbe/evad027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaenepoel A, Li Z, Lohaus R, Van de Peer Y. Finding evidence for whole genome duplications: a reappraisal. Mol Plant. 2019:12(2):133–136. 10.1016/j.molp.2018.12.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The presented genome is deposited in the NCBI (Genome assembly ID CAF_SchTab_1.0, Bioproject ID PRJNA1055192, and Biosample ID SAMN38984562). The raw reads used to generate this genome assembly are stored at Sequence Read Archive (SRA) (ONT long-reads: SRR27340954; Illumina short reads: SRR27340955). The complete set of genome annotation files in gff3 format, including coding sequences, ncRNA sequences, and repeat sequences, are shared at https://figshare.com/articles/dataset/Annotation_files_for_the_chromosome-level_genome_assembly_of_soft-stem_bulrush_i_i_i_Schoenoplectus_tabernaemontani_i_/25367605.